Abstract
Aromatase (CYP19) is a critical enzyme for estrogen biosynthesis, and aromatase inhibitors (AIs) are established endocrine therapy for post-menopausal women with breast cancer. DNA samples were obtained from 52 women pre- and post-AI treatment in the neoadjuvant setting. 82 breast cancer and 19 normal breast samples were resequenced to test the hypothesis that single nucleotide polymorphisms (SNPs) in the CYP19 gene might contribute to response to neoadjuvant AI therapy. There were no differences in CYP19 sequence between tumor and germline DNA in the same patient. Forty-eight CYP19 SNPs were identified, with four being novel when compared with previous resequencing data. Genotype-phenotype association studies performed with levels of aromatase activity, estrone, estradiol and tumor size pre- and post-AI treatment indicated that two tightly linked SNPs, rs6493497 and rs7176005 in the 5’-flanking region of CYP19 exon 1.1, were significantly associated with a greater change in aromatase activity after AI treatment. A follow-up study in 200 women with early breast cancer treated with adjuvant anastrozole showed that these same two SNPs were also associated with higher plasma estradiol levels pre- and post-AI treatment. Electrophoretic mobility shift and reporter gene assays confirmed the potential functional effects of these two SNPs on transcription regulation. These studies provide insight into the role of common genetic polymorphisms in CYP19 in variation in response to AIs by breast cancer patients.
Keywords: Aromatase, aromatase inhibitors, CYP19, gene resequencing, SNP, functional genomics, genotype-phenotype association
Introduction
In post-menopausal women, the major source of estrogen is the peripheral synthesis of estrone and estradiol by the cytochrome P450 enzyme aromatase (CYP19A1). The third generation aromatase inhibitors (AIs), anastrozole, exemestane, and letrozole, are potent and specific inhibitors of aromatase (1, 2) and have become established for the treatment of metastatic disease (3) and as adjuvant therapy in early breast cancer (4). These three AIs have been studied in the neo-adjuvant setting by two of the authors (WRM, JMD) and, in these 59 patients, 46 (78%) had a clinically determined major response, defined as >50% reduction in tumor volume, 8 (14%) had a 25-50% reduction in tumor volume and 5 (8%) had <25% reduction (5).
Selected CYP19 polymorphisms have been investigated for their possible association with the therapeutic efficacy of aromatase inhibitors and with sex hormone levels or risk for estrogen-dependent diseases (6-17). Although the results of those studies are controversial, they suggest an important role for genetic variation in aromatase function and, as a result, in estrogen production. Previously, we performed a CYP19 resequencing study with 240 germline DNA samples from 4 ethnic groups to identify 88 common SNPs and 44 common haplotypes (18). In this study, we investigated whether a difference in CYP19 sequence between breast tumor and germline DNA might exist and whether SNPs in tumor CYP19 might have a significant affect on response to AI therapy. Specifically, we obtained DNA samples isolated from breast tumor and normal breast tissue from 52 Edinburgh women treated with AIs in a neoadjuvant setting for which in vivo assays had been performed to measure levels of aromatase activity, estrone (E1) and estradiol (E2) in the tumors as well as tumor size before and after AI treatment (5). Gene resequencing was performed with DNA from both tumor and normal breast tissue pre- and post-AI treatment. A genotype-phenotype association study was then performed with the resequenced SNPs and all of the in vivo phenotypes. Functional genomic studies and a follow-up study were also performed to confirm observations made during the initial genotype-phenotype association study.
Material and Methods
Clinical samples and phenotype data
Both tumor and normal breast tissue samples were obtained from patients previously described in detail (5). Specifically, women with ER-positive (defined as >20 fmol/mg cytosol protein) primary breast cancer larger than 3 cm (stage T2,3,4b, N0,1, M0) and no prior treatment or concurrent treatment with hormone preparations were recruited to a neoadjuvant study to assess levels of E1, E2 and aromatase activity in the tumor, as well as tumor size pre- and post-AI treatment. All patients had received either letrozole (2.5mg or 10mg daily), anastrozole (1mg or 10mg daily) or exemestane (25mg daily). Patients in the study from whom the specimens were obtained underwent tumor measurement before, at monthly intervals and at the end of the 3 months of AI therapy. In addition, patients were given an infusion of 3H-androstenedione and 14C-estrone before and at the end of the 3-month treatment period, with blood, tumor and non-malignant breast tissue taken immediately after the infusion. That original study was designed to determine the effect of neoadjuvant AI therapy on the tumor – while the present study utilize those data to determine genotype-phenotype associations for CYP19 polymorphisms.
DNA samples were isolated from 52 patients, including 19 from normal and 82 from tumor tissue. 13 were normal-tumor pairs obtained from the same individuals. Tumor DNA included pre- and post-AI treatment samples. After quality control for the gene resequencing results and merging with clinical phenotypes, 77 DNA samples from 45 individuals were available for use in the genotype-phenotype studies. Characteristics of these 45 patients are listed in Supplementary Table 1. This clinical protocol was reviewed and approved by the Lothian Research Ethics Committee, Edinburgh, UK and all participants provided written consent. The genotyping study reported subsequently was reviewed and approved by the Mayo Institutional Review Board.
Follow-up study sample sets
200 DNA samples were isolated from women treated with 1mg anastrozole per day in an ongoing Mayo Clinic-MD Anderson collaborative clinical pharmacogenomic study. Plasma samples were collected from these women to measure levels of E1 and E2 as well as anastrozole concentrations. E1 and E2 levels pre- and post-treatment with anastrozole were measured by a gas chromatographic negative ionization tandem mass spectrometry (GC/NCI/MS/MS) and liquid chromatographic electrospray tandem mass spectrometry (LC/ESI/MS/MS) bioanalytical method at Taylor Technology (Princeton, NJ). Anastrozole and anastrozole metabolite levels were measured by an LC/MS/MS assay performed at Indiana University. This protocol was reviewed and approved by the Mayo Clinic and MD Anderson Institutional Review Boards, and all participants provided written consent.
CYP19 gene resequencing
Resequencing was performed using primers and PCR conditions listed in Supplementary Table 2. Resequenced regions included all coding exons, noncoding upstream exons, intron-exon splice junctions, 1000 bp of 5’–flanking regions (5’-FRs) for each of the noncoding exons and the 3’-untranslated region (3’-UTR). The primers and PCR conditions differed slightly from our previous resequencing study using 240 ethnically defined DNA samples (18) in that we sequenced longer regions than in the previous study. Because of this difference, we also resequenced additional regions included in the present study using the same 60 Caucasian DNA samples used in the previous study to allow us to directly compare the resequencing results between germline DNA from healthy individuals and tumor DNA from breast cancer patients. Amplicons were sequenced on both strands with an ABI 3730 DNA sequencer. Independent amplifications were performed for samples in which a SNP was observed only once or for samples with ambiguous chromatograms. The chromatograms were analyzed using Mutation Surveyor (SoftGenetics, State College, PA).
Cell culture and transfection
A mammalian expression construct was created for Arg128 by performing site-directed mutagenesis using the WT construct described previously (18) as template for circular PCR. Primers used to perform mutagenesis are listed in Supplementary Table 3. COS-1 cells obtained from ATCC (American Tissue Culture Center, Manassas, VA) were transfected with the WT and variant CYP19 constructs as well as empty vector as control using the TranFast reagent (Promega, Madison, WI). The pSV40-β-galactosidase vector encoding β-galactosidase (Promega, Madison, WI) was used as a control for transfection efficiency.
Aromatase enzyme assay and inhibition study
Aromatase activity was determined as described previously (18-20). Specifically, microsomes were isolated from COS-1 cells transfected with WT, Arg128 and empty vector together with β-galactosidase. Microsomal preparations were then used to perform the aromatase enzyme assay using [1β3H] androst-4-ene-3,17-dione (NEN Life Science Products, Boston, MA) as substrate (18). Aromatase activity was assayed by measuring the release of 3H2O from radio-labeled substrate. Reactions were carried out for 45 minutes at 37°C with an NADPH regeneration system in a final 100 μl volume as described previously. Blanks were samples that lacked enzyme. For substrate kinetic studies, substrate concentrations ranged from 0 to 80nM. For inhibition studies, assays were performed in the presence of three concentrations of letrozole (0.2, 0.4 and 0.8nM). Km and Kis values were then determined.
Electrophoresis mobility shift assay (EMSA)
Nuclear extracts from SKBR03 cells (ATCC) were isolated and protein was quantified with the method of Bradford (21). Biotin labeled sense and anti-sense oligonucleotides were designed to contain WT and variant sequences for the rs6493497 (G/A) and rs7176005 (C/T) SNPs. Probe sequences are listed in Supplementary Table 3. EMSAs were performed with the LightShift™ Chemiluminescent EMSA Kit (Pierce, Rockford, IL). A 400-fold excess of unlabeled probe was added for the competition assays.
Reporter gene assay
Reporter gene constructs were created by PCR amplification of a 1000 bp region surrounding the two SNPs using Coriell DNA as template. The WT PCR product was cloned into pGL3 Basic vector (Promega, Madison, WI) followed by mutagenesis to obtain the variant constructs. Primers used to perform mutagenesis are listed in Supplementary Table 3. DNA sequences were verified by sequencing both strands. Vector without an insert was used as a control. SKBR03 cells were transfected with WT and variant constructs together with a pRL-TK DNA construct encoding Renilla luciferase as a control for transfection efficiency. Cells were then lyzed and luciferase activity was measured using a dual luciferase activity assay with a TD-20/20 Luminometer (Turner Designs, Sunnyvale, CA). Results were reported as the ratio of firefly luciferase to Renilla luciferase light units, and all values were expressed as a percentage of the activity of the WT construct. All assays were performed in triplicate.
Taqman assay
In a follow-up study, SNPs rs6493497 (G/A) and rs7176005 (C/T), upstream of CYP19 exon 1.1, were genotyped using 200 germline DNA samples from breast cancer patients being treated with adjuvant anastrozole. Taqman assays were performed according to the manufacturer's instructions. Positive (Coriell DNA with known genotypes) and negative controls (lacking DNA) were included in the assays.
Statistical methods
SNPs obtained during the gene resequencing study were subjected to quality control to remove those with minor allele frequencies (MAFs) <5%, SNPs that deviated significantly from Hardy-Weinberg equilibrium (HWE) (P<0.001) or with call rates <95%. Due to skewness of the phenotypes, the van der Waerden rank normal score transformation was performed for all phenotypes. Phenotypes were compared among anastrozole, exemestane, and letrozole using a Monte Carlo approximated exact Krustal Wallis test. SNP associations with transformed phenotypes were assessed using ordinary regression, treating genotype effect as the count of minor alleles (i.e., 1df or trend test) in all treatment groups, as well in a subset analysis by treatment group. For the genotypic analyses, p-values were corrected for multiple testing by a permutation step-up procedure (22). Linkage disequilibrium among CYP19 polymorphisms was determined by calculating D' and R2 values for all possible pairwise combinations of polymorphisms, and the data were plotted using Haploview software (23). Haplotypes were estimated for the markers passing QC and compared among phenotypes using score tests from the haplo.stats software (http://mayoresearch.mayo.edu/mayo/research/schaid_lab/software.cfm) (24). For the follow-up study, the relationship between genotypes and pre- and post-drug E1 and E2 concentrations were modeled using a negative bimodal generalized linear model due to the extreme skewness of the phenotypes (25, 26). Models of post-treatment E1 and E2 were also adjusted for anastrozole levels. Differences in E1 and E2, pre- to post-treatment, were left on their original scale. Ordinary regression was used to assess the association of phenotypes and genotypes treated as the count of minor alleles (i.e. 1df).
Differences in enzyme activity, protein levels and luciferase activities were tested using Student's T-test within the software prism. Ki and Km values were calculated with the Prism Program.
Results
CYP19 gene resequencing
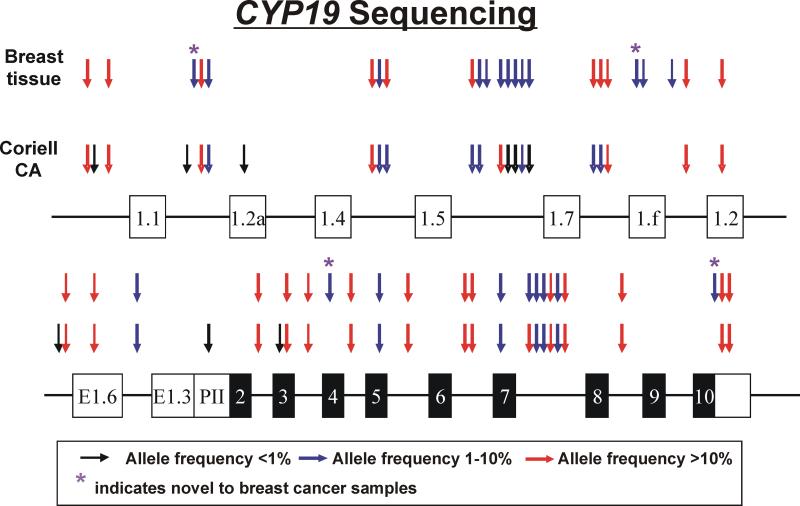
CYP19 resequencing identified 48 SNPs (Figure 1 and Supplementary Table 4). No differences were observed between normal germline and tumor DNA. Since one reaction covering part of the 5’-FR of exon 1.7 contained two indels, one located 1051 bp and the other 1000 bp upstream of exon 1.7, respectively, we could not determine the allele frequencies of the indels or of a SNP at (−787) upstream of exon 1.7. We also could not determine the genotype of an indel and VNTR (variable number of tandem repeats) in intron 4 that were reported previously. While using the 60 CA DNA samples to resequence the additional regions that were not covered in our previous study (18), we identified 11 additional SNPs in Caucasians in addition to the original 37 SNPs identified during our previous study (Figure 1, Supplementary Table 4). Four SNPs, including the A383G that resulted in a 128His>Arg amino acid alteration, were observed only in breast tissue, including both normal and tumor tissue (Figure 1 and Supplementary Table 4).
Figure 1.
Human CYP19 gene structure and polymorphisms. The figure shows a schematic representation of the human CYP19 gene. Exons are represented as rectangles, with black rectangles indicating the open reading frame (ORF) and white indicating untranslated regions (UTRs). Arrows indicate the locations of polymorphisms, with colors representing minor allele frequencies. Red arrows indicate polymorphisms with frequencies of >10%, while blue and black arrows indicate polymorphisms with frequencies from 1% to 10% and of <1%, respectively. “*” represents SNPs that were “novel” to this study.
Functional characterization of His128Arg
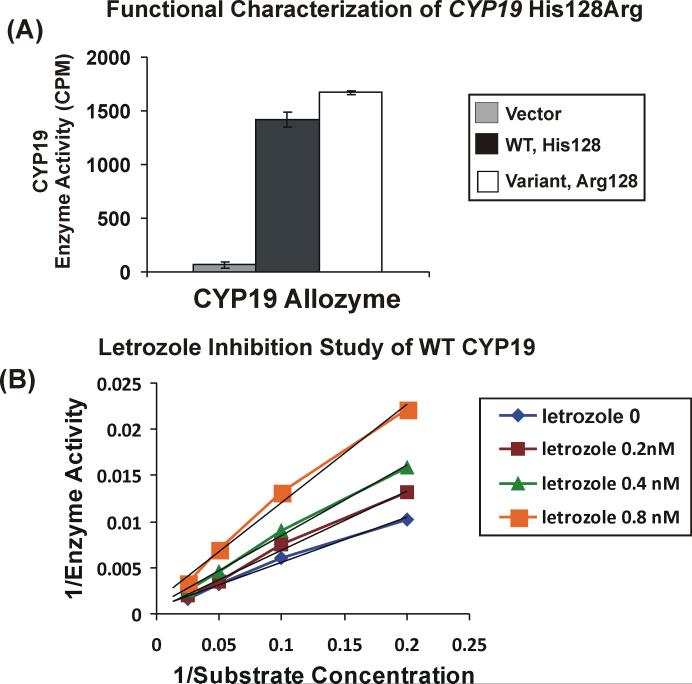
Since the His128Arg was only observed in breast tumor and normal breast germline DNA at a 1% allele frequency and had not been observed in the 60 Caucasian subjects included in our previous resequencing study, we performed functional genomic studies with that SNP. Microsomal preparations from COS-1 cells transiently transfected with WT, Arg128 and empty vector were used to perform enzyme assays with [1β3H] androst-4-ene-3,17-dione as substrate. No significant difference in aromatase enzyme activity was observed between the WT and Arg128 allozymes (Figure 2A). Substrate kinetic studies were also performed with 6 different concentrations of [1β3H] androst-4-ene-3,17-dione, 2.5, 5, 10, 20 40 and 80nM. Representative substrate kinetic studies for WT and Arg128 showed that apparent Km values for the WT and Arg128 allozymes were 81 ± 4 and 164 ± 32nM, respectively. These values did not differ significantly (P=0.08). We also performed inhibition studies with increasing concentrations of letrozole (0.2, 0.4 and 0.8nM). Double inverse plots for the WT inhibition study with letrozole is shown in Figure 2B. The Kis for WT was 1.6nM ± 0.11 and that for Arg128 was 1.6nM ± 0.27 (P=0.9). Western blot analysis using antibody directed against aromatase showed no difference in expression (data not shown).
Figure 2.
Functional characterization of the CYP19 Arg128 variant allozyme. (A). Enzyme activity assay. CYP19 enzyme activity was measured with microsomal preparations from cells transfected with “empty” vector, WT and Arg128 constructs using [1β3H] androst-4-ene-3,17-dione as substrate. Error bars are mean ± SEM for 3 independent experiments. (B). Representative letrozole inhibition study of WT CYP19. Double inverse plots were created using 6 concentrations of [1β3H] androst-4-ene-3,17-dione in the presence of increasing concentrations of letrozole (0-0.8nM).
Genotype-phenotype association study
In order to identify CYP19 SNPs in breast tumor tissue that might contribute to variation in response to AI treatment in women with ER(+) breast cancer, we performed genotype-phenotype studies with our resequencing data using tumor DNA together with in vivo phenotypes that had been obtained in a neoadjuvant setting, as described previously (5). DNA samples from 45 patients were used in this analysis. Supplementary Table 1 lists the demographic and clinical characteristics of these patients. These patients were treated with three different AIs, with information on aromatase activity before and after AI treatment recorded for 45 patients and tumor size pre- and post-AI treatment for 44 patients. However, only the 30 patients treated with letrozole and anastrozole had information on levels of E1 and E2 pre- and post-treatment. That was because exemestane is a steroid analogue and interferes with the assays used to measure E1 and E2 (27). As expected, most patients treated with AIs had decreases in E1, E2 and tumor size. However, there was variation in changes in these phenotypes before and after AI treatment (Supplementary Figure 1). Before performing the genotype-phenotype association study, we determined the effect of the three treatments on the phenotypes of interest. No significant differences for phenotypes were observed among drugs. Since the effects of different AI drugs on the phenotypes were minimal, statistical analysis was completed on the combined data for all three AIs.
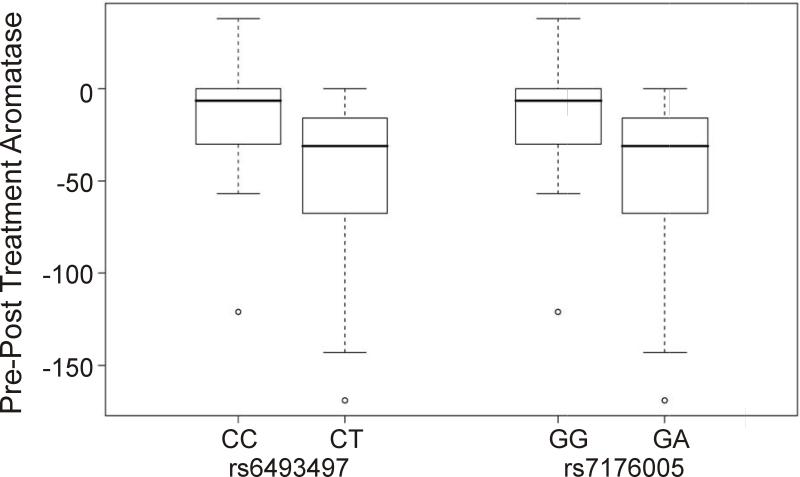
Genotype-phenotype association analysis was then performed using the SNPs identified by resequencing CYP19 in breast tumor DNA samples from 45 patients. After quality control, 27 SNPs were used in the analysis. Table 1 lists the phenotypes studied, including changes in E1, E2, aromatase activity and tumor size as well as each of these phenotypes measured before and after AI treatment. Two SNPs, one located at 144 bp (rs6493497) and the other located at 588 bp (rs7176005) 5’-upstream of exon 1.1, the major “placental” exon 1, were significantly associated with change in aromatase activity before and after AI treatment (nominal P=0.0063, P=0.039 after adjustment for multiple testing) (Table 2A and Figure 3). Specifically, those SNPs result in greater change in aromatase activity pre- and post-AI, which indicated that samples containing these two SNPs had greater inhibition of aromatase activity. These same SNPs were also associated with change in E1 level (P=0.038) and with higher aromatase activity at baseline (P=0.028), although these differences were not significant after correction for multiple comparisons (Table 2A). Rs6493497 and rs7176005 were in LD (R2=1), with observed MAFs of 17% in these samples (Supplementary Figure 2). Another two linked SNPs (R2=1), one located (−690) and the other (−725) upstream of exon 1.f, were significantly associated with post-drug aromatase activity, with P=0.022 adjusted for multiple testing. Marginally, the SNP at (−628) upstream of 5'-FR for exon 1.5 was also associated with change in aromatase (P=0.06 after adjustment for multiple testing). We also performed haplotype analysis using the resequenced SNPs, but none of the haplotypes showed a significant association with the phenotypes of interest after correction for multiple testing (data not shown).
Table 1.
Summary of clinical and in vivo phenotypes for breast cancer patients. These phenotypes were determined as part of a previous study (5).
| Phenotype | Pre-treatment | Post-treatment | Post-Pre |
|---|---|---|---|
| Tumor volume | |||
| N | 44 | 44 | 44 |
| Mean (SD) |
8.5 (11.00) |
2.2 (2.38) |
−6.4 (9.08) |
| Aromatase activity (fmol/mg protein/h) | |||
| N | 45 | 45 | 45 |
| Mean (SD) | 33.9 (41.80) | 6.1 (13.16) | −27.8 (40.71) |
| Median |
23.0 |
0.0 |
−21.0 |
| E1 Level (dpm/g) | |||
| N | 30 | 30 | 30 |
| Mean (SD) | 1.9 (2.79) | 0.4 (0.40) | −1.5 (2.73) |
| Median |
0.7 |
0.3 |
−0.5 |
| E2 Level (dpm/g) | |||
| N | 30 | 30 | 30 |
| Mean (SD) | 1.5 (1.27) | 0.3 (0.34) | −1.2 (1.25) |
| Median | 1.2 | 0.3 | −0.8 |
Table 2A.
Results of genotype-phenotype correlation study for rs7176005 and rs6493497.
| Phenotype | Marker | Pearson r | Unadjusted P-value | Adjusted P-value |
|---|---|---|---|---|
| Change in aromatase activity | 5'-FR of Exon 1.2 (−588) rs7176005 |
−0.40 | 0.006 | 0.039 |
| |
5'FR of Exon 1.1 (−144) rs6493497 |
−0.40 |
0.006 |
0.039 |
| Change in E1 | 5'-FR of Exon 1.2 (−588) rs7176005 |
−038 | 0.038 | 0.180 |
| |
5'FR of Exon 1.1 (−144) rs6493497 |
−0.38 |
0.038 |
0.180 |
| Pre-treatment aromastase activity | 5'-FR of Exon 1.2 (−588) rs7176005 |
0.33 | 0.028 | 0.151 |
| 5'FR of Exon 1.1 (−144) rs6493497 |
0.33 | 0.028 | 0.151 |
Figure 3.
Genotype-phenotype association study. Quartile box plot was plotted between the genotypes of the two SNPs, rs6493497 (−588) and rs7176005 (−144), and change in aromatase activity measured in tumor tissue pre- and post-AI treatment.
Since rs6493497 and rs7176005 were associated with multiple phenotypes that were related to aromatase activity and aromatase inhibition, we also performed a follow-up study by genotyping these two SNPs using an additional 200 DNA samples isolated from post-menopausal women with breast cancer who were treated with 1.0 mg anastrozole per day in the adjuvant setting. Plasma samples had been collected before and after the initiation of anastrozole treatment for the measurement of E1 and E2 levels as well as plasma anastrozole concentrations, although tissue aromatase enzyme activity was not available for these patients. The MAFs for rs6493497 and rs7176005 were 12% and 13%, respectively, in these 200 samples. The genotype-phenotype association study for these 200 subjects was performed using levels of E1, E2 and change in E1 and E2 before and after anastrozole as phenotypes. Both SNPs were significantly associated with pre- (P=0.028 for rs6493497 and 0.057 for rs7176005) and post-drug E2 (P=0.0002 for both SNPs) concentrations after adjustment for plasma anastrozole concentration (Table 2B). The average plasma concentration of anastrozole in these 200 subjects was 32.2 ng/ml, with a range from 0.0 to 98.8 ng/ml. Specifically, with each added minor allele, rs6493497 and rs7176005 were associated with 13- and 11-fold increases in post-drug E2 level, respectively. We will discuss the possible implications of the genotype-phenotype association results in the Discussion.
Table 2B.
Effect of two SNPs on plasma estrogen levels pre- and post-anastrazole treatment for 200 post-menopausal patients recruited for the Mayo pharmacogenomics study.
| Phenotype | Marker | Multiplicative effect of 1 rare allele increase in phenotype | 95% Confidence interval | P-value for marker effect | P-value for anastrozole effect |
|---|---|---|---|---|---|
| Pre-Estrone | rs6493497 | 1.20 | (1.00,1.45) | 0.055 | --- |
| |
rs7176005 |
1.14 |
(0.95,1.36) |
0.15 |
--- |
| Pre-Estradiol | rs6493497 | 1.31 | (1.03,1.67) | 0.028 | --- |
| |
rs7176005 |
1.24 |
(0.99,1.56) |
0.057 |
--- |
| Post- Estrone* | rs6493497 | 3.38 | (0.79,14.36) | 0.099 | 0.63 |
| |
rs7176005 |
3.02 |
(0.78,11.68) |
0.110 |
0.63 |
| Post- Estradiol* | rs6493497 | 12.58 | (3.39,46.68) | 0.0002 | 0.93 |
| rs7176005 | 10.88 | (3.12,37.94) | 0.0002 | 0.80 |
Adjusted for anastrozole plasma level.
Functional characterization of rs6493497 and rs7176005
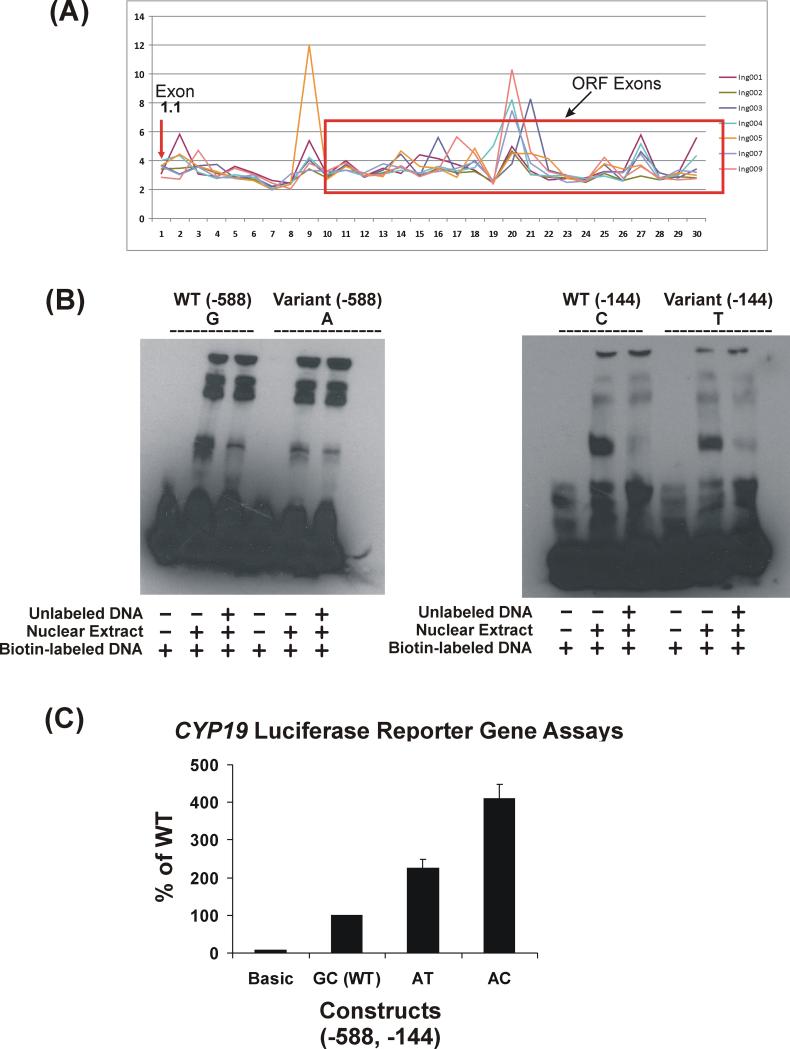
To further understand the functional effect of the two significant SNPs located in the 5’-FR of CYP19 exon1.1, we performed a series of functional genomic studies. These SNPs are located in the major “placental” promoter, exon1.1, so we first attempted to determine whether this exon was expressed in breast tumor tissue using exon array data obtained with seven RNA samples isolated from breast tumor tissue, a subset of the tumor samples included in this study. Figure 4A shows these exon array data and indicates that, although the expression of this exon in breast tumor tissue was low, as shown by the arrow, it was clearly expressed. We then performed EMSA to test for possible DNA-protein binding within the regions containing these two SNPs using nuclear extract from the SKBR03 cells. SKBR03 cells have WT sequence for both of these SNPs (Figure 4B). We chose this cell line since a previous study performed by Kinoshita and Chen demonstrated that aromatase is highly expressed in these cells and that estrogen can upregulate aromatase expression through CYP19 promoter 1.1 (28). EMSA showed that the WT sequence at position (−588) displayed greater DNA-protein binding than did the variant sequence. This effect was less striking for the SNP at position (−144), although binding for the WT sequence was slightly stronger than for the variant (Figure 4B). Reporter gene assay indicated that the AT and AC combinations both displayed increased transcription activity, with AC having the most significant effect (Figure 4C), i.e., that the effect on transcription was most striking for the (−144) SNP. The TC combination had not been observed in any of the patients. Therefore, we did not study that haplotype.
Figure 4.
Functional characterization of CYP19 rs6493497 (−588) and rs7176005 (−144). (A) Exon array analysis of CYP19 expression in 7 breast tumor samples. Exon array probe sets (1-30) for CYP19 are plotted against expression levels. The arrow indicates probe sets for exon 1.1. The red rectangular box indicates the coding region of the gene. The cutoff for the expression level is 3. (B) EMSA of SNPs at (−588) and (−144) upstream of exon 1.1. EMSA were performed using biotin-labeled probes containing WT or variant sequences using nuclear extract from SKBR03 Cells. Competition reactions were performed with 400-fold excess of unlabeled probes. (C) Reporter gene assays. Reporter gene constructs were created for WT (GC) and the observed haplotypes (AT and AC) for the rs6493497 and rs7176005 SNPs. Luciferase activity was corrected for Renilla luciferase activity and is expressed as a percent of the WT activity. Values are mean ± SEM for 3 independent experiments.
Discussion
Breast cancer remains the most common cancer among women, and the majority of post-menopausal women have tumors that are ER(+) and, as a result, are candidates for endocrine therapy (29). Over the past decade, AIs have become established in both the metastatic and adjuvant settings and are a main focus of research in the prevention setting in post-menopausal women at high risk of developing breast cancer (3, 4, 30). Aromatase (CYP19) catalyzes the conversion of androgens to estrone (E1) or estradiol (E2) (31). Therefore, inhibition of aromatase has become a mainstream treatment of post-menopausal women with ER(+) breast cancer. Third generation aromatase inhibitors (AIs) include the steroid derivative, exemestane, and the nonsteroidal agents, letrozole and anastrozole. However, response to treatment with these drugs varies widely, ranging from lack of efficacy to severe side effects (32, 33). One possible explanation is that genetic variation in the genes encoding the drug target, or drug-metabolizing enzymes, might contribute to this variation in response. Previous resequencing studies of CYP19, identified 88 SNPs using 240 ethnically-defined germline DNA samples (18). In the current study, we expanded our previous resequencing effort to include DNA isolated from breast tumor and normal tissue to test the hypothesis that genetic variation in CYP19 might play an important role in variation in aromatase enzyme activity, estrogen production and clinical response. Our study took advantage of a unique sample set collected at the University of Edinburgh (5). These samples were collected in a neoadjuvant setting, with information on in vivo E1 and E2 levels, aromatase activity in the tumor and tumor size pre- and post-AI treatment.
Resequencing of CYP19 in these samples identified a total of 48 SNPs, including 4 novel SNPs as compared with previous resequencing efforts (Figure 1 and Supplementary Table 4). Functional characterization of a novel nonsynonymous cSNP, Arg128, failed to show an effect on aromatase enzyme activity, apparent Km value or Ki values with letrozole (Figure 2). However, genotype-phenotype association studies performed using 27 SNPs identified during resequencing with in vivo and clinical data for 45 patients as phenotypes, identified two tightly linked SNPs, rs6493497 and rs7176005, that were significantly associated with a greater change in aromatase activity before and after AI treatment (Table 2A and Figure 3), with adjusted P values=0.038 for both SNPs. These same SNPs were also moderately associated with change in E1 level and with baseline aromatase activity in these tumor samples. These two SNPs were associated with higher basal aromatase activity, compatible with the results of our luciferase reporter gene assay which showed high activity for the variant SNPs, as well as greater inhibitory effect of AIs, which might be due to the elevated baseline level of aromatase activity (Figure 3 and Figure 4B and C). However, these two SNPs were not associated with levels of either E1 or E2 pre- or post-AI treatment in tumor tissue, which could be due to the small sample size since E1 and E2 levels were only available for the 30 samples from patients treated with letrozole or anastrozole.
We next performed a follow-up study with an additional 200 DNA samples obtained from women with breast cancer who had been treated with 1.0mg anastrozole. We had measured plasma E1 and E2 concentrations as well as anastrozole levels in these patients before and after AI treatment. These phenotypes were used to perform a genotype-phenotype correlation study to determine whether the two SNPs identified in the Edinburgh patients might be associated with plasma estrogen concentrations. Although the samples in the follow-up study were germline DNA, we hypothesized that, if the SNPs had functional effects on aromatase activity and inhibition by AIs, they might have similar effects on plasma estrogen levels. We performed the follow-up study with the two SNPs independently in order to avoid multiple comparisons, since they were tightly linked. The results showed that both SNPs were significantly associated with plasma E2 level before and after anastrozole treatment (Table 2B), adjusted for plasma drug concentrations, a potential confounding variable. We observed this association only with E2 and not E1, which might be due to different precursors for E1 and E2, with E1 being formed from androstenedione and E2 from testosterone. Both SNPs were associated with higher E2 levels, particularly after treatment with anastrozole (Table 2B), which could be related to our observations with the tumor DNA in which the two SNPs were moderately associated with higher aromatase activity at baseline. Although inhibition was greater with samples harboring these SNPs in tumor tissue (Figure 3), this might not result in lower estrogen levels after AI treatment because of the higher basal aromatase activity for samples with these two SNPs.
We next determined how these two SNPs might affect aromatase function. We performed reporter gene and EMS assays which demonstrated that the two SNPs showed different DNA protein binding patterns for WT and variant sequences – particularly for the (−588) SNP – and that they displayed different transcriptional activity when compared with the WT sequence (Figure 4B and C). These functional studies provided additional evidence that these SNPs might result in increased aromatase transcription and, thus, higher aromatase activity, resulting in higher estrogen production even though the inhibition of aromatase activity was greater in the presence of the SNPs, which might be due to the higher aromatase expression (more drug target for AIs). It would be interesting to determine whether tumor samples containing these SNPs have different CYP19 expression than WT. However, because the quantity of tumor tissue was very limited, we were unable to test that hypothesis. This possibility should be tested in future studies. We did not observe an association with tumor size or change in tumor size before and after AIs, nor did we observe an association with E1 or E2 pre- and post-AI treatment in tumors, which might be due to the small sample size studied. However, the fact that rs6493497 and rs7176005 in the 5’-FR of exon 1.1 of CYP19 were associated with aromatase activity and also displayed an association with plasma E2 levels before and after AI treatment suggests an important functional role for these two SNPs in variation in response to AI therapy. It also raises the possibility that these same SNPs might play a role in risk for breast cancer, since estrogen is a major risk for breast cancer. All of these possibilities need to be tested further in further larger independent patient cohorts.
Supplementary Material
Acknowledgements
This work was supported in part by National Institutes of Health (NIH) grants U01 GM61388 (The Pharmacogenetics Research Network), P50 CA116201 (Mayo Clinic Breast Cancer SPORE), R01 CA138461 and a PhRMA Foundation “Center of Excellence in Clinical Pharmacology” Award.
References
- 1.Geisler J, Haynes B, Anker G, Dowsett M, Lonning PE. Influence of letrozole and anastrozole on total body aromatization and plasma estrogen levels in postmenopausal breast cancer patients evaluated in a randomized, cross-over study. J Clin Oncol. 2002;20:751–7. doi: 10.1200/JCO.2002.20.3.751. [DOI] [PubMed] [Google Scholar]
- 2.Geisler J, King N, Anker G, et al. In vivo inhibition of aromatization by exemestane, a novel irreversible aromatase inhibitor, in postmenopausal breast cancer patients. Clin Cancer Res. 1998;4:2089–93. [PubMed] [Google Scholar]
- 3.Ingle JN, Suman VJ. Aromatase inhibitors for therapy of advanced breast cancer. J Steroid Biochem Mol Biol. 2005;95:113–9. doi: 10.1016/j.jsbmb.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 4.Ingle JN, Dowsett M, Cuzick J, Davies C. Aromatase inhibitors versus tamoxifen as adjuvant therapy for postmenopausal women with estrogen receptor positive breast cancer: meta-analyses of randomized trials of monotherapy and switching strategies. Cancer Res. 2009;69(2 Suppl S):66S. [Google Scholar]
- 5.Miller WR, Dixon JM. Local endocrine effects of aromatase inhibitors within the breast. J Steroid Biochem Mol Biol. 2001;79:93–102. doi: 10.1016/s0960-0760(01)00148-0. [DOI] [PubMed] [Google Scholar]
- 6.Watanabe J, Harada N, Suemasu K, Higashi Y, Gotoh O, Kawajiri K. Arginine-cysteine polymorphism at codon 264 of the human CYP19 gene does not affect aromatase activity. Pharmacogenetics. 1997;7:419–24. doi: 10.1097/00008571-199710000-00014. [DOI] [PubMed] [Google Scholar]
- 7.Kristensen VN, Harada N, Yoshimura N, et al. Genetic variants of CYP19 (aromatase) and breast cancer risk. Oncogene. 2000;19:1329–33. doi: 10.1038/sj.onc.1203425. [DOI] [PubMed] [Google Scholar]
- 8.Probst-Hensch NM, Ingles SA, Diep AT, et al. Aromatase and breast cancer susceptibility. Endocr Relat Cancer. 1999;6:165–73. doi: 10.1677/erc.0.0060165. [DOI] [PubMed] [Google Scholar]
- 9.Siegelmann-Danieli N, Buetow KH. Constitutional genetic variation at the human aromatase gene (Cyp19) and breast cancer risk. Br J Cancer. 1999;79:456–63. doi: 10.1038/sj.bjc.6690071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haiman CA, Hankinson SE, Spiegelman D, et al. A tetranucleotide repeat polymorphism in CYP19 and breast cancer risk. Int J Cancer. 2000;87:204–10. [PubMed] [Google Scholar]
- 11.Bershtein LM, Imianitov EN, Suspitsyn EN, et al. A polymorphism study of the CYP19 gene in endometrial cancer patients. Vopr Onkol. 2000;46:302–5. [PubMed] [Google Scholar]
- 12.Baxter SW, Choong DY, Eccles DM, Campbell IG. Polymorphic variation in CYP19 and the risk of breast cancer. Carcinogenesis. 2001;22:347–9. doi: 10.1093/carcin/22.2.347. [DOI] [PubMed] [Google Scholar]
- 13.Masi L, Becherini L, Gennari L, et al. Polymorphism of the aromatase gene in postmenopausal Italian women: distribution and correlation with bone mass and fracture risk. J Clin Endocrinol Metab. 2001;86:2263–9. doi: 10.1210/jcem.86.5.7450. [DOI] [PubMed] [Google Scholar]
- 14.Miyoshi Y, Ando A, Hasegawa S, et al. Association of genetic polymorphisms in CYP19 and CYP1A1 with the oestrogen receptor-positive breast cancer risk. Eur J Cancer. 2003;39:2531–7. doi: 10.1016/j.ejca.2003.08.017. [DOI] [PubMed] [Google Scholar]
- 15.Thyagarajan B, Brott M, Mink P, et al. CYP1B1 and CYP19 gene polymorphisms and breast cancer incidence: no association in the ARIC study. Cancer Lett. 2004;207:183–9. doi: 10.1016/j.canlet.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 16.Dunning AM, Dowsett M, Healey CS, et al. Polymorphisms associated With circulating sex hormone levels in postmenopausal women. J Natl Cancer Inst. 2004;96:936–45. doi: 10.1093/jnci/djh167. [DOI] [PubMed] [Google Scholar]
- 17.Lloveras B, Monzo M, Colomer R, et al. Letrozole efficacy is related to human aromatase CYP19 single nucleotide polymorphisms (SNPs) in metastatic breast cancer patients. Journal of Clinical Oncology, 2004 ASCO Annual Meeting Proceedings (Post-Meeting Edition) 2004;22:507. [Google Scholar]
- 18.Ma CX, Adjei AA, Salavaggione OE, et al. Human aromatase: gene resequencing and functional genomics. Cancer Res. 2005;65:11071–82. doi: 10.1158/0008-5472.CAN-05-1218. [DOI] [PubMed] [Google Scholar]
- 19.Brueggemeier RW, Richards JA, Joomprabutra S, Bhat AS, Whetstone JL. Molecular pharmacology of aromatase and its regulation by endogenous and exogenous agents. J Steroid Biochem Mol Biol. 2001;79:75–84. doi: 10.1016/s0960-0760(01)00127-3. [DOI] [PubMed] [Google Scholar]
- 20.Hahn EF, Fishman J. Immunological probe of estrogen biosynthesis. Evidence for the 2 beta-hydroxylative pathway in aromatization of androgens. J Biol Chem. 1984;259:1689–94. [PubMed] [Google Scholar]
- 21.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem. 1976;72:248–154. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 22.Westfall RDT Peter H., Rom Dror, Wolfinger Russell D., Hochberg Yosef. Multiple Comparisons and Multiple Tests. Using the SAS System. SAS Institute; Cary, USA: 2000. [Google Scholar]
- 23.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 24.Schaid D J, Rowland C M, Tines D E, Jacobson R M, Poland G A. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–34. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agresti A. Categorical Data Analysis. John Wiley & Sons, Inc.; Hoboken, NJ: 2002. [Google Scholar]
- 26.Wedderburn RWM. Quasi-likelihood functions, generalized linear models, and the Gauss-Newton method. Biometrika. 1974;61:439–47. [Google Scholar]
- 27.Johannessen DC, Engan T, Di Salle E, et al. Endocrine and clinical effects of exemestane (PNU 155971), a novel steroidal aromatase inhibitor, in postmenopausal breast cancer patients: a phase I study. Clin Cancer Res. 1997;3:1101–8. [PubMed] [Google Scholar]
- 28.Kinoshita Y, Chen S. Induction of aromatase (CYP19) expression in breast cancer cells through a nongenomic action of estrogen receptor alpha. Cancer Res. 2003;63:3546–55. [PubMed] [Google Scholar]
- 29.Anderson WF, Chatterjee N, Ershler WB, Brawley OW. Estrogen receptor breast cancer phenotypes in the Surveillance, Epidemiology, and End Results database. Breast Cancer Res Treat. 2002;76:27–36. doi: 10.1023/a:1020299707510. [DOI] [PubMed] [Google Scholar]
- 30.Ingle JN. Endocrine therapy trials of aromatase inhibitors for breast cancer in the adjuvant and prevention settings. Clin Cancer Res. 2005;11:900s–5s. [PubMed] [Google Scholar]
- 31.Simpson ER, Mahendroo MS, Means GD, et al. Aromatase cytochrome P450, the enzyme responsible for estrogen biosynthesis. Endocr Rev. 1994;15:342–55. doi: 10.1210/edrv-15-3-342. [DOI] [PubMed] [Google Scholar]
- 32.Forbes JF, Cuzick J, Buzdar A, Howell A, Tobias JS, Baum M. Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 100-month analysis of the ATAC trial. Lancet Oncol. 2008;9:45–53. doi: 10.1016/S1470-2045(07)70385-6. [DOI] [PubMed] [Google Scholar]
- 33.Coates AS, Keshaviah A, Thurlimann B, et al. Five years of letrozole compared with tamoxifen as initial adjuvant therapy for postmenopausal women with endocrine-responsive early breast cancer: update of study BIG 1-98. J Clin Oncol. 2007;25:486–92. doi: 10.1200/JCO.2006.08.8617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.