Abstract
Patterns in shape and property distributions on the surface of binding sites are often conserved across functional proteins without significant conservation of the underlying amino-acid residues. To explore similarities of these sites from the viewpoint of a ligand, a sequence and fold-independent method was created to rapidly and accurately compare binding sites of proteins represented by property-mapped triangulated Gauss-Connolly surfaces. Within this paradigm, signatures for each binding site surface are produced by calculating their property-encoded shape distributions (PESD), a measure of the probability that a particular property will be at a specific distance to another on the molecular surface. Similarity between the signatures can then be treated as a measure of similarity between binding sites. As postulated, the PESD method rapidly detected high levels of similarity in binding site surface characteristics even in cases where there was very low similarity at the sequence level. In a screening experiment involving each member of the PDBBind 2005 dataset as a query against the rest of the set, PESD was able to retrieve a binding site with identical E.C. (Enzyme Commission) numbers as the top match in 79.5% of cases. The ability of the method in detecting similarity in binding sites with low sequence conservations were compared with state-of-the-art binding site comparison methods.
Keywords: similarity, structure, function, 3D recognition, cross-reactivity, virtual screening, drug discovery
Introduction
Methods for protein binding site characterization and quantitative comparison have been of interest to the scientific community for at least a decade1–12, primarily because chemical environments and binding site shapes are strong determinants of ligand selectivity. However, it still remains difficult to accurately predict protein-ligand interactions. With each method having its limitations, some of which are shown here, having several methods at disposal helps an investigator to choose the most relevant one for the task at hand.
Reliable identification of the similarities between binding sites across families of proteins is crucial for predicting protein function and suggesting potential ligand cross-reactivity13. In the context of practical drug discovery efforts, quantitative characterization of similarities between binding sites occupied by structurally dissimilar ligands can aid in reducing the chance of ligand cross-reactivity that lead to unwanted side-effects. Crystal structures of ligand bound complexes are being solved at an ever-increasing pace14, enabling alternate ligand and ligand substructures to be also identified from the structural similarity of ligand bound active sites13.
Existing structure-based methods for performing quantitative binding site comparisons use two broad representations of the binding site: Type 1, which uses atom, pseudo-center or residue-based representations1–5 and Type 2 that use property-mapped molecular surface-based representations6–8. In the former case, geometric hashing is the algorithm of choice for similarity detection, although statistical and graph-theoretical approaches have also been successfully utilized in this role. These Type 1 methods are designed to detect similarity in residue, residue type or atom composition, and work well where sequence, sequence type, motif or atomic positions are well conserved. However, as has been shown here and in earlier works9, 15, 16, protein binding sites showing no significant conservation in residue or atomic composition and orientation or fold can show remarkable selectivity towards a common ligand. For example, low conservation of amino acids is known to occur in estradiol and adenine binding sites1, 16. In such cases, the property distribution on the binding site surface and shape of the site are the determining factors for ligand selectivity.
There are far fewer methods reported that utilize Type 2 representations than Type 1, primarily because of the computational complexity associated with comparing large densely triangulated surfaces17. For example, Kinoshita et al. 6, 7 have developed a clique-detection based method for binding site comparison, but the algorithm is NP-complete and is therefore difficult to deploy for large scale virtual screening studies. Another method utilizing 3D-Zernike descriptors that is capable of rapidly comparing binding sites has also been recently proposed8. However, this does not simultaneously encode positive and negative values in surface properties but rather combines them linearly – a characteristic that leads it to produce additional false positives in cases discussed here (See Results and Discussion). It also requires scaling of surfaces as a pre-processing step. Consequently, a new methodology is needed that addresses existing limitations while staying within the robust Type 2 (surface based) similarity paradigm. To this end, we report a novel method known as pair-wise “Property-Encoded Shape Distributions” (PESD) which was inspired by the concept of Shape Distributions as originally proposed by Osada et al. 18. By coupling key features of shape distributions with property information a new method was created that yields a shape-property hybrid signature encoding both shape and chemical environment of the binding site surface as a string of numbers. Unlike the 3D Zernike method, PESD simultaneously captures the spatial relationships of multiple surface property distributions for both positive and negative valued properties into a single signature and does not require scaling of surfaces. Spatial relationships of multiple surface properties such as hydrophobicity, polar surface area and hydrogen-bonding donor/acceptor features can thus be simultaneously encoded into a single compact signature. The resulting signature is independent of alignment, translation and rotation and can be rapidly computed, enabling high throughput comparisons of shape and property distributions of binding sites to be made in a computationally efficient and effective manner.
Following a description of the PESD method below, we study the ability of PESD in picking binding sites of identical functions, from a large set of binding sites. With illustrative examples, we also show that PESD can detect conservations in both shape and property distributions across binding sites when no significant similarity exists at the fold, residue or atomic level. Next, in a classification experiment we compare the results of PESD similarity analyses on a set of binding sites from TIM β/α barrel containing proteins with those obtained using 3D-Zernike descriptors and use example cases to highlight how the PESD method can overcome the limitations of the 3D-Zernike method. Finally we show how simple binding site comparison using PESD can be employed to predict cross-reactivity of bound ligands.
Materials and Methods
Dataset and protein preparation
Binding sites from a set of 40 proteins (Dataset 1) of low pair-wise sequence identity binding to a single Nicotinamide Adenine Dinucleotide (NAD), Adenosine tri-phosphate (ATP), heme or steroid molecule from Morris et al. 19 were clustered to determine the optimum weights as outlined in the “parameter optimization” step.
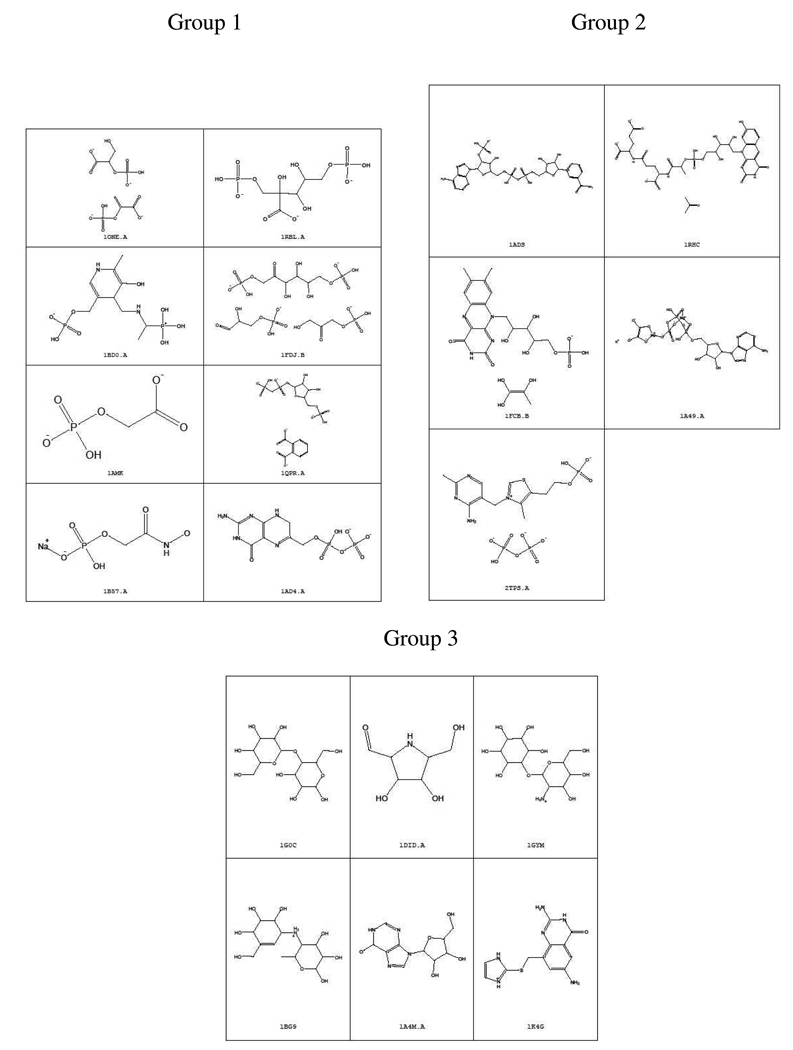
To compare the performance of PESD with 3D-Zernike descriptors, a second dataset (Dataset 2) consisting of 19 binding sites from 19 TIM β/α barrel proteins from the work of Sael et al. 8, was included. In this work, each protein was representative of a family as classified by Nagano et al. 15. Binding sites in this dataset included sites with both single and multiple ligands– see Figure 7. For dataset 1 and dataset 2, side chain ionization states were adjusted to reflect a pH of 7.0 using PROPKA 20.
Figure 7.
Structures of ligands and their corresponding receptor PDB ids for the three binding site groups identified by PESD.
The dataset for virtual screening (Dataset 3) had 1257 complexes, culled from the original set of 1296 complexes in PDBBind v2005.121 after a run of Protonate3D22 in MOE23 for pH dependent protonation state assignment using a SVL script and was used in our earlier works24, 25. The pH values were extracted directly from the PDB14 files of the respective complexes. For structure files not having a pH value, a default pH of 7.0 was used. The electrostatics cutoff was set at 12 Å; all other parameters were set to default. To reduce the computational resources used in preprocessing protein structures, a “sliding scale” in the inclusion of waters on structure optimization was adopted; if the protein structure was of significantly high resolution (less than 2.2 Å resolution and an r-factor of less than 0.29) and was small enough (less than 6000 daltons), all waters were included in subsequent optimization. For all other structures, waters were included only if they were located less than 3.8 Å from the ligand.
Binding site surface generation
All molecular modeling steps were performed in MOE on protein crystal structures. The binding site was defined as all protein atoms within 4.5 Å of any atom of bound ligands in the site.
For each site, two property mapped Gauss-Connolly molecular surfaces were computed26, 27: One with default MOE Active LP (ALP) color coding and other with default MOE Electrostatics (EP) color coding. Color-mapped surfaces generated in MOE are triangulated meshes with each vertex of a triangle having a color-coded property value. The Gauss-Connolly surface is a smooth approximation of the Connolly surface obtained by a sum-of-Gaussians density derived from the atomic coordinates. The ALP surface was encoded by the three colors whose varying shades represented magnitudes of mildly polar, hydrophobic and hydrogen-bonding regions, while the EP surface is encoded with a screened (localized) electrostatic potential represented by another set of three colors, the various shades of which covered electrostatic potential values in the range [−35,35 kcal/mol]. Potential values occurring outside the range were clamped to lie inside the range. The Amber99 force field was used for calculating partial charges in Dataset 1 and 2, while the charges assigned by Protonate3D for Dataset 3 were used as such.
Property-Encoded Shape Distributions
Shape Distributions18 are defined as rotation and translation invariant probability density functions that capture the geometry of a 3D object. An example of this approach is the D2 shape distribution which can be obtained by binning distances of two randomly selected points on the surface of a triangulated three-dimensional mesh. In the PESD method, we make use of a D2 shape distribution where each vertex of the triangulated surface is represented not only by its Cartesian coordinates, but also by a surface property-based “color” code in RGB format representing the magnitude of the surface properties found there. To accomplish this, two points are randomly chosen on a property-encoded surface, after which the Euclidean distance between the two points is calculated, and their color codes are recorded. We employed a coarse-grained binning scheme that utilizes 19 distance bins (24 in the case of screening of Dataset 3 to ensure adequate resolution within poly-peptide binding sites), each 1 Å wide, while the 20th bin (25th bin) records all distances greater than 19 Å (24 Å). Each distance bin is further divided into 4096 sub-bins. For each vertex point the R, G and B values are transformed into integers 1 to 4, producing a coarse-grained “color” (property) space of 64 values. This makes the number of possible combinations of colors for a pair of points (43 × 43 = 4096), each combination being represented by an individual sub-bin. In each case, a total of 50,000 (100,000 in the case of screening Dataset 3) pairs of random points are selected on the surface. The value of each sub-bin, chosen by the combination of color codes of the two points and the distance between them, is incremented by one unit for each pair selected in the random process. When a representative sample of binding site point pairs has been collected, the resulting distribution is dubbed a Property-Encoded Shape Distribution (PESD) signature where the value of each sub-bin is proportional to the probability of a color (or property) being present at a certain distance to another color on the surface of the object (in this case, a binding site). To eliminate bias in surface point selection, the procedure of Osada et al. was utilized18. Within this scheme, the area of each triangle on the surface is calculated and stored as an array of cumulative areas. A number between 0 and the total area is then randomly chosen, and the triangle corresponding to the array of cumulative areas containing that value is selected. A co-planar point within this triangle is then randomly selected using as shown in Equation (1), where r1 and r2 are random numbers and A, B and C are vertices of the selected triangle:
| (1) |
The color of the selected point is then set equal to the color code of the nearest vertex of the triangle.
Signature generation
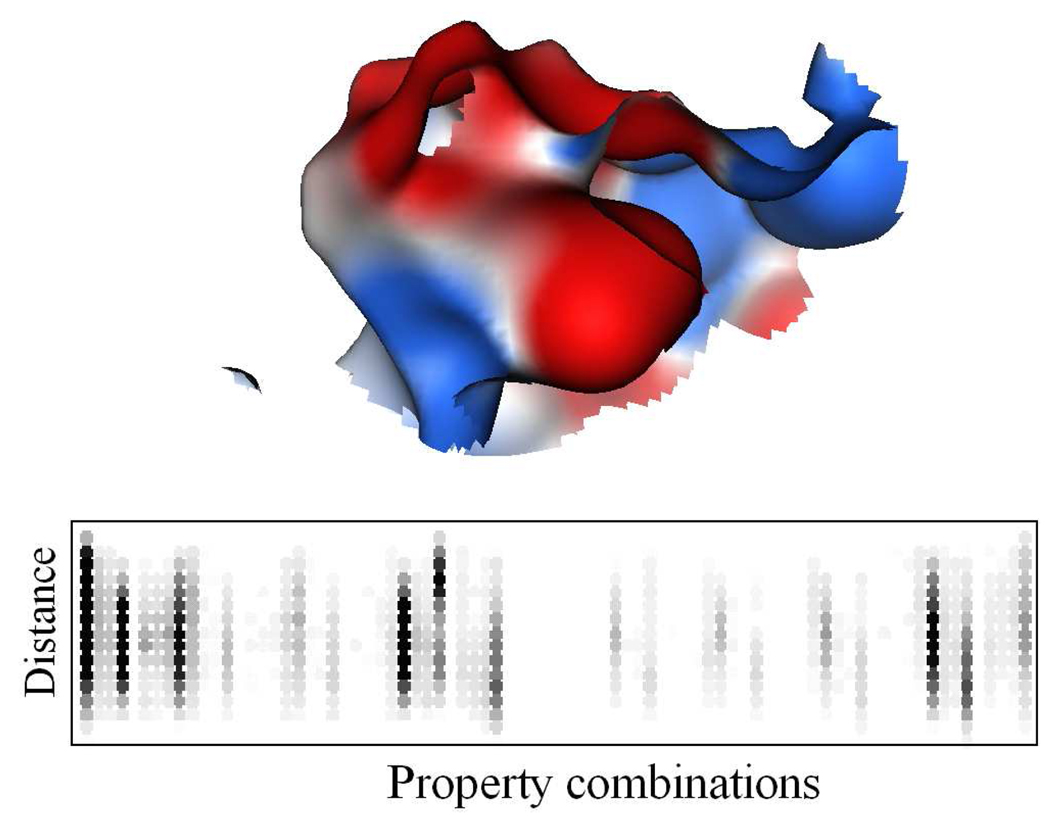
PESD signatures were calculated separately for both EP and ALP surfaces. As a result of coarse-graining, the EP color space was represented by 9 colors giving 81 color/property combinations, while the numbers for ALP surface stood at 14 colors and 196 color/property combinations. A representative EP surface and the graphical representation of the corresponding PESD signature are shown in Figure 1. Darker circles indicate greater magnitude of bin values. PESD property combinations change by columns and point-pair distances increase by rows from top to bottom.
Figure 1.
Electrostatic potential mapped binding site surface of protein neuraminidase (PDB: 1f8b) from influenza virus and the graphical representation of the corresponding PESD signature below it. Each circle represents a bin. Empty bins are white while bins containing non-zero values are represented by various sheds of grey. Any bin having value ≥ 400 is drawn black. PESD property combinations change by columns and point-pair distances increase by rows from top to bottom.
Signature Comparison
The performance of three standard dissimilarity measures, the L1 and L2 norms and the χ2 statistic, was evaluated in their ability to correctly classify binding sites in Dataset 1. The dissimilarity measure d between two signatures H and K, encoding a particular set of properties and having i elements, is shown in Equation 2 with r=1 and with r=2 respectively for the L1 and L2 norms, and in Equation 3 for the χ2 statistic.
| (2) |
| (3) |
Parameter Optimization
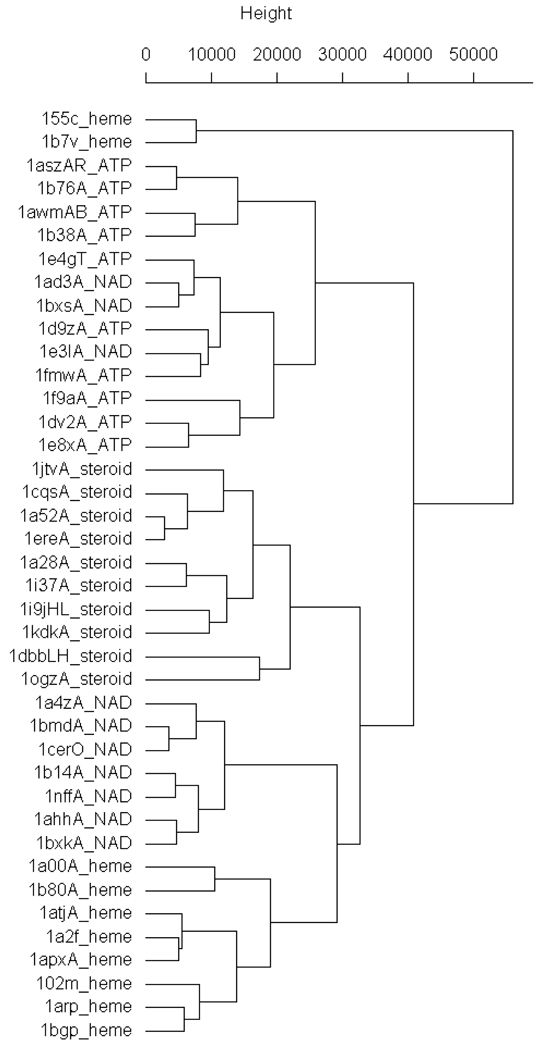
The EP and ALP signatures were combined linearly with weights that were varied until a well-differentiated cluster was obtained for Dataset 1 that corresponded with the nature of the bound ligands. The L1 norm and the χ2 statistic with the weights 1 and 0.7 for respective EP and ALP signatures (Equation 4) produced the best set of clusters (Figure 2) with just three misclassifications (mixing of ATPs and NADs). Two heme binding sites appeared in a separate cluster from the rest as these two sites completely engulfed the ligand (not solvent exposed like the rest) giving them unique shapes. The χ2 statistic produced higher relative differences between clusters of similar binding sites and lower differences within clusters; hence, this measure was applied for all further comparison of binding sites.
| (4) |
Figure 2.
Complete linkage clustering of Dataset 1. Entries in dataset 1 are represented by a four letter PDB code followed by a letter representing the chain on which the binding site is located followed by the type of ligand.
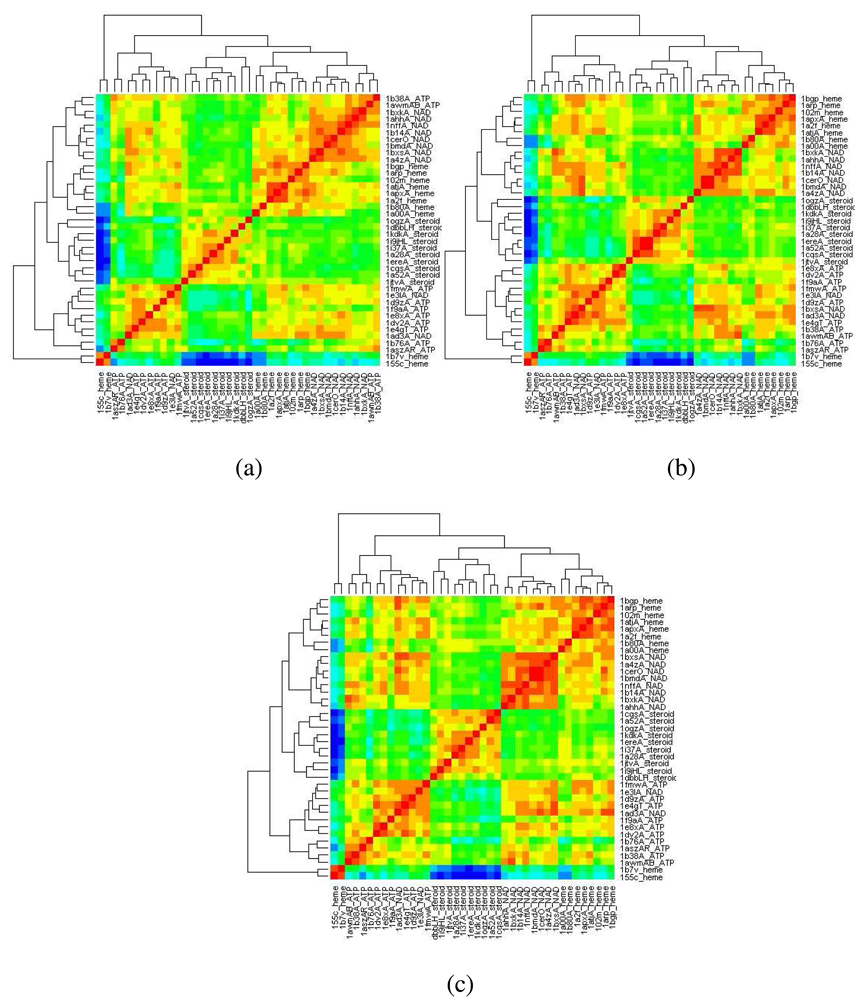
The effect of bin width on classification accuracy was also investigated with bin widths of 0.5 Å, 1 Å and 2 Å. The five primary clusters remained stable for all three bin widths but the difference between different classes decreased on moving from fine to coarse bin width. The trend is shown as heat maps in Figure 3 with overall temperature progressively increasing from Figures 3a to 3c, indicating that a fine bin would be more suitable for in-class comparison. The 1 Å bin was finally chosen to allow for local fluctuations in binding site shape within a class, crystallographic errors and minimal time for distance computation while maintaining reasonable distance between clusters of different classes.
Figure 3.
Heat maps of clustering on Dataset 1 with (a) 0.5 Å bin, (b) 1 Å bin and (c) 2 Å bin PESD signatures.
Clustering
Hierarchical clustering was done on the final combined signature distances using the complete linkage clustering method in R 28.
Results
Computation Speed
Signature generation took about two to four minutes per surface on an Intel 8 core 2.66 GHz based computer. A large part of the time was spent in searching the array of cumulative areas. Segmentation of the array and an introduction of lookup table for the search reduced the computation time to typically 33 seconds per surface. This optimized code is also available for download from our website. Binding site comparison with pre-computed signatures took 0.0056 second per pair-wise comparison on an average.
Virtual Screening
Ability to recall a binding site having the same Enzyme Commission (E.C.) numbers as the query was studied by creating a subset of Dataset 3, such that at least two entries had the same E.C. numbers29. This resulted in a set of 881 binding sites. The E.C. numbers are specific to a certain protein function29 and are not fold dependent. The ability to positively recall a binding site having the same E.C. numbers, in the top rank, by using each member of the 881 binding site set as a query against 1256 other complexes of Dataset 3 was found in 79.5% of the cases (700 out of 881 cases). The results are shown in Table I with different criteria for a positive recall.
Table I.
Ability of PESD to return a binding site with the same E.C. numbers in the top ranks of matched sites.
| Top | 1 | 3 | 1% | 2% | 5% |
|---|---|---|---|---|---|
| Positive (%) | 79.5 | 85.1 | 87.9 | 89.7 | 92.5 |
Case studies
Specific examples are provided to show how the PESD method can correctly identify similarities between binding sites in cases that are missed by state-of-the art binding site comparison algorithms.
Inositol phosphate bound sites
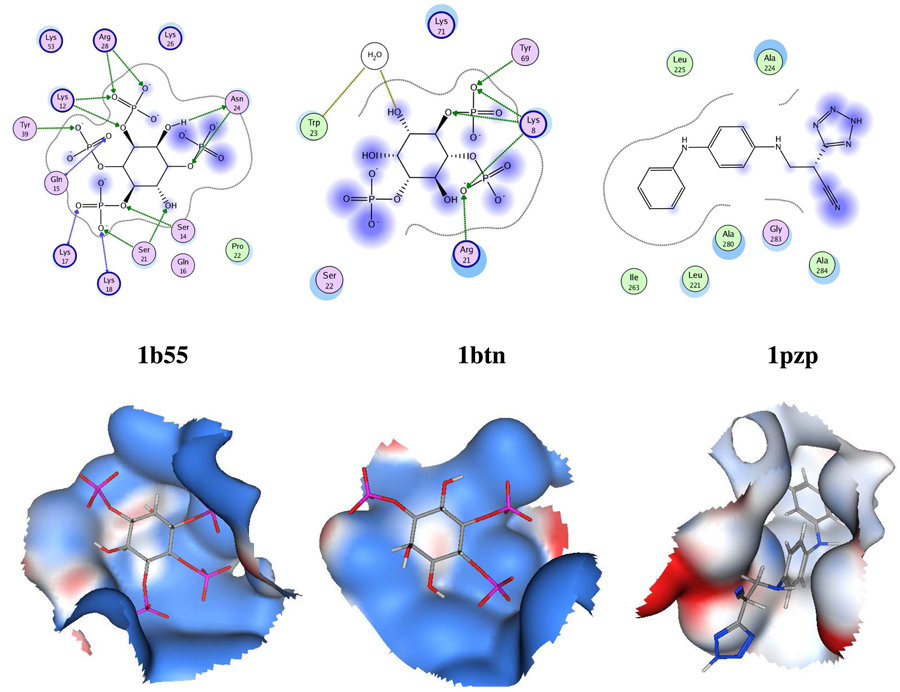
On screening Dataset 3 with the binding site of 1b55 (Pleckstrin Homology (PH) domain of Bruton's tyrosine kinase) in PESD, 1btn (beta-spectrin PH domain) appeared in rank 7. Although the ligand (Inositol 1,3,4,5-tetrakisphosphate) of 1b55 also binds on to the receptor in 1btn30, only three hydrogen bonding residues are conserved (Lysine, Arginine and Tyrosine) in different orientations. In spite of the residue conservation being low, the chemical environment (as depicted by the Electrostatic Potential mapped surfaces in Figure 4) and the shapes of the binding sites in 1b55 and 1btn are similar, which are effectively captured by PESD. Recent methods for binding site comparison such as the Multibind1 and SitesBase5 methods do not find significant similarities between the two sites. The highest similarity score for Multibind (detects similarity by local multiple alignment of binding sites represented as pseudocenters) for example, is 23.2074 which is in fact lower than the Multibind similarity score between 1b55 and an unrelated site from TEM-1 Beta-Lactamase (1pzp, score = 23.849). In PESD, 1pzp appeared in rank 1250 (lower than top 99%) which indicated the dissimilarity between the two sites. SitesBase, which looks for similarities at the levels of atom type and atom position by geometric hashing, found no significant similarities between 1b55 and 1btn and did not report any score.
Figure 4.
Binding site surfaces and schematic diagrams of binding sites of proteins 1b55, 1btn and 1pzp.
1btn at rank 7 is highlighted to show the advantage of the PESD method over other binding site comparison algorithms in a low sequence similarity scenario. Functionally relevant hits were also found in other top ranks. For example, PH domains appeared also in ranks 1 (1w1g), 2 (1mai) and 3 (1w1d), all bound to inositol phosphates. Similar to the query, 1w1g and 1w1d are kinases having a PH domain while 1mai is a phospholipase. Both SitesBase and Multibind detected similarities (with scores of 15 and 31 respectively) between the binding sites in 1b55 and 1w1g. These sites however had higher sequence similarity: 4 (or 5 hydrogen bonding residues common between the two sites if lysine and arginine are considered equivalent). In ranks 4, 5 and 6, were binding sites of 1pgp, 1x8t and 1x8r which did not have a PH domain, but were all bound to ligands that were polyolic phosphates (cf. inositol phosphates). Functionally, 1x8t and 1x8r were transferases (as was the query protein) and 1pgp was an oxidoreductase. Ranks 8 (1bq4 - phosphoglycerate mutase) and 10 (1cru - glucose dehydrogenase) were binding sites of cyclic polyanionic ligands with several carboxylic acid groups arranged in a way similar to phosphates in inositol phosphates, while rank 9 was 1fao (DAPP1/PHISH PH domain) that was bound to inositol 1,3,4,5-tetrakisphosphate. An interesting functional relationship in fact exists between bound ligand of phosphoglycerate mutase in 1bq4 and inositol phosphate: Both the bound ligand (benzene hexacarboxylic acid) and inositol hexakisphosphate inhibit phosphoglycerate mutase comparably31.
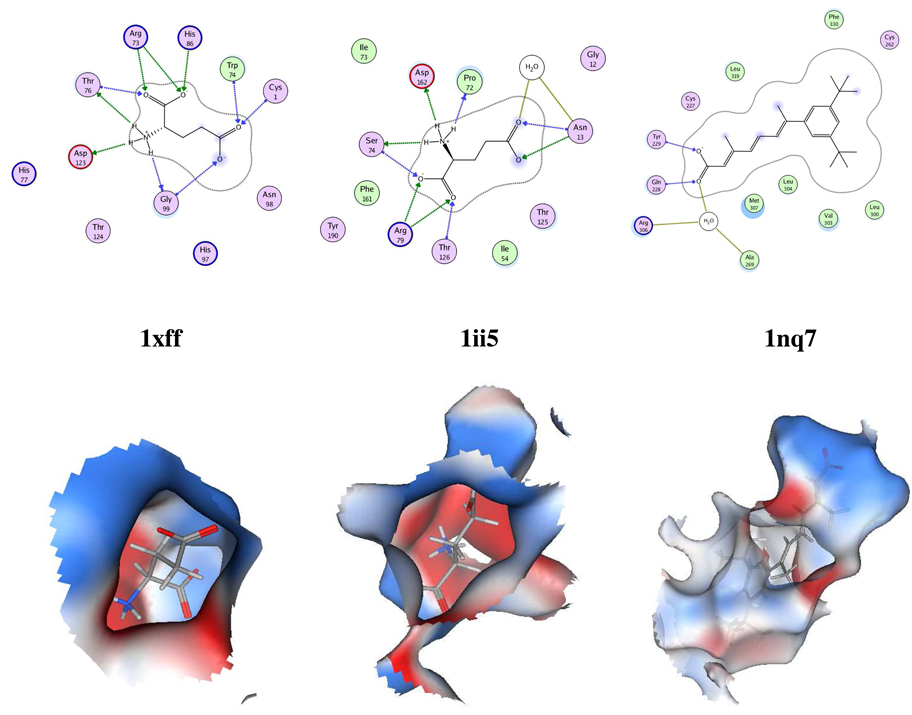
Glutamate bound sites
Another example where low conservation in amino acid composition and orientation is detected by PESD is in the case of binding sites of glucosamine 6-phosphate synthase (1xff, fold: Ntn hydrolase-like) and GluR0, a glutamate receptor ion channel (1ii5, fold: Periplasmic binding protein-like II). Arginine, aspartic acid and threonine are the three hydrogen-bonding amino acids conserved in the two sites (Figure 5). SitesBase did not find significant similarity between the two sites and Multibind similarity score for the two sites is a low 17.3517. The Multibind similarity score between two unrelated sites, 1xff and 1nq7 (orphan nuclear receptor RORbeta) is in fact 17.494. The binding site of 1ii5 appeared within the top 4.1% (rank 51) on screening Dataset 3 by 1xff in PESD, whereas the binding site of 1nq7 appeared only at 98.3% (rank 1234). Here we see that irrespective of binding sites being from proteins with different folds, PESD ranked them as similar when the chemical distributions and shapes of the binding sites were similar.
Figure 5.
Binding site surfaces and schematic diagrams of binding sites of proteins 1xff, 1ii5 and 1nq7.
Similar to the previous case study, functionally relevant hits were also found in ranks higher than 51. For example, among the top 10 ranks, the binding site of glutamine binding protein (1wdn) appeared in rank 2 and glutamate receptors in ranks 5 and 6. The query protein 1xff hydrolyses glutamine to glutamic acid and hence recognizes glutamine. A cysteine bound site appeared in rank 3 (1ssq). Other high ranking PESD hits (1ebg, 1kv5 and 1ik4) included binding sites of highly polar acyclic ligands with terminal acidic groups (cf. glutamic acid) and some sugar binding sites (1bap and 1drj). Of particular interest is rank 1 which was a cysteine bound site of a cysteine transporter (PDB: 1xt8). Although known to bind to amino acids similar to glutamine namely cysteine and serine (neutral and polar), the protein closely resembles glutamine-binding protein in terms of sequence32.
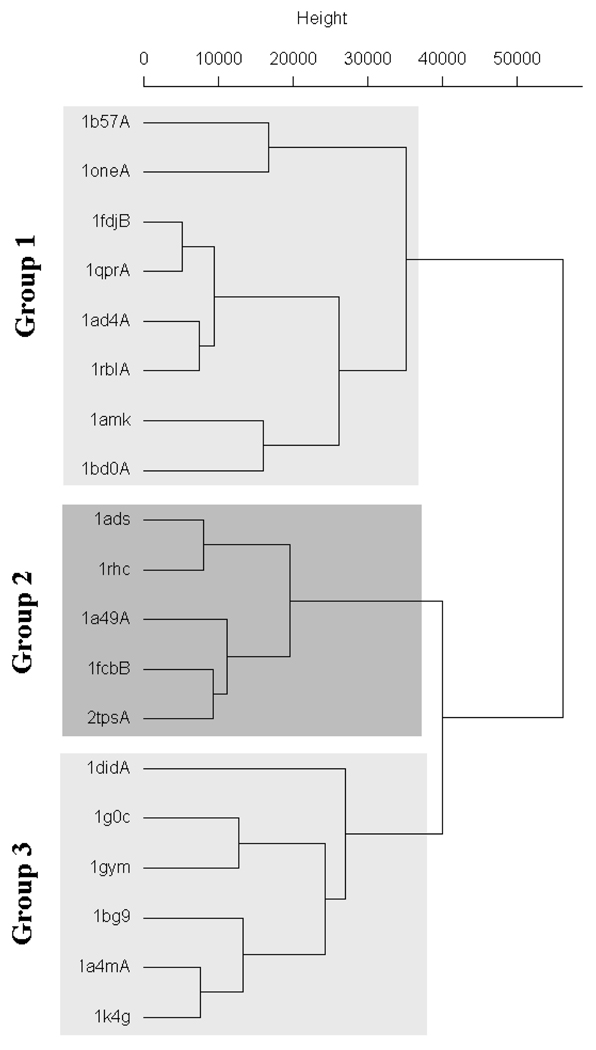
Comparison with 3D Zernike Descriptors
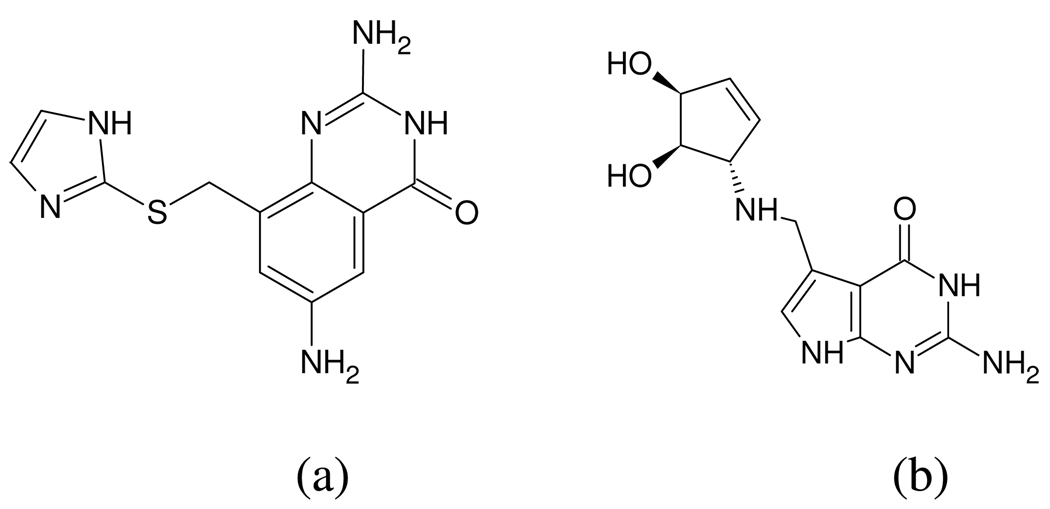
3D Zernike descriptors also encode surface mapped properties as strings of numbers. However, the method did not give as well defined clusters as PESD on the same dataset used by Sael et al.8 In all, PESD signatures were calculated for binding sites of 19 TIM β/α barrel proteins and clustered using the complete linkage hierarchical clustering method (Figure 6). Three groups of clusters were obtained at a chi-squared distance threshold of 37,000. Group 1 consisted of sites binding to smaller sized molecules with terminal phosphate groups. Group 2 comprised of nucleotide and coenzyme binding sites and all of Group 3 comprised of polyol binding sites. The structures of the ligands belonging to each group are shown in Figure 7. Results obtained from 3D Zernike descriptor based clustering by Sael et al.8 which uses only EP surfaces for descriptor generation, does not show such a clear demarcation of bound ligand characteristics among cluster groups. 3D Zernike descriptors had also given three well differentiated groups; however, 1b57 and 1fdj appear in different groups even though they are binding sites of the same protein that are bound to structural analogs. These sites appear together in group 1 in Figure 6. Also, binding sites of enzymes 1fcb and 1rhc appear in different groups in the 3D Zernike descriptors based clustering but appear together in a single group in the PESD results shown in Figure 6. Both these enzymes are dehydrogenases which bind to structurally related coenzymes FMN and F420. Third, the substrate binding site of tRNA-Guanine-transglycosylase (TGT) 1k4g is grouped together with other polyol binding sites in Figure 6 although the bound ligand, which mimics a diol substrate (Figure 8) 33, does not have any hydroxyl groups itself. In the work of Sael et al. the binding site of 1k4g has the polyol binding site of 1g0c as its nearest neighbor but is clustered in group 1 with binding sites of phosphorylated ligands (1one and 1ad4).
Figure 6.
Complete linkage clustering of PESD signatures of the binding sites of 19 TIM β/α barrel proteins showing three groups formed at a chi-squared distance of ~37000.
Figure 8.
(a) Designed ligand (PDB: 1k4g) (b) Natural ligand of tRNA-Guanine-transglycosylase.
Application in cross-reactivity prediction
Binding site comparisons can pin-point sites which are similar in their ability to bind onto the same ligand. For example in the case of the screen with 1b55, 1btn appeared in rank 7. It is experimentally known that the ligand of 1b55 (Inositol 1,3,4,5-tetrakisphosphate) binds onto the binding site in 1btn, even though in the crystal structure 1btn is bound to D-myo-inositol-1,4,5-triphosphate 30. Similarly, a screen with the orphan nuclear receptor RORbeta 1nq7, resulted in 1rdt in rank 2. The ligand bound to the 1nq7 is an analog of all-trans retinoic acid whereas 1rdt is a retinoic acid receptor bound to an agonist. Although the two proteins are bound to different ligands the underlying cross-reactive relationship (both bind to all-trans retinoic acid34, 35) is established by PESD similarity analysis. A way to look at the ability of a binding site comparison algorithm to predict cross-reactivity is to screen with a promiscuous ligand binding site such as ADP or ATP binding sites. Screening with an ATP binding site should not only return binding sites of the same functional class of proteins but also ATP binding sites from other classes. We see a similar recall with PESD: With the ATP binding site in Cystic fibrosis transmembrane conductance regulator (CFTR) nucleotide-binding domain one (PDB: 1r0x) as a query, PESD ranked ATP binding sites in ranks 2, 3 and 9 in the top 10 hits among other nucleotide binding sites from a variety of proteins. Two of the ATP binding sites were from kinases (1b38 in rank 2, 1b39 in rank 9) and one from a synthetase (1t3t in rank 3).
PESD can identify cross-reacting bound ligands even when ligand similarity is low. On screening with 1fmo (adenosine bound to cAMP-dependent protein kinase), 1nvs (staurosporine analog bound to checkpoint kinase Chk1) appeared in rank 11 or within top 0.9% of the database. The Tanimoto coefficient (TC) of the MACCS structural keys36, 37 implemented in MOE of the two ligands is low at 0.5 but it is known that staurosporine binds onto cAMP-dependent protein kinase (1fmo)38. The ROCS39 Shape Tanimoto (0.501) and combo scores (0.690) are also low with the default rank by combo option. However, on screening ligands of Dataset 3 with rank by Tversky(q) score (accounting for subshape match with respect to query ligand) option in ROCS, 1nvs appeared in rank 36 (score=0.896) or within top 2.9% of the database (still 2% lower than PESD).
Discussion
Since binding sites can be represented as a set of points or a surface, several algorithms have been adopted from the field of computer vision into computational analysis of ligand binding sites, with very few reported approaches for comparing surface representation of binding sites17. In this work we have shown that for relatively small sized sites with low sequence conservation (such as 1b55 and 1btn), comparison of surfaces with PESD can lead to correct identification of functional similarity (binding to the same ligand). Geometric hashing and graph matching methods show higher accuracy with increasing number of matched coordinates (more plausible with larger sites), that is, the matching algorithms are “growth” based. Since PESD is not “growth” based, it is especially suitable for detecting surface similarity when underlying conservation in orientation and type of sequences is low.
The PESD method was able to correctly predict the true functional class (E.C. number) for a given query from the best matched binding site in 79.5% of the cases. It is of interest to look at why in 20.5% of the cases this did not happen. Two main reasons for this were: 1) A protein was bound to two functionally different molecules in separate sites and 2) relevant binding sites had large size differences and only partial shape similarity40. The first reason actually bodes well for PESD, since we would not expect PESD to find similarities between functionally different sites of unrelated ligands. An example is the glutaminase domain (1xff bound to glutamate) and isomerase domain (1moq bound to glucosamine 6-phosphate) of glucosamine 6-phosphate synthase. These two sites being the only ones with E.C. numbers 2.6.1.16 in Dataset 3, PESD is unable to assign the correct E.C. number to 1xff. The second reason is a limitation of any global similarity search algorithm such as PESD which is unable to detect partial local similarities when sites being compared differ greatly in size or when there is only partial shape similarity arising out of head to tail overlap of binding sites or large flexible ligands adapting to sites of significantly different shapes40. In the case of binding sites of inosine-5'-monophosphate dehydrogenase in 1lrt and 1me7 41,42, the binding site of the ligand beta-methylene-thiazole-4-carboxyamide-adenine dinucleotide (TAD) in 1lrt overlaps only partially with the binding site of the smaller ligand ribavirin monophosphate in 1me7 in a head to tail fashion. Here, however, the two ligand molecules are also functionally different. While ribavirin is a substrate mimicking inhibitor, TAD is an analog of the coenzyme NAD+. The substrate gets oxidized in an uninhibited enzyme while NAD+ gets reduced. PESD is unable to find a similarity between the two binding sites and assign the correct E.C. numbers to 1lrt since all other sites with the same E.C. numbers in Dataset 3 were substrate binding sites. In spite of the limitation of PESD with respect to partial matching, within the class of global search algorithms, we note that the PESD method gave better resolved clusters than 3D-Zernike descriptors for Dataset 2 and the spherical harmonics based method of Thornton et al.19 and the PocketMatch algorithm of Yeturu et al.43 for Dataset 1.
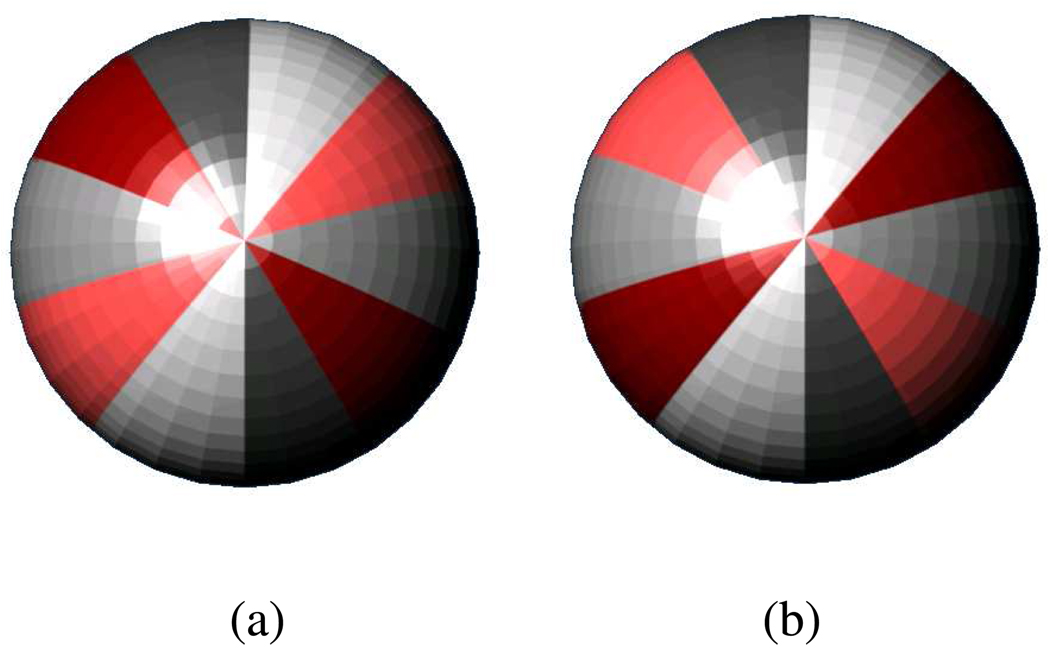
Unlike the surface based 3D-Zernike signatures, PESD is capable of simultaneously encoding spatial relationships of both positive and negative values as a single signature. This prevents misclassification in cases similar to Figure 9. Analogous to colored sphere models described in Sael et al.8, the various shades of grey on spheres in Figures 9a and 9b represent different magnitudes of a single property on a protein surface with a positive sign, whereas the different shades of red represent varying magnitudes with a negative sign of the same property. The Figure 9a and 9b are different in that the order of variation in magnitude of negative property values is reverse with respect to the order of positive property values. Because 3D Zernike descriptor encodes each positive and negative property values separately and then combines them linearly, it will be unable to distinguish between the two cases, and would result in a false positive. PESD on the other hand, is capable of differentiating between the two as the distances between the different shades of colors change in Figure 9a and 9b that is essentially captured by the PESD signatures. Also, the PESD method can create a single signature capturing spatial relationships of multiple surface properties in cases where a single surface mesh has multiple properties or colors mapped onto each vertex. An increase in the number of columns or property combinations in the PESD signature accommodates such cases. For the 3D Zernike descriptors the increased information can only be captured as a linear combination of individual property distributions as in the case of positive and negative values; hence the inter-property spatial relationship is lost. Since 3D Zernike descriptors are inherently scale invariant, a prescreening step to filter out unequal sizes is required to achieve optimal performance44, 45. Since PESD is not scale invariant, the prescreening step is not required.
Figure 9.
The different shades of grey represent different magnitudes of a single property with a positive sign and different shades of red represent varying magnitudes of the same property with a negative sign. The Figures 9a and 9b are different in that the order of variation in magnitude of negative properties is reverse with respect to the order of variation of positive properties.
The suggested uses13 of binding site comparison algorithms are in 1) analysis of protein function, 2) finding alternate ligand and ligand substructures for a binding site and 3) cross-reactivity prediction. We have shown that PESD is applicable to all three areas. With PESD it is possible to find alternate ligand structures from ligands bound to similar binding sites even when the bound ligands are “dissimilar” (having low global similarity score in ligand 2D and 3D similarity analysis algorithms). This was seen in the screening of Dataset 3 with 1fmo bound to adenosine. Thus, where bound crystal structures are available, PESD can be used as a powerful tool for virtual screening in conjunction with ligand based similarity search methods where the high ranking sites or bound ligands can be candidates for further experimental verification.
Conclusions
Given that conservation in atomic or residue arrangements cannot be the only criteria for inferring ligand preferences and hence elucidating function in proteins, we have validated using the PESD method that conserved shape and property distributions on binding site molecular surfaces can account for functional binding site characteristics. PESD is able to create functionally relevant classifications of binding sites from property-mapped binding site surfaces and overcome many known limitations of currently used methods. For the latter purpose, we have evaluated the performance of PESD on datasets of recently published methods8, 19, 43.
In PESD, simultaneous stochastic binning of pair-wise properties and distances provides a probability measure that two properties are present at a certain distance within a binding site, and acts as a site-specific signature. The PESD technique was shown to work on a surface representation extracted from all atoms of a protein, is fold and sequence independent, is alignment free and does not require any reference points - unlike moment invariants46 and spherical harmonics based methods19. PESD is extremely fast, allowing thousands of comparisons to be made in a matter of seconds from pre-computed signatures. The current method is designed for global, and not partial matching. Segmenting a binding site and comparing the segments can overcome this limitation, and will be the subject of future research. In addition, surface representations other than the Gauss-Connolly surface used in the current study are also being investigated.
In summary, PESD has been shown to be a fast and effective shape-property hybrid signature capable of capturing global similarities of binding sites. Its speed and alignment invariance assures that it could become be a valuable virtual screening tool for drug discovery and structural bioinformatics.
Acknowledgements
This work was supported by the National Institutes of Health, Grant number 1P20 HG003899 “Establishment of the Rensselaer Exploratory Center for Cheminformatics Research – RECCR” and was conducted in the RPI Center for Biotechnology and Interdisciplinary Studies (CBIS). The authors would like to acknowledge Dr. N. Sukumar and Dr. Dominic Ryan for assistance and valuable discussions.
Footnotes
Work was done at Department of Chemistry & Chemical Biology / RECCR Center, Center for Biotechnology and Interdisciplinary Studies, Rensselaer Polytechnic Institute, Troy, NY, USA
Availability: Perl and Visual Basic source codes along with instructions are available at http://breneman.chem.rpi.edu/PESD
Contributor Information
Sourav Das, Department of Chemistry & Chemical Biology Rensselaer Polytechnic Institute 110-8th Street Troy, NY 12180.
Arshad Kokardekar, Department of Chemistry & Chemical Biology Rensselaer Polytechnic Institute 110-8th Street Troy, NY 12180.
Curt M. Breneman, Department of Chemistry & Chemical Biology / RECCR Center, Rensselaer Polytechnic Institute, 110-8th Street, Center for Biotechnology and Interdisciplinary Studies, Troy, NY 12180, Phone Number: 518-276-2678, Fax Number: 518-276-4887, brenec@rpi.edu
References
- 1.Shulman-Peleg A, Shatsky M, Nussinov R, Wolfson HM. MultiBind and MAPPIS: webservers for multiple alignment of protein 3D-binding sites and their interactions. Nucleic Acids Res. 2008;36:W260–W264. doi: 10.1093/nar/gkn185. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamdan Y, Wolfson HJ. Geometric hashing: A general and efficient model-based recognition scheme; Proceedings of the IEEE International Conference on Computer Vision; IEEE Press; 1988. pp. 238–249. [Google Scholar]
- 3.Nussinov R, Wolfson HJ. Efficient detection of three-dimensional structural motifs in biological macromolecules by computer vision techniques. Proc. Natl. Acad. Sci. USA. 1991;88:10495–10499. doi: 10.1073/pnas.88.23.10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachar O, Fischer D, Nussinov R, Wolfson H. A computer vision based technique for 3-D sequence independent structural comparison. Protein Eng. 1993;6:279–288. doi: 10.1093/protein/6.3.279. [DOI] [PubMed] [Google Scholar]
- 5.Gold ND, Jackson RM. SitesBase: a database for structure-based protein–ligand binding site comparisons. Nucleic Acids Res. 2006;34:231–234. doi: 10.1093/nar/gkj062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kinoshita K, Furui J, Nakamura H. Identification of proteins functions from a molecular surface database, eF-site. J. Struct. Funct. Genomics. 2001;2:9–22. doi: 10.1023/a:1011318527094. [DOI] [PubMed] [Google Scholar]
- 7.Kinoshita K, Nakamura H. Identification of protein biochemical functions by similarity search using the molecular surface database ef-site. Protein Sci. 2003;12:1589–1595. doi: 10.1110/ps.0368703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sael L, La D, Li B, Rustamov R, Kihara D. Rapid comparison of properties on protein surface. Proteins. 2008;73:1–10. doi: 10.1002/prot.22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moodie SL, Mitchell JB, Thornton JM. Protein recognition of adenylate: an example of a fuzzy recognition template. J. Mol. Biol. 1996;263:486–500. doi: 10.1006/jmbi.1996.0591. [DOI] [PubMed] [Google Scholar]
- 10.Artymiuk PJ, Poirrette AR, Grindley HM, Rice DW, Willett P. A graph-theoretic approach to the identification of three-dimensional patterns of amino acid side-chains in protein structures. J. Mol. Biol. 1994;243:327–344. doi: 10.1006/jmbi.1994.1657. [DOI] [PubMed] [Google Scholar]
- 11.Schmitt S, Kuhn D, Klebe G. A new method to detect related function among proteins independent of sequence and fold homology. J. Mol. Biol. 2002;323:387–406. doi: 10.1016/s0022-2836(02)00811-2. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z, Grigorov MG. Similarity networks of protein binding sites. Protein Struct. Funct. Bioinform. 2006;62:470–478. doi: 10.1002/prot.20752. [DOI] [PubMed] [Google Scholar]
- 13.Shulman-Peleg A, Nussinov R, Wolfson HJ. Recognition of Functional Sites in Protein Structures. J. Mol. Biol. 2004;339:607–633. doi: 10.1016/j.jmb.2004.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, Fagan P, Marvin J, Padilla D, Ravichandran V, Schneider B, Thanki N, Weissig H, Westbrook JD, Zardecki C. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagano N, Orengo CA, Thornton JM. One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J. Mol. Biol. 2002;321:741–765. doi: 10.1016/s0022-2836(02)00649-6. [DOI] [PubMed] [Google Scholar]
- 16.Denessiouk KA, Rantanen VV, Johnson MS. Adenine recognition: a motif present in ATP-, CoA-, NAD-, NADP-, and FAD-dependent proteins. Proteins: Struct. Funct. Gen. 2001;44:282–291. doi: 10.1002/prot.1093. [DOI] [PubMed] [Google Scholar]
- 17.Kahraman A, Thornton JM. Methods to characterize the structure of enzyme binding sites. In: Schwede T, Peitsch MC, editors. Computational Structural Biology. Singapore: World Scientific Press; 2008. pp. 189–221. [Google Scholar]
- 18.Osada R, Funkhouser T, Chazelle B, Dobkin D. Shape Distributions. ACM Trans. Graph. 2002;21:807–832. [Google Scholar]
- 19.Morris RJ, Najmanovich RJ, Kahraman A, Thornton JM. Real spherical harmonic expansion coefficients as 3D shape descriptors for protein binding pocket and ligand comparisons. Bioinformatics. 2005;21:2347–2355. doi: 10.1093/bioinformatics/bti337. [DOI] [PubMed] [Google Scholar]
- 20.Li H, Robertson HD, Jensen JH. Very fast empirical prediction and rationalization of protein pKa values. Proteins: Struct. Funct. Bioinf. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 21.Wang R, Fang X, Lu Y, Wang S. The PDBbind Database: Collection of Binding Affinities for Protein-Ligand Complexes with Known Three-Dimensional Structures. J. Med. Chem. 2004;47:2977–2980. doi: 10.1021/jm030580l. [DOI] [PubMed] [Google Scholar]
- 22.Labute P. Protonate3D: Assignment of ionization states and hydrogen coordinates to macromolecular structures. Proteins: Struct., Funct., Bioinf. 2009;75:187–205. doi: 10.1002/prot.22234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molecular Operating System (MOE), version 2007.09. Montreal, QC: Chemical Computing Group; 2007. [Google Scholar]
- 24.Ryan MD, Hepburn T, Sukumar N, Das S, Breneman CM. TAE Augmented scoring functions: Two approaches, atom and surface based. Abstracts of Papers, 234th ACS National Meeting; August 19–23 2007; Boston, MA, United States. 2007. COMP-42. [Google Scholar]
- 25.Das S, Breneman CM, Ryan MD. TAE Augmented Scoring Functions: Application to Enzymatic and Non-enzymatic proteins. Abstracts of Papers, 235th ACS National Meeting; April 6–10 2008; New Orleans, LA. 2008. COMP-121. [Google Scholar]
- 26.Santavy M, Labute P. Electrostatic Fields and Surfaces in MOE. [accessed Oct 16, 2009];J. Chem. Comput. Group. 1998 [Online], http://www.chemcomp.com/journal/grid.htm.
- 27.Labute P. An Integrated Application in MOE for the Visualization and Analysis of Protein Active Sites with Molecular Surfaces, Contact Statistics and Electrostatic Maps. [accessed Oct 16, 2009];J. Chem. Comput. Group. 2006 [Online], http://www.chemcomp.com/journal/f_surfmap.htm.
- 28.Ripley BD. The {R} project in statistical computing. MSOR Connections. Newsletter of the LTSN Maths, Stats & OR Network. 2001;1:23–25. [Google Scholar]
- 29.Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB). Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the nomenclature and classification of enzymes by the reactions they catalyse. [accessed Oct 16, 2009]; http://www.chem.qmul.ac.uk/iubmb/enzyme/
- 30.Hyvönen M, Macias MJ, Nilges M, Oschkinat H, Saraste M, Wilmanns M. Structure of the binding site for inositol phosphates in a PH domain. EMBO J. 1995;14:4676–4685. doi: 10.1002/j.1460-2075.1995.tb00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rigden DJ, Walter RA, Phillips SEV, Fothergill-Gilmore LA. Polyanionic inhibitors of phosphoglycerate mutase: combined structural and biochemical analysis. J. Mol. Biol. 1999;289:691–699. doi: 10.1006/jmbi.1999.2848. [DOI] [PubMed] [Google Scholar]
- 32.Müller A, Thomas GH, Horler R, Brannigan JA, Blagova E, Levdikov VM, Fogg MJ, Wilson KS, Wilkinson AJ. An ATP-binding cassette-type cysteine transporter in Campylobacter jejuni inferred from the structure of an extracytoplasmic solute receptor protein. Mol. Microbiol. 2005;57:143–155. doi: 10.1111/j.1365-2958.2005.04691.x. [DOI] [PubMed] [Google Scholar]
- 33.Meyer EA, Brenk R, Castellano RK, Furler M, Klebe G, Diederich F. De novo design, synthesis, and in vitro evaluation of inhibitors for prokaryotic tRNA-guanine transglycosylase: a dramatic sulfur effect on binding affinity. Chem. Bio. Chem. 2002;3:250–253. doi: 10.1002/1439-7633(20020301)3:2/3<250::AID-CBIC250>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 34.Stehlin-Gaon C, Willmann D, Zeyer D, Sanglier S, Van Dorsselaer A, Renaud J, Moras D, Schule R. All-trans retinoic acid is a ligand for the orphan nuclear receptor ROR[beta] Nat. Struct. Mol. Biol. 2003;10:820–825. doi: 10.1038/nsb979. [DOI] [PubMed] [Google Scholar]
- 35.Haffner CD, Lenhard JM, Miller AB, McDougald DL, Dwornik K, Ittoop OR, Gampe RT, Xu HE, Blanchard S, Montana VG, Consler TG, Bledsoe RK, Ayscue A, Croom D. Structure-based design of potent retinoid X receptor alpha agonists. J. Med. Chem. 2004;47:2010–2029. doi: 10.1021/jm030565g. [DOI] [PubMed] [Google Scholar]
- 36.MACCS Structural Keys. San Ramon, CA: Symyx Software; [Google Scholar]
- 37.Durant JL, Leland BA, Henry DR, Nourse JG. Reoptimization of MDL keys for use in drug discovery. J. Chem. Inf. Comput. Sci. 2002;42:1273–1280. doi: 10.1021/ci010132r. [DOI] [PubMed] [Google Scholar]
- 38.Fabian MA, Biggs WH, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, Ford JM, Galvin M, Gerlach JL, Grotzfeld RM, Herrgard S, Insko DE, Insko MA, Lai AG, Lelias J, Mehta SA, Milanov ZV, Velasco AM, Wodicka LM, Patel HK, Zarrinkar PP, Lockhart DJ. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat. Biotech. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 39.Rush TS, III, Grant JA, Mosyak L, Nicholls A. A shape-based 3-D scaffold hopping method and its application to a bacterial protein-protein interaction. J. Med. Chem. 2005;48:1489–1495. doi: 10.1021/jm040163o. [DOI] [PubMed] [Google Scholar]
- 40.Kahraman A, Morris RJ, Laskowski RA, Thornton JM. Shape Variation in Protein Binding Pockets and their Ligands. J. Mol. Biol. 2007;368:283–301. doi: 10.1016/j.jmb.2007.01.086. [DOI] [PubMed] [Google Scholar]
- 41.Gan L, Petsko GA, Hedstrom L. Crystal structure of a ternary complex of Tritrichomonas foetus inosine 5'-monophosphate dehydrogenase: NAD+ orients the active site loop for catalysis. Biochemistry. 2002;41:13309–13317. doi: 10.1021/bi0203785. [DOI] [PubMed] [Google Scholar]
- 42.Prosise GL, Wu JZ, Luecke H. Crystal structure of Tritrichomonas foetus inosine monophosphate dehydrogenase in complex with the inhibitor ribavirin monophosphate reveals a catalysis-dependent ion-binding site. J. Biol. Chem. 2002;277:50654–50659. doi: 10.1074/jbc.M208330200. [DOI] [PubMed] [Google Scholar]
- 43.Yeturu K, Chandra N. PocketMatch: A new algorithm to compare binding sites in protein structures. BMC Bioinformatics. 2008;9:543–559. doi: 10.1186/1471-2105-9-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Novotni M, Klein R. 3D Zernike descriptors for content based shape retrieval. ACM symposium on solid and physical modeling; Proceedings of the 8th ACM symposium on Solid modeling and applications; ACM; New York. 2003. [Google Scholar]
- 45.Sael L, Li B, La D, Fang Y, Ramani K, Rustamov R, Kihara D. Fast protein tertiary structure retrieval based on global surface shape similarity. Proteins. 2008;72:1259–1273. doi: 10.1002/prot.22030. [DOI] [PubMed] [Google Scholar]
- 46.Sommer I, Müller O, Domingues FS, Sander O, Weickert J, Lengauer T. Moment invariants as shape recognition technique for comparing protein binding sites. Bioinformatics. 2007;23:3139–3146. doi: 10.1093/bioinformatics/btm503. [DOI] [PubMed] [Google Scholar]