Abstract
Despite extensive research efforts to characterize peripheral regulatory T cells (Treg) expressing transcription factor Foxp3, their subset complexity, phenotypic characteristics, TCR repertoire and antigen specificities remain ambiguous. Here, we identify and define two subsets of peripheral Treg cells differing in Foxp3 expression level and TCR repertoires. Treg cells expressing a high level of Foxp3 and TCRs not utilized by naive CD4+ T cells present a stable suppressor phenotype and dominate the peripheral Treg population in unmanipulated mice. The second Treg subset, expressing a lower level of Foxp3 and utilizing TCRs shared with naive CD4+ T cells constitutes a small fraction of all Treg cells in unmanipulated mice and enriches Treg population with the same antigen specificities as expressed by activated/effector T cells. This Treg subset undergoes extensive expansion during response to antigen when it becomes a major population of antigen-specific Treg cells. Thus, Treg cells expressing TCRs shared with naive CD4+ T cells have a flexible phenotype and may downregulate Foxp3 expression which may restore immune balance at the conclusion of immune response or convert these cells to effector T cells producing inflammatory cytokines.
Keywords: T cells, Tolerance/Suppression/Anergy, Autoimmunity, Transcription Factors
Regulatory CD4+ T cells (Treg) expressing the transcription factor Foxp3 represent a major population of suppressor cells maintaining homeostasis of the immune system (1). The origin, subset composition and functional properties of Treg cells residing in peripheral lymphoid organs of healthy mice are not well known. Previously, two major subsets of Treg cells, natural (nTreg) and adoptive (a Treg) were defined based on whether their suppressor function is acquired during normal T cell development or following TCR stimulation in peripheral tissues or in vitro. Hence, nTreg cells, that have intrinsic suppressor function, arise and mature in the thymus, and a Treg cells are generated from naive CD4+ T cells extrathymically, particularly in conditions of sub-optimal antigen exposure (2–5). Of the two subsets, the nTreg population has undergone the most characterization. However, a recent report showed that thymus-derived nTreg cells may be further subdivided into two functional subsets, which brings into question whether the nTreg population is homogenous (6). In addition, the relative contribution of nTreg and a Treg cells to the peripheral pool of Treg cells in healthy mice remains controversial.
It has been shown that a Treg cells are efficiently generated in vitro upon antigen stimulation of naive cells in the presence of TGF-β and Il-2 and that their suppressor function is similar to that of nTreg cells based on in vitro tests (7, 8). Though, the extent to which these subsets are equivalent in vivo is not known considering their function may be modified by differing homing capacity, antigen specificity and ability to expand in response to antigen and/or inflammatory cytokines. Further shown was the capacity of in vitro generated a Treg cells to suppress antigen-induced T cell activation in cell culture and to protect mice from the development of autoimmune diseases in vivo. However, in those experiments, most cells quickly lost Foxp3 expression in the recipient mice (9, 10).
In vivo upregulation of Foxp3 and conversion to a Treg cells happens during homeostatic expansion, particularly in recipients expressing systemic antigen. Foxp3 can also be upregulated by stimulation with low doses of cognate antigen (4, 5, 11). Some studies have found that peripheral conversion of effector CD4+ T cells (Teff) may also occur in a steady-state, particularly in organs like the respiratory tract and intestine subject to continuous antigen stimulation (12–14). Still another study has shown that in situ conversion to a Treg cells has only a marginal contribution to the expansion of Treg cells in acute inflammation (15). Similarly, analysis of monoclonal T cells specific for islet self-antigen has led some to conclude that peripheral conversion occurs infrequently (16). The conflicting data reported thus far emphasize the need for an experimental model that would allow for further dissection of the peripheral population of Treg cells in the steady state and during response to antigen.
Foxp3 is a transcription factor that regulates the expression of genes involved in the control of multiple cellular and immune functions of CD4 lymphocytes (17, 18). Some data suggest that Foxp3 acts in a dose-dependent manner instead of being a binary switch between Teff and Treg phenotypes (19). Thus, the differences in Foxp3 expression may form the basis for the heterogeneity of the Treg population. Complete demethylation at the Foxp3 locus has been found to be necessary for stable expression of the Foxp3 gene (20). Incomplete demethylation of the Foxp3 locus leads to transient expression of Foxp3 observed in a Treg cells, as well as expression of Foxp3 in activated human T cells that did not have suppressor function (21). Thus, the stability of the Treg phenotype correlates with molecular features at the Foxp3 locus. Weak signaling through the TCR, also responsible for the maintenance of T cells in peripheral tissues, creates conditions for opening chromatin structure at the Foxp3 gene (22). In summary, molecular studies suggest that the level of Foxp3 expression in peripheral CD4+ T cells may vary depending on the environmental signals received through the TCR and could be further modulated by cytokines like TGF-β.
Here, we utilize a new mouse model (Foxp3GFP mice) where we analyze Treg cells in unmanipulated and immunized mice to gain further characterize their properties. This analysis shows that the peripheral population of Treg cells consists of two major subsets that can be discriminated by the level of Foxp3 expression and by the non-overlapping TCRs they express. The Treg subset expressing high levels of Foxp3 presents a stable phenotype of suppressor cells. Treg cells expressing lower levels of Foxp3 express the same TCRs as naive cells suggesting that they represent Teff cells at various stages of differentiation towards becoming a Treg cells. However, some cells within this subset can be diverted into Teff cells when appropriately stimulated. Despite being only a small component in a steady state, this subset expanded quicker than Treg cells expressing an exclusive set of TCRs upon stimulation, thus playing an important role in maintaining the balance between tolerance and immunity.
We propose that peripheral Treg cells expressing an exclusive set of TCRs, not found in the population of naive CD4+ T cells, form the bulk of nTreg population. In contrast, Treg cells sharing the same set of TCRs as naive cells dominate the a Treg subset. Our data also emphasize the importance of thymic selection which endows a developing CD4+ T cell with distinctive properties to become a Treg cell or to function as a Treg or a Teff cell depending on the expression of a particular TCR.
Materials and Methods
Mice
Mice expressing the Foxp3GFP reporter transgene were produced by pronuclear injection of the modified BAC construct encompassing Foxp3 locus into C57BL6 oocytes (submitted). DNA fragment encoding GFP followed by the polyadenylation signal was introduced in frame with Foxp3 translation initiation site into exon 1 of the Foxp3 gene. Foxp3GFP mice expressing a limited TCR repertoire were produced by crossing TCRmini and Foxp3GFP mice (23). Mice were housed under specific pathogen-free conditions and used according to the guidelines of the Institutional Animal Care and Use Committee of the Medical College of Georgia.
Cell purification, flow cytometry and cell sorting
Single-cell suspensions were prepared from lymph nodes by mechanical disruption and cells were stained with antibodies available commercially (eBioscience or BD Biosciences). Cells were analyzed using FACSCanto flow cytometer (Becton Dickinson) and FACSDiva or WinList software. Cells were sorted on a MoFlo cell sorter (Cytomation). Purity of sorted populations exceeded 98.5%. For some experiments, CD4+ T cells were negatively sorted using a commercial kit and an AutoMACS magnetic cell sorter (Miltenyi, Auburn, CA).
Single-cell RT-PCR and TCR sequencing
Single cells from various CD4+ populations were sorted for TCR sequencing as described previously (23). Brachial, axillary and inguinal lymph nodes isolated from two 6 week old TCRmini-Foxp3GFP mice were combined for cell sorting. We analyzed 701 TCRs from naive CD44−CD62L+Foxp3GFP−, 341 from Foxp3GFPlo, 366 from Foxp3GFPhi and 298 from activated CD44+CD62L−Foxp3GFP− T cell subsets of unmanipulated mice. Another two TCRmini-Foxp3GFP mice were analyzed in a similar experiment (data not shown). For analysis of CD4+ T cell subsets in mice undergoing response to antigen we sorted the same CD4+ T cell populations from popliteal lymph nodes draining antigen injection site of two TCRmini-Foxp3GFP mice immunized with Ep63K peptide and CFA. We analyzed 215 TCRs from naive CD44−CD62L+Foxp3GFP−, 144 from Foxp3GFPlo, 64 from Foxp3GFPhi and 254 from activated CD44+CD62L−Foxp3GFP− T cell subsets of immunized mice. DNA sequencing was done in the DNA sequencing core facility at the University of Illinois.
Proliferation assay
Lymph node proliferation assays was performed with total population of lymph node cells from TCRmini mice (4×105 cells/well) incubated with Ep63K peptide (■, ●) (0.1, 1 and 10 µM) in round-bottom 96-well plates. IgGVH(59–74) peptide (□, ○)(0.1, 1 and 10 µM) was used as a control. Proliferation responses were measured by adding 1 μCi/well of 3H-thymidine on day 3 of a 4-day culture.
Inhibition assay
Sorted CD4+Foxp3GFP− cells (5×104/well) were incubated on a 96-well plate with irradiated splenocytes (5×104/well, 3000 Rad) and soluble anti-CD3ε (5 µg/ml). Various numbers of sorted CD4+Foxp3GFP+ cells (1–5×104/well) were added. Cells were sorted using MoFlo sorter. After culturing cells for 3 days, proliferation was measured by adding 1 μCi/well of 3H-thymidine. The percent of cells stimulated was calculated by dividing the proliferation reading from a particular well by the reading from a well with only Foxp3GFP− cells.
In vitro cultures of Treg cells
Flow cytometer sorted CD4+Foxp3GFP−, CD4+Foxp3GFPlo and CD4+Foxp3GFPhi cells were stimulated for 2 days with plate-bound anti-CD3ε/anti-CD28 antibodies alone or in the presence of TGFβ (3 ng/ml). Foxp3GFP expression in cultured cells was assessed by flow cytometry. To examine the TCR repertoire studies of Treg cells that retain or lose Foxp3 expression CD4+Foxp3GFP+ cells were stimulated for 5 days with plate-bound anti-CD3ε/anti-CD28 antibodies in the presence of Il-2 (50 u/ml). Single CD4+Foxp3GFP+ and CD4+Foxp3GFP− cells were sorted for TCR sequencing as described above.
Adoptive transfer
Donor cells for adoptive transfer were isolated by flow cytometry sorting of CD4+Foxp3GFPlo or CD4+Foxp3GFPhi cells from Ly5.1+Foxp3GFP mice and CD4+Foxp3GFP− cells from Ly5.1−Foxp3GFP mice. CD4+Foxp3GFPlo or CD4+Foxp3GFPhi cells (105/mouse) were cotransferred i.v. with CD4+Foxp3GFP− (7.5×105/mouse) cell into recipient TCRα chain knockout mice. To determine the stability of Foxp3 expression, recipient mice were analyzed 4 weeks after adoptive transfer. Weight of the recipient animals was measured over a period of 17 weeks to determine suppressive capacity of Foxp3GFPlo or Foxp3GFPhi cells. To show upregulation of Foxp3 by peripheral naive CD4+ T cells lymphoreplete Ly5.1− TCRmini-Foxp3GFP mice received i.v. transfer of 3×106 Foxp3GFP− cells sorted by flow cytometry from C57BL6 Ly5.1+ Foxp3GFP mice. Lymph node cells of recipient mice were analyzed by flow cytometry 8 days after transfer.
RT-PCR for Foxp3 transcript
Lymph node cells were isolated from Foxp3GFP transgenic mice. RNA was isolated from sorted CD4+Foxp3GFP− and CD4+FoxpGFP+ cells expressing increased levels of the GFP reporter (103 cells/sample). cDNA was produced using the Superscript III CellsDirect cDNA synthesis system (Invitrogen) according to the manufacturer’s instructions. β-actin was used to normalize cDNA quantities. Foxp3 cDNA was amplified with the sense primer 5’ATCCAGCCTGCCTCTGACAAGAACC 3’ and the reverse primer 5’ GGGTTGTCCAGTGGACGCACTTGGAGC 3’. These primers distinguish between the amplification product of the endogenous Foxp3 gene (401 bp) and the transgenic transcript (1357 bp). PCR products were resolved on agarose gels, with the gel being scanned. The resulting gel image was analyzed and DNA bands were quantitated using ImageQuant software (Molecular Dynamics). Relative band intensity was expressed in arbitrary units.
Western blotting
Foxp3 protein was detected in sorted (105 cells/sample) CD4+Foxp3GFP−, CD4+Foxp3GFPlo and CD4+Foxp3GFPhi cells. Cells were lysed in the gel-loading buffer and resolved on 10% polyacrylamide gel. Proteins were transferred onto a PVDF membrane (Millipore). Membranes were probed with anti-Foxp3 antibody eBio7979 (eBioscience) followed by goat anti-mouse polyclonal antibody coupled with horseradish peroxidase (BioRad). Membranes were developed with ECL chemiluminescence kit (Amersham) according to the manufacturer’s instructions.
Cytokine and transcription factor detection by RT-PCR
Production of cytokines in vitro by sorted CD4+Foxp3GFP−, CD4+Foxp3GFPlo and CD4+Foxp3GFPhi T cells was assessed by RT-PCR. Cells were sorted onto 96-well plates (5×104/well) coated with anti-CD3 (10 µg/ml) and anti-CD28 (1 µg/ml) antibodies and cultured under neutral, Th1, Th2 or Th17 conditions for detection of Il-2, IFNγ, Il-4, Il-17 respectively. After 3 days, cells were collected and RNA was isolated with an RNeasy Mini Kit (Qiagen) and reverse transcribed using a Superscript kit (Invitrogen) according to the manufacturer’s instructions. cDNA prepared from cells directly after sorting served as a negative control. β-actin was used to normalize cDNA quantities and was amplified with the sense primer 5’CCTTCTACAATGAGCTGCGTGTGGC3’ and antisense primer 5’CATGAGGTAGTCTGTCAGGTCC3’. Cytokine cDNA was amplified using the following primers: Il-2, sense: 5’CCTTGCTAATCACTCCTCACA’, antisense: 5’GAGCTCCTGTAGGTCCATCA3’, Il-4 sense: 5’CAAGGTGCTTCGCATATTTT3’, antisense: 5’ATCCATTTGCATGATGCTCT3’, Il-17, sense: 5’AGGCCCTCAGACTACCTCAA3’, antisense: 5’CAGGATCTCTTGCTGGATGA3’, IFN-γ, sense: 5’AGTGGAGCAGGTGAAGAGTG3’, antisense: 5’TTCGGAGAGAGGTACAAACG3’. Transcription factor cDNAs were amplified with the following primers: GATA-3, sense: 5’CTCCTTTTTGCTCTCCTTTTC3’, antisense: 5’AAGAGATGAGGACTGGAGTG3’, T-bet, sense: 5’TTCCCATTCCTGTCCTTCACCG3’, antisense: 5’CTGGAAGGTCGGGGTAGAAACG3’, RORγt: sense: 5’GCACCCGCTGAGAGGGCTTCACC3’, antisense: 5’CTGCACTTCTGCATGTAGACTGTCCC3’. cDNA prepared from cells known to produce a particular cytokine or transcription factor was used as a positive control.
Immunization
TCRmini-Foxp3GFP mice were immunized s.c. in the footpad with 1:1 emulsified mixture of Ep63K peptide (5 mM sol. in PBS) and CFA. Popliteal lymph nodes were removed after 7 days and analyzed by flow cytometry. Single cells from various CD4+ T cell subsets were sorted for TCR repertoire studies.
Statistical analysis
The similarity indices, Shanon means and overlap estimators were used as described previously (23). Additional information can be found in the help files for the EstimateS software (http://viceroy.eeb.uconn.edu/estimates).
Testing for symmetry in protein sequence distribution among naive (N) and Foxp3GFP+ (G) cells was done using two approaches. In both of them we consider binomial counts Xi, i=1…m, of the protein sequences in N group for ni trials (hence G group count is simply ni - Xi). We restricted our attention to proteins sequences where ni>l>0 for some fixed integer l.
In the first approach the probability of a random sequence distribution, given the experimental data about TCR distribution, was calculated. We made an assumption that Xi’s are independent with binomial distribution with parameters ni, pi (Xi ~binom (ni, pi)) where pi has an unknown distribution of Fp (pi ~ Fp). We tested the hypothesis H0 of between-groups-symmetry i.e., H0: p=dq (or Fq(1 - x) = 1 - Fp(x)), where q = 1 - p. Using the fact that under the null hypothesis we have P(k zeros in N) = P(k zeros in G), we defined a test statistic for H0 given by: D0 = (number of zeros in N) – (number of zeros in G). It is symmetric on H0, and the p-value was calculated as p-val ≈ #(| D0perm | > |D0obs |)/B where, # is the number of elements for which expression in the parenthesis is true, D0perm is the permutation distribution of D0, that is the approximate distribution (conditional on the data) of D0, recalculated many times (say B) from pairs of counts from data sets N, G but in which the labels were randomly scrambled. For the experimental data where we have taken l = 4, we obtained D0perm = 13 and as p-val = 0.01.
In the second approach, model of a symmetric distribution of TCRs between N and G cells was compared with a model of an asymmetric distribution using a Bayesian approach. Here we assumed that Xi’s are independent with Xi ~binom(ni, pi) where pi ~ βeta(α, β). We compared the models of between-groups-symmetry versus no symmetry i.e., M0: α = β vs. M1: α ≠ β. Assuming m independent priors model, we computed the Bayesian factor (posterior odds) as:
where B(α, β) with (α, β > 0) is a beta function. Using the empirical Bayes ideas we calculated α0 α1, β1 from the data using moment estimates. Noticing that:
we have Denoting and equating the empirical models to the theoretical ones we got
These relations gave a set of estimates α0, α1 and β1 in terms of estimators μ0, μ1, μ2 as follows:
Using l = 4 we obtained log BF(M0, M1) = −9.64. This indicates that the model M0 (symmetry between H and G) is very unlikely against model M1 (no symmetry). This result is consistent with previously performed non-parametric analysis.
To show that the ratio of Foxp3GFPlo versus Foxp3GFPhi cells is increased in the draining (D) versus control (C) lymph nodes in immunized mice, we tested the following hypothesis: H0: P(pD > pC) = ½ versus H0: P(pD > pC) > ½, where pD is the fraction of Foxp3GFPhi cell population in Foxp3GFPlo cells in the draining lymph node and pC is the fraction of Foxp3GFPhi cell population in Foxp3GFPlo cells in the control lymph node. Based on the data, we obtained that the increase in the draining lymph nodes versus control lymph node is significant (p-val = 0.0078).
Permutation tests were used to determine if there are differences in the numbers of cells isolated from lymph nodes or the percentages of CD4+ T cells in blood of Foxp3GFP and TCRmini-Foxp3GFP mice. In the first case we tested the hypothesis that the average numbers of cells in the lymph nodes of Foxp3GFP and TCRmini-Foxp3GFP mice were similar. We obtained that the differences are not statistically significant (p-val =0.258). In the second case the null hypothesis tested that the percentages of CD4+ T cells in blood of Foxp3GFP and TCRmini-Foxp3GFP mice before and after transfer were the same also could not be rejected (p-val =0.37 and 0.18, respectively). To test if adoptive transfer of GFP− cells resulted in an increase in the percentage of CD4+ T cells in recipient mice we performed a sign test. We tested the null hypothesis that the percentages before and after transfer are equal (H0: P(ppost>ppre)=½) versus an alternative outcome that the percentages after transfer are higher (H1: P(ppost>ppre) >½). The p-val =0.6875 indicates that the null hypothesis can not be rejected.
Results
Peripheral Treg cells express a wide range of Foxp3 levels
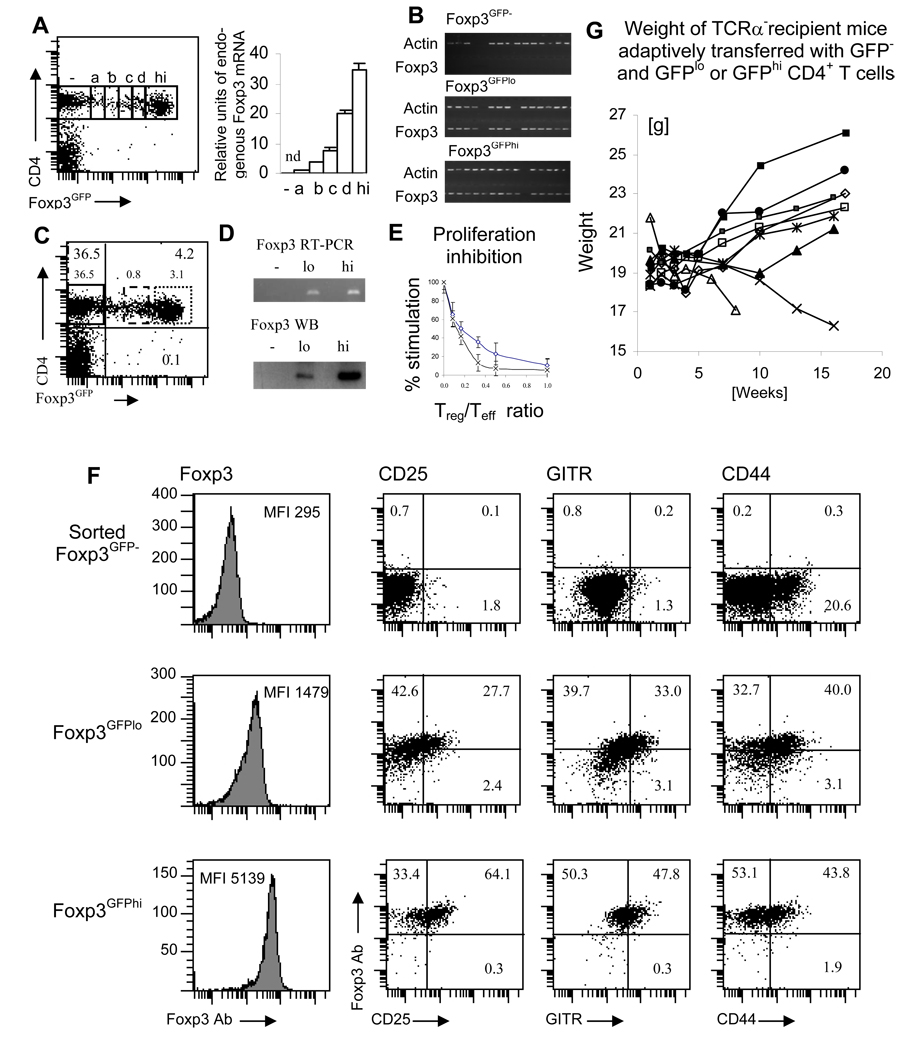
We analyzed peripheral Treg cells in Foxp3GFP mice, who express the GFP reporter driven by the Foxp3 regulatory sequences to investigate the diversity of Foxp3 expression (submitted). The GFP reporter followed by the polyA signal sequence was inserted into exon 1 of the Foxp3 gene on a BAC transgene. The endogenous Foxp3 locus was not modified ensuring proper stability and transcriptional control of the Foxp3 locus. The design of the transgenic construct does not allow for the expression of native or truncated Foxp3 protein. These transgenic mice expressed a broad range of the reporter protein that allowed for the separation of cells based on the level of the reporter GFP. Extensive analyses established that GFP upregulation is tantamount to the expression of the endogenous Foxp3 gene and that the intensity of the GFP fluorescence is proportional to the level of Foxp3 transcript (Fig. 1A). To further show that expression of the Foxp3GFP reporter is induced only in cells expressing endogenous Foxp3, we sorted single cells from CD4+ Foxp3GFP−, Foxp3GFPlo and Foxp3GFPhi populations and amplified their transcripts (Fig. 1B). Next, to determine the amount of Foxp3 transcript and protein, sorted CD4+ Foxp3GFP−, Foxp3GFPlo and Foxp3GFPhi cells were lysed and their Foxp3 expression was quantitated by RT-PCR and Western blotting (Fig. 1C, D). Alternatively, cells were stained with antibody specific for Foxp3 and analyzed by flow cytometry (Fig. 1F). Additionally, we analyzed the conversion of Teff cells into a Treg cells induced in vitro in the presence of TGF-β and Il-2. After 48 hours, all sorted Foxp3GFP+ cells expressed endogenous Foxp3 transcripts and cells remaining Foxp3GFP− cells were devoid of Foxp3 transcripts. In summary, expression of the Foxp3GFP reporter reliably identifies cells expressing native Foxp3 and the level of expression correlates with expression of the Foxp3 gene. Thus, peripheral Treg cells differ in their level of Foxp3 expression.
FIGURE 1.
Analysis of CD4+ Treg cell subsets defined by Foxp3 expression level. (A) The level of the Foxp3 transcript is proportional to the expression of the GFP reporter. Foxp3GFP− and Foxp3GFP+ CD4+ T cells expressing various levels of GFP were sorted and Foxp3 transcript was quantitated by RT-PCR with primers specific for endogenous Foxp3. Sorting gates are shown as rectangles on the histogram. The plot shows relative intensities of DNA bands after scanning the gel image (nd – not detectable). The Foxp3 mRNA level in the cell subset labeled “a” was set as “1”. Experiment was repeated two times. (B) GFP reporter is expressed only in cells expressing endogenous Foxp3 transcript. Single cell analysis of the Foxp3 expression in populations of Foxp3GFP− and Foxp3GFPlo and Foxp3GFPhi CD4+ T cells (sorting gates are shown in C). Actin and Foxp3 were amplified in sorted single cells. (C) Flow cytometry analysis of peripheral lymph node cells stained with CD4. Gates used to sort CD4+ Foxp3GFP− (continuous line), Foxp3GFPlo (broken line) and Foxp3GFPhi (dotted line) Treg cells are shown. (D) Foxp3GFPlo and Foxp3GFPhi Treg cells express low and high levels of Foxp3 transcript and protein. RT-PCR and Western blot analysis of Foxp3 expression in sorted Foxp3GFP−, Foxp3GFPlo and Foxp3GFPhi Treg cells. Experiment was repeated two times. (E) Foxp3GFPlo (open rectangles) and Foxp3GFPhi (crosses) Treg cells suppress proliferation of effector T cells. Typical experiment of three is shown. (F) Foxp3GFPlo and Foxp3GFPhi Treg cells differ in the expression of cell surface markers. Flow cytometry analysis of intracellular Foxp3 expression and cell surface expression of CD25, GITR and CD44. Analysis gates are shown in C. Numbers represent percentage of cells in each quadrant. Representative experiment of three is shown. (G) Weight of the TCRα− recipients receiving adoptive transfer of Foxp3GFPlo (□, ◊, Δ, x, *) and Foxp3GFPhi (■, ▪, ▲, ●) cells and co-transferred with Foxp3GFP− cells into TCRα− mice. Each line represents individual mouse. All recipients receiving FoxpGFPhi cells remained healthy while two (marked by x, Δ) of the recipients of Foxp3GFPlo cells succumbed to wasting disease.
Phenotypic and functional heterogeneity of peripheral Treg cells correlates with the level of Foxp3 expression
We have analyzed the expression of surface markers on CD4+ Foxp3GFP−, Foxp3GFPlo and Foxp3GFPhi cells to further characterize the population of peripheral Treg cells (Fig. 1F). Only Foxp3GFPhi cells expressed high levels of CD25 and GITR, molecules characteristic of Treg cells. The high level of CD25 observed in Foxp3GFPhi cells is consistent with the dependence of Treg cells on exogenous Il-2 for peripheral survival (24). The population of Foxp3GFPlo cells was enriched in cells with little or no CD25 expression, consistent with an earlier report that some Foxp3+ cells do not express CD25 (25). Low CD25 expression on Foxp3GFPlo cells suggests that this cell population may be Il-2 independent. The heterogeneity of Foxp3GFP+ cells shown by flow cytometry analysis suggested that cells expressing various levels of Foxp3 may have differential suppressor capacity depending on the level of Foxp3 expression (26, 27). To determine if both subsets of Foxp3+ cells possess suppressor function we set up an in vitro inhibition assay with Foxp3GFPlo and Foxp3GFPhi cells. Foxp3GFPhi cells had robust suppressor function and a small number of these cells was able inhibit the proliferation of Teff cells (Fig. 1E). However, in order for Foxp3GFPlo cells to achieve the same level of inhibition, more cells were required (Fig. 1F). Similarly, in vivo experiments where Foxp3GFPhi or Foxp3GFPlo cells, along with Foxp3GFP− cells, were co-transferred into lymphopenic mice the same results were observed. Foxp3GFPhi cells protected all recipient mice from autoimmune disease whereas some recipients of Foxp3GFPlo cells succumbed to autoimmune disease (Fig. 1G).
Stability of Foxp3GFPlo and Foxp3GFPhi Treg cell phenotype
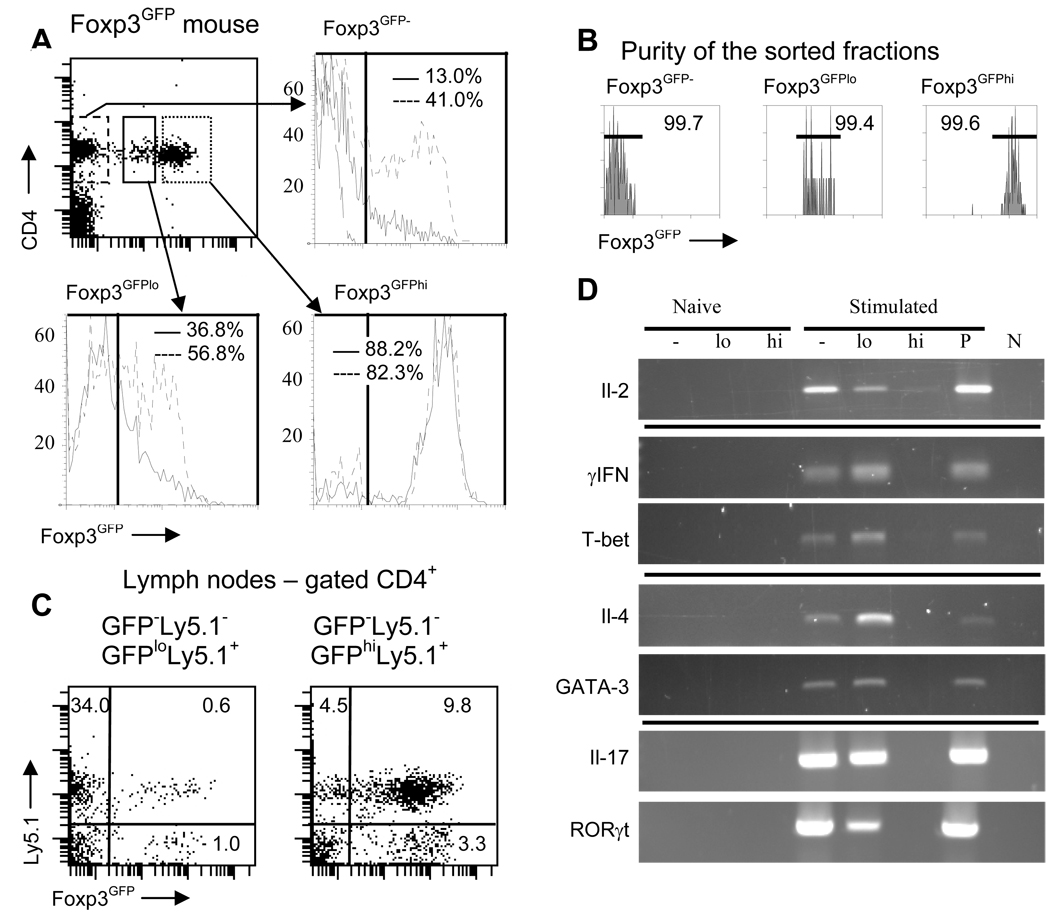
To examine the stability of Foxp3 expression in Foxp3GFPlo and Foxp3GFPhi Treg cells we stimulated cells sorted from relevant populations with plate-bound anti-CD3/anti-CD28 either alone or in the presence of TGF-β (Fig. 2A). Flow cytometry analysis conducted after 2 days of in vitro culture showed a dramatic difference in the stability of Foxp3 expression between Foxp3GFPlo and Foxp3GFPhi cells. While almost all Foxp3GFPhi cells retained Foxp3 expression, Foxp3GFPlo cells lost, retained or upregulated Foxp3 expression. This result and heterogeneous expression of cell surface markers demonstrates that the Foxp3GFPlo subset most likely includes cells at various stages in the developmental pathway to becoming Treg cells. Up- and downregulation of the Foxp3 that occurs in stimulated Foxp3GFPlo cells indicates that commitment towards the Treg phenotype is, at least initially, reversible. High purity of sorted populations and short incubation period exclude a possibility that changes in the Foxp3 expression result from preferential survival or expansion of contaminating cells (Fig. 2B).
FIGURE 2.
Stability of the Treg phenotype of Foxp3GFPlo and Foxp3GFPhi cells. (A) Flow cytometry analysis of Foxp3GFP expression in in vitro cultured sorted CD4+ Foxp3GFP− (dashed line), Foxp3GFPlo (continuous line) and Foxp3GFPhi (dotted line) cells (upper left panel). Cells were stimulated with plate-bound anti-CD3/anti-CD28 antibodies either alone (continuous line) or in the presence of TGFβ and Il-2 (broken line). Foxp3GFP expression in unstimulated CD4+ Foxp3GFP− is shown on the upper right panel (dashed line). Numbers in each plot indicate percentage of Foxp3GFP+ cells. Experiment was repeated three times. (B) Purity of the sorted populations shown in (A). Histograms showing purity of sorted fractions were analyzed on flow cytometer and sorting was done on a cell sorter so the x-axes of the dot plot and histograms have different scale. (C) Foxp3GFP expression in adoptively transferred Foxp3GFPlo (left panel) and Foxp3GFPhi (right panel) cells. Ly5.1+CD4+ Foxp3GFPlo or Ly5.1+Foxp3GFPhi cells were co-transferred with Ly5.1−CD4+Foxp3GFP− cells into TCRα chain knockout mice and recipient mice were analyzed after 4 weeks. Gates used for sorting donor populations are shown in (A). At least three recipient mice were analyzed for each adoptive transfer. (D) Cytokine and transcription factor expression in unstimulated or in vitro stimulated populations of Foxp3GFP−, Foxp3GFPlo and Foxp3GFPhi cells. Sorted cells were lysed directly or were stimulated with plate-bound anti-CD3/anti-CD28 antibodies alone for Il-2 detection or in conditions favoring differentiation of Th1, Th2 or Th17 cells. Population of total CD4+ T cells stimulated like experimental samples served as positive control (P). Cytokine expression was analyzed by RT-PCR. PCR reaction without added template was used as a negative control (N).
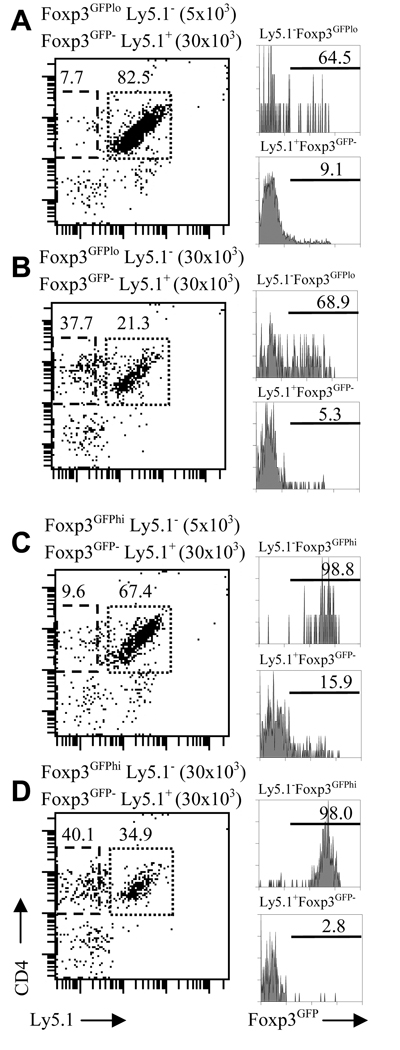
We analyzed Foxp3GFP−, Foxp3GFPlo and Foxp3GFPhi cells in a co-culture assay to further investigate Foxp3 expression in CD4+ T cells subject to activation through the antigen receptor complex in the presence of antigen presenting cells (Fig. 3). Foxp3GFP− cells, expressing an allele-specific marker (Ly5.1+) were stimulated in vitro with soluble anti-CD3 antibody in the presence of a low or high number of Foxp3GFPlo or Foxp3GFPhi cells (both Ly5.1−) and irradiated splenocytes. After 3 days, cells were analyzed for Foxp3 expression by gating on CD4 and Ly5.1 markers. The majority of Foxp3GFPlo (Fig. 3A, B) and almost all Foxp3GFPhi cells (Fig. 3C, D) retained Foxp3 expression. Twice as many Foxp3GFPlo cells retained Foxp3 expression when stimulated in the presence of irradiated splenocytes, in contrast to stimulation with plate bound anti-CD3/anti-CD28 antibodies (Fig. 2A, Fig. 3A, B). These results suggest that interactions with antigen presenting cells, involving accessory signaling molecules, may be particularly important for preserving Treg suppressor function.
FIGURE 3.
Regulation of Foxp3 expression in Foxp3GFP−, Foxp3GFPlo and Foxp3GFPhi cells in in vitro co-culture assays. Foxp3GFP− Teff cells (Ly5.1+, 30×103/well) were stimulated in vitro with soluble anti-CD3 antibody in the presence of low (5×103/well, A, C) or high (30×103/well, B, D) number of Foxp3GFPlo (A, B) or Foxp3GFPhi (C, D) Treg cells (both Ly5.1−) and irradiated splenocytes from TCRα− mice. Panels on the left show CD4 and Ly5.1 expression on cultured cells and gates used to define Ly5.1+ Teff or Ly5.1− Treg cells. Histograms on the right show Foxp3 expression in Foxp3GFPlo (A, B) and Foxp3GFPhi (C, D) cells (upper histogram in each pair) and in Teff cells (lower histograms in each pair).
A substantial fraction of Foxp3GFP− cells stimulated in the presence of a low number of Foxp3GFPlo or Foxp3GFPhi cells upregulated Foxp3 (9.1 and 15.9% respectively) (Fig. 3A, C). This outcome was not a result of contamination of Ly5.1+Foxp3GFP− cells with Foxp3GFP+ cells during cell sorting since the same preparation of Foxp3GFP− cells stimulated in the presence of a high number of Foxp3GFPhi cells did not contain Foxp3GFP+ cells. The fraction of cells upregulating Foxp3 was inversely proportional to the number of Foxp3GFP+ cells in culture implying that activation (blocked by a large proportion of Foxp3+ cells) is necessary for Foxp3 expression. In conclusion, studies of CD4+ T cell activation show that some Teff cells are prone to express Foxp3 after TCR stimulation even in the absence of exogenously added TGF-β.
To investigate the stability of in vivo Foxp3 expression, we analyzed Foxp3GFPlo and Foxp3GFPhi cells transferred into lymphopenic recipients (Fig. 2C). The respective cell populations, expressing Ly5.1, were sorted and co-transferred with Foxp3GFP− cells. Most Foxp3GFPhi cells preserved Foxp3 expression while almost all transferred Foxp3GFPlo cells downregulated Foxp3 expression. Adoptive transfer experiments also demonstrate that some Foxp3GFP− cells upregulate Foxp3 expression in a lymphopenic environment as reported (28). In conclusion, FoxpGFP− and Foxp3GFPlo cells dynamically regulate Foxp3 expression when activated by antigen or when transferred into recipient animals.
Reversible effector functions of the Foxp3GFPlo cell subset
Next, to gain insight into their potential for differentiation, we investigated the ability of Foxp3GFPlo and Foxp3GFPhi subsets to produce inflammatory cytokines. Sorted cells were stimulated in vitro in conditions promoting the generation of Th0, Th1, Th2 or Th17 cells. Cytokine production and expression of transcription factors T-bet, GATA-3 and RORγt was assessed by RT-PCR (Fig. 2D). While Foxp3GFPhi cells were not able to produce inflammatory cytokines and did not express Th lineage-specific transcription factors, Foxp3GFPlo cells produced Il-2, IFN-γ, Il-4 and Il-17. Predicting the extent to which Th cell generation happens in vivo is difficult, but the functions of at least some Treg cells might be modulated by local exposure to antigen and/or cytokines allowing for quick alterations between the Treg/Teff balance in situ. In summary, the level of Foxp3 expression defines two distinct subsets of Treg cells in normal mice. Foxp3GFPhi cells possess a stable suppressor phenotype exhibited by a constant level of Foxp3 expression and the lack of inflammatory cytokine production. In contrast, Foxp3lo cells have the capacity to up- or downregulate Foxp3, when activated by antigen and can be induced to produce inflammatory cytokines. In summary, though we do not know the source of both subsets of peripheral Treg cells their properties resemble nTreg and a Treg cells described in earlier reports.
Analysis of the TCR repertoires expressed by Foxp3GFPlo and Foxp3GFPhi cells provides insight into the origin of peripheral Treg cell heterogeneity
Considering that the phenotype of Foxp3GFPlo cells resembles a Treg cells, it was tempting to speculate that they represent or, at least, are enriched in Teff cells that upregulated Foxp3 expression (10, 13). So far, no surface markers discriminating between Teff cells induced to become a Treg cells and nTreg cells exist. Previous studies have shown a very limited TCR repertoire overlap between Teff and Treg cells (23, 29, 30). However, it was not known if the TCR repertoire correlates with the level of Foxp3 expression. To identify TCRs expressed by the Foxp3GFPlo and Foxp3GFPhi Treg subsets, we have analyzed these populations in TCRmini mice crossed to Foxp3GFP mice (TCRmini-Foxp3GFP mice) (23). TCRmini mice harbor a mini-repertoire of TCR α chains encoded by Vα2.9 and Jα26 (or Jα2) associated with one rearranged TCR Vβ14 chain. Foxp3GFP+ cells accounted for 0.5% of single positive CD4+ thymocytes and about 3.5–4% of all CD4+ T lymphocytes in peripheral lymph nodes. The TCR repertoire of TCRmini-Foxp3GFP mice was analyzed by sorting single CD4+ T cells from naive CD44−CD62L+Foxp3GFP−, activated/memory CD44+CD62L−Foxp3GFP− (Tm) and Foxp3GFPlo and Foxp3GFPhi Treg subsets (Fig. 4). CDR3 regions of TCR α chains were amplified from single cells and sequenced. T cells used in all experiments were isolated from brachial, axillary and inguinal lymph nodes to minimize a possibility that TCRs repertoires are affected by antigens derived from gut or lung flora.
FIGURE 4.
Flow cytometry analysis of lymph node CD4+ T cells from TCRmini-Foxp3GFP mice. The fraction of CD4+ T cells expressing Foxp3GFP and expression of CD44 and CD62L on Foxp3GFP− cells is shown. Gates used to define Foxp3GFP− cells and subsets of naive CD44−CD62L+ and activated CD44+CD62L− cells as well as Foxp3GFPlo and Foxp3GFPhi Treg cells for TCR repertoire studies are shown as rectangles. Figure shows representative data of at least five mice analyzed.
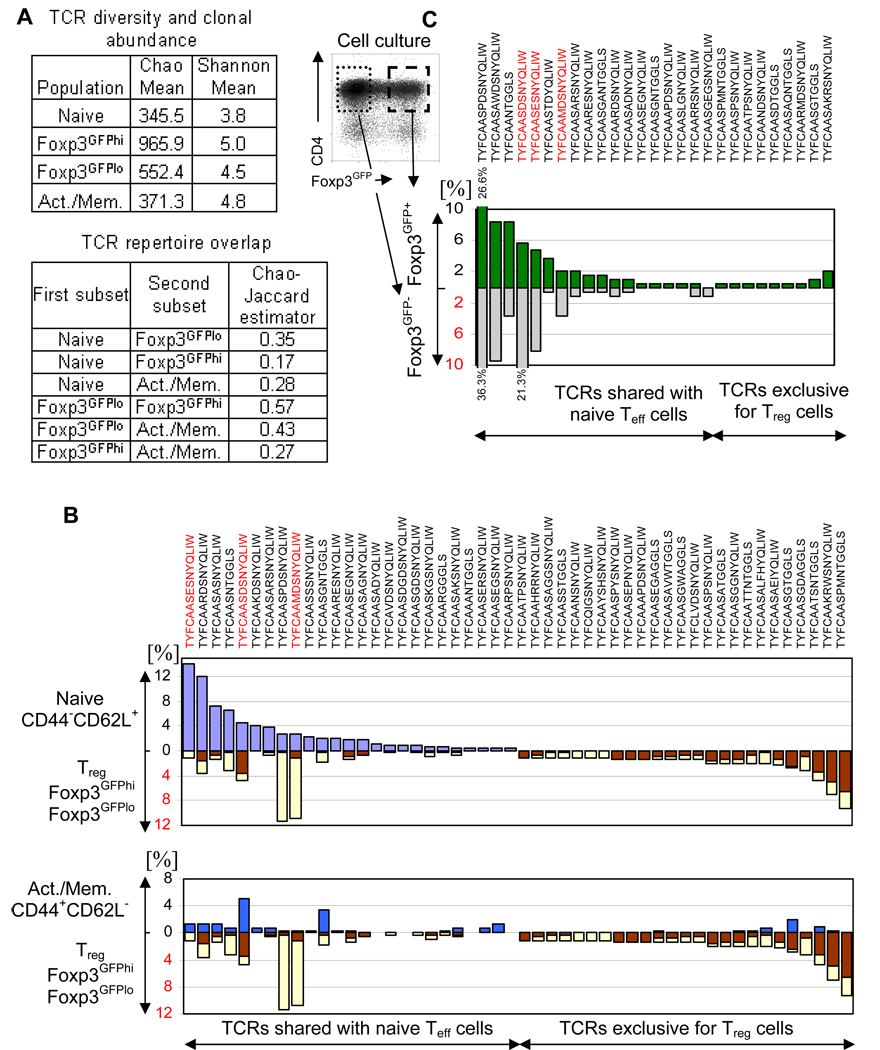
The diversity of TCRs within populations and the overlap between populations were assessed using previously published estimators (Fig. 5A)(23, 31). Naive cells have the lowest diversity (Chao mean) and highest fraction of the TCR repertoire encoded by repetitive clones (lowest Shannon mean). We analyzed the TCR repertoire of naive cells first (Fig. 5B). A significant proportion of TCRs that were common in the naive population were also found in populations expressing Foxp3 and in Tm cells. TCRs characteristic of naive cells constitute 36.4% of TCRs expressed by Foxp3GFPlo cells and 13.4% of TCRs expressed by Foxp3GFPhi cells, consistent with values obtained using Chao-Jaccard estimators (0.35 and 0.17 respectively). Second, we focused on the reciprocal TCR subset, not expressed by naive cells. These TCRs were found to be most frequent in Foxp3GFPhi cells, less frequent in Foxp3GFPlo cells and rare in the Tm population. The repertoire overlap between Teff and Foxp3GFPlo cells was more extensive than between Teff and Foxp3GFPhi cells, suggesting that Treg cells expressing TCRs shared with naive cells, on average, express lower levels of Foxp3 than Treg cells expressing an exclusive set of TCRs. Figure 5B lists the 25 most abundant clones from each population and their frequencies in all analyzed populations. Remarkably, a significant proportion (15 of 25) of TCRs expressed by naive cells was also found in the Foxp3GFP+ population (Foxp3GFPlo or Foxp3GFPhi cells). Large differences in the fraction of Foxp3GFP+ cells existed for T cells expressing individual TCRs. Some abundant naive T cell clones had only a small fraction of Foxp3GFP+ cells while other, less abundant clones, had a large fraction of Foxp3GFP+ cells. This pattern of receptor use implies that the presence of Foxp3GFP+ cells is not a transgenic artifact, affecting all clones equally, but rather that antigen specificity might be important in the recruitment of naive cells into the Foxp3+ population.
FIGURE 5.
Analysis of the TCR repertoire in TCRmini-Foxp3GFP mice. (A) Estimation of the TCR repertoire diversity and clonal abundance (upper table) and TCR repertoire overlap (lower table) of naive and activated/memory Teff cells and Foxp3GFPlo and Foxp3GFPhi Treg cells. (B) Comparison of the frequencies (%) of most abundant TCRs in naive Teff (purple bars) and Treg (brown bars – FoxpGFPhi Treg, yellow bars – Foxp3GFPlo Treg) subsets (upper panel) and in activated/memory (blue bars) subset (lower panel). (C) Partitioning of the most abundant TCRs sequenced from in vitro stimulated Foxp3GFP+ cells (combined Foxp3GFPlo and Foxp3GFPhi cells) between T cells that retained and lost Foxp3 expression. Percentages of a particular TCR in Foxp3GFP− and Foxp3GFP+ subsets are shown on the plot (numbers above and below bars show percentages for the most abundant clones). Sorted Foxp3GFP+ cells were stimulated in vitro. After 5 days, single cells were sorted (sorting gates are shown on the plot) into Foxp3GFP+ and Foxp3GFP− subsets and TCRα chains were amplified and sequenced. Sequences of the TCR α chain CDR3 regions are shown in (B) and (C), red sequences mark T cell clones specific for Ep63K peptide.
Since we observed a larger TCR repertoire overlap than reported, we used two statistical approaches to obtain an objective measure of the significance of the bimodal distribution of receptors between Teff and Treg cell populations. In the first approach, we performed a non-parametric statistical test based on randomization. Using experimental data, we tested the hypothesis that the distribution of TCRs between naive and Foxp3GFP+ (combined Foxp3GFPlo and Foxp3GFPhi cells) T cell subsets is symmetric. This approach found significant deviation from symmetry (p=0.01). In another approach, a model of symmetric distribution of TCRs between naive and Foxp3GFP+ populations was compared with a model of asymmetric distribution between populations using a Bayesian approach. The Bayes factor (ratio of marginal likelihoods for both models) for this analysis was 0.00006517, indicating that an asymmetric distribution is greatly favored over a symmetric distribution. The details of both statistical approaches are described in the Materials and Methods section. The analysis of the TCR distribution reveals that the overlap of repertoires between naive Teff and Treg cells results from contribution of Teff cells to Treg subset (but not vice versa) by upregulation of Foxp3 expression in naive cells. Consequently, the diversity of the Foxp3GFPlo population is higher than the diversity of naive population, consistent with the Chao-Jaccard estimator (Fig. 5A).
Treg cells sharing TCRs with naive cells downregulate Foxp3 in in vitro culture
To determine if the stability of Foxp3 expression and hence the suppressor phenotype depends on the TCR expressed by a particular Treg cell we have stimulated in vitro Foxp3GFP+ cells sorted from TCRmini-Foxp3GFP mice. We analyzed the TCR repertoires of cells that lost or preserved Foxp3 expression (Fig. 5C). Cells that downregulated Foxp3 selectively utilized TCRs shared with the naive population while Treg cells that preserved Foxp3 expression, utilized both TCRs shared with naive T cells, and those found exclusively in Treg cells. In addition, the fraction of Treg cells sharing TCRs with naive T cells was greatly expanded in the subset that remained Foxp3GFP+ at the conclusion of the culture. These cells have a proliferative advantage over Treg cells utilizing the exclusive set of TCRs. In conclusion, the stability of a suppressor phenotype and the ability to expand correlates with the TCR expressed by a particular Treg cell.
Naive CD4+ T cells upregulate Foxp3 in lymphoreplete mice to become a Treg cells
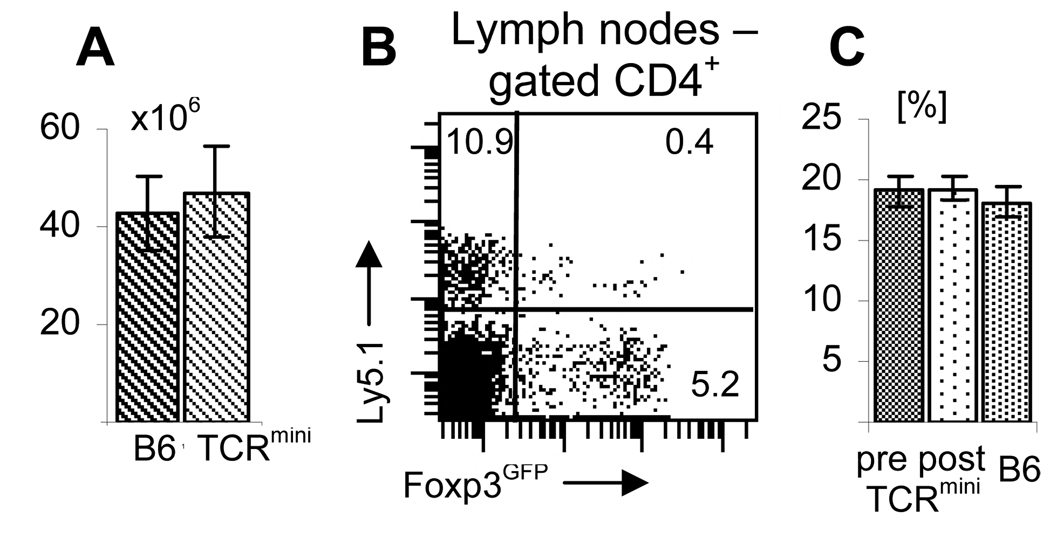
Identical TCRs and the ability to dynamically regulate Foxp3 expression shown in in vitro studies strongly suggest that Foxp3GFP+ cells expressing TCRs shared with naive cells can arise from Foxp3GFP− cells in peripheral tissues. To demonstrate that such a process occurs in lymphoreplete animals, purified CD4+ Foxp3GFP− cells from the wild type Ly5.1+ Foxp3GFP mice were adoptively transferred into Ly5.1− TCRmini-Foxp3GFP mice. Both mice are on the C57BL6 genetic background, have normal number of T lymphocytes and express the same class II MHC/peptide complexes that mediate thymic selection and peripheral maintenance of their T cells (Fig. 6A). Successful engraftment of transferred cells also implies that genetic differences between recipients and donors are minimal, if any. Since the original rearranged TCR that was used to generate DNA constructs to produce TCRmini mice was MHC class II restricted these mice have larger fraction of CD4+ T cells in peripheral lymph nodes than Foxp3GFP mice excluding a possibility of a lymphopenic CD4+ compartment (23, 32). We observed that a significant proportion of transferred Foxp3GFP− cells (3.5%) upregulated Foxp3 when examined 8 days after transfer (Foxp3GFP+ cells constituted 5.8% of recipient CD4+ T cells)(Fig. 6B). The adoptive transfer experiment provides evidence that Foxp3GFP+ cells may be generated in peripheral tissues of lymphosufficient mice from naive CD4+ T cells that upregulate Foxp3. Mixing two populations of CD4+ T cells expressing diverse, polyclonal TCR repertoires most likely leads to interclonal competition. This process may involve some degree of clonal expansion and contraction, however it does not lead to a homeostatic expansion of the CD4+ T cell compartment since the proportion of the CD4+ T cells in the peripheral blood and lymph nodes did not increase (Fig. 6C, data not shown). Wide differences in the level of Foxp3 expression by transferred cells suggest that, in fact, upregulation of Foxp3 may depend on the strength of interaction between the TCR and MHC/peptide complexes.
FIGURE 6.
Foxp3 is upregulated in Foxp3GFP− cells in peripheral lymph nodes of lymphoreplete mice. (A) TCRmini-Foxp3GFP and wild type C57BL6 Foxp3GFP mice have similar number of cells in peripheral lymph nodes. The plot shows average number of lymph node cells isolated from axillary, brachial and inguinal lymph nodes. Twelve mice in each group was analyzed. (B) CD4+ Foxp3GFP− cells upregulate Foxp3GFP expression when transferred into lymphoreplete TCRmini-Foxp3GFP mice. Flow cytometer sorted CD4+ Foxp3GFP− cells (3×106/mouse) from Ly5.1+Foxp3GFP mice expressing wild type TCR repertoire were transferred into Ly5.1− TCRmini-Foxp3GFP expressing restricted TCR repertoire. Recipient mice were analyzed 8 days after transfer. Three recipient mice were analyzed. (C) CD4+ T cells in TCRmini mice reconstituted with sorted CD4+ Foxp3GFP− cells do not undergo homeostatic expansion. Percentage of CD4+ T cells in peripheral blood of recipient mice is shown before (left column) and after (middle column) adoptive transfer. Three recipient mice were examined. For comparison percentage of CD4+ T cells in peripheral blood of Foxp3GFP mice expressing wild type TCR repertoire is shown (right column).
Different clonal dynamics of Treg cells expressing TCRs shared with Teff cells or expressing a Treg restricted repertoire of TCRs during immune response
TCRmini-Foxp3GFP mice were immunized with Ep63K peptide in CFA to determine how both Treg subsets contribute to the immune response. The Ep63K peptide is a cognate antigen for the rearranged TCR that was a prototype for TCR α and β chain constructs used to produce TCRmini mice (32). As a result, the population of CD4+ T cells in TCRmini mice is enriched in cells specific for Ep63K without prior immunization (Fig. 7A). This makes it easier to find Ep63K-specific clones in TCR libraries prepared from various CD4+ T cell subsets. Figure 7B lists CDR3 sequences of α chains of TCRs specific for Ep63K found in TCRmini mice and the frequency of specific Teff cells in naive population. All sequences were obtained from CD4+ T cell hybridomas produced by immunization of TCRmini-Foxp3GFP mice with Ep63K peptide/CFA and their specificity was confirmed by an in vitro Il-2 production assay.
FIGURE 7.
Analysis of the TCR repertoire in TCRmini-Foxp3GFP mice immunized with Ep63K peptide. (A) Naive TCR repertoire in TCRmini-Foxp3GFP mice is enriched in CD4+ cells specific for Ep63K peptide. Lymph node cells from unmanipulated mice proliferate in response to Ep63K peptide (●, ■) but not to a control peptide derived from IgGVH (○, □). Two TCRmini mice were analyzed. (B) Amino-acid sequences of TCRα chain CDR3 regions of Ep63K-specific CD4+ T cell hybridomas obtained from TCRmini-Foxp3GFP mice. (C) Flow cytometry analysis of the draining lymph node cells from TCRmini-Foxp3GFP mice immunized with Ep63K and CFA. CD4+ T cells expressing Foxp3GFP are shown. Gates used to define Foxp3GFP− cells (continuous black line) and subsets of naive CD44−CD62L+ (yellow continuous line) and activated CD44+CD62L− cells (red dotted line) as well as Foxp3GFPlo (dashed line) and Foxp3GFPhi Treg cells (dotted line) for TCR repertoire studies are shown as rectangles. (D) Frequencies (%) of the 25 most abundant T cell clones from activated subset (blue bars) are shown with the frequencies of the Treg clones expressing the same TCR. FoxpGFPhi (brown bars) and Foxp3GFPlo Treg (yellow bars) subsets (upper panel) are shown. (E) Frequencies (%) of the most abundant Foxp3GFP+ Treg clones (combined Foxp3GFPlo and Foxp3GFPhi clones) in the draining lymph nodes of immunized (grey bars) TCRmini-Foxp3GFP mice are compared with frequencies of the same TCRs in Treg subset of unmanipulated mice (light blue bars). TCRs shared with Teff cells and exclusively expressed by Treg cells are indicated by the arrows under the plot. Sequences of the TCRα chain CDR3 regions are shown in (D) and (E), red sequences mark T cell clones specific for Ep63K peptide. (F) Clonal abundance (%) of Ep63K-specific T cell clones in naive (purple bars) and activated (blue bars) subsets of Teff cells and Foxp3GFPlo (yellow bars) and Foxp3GFPhi (brown bars) Treg cells in control TCRmini-Foxp3GFP mice (upper panel) and mice immunized with Ep63K peptide (lower panel). Names of hybridomas expressing a particular TCRα chain are shown above upper panels.
Populations of naive, Tm cells and Foxp3GFPlo and Foxp3GFPhi Treg cells in TCRmini-Foxp3GFP mice were examined by flow cytometry one week after immunization (Fig. 7C). A population of CD4+ T cells isolated from draining lymph nodes had an elevated fraction of activated cells. The Foxp3GFP+ cell subset was not increased compared to control mice. However the fraction of Foxp3GFPlo cells was increased relative to Foxp3GFPhi cells. In all immunized mice (n=7) the Foxp3GFPlo/Foxp3GFPhi ratio was higher in the draining than in the control lymph nodes (0.25 and 0.21, 0.4 and 0.32, 0.33 and 0.3, 0.17 and 0.12, 0.23 and 0.2, 0.22 and 0.17, 0.15 and 0.11). The first number represents Foxp3GFPlo/Foxp3GFPhi ratio in the draining lymph nodes followed by the ratio in the control lymph nodes of the same mouse. This result shows that increased Foxp3GFPlo/Foxp3GFPhi ratio in the draining lymph nodes is highly significant (p-val=0.0078). An increased proportion of the Foxp3GFPlo population most likely suggests that some of naive CD4+ T cells activated by antigen upregulate Foxp3. This interpretation is further supported by the analysis of the TCR repertoire (see below).
We analyzed the repertoires of the naive, activated Teff, Foxp3GFPlo and Foxp3GFPhi Treg subsets to gain insight into the clonal dynamics of CD4+ T lymphocytes. This analysis showed that naive cells in healthy and immunized mice were very similar. Of the 15 most frequent TCRs in unprimed mice, 13 were also found to be the most frequent in the naive population of immunized mice (data not shown). In contrast, comparison of the TCR repertoires of activated populations from unprimed and immunized mice showed a dramatic expansion of Ep63K-specific T cell clones. In particular, clones #274 and #7, that are rare in naive mice, were greatly expanded, suggesting that they represent high-affinity TCRs specific for the Ep63K peptide (Fig. 7D). These clones accounted for 0.1 and 0.3% of naive cells in control mice and for 0.9 and 1.4% of naive cells in immunized mice (Fig. 7F). Clones #274 and #7 were not found among the activated cells in control mice but their fraction increased to 30.7% of all activated cells (19.3 and 11.4%, respectively) in immunized mice (Fig. 7D, F). Notably, the frequencies of Ep63K-specfic clones #31, #36 and #26 in the populations of naive and activated T cells were similar in control and immunized mice. These clones most likely represent low affinity Ep63K-specific clones that did not undergo efficient clonal expansion and though their absolute numbers increased in immunized mice, due to expansion of activated cells, their proportion among activated cells remained similar in healthy and in immunized mice (6.6% and 4.8% respectively). Thus, the immune response to peptide antigen in TCRmini-Foxp3GFP mice follows the paradigm of clonal selection and interclonal competition driven by TCR affinity for antigen (33).
Analysis of the TCR repertoires of Foxp3GFPlo and Foxp3GFPhi Treg subsets in immunized mice showed that Ep63K-specific clones were among the most abundant clones in the population of Treg cells (Fig. 7D, E). Surprisingly, these clones utilized the same TCRs as Teff cells. The proportion of three Ep63K-specific clones (#31, 274, 7) was greatly increased in Foxp3GFP+ cells (15.9%) while two other clones (#26, 36) constituted a similar fraction of Treg cells in immunized and control mice (5.8%)(Fig. 7F). This finding shows that expansion of antigen-specific Teff cells is accompanied by parallel expansion of Treg cells with the same antigen specificity even in conditions of acute inflammation.
In contrast to expanded Treg clones expressing TCRs found in Teff cells, none of the Treg clones utilizing an exclusive set of TCRs was greatly expanded in immunized mice. The three most frequent clones in this subset were represented in control and immunized mice indicating that their abundance is not a result of antigen stimulation. All other clones constituted 1.4% (one clone) or less of the total Treg population (Fig. 7E). The lack of highly abundant clones may be due to the lack of Ep63K-specific clones but this possibility seems unlikely since the TCR repertoire of FoxpGFPhi cells was estimated to be more diverse than that of naive T cells. Another possibility is that Treg cells expressing an exclusive set of TCRs or expressing TCRs shared with naive cells are differentially regulated and clones belonging to the former subset do not expand in the inflammatory environment. Finally, since we do not know the sequences of Ep63K-specific Treg cells expressing an exclusive set of TCRs, we can not exclude that some of Treg clones found in immunized mice are in fact expanded clones specific for Ep63K. However, since none of these Treg clones accounted for a substantial fraction of the Treg subset, we conclude that Treg cells expressing an exclusive set of TCRs have much lower expansion dynamics than antigen-specific Treg cells expressing the same TCRs as Teff cells (Fig. 7F). This interpretation is consistent with our attempts to generate Ep63K-specific clones from Treg cells by using dendritic cells that express a covalent complex AbEp63K to stimulate either CD4+CD25+ cells or the total population of Foxp3GFP+ T cells. This approach produced only clones that expressed the same TCRs found in Ep63K-specific Teff cells (data not shown).
Discussion
To gain insight on the heterogeneity of the peripheral population of Treg cells, we used our Foxp3GFP mouse model and identified two distinct subsets of Treg cells differing in the level of Foxp3 expression. We have correlated these differences with Foxp3 stability, suppressor function, cytokine production and the TCR repertoires they express. Foxp3 was stably expressed in Foxp3hi cells expressing an exclusive set of TCRs. In contrast, within the Foxp3lo population expressing TCRs shared with naive T cells, Foxp3 expression was either up or downregulated when stimulated through the TCR.
While both Foxp3hi and Foxp3lo Treg cells can inhibit lymphocyte proliferation, more Foxp3lo than Foxp3hi cells was required to achieve similar inhibition. This finding is consistent with a previous report showing that Teff cells upregulating Foxp3 in vivo acquire suppressor functions (4). While some Foxp3lo cells downregulated Foxp3 when stimulated, other cells retained or upregulated Foxp3, demonstrating that this cell subset contains genuine suppressor Treg cells and not just cells that transiently upregulated Foxp3. In addition, suppressor function of Foxp3lo cells was similar to the suppressor function of Treg cells expressing the same level of Foxp3 and generated in vitro from sorted Foxp3− cells (data not shown). In conclusion, considering that the level of Foxp3 expression correlates with suppressor function, decreased suppressor activity of Foxp3lo subset is likely due to lower activity of individual Treg cells (34).
Despite similarities in suppressor function our studies have found marked differences between Foxp3hi and Foxp3lo Treg cells in cytokine production. Foxp3lo but not Foxp3hi, cells have the capacity to become Teff cells expressing Th1, Th2 or Th17 cytokines upon the appropriate stimulation in cell culture. Cells producing inflammatory cytokines could either represent conversion of a significant fraction of Foxp3lo cells or expansion of a very small number of these cells. Regardless, the data suggest that precursors of multiple T cell lineages are present in this population. We favor the interpretation that Foxp3lo cells that produce inflammatory cytokines represent Teff cells that have upregulated Foxp3 in order to become Treg cells but are still in a reversible phase of the differentiation process. This finding shows that Foxp3 expression does not “lock” a T cell into a regulatory phenotype and highlights the importance of other genes needed to achieve a stable Treg phenotype (35). Alternatively, a proportion of Foxp3lo cells may represent a recently identified, persistent T cell subset that expresses both Foxp3 and RORγt, that has the capacity to augment or suppress immune response depending on the outcome of cytokine/antigen stimulation (36). In our case cells co-expressing Foxp3 and RORγt could constitute only a minority of all Treg cells varying between different organs, since the RORγt transcript was not detected when we analyzed its expression in sorted Foxp3lo cells.
In our mouse model, when we combine the Foxp3lo and Foxp3hi subsets, we estimate that Treg cells bearing TCRs identical with Foxp3GFP− cells constitute about 20% of all Treg cells in unmanipulated mice, consistent with an earlier report (23). Our analysis of the TCR repertoires of these subsets show that almost 2/3 of Foxp3GFP− cells bear TCRs that can be found within the Foxp3lo and Foxp3hi TCR repertoire, more so within the Foxp3lo population. However, the remaining 1/3 of Foxp3GFP− TCRs could not be found within the Treg population, these are exclusive to the naive repertoire. Similarly, the Foxp3hi cells bear many TCRs not found within the naive repertoire. In conclusion, Foxp3GFP− and Foxp3hi cells represent populations with almost non-overlapping, exclusive TCR repertoires while Foxp3lo cells harbor a complex, mixed population expressing TCRs found either within the naive or Treg cell repertoires.
We propose that sustained generation of Treg cells is mediated by transient interactions of naive Teff with self-antigens and/or with exogenous antigens, in organs like the gut or respiratory tract, and these cells constitute a lasting component of Treg cells in unmanipulated mice (13, 14). The fraction of lymphocytes expressing a particular TCR and co-expressing Foxp3 varied greatly between different T cell clones indicating that the extent of Foxp3 upregulation by cells expressing the same TCR may be determined by the specificity of the TCR for self-peptide/MHC complexes. The process of recruiting Teff cells into the pool of Treg based on the local availability of self-antigens could have an important contribution to the preservation of tissue-specific tolerance, complementing the activity of tissue-specific nTreg cells.
Our studies of Foxp3lo cells sharing TCRs with naive T cells seem highly relevant for revealing the contribution of a Treg cells, expressing exactly the same set of TCRs, to immune regulation. Experiments investigating the contribution of a Treg to the peripheral population of Treg cells have used adoptive transfer to show conversion of Foxp3− into Foxp3+ T cells. Transferred Foxp3+ cells were easily found in lymphopenic animals but were barely detectable in lymphoreplete animals leading to the conclusion that conversion plays only marginal role in forming the population of peripheral Treg cells, even in organs subject to continuous stimulation with exogenous antigens (11, 37). One reason that may account for the failure to observe a larger fraction of Foxp3+ cells originating from transferred Foxp3− effector cells is that analysis of the TCR repertoire shows that many naive T cell clones do not have a fraction that are Foxp3+, while for other clones, Foxp3+ cells account for only a small fraction of cells in unmanipulated mice. This observation may explain why T cells that express some transgenic TCRs may not undergo efficient conversion to Foxp3+ cells (16).
A process of converting naive T cells into a Treg cells may require multiple differentiation steps and be completed only by a fraction of cells that initially upregulated Foxp3. Conversion process may be inhibited by Treg cells present in lymphosufficient recipient mice. Our in vitro activation studies have shown that the fraction of activated Teff cells that upregulated Foxp3 was inversely proportional to the number of preexisting Treg cells, suggesting that a mechanism may exist that allows Treg cells to constitute only a certain fraction of all CD4+ T cells. Finally, the proportion of cells upregulating Foxp3 may be established when thymus-derived naive cells populate lymphoid organs, shortly after birth, and may be associated with clonal expansion in peripheral organs.
Our adoptive transfer studies suggest that upregulation of the Foxp3 expression by peripheral naive CD4+ T cells occurs in lymphoreplete mice. When two populations of CD4+ T cells expressing diverse, polyclonal TCR repertoires are mixed a new hierarchy of clonal abundance is established as a result of clonal competition for the MHC/peptide ligands (38, 39). While, in normal mice sudden changes in the TCR repertoire available to compete for MHC/peptide complexes are unlikely to occur, interclonal competition among T cell clones likely results from constant alterations of peptides presented by class II MHC. Such alterations are due to changes in proteins expressed in peripheral tissues at different differentiation stages or in the course of cellular responses to external stimuli. In summary, adoptive transfer experiment shows that in lymphoreplete mice Foxp3GFP+ cells in peripheral organs may originate from naive CD4+ T cells that upregulate Foxp3 expression and become a Treg cells.
Treg cells involved in the immune response have been found in sites of chronic inflammation caused by foreign antigens derived from microorganisms, allergens or self-antigens (40–43). Expansion of Treg cells has also been reported during acute primary immune response (15). However, the cellular origin and antigen specificity of Treg cells found in sites of inflammation are still controversial. Analysis of TCRmini-Foxp3GFP mice immunized with peptide antigen and CFA revealed dramatic expansion of antigen-specific Teff cells. Surprisingly, this population of Foxp3+ Treg cells had a significant component (21.7%) of cells expressing the same TCRs as antigen-specific Teff cells. Since most expanded antigen-specific Teff clones are rare in normal mice, we can not determine if antigen-specific Treg cells in immunized mice originate from pre-existing Foxp3+ cells or are converted from activated cells. However, considering that antigen-specific Foxp3+ T cells constitute a substantial subset of all Treg cells, they must have undergone dramatic expansion similar to the expansion of Teff cells. In contrast, the population of Treg cells expressing an exclusive set of TCRs contained much less abundant clones. This observation may indicate that the population of Treg expressing an exclusive set of TCRs contains only low affinity antigen-specific clones that do not undergo efficient expansion when recruited into an inflammation site.
In summary, Treg clones with stable suppressor function and exclusive TCRs that dominate peripheral Treg population in unmanipulated mice are complemented by clones with a flexible phenotype expressing the same TCRs as Teff cells responding to antigen. Since Treg cells need to be activated in an antigen-specific manner to exert antigen-nonspecific suppression, newly generated a Treg cells play an important role in limiting immune response to antigen and protecting against excessive inflammation and tissue damage. This scenario might be particularly relevant in organs exposed to continuous stimulation with microbial antigens and could contribute to local tolerance (13, 14, 44). This does not exclude a possible contribution of Treg cells expressing an exclusive set of TCRs to the immune response to exogenous antigens, though in our experimental model we did not observe extensive clonal expansion of this subset. It is tempting to speculate that a Treg cells generated during the immune response lose Foxp3 expression as a mechanism to restore immune balance once an antigen challenge is removed. In conclusion, we postulate that Treg cells sharing antigen specificity with Teff cells are an important component of the immune regulation in the course of response to antigen.
Acknowledgements
We thank Dr. T. Denning and Dr. L. Ignatowicz and D. Daniely for critically reading the manuscript.
Footnotes
This work was supported by NIH grants R01 CA107349-01A1 to P.K. and 1R01DE019243-01 to G.A.R.
Disclosures
The authors have no conflicting financial interests.
References
- 1.Sakaguchi S, Sakaguchi N, Shimizu J, Yamazaki S, Sakihama T, Itoh M, Kuniyasu Y, Nomura T, Toda M, Takahashi T. Immunologic tolerance maintained by CD25+ CD4+ regulatory T cells: their common role in controlling autoimmunity, tumor immunity, and transplantation tolerance. Immunol. Rev. 2001;182:18–32. doi: 10.1034/j.1600-065x.2001.1820102.x. [DOI] [PubMed] [Google Scholar]
- 2.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 3.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- 4.Kretschmer K, Apostolou I, Hawiger D, Khazaie K, Nussenzweig MC, von BH. Inducing and expanding regulatory T cell populations by foreign antigen. Nat. Immunol. 2005;6:1219–1227. doi: 10.1038/ni1265. [DOI] [PubMed] [Google Scholar]
- 5.Knoechel B, Lohr J, Kahn E, Bluestone JA, Abbas AK. Sequential development of interleukin 2-dependent effector and regulatory T cells in response to endogenous systemic antigen. J. Exp. Med. 2005;202:1375–1386. doi: 10.1084/jem.20050855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito T, Hanabuchi S, Wang YH, Park WR, Arima K, Bover L, Qin FX, Gilliet M, Liu YJ. Two functional subsets of Foxp3+ regulatory T cells in human thymus and periphery. Immunity. 2008;28:870–880. doi: 10.1016/j.immuni.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25− regulatory T cells TGF-beta induction of trasnscription factor Foxp3. J. Exp. Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidson TS, DiPaolo RJ, Andersson J, Shevach EM. Cutting Edge: IL-2 is essential for TGF-beta-mediated induction of Foxp3+ T regulatory cells. J. Immunol. 2007;178:4022–4026. doi: 10.4049/jimmunol.178.7.4022. [DOI] [PubMed] [Google Scholar]
- 9.You S, Leforban B, Garcia C, Bach JF, Bluestone JA, Chatenoud L. Adaptive TGF-beta-dependent regulatory T cells control autoimmune diabetes and are a privileged target of anti-CD3 antibody treatment. Proc. Natl. Acad. Sci. USA. 2007;104:6335–6340. doi: 10.1073/pnas.0701171104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selvaraj RK, Geiger TL. A kinetic and dynamic analysis of Foxp3 induced in T cells by TGF-beta. J. Immunol. 2007;178:7667–7677. doi: 10.4049/jimmunol.178.12.7667. [DOI] [PubMed] [Google Scholar]
- 11.Liang S, Alard P, Zhao Y, Parnell S, Clark SL, Kosiewicz MM. Conversion of CD4+ CD25− cells into CD4+ CD25+ regulatory T cells in vivo requires B7 costimulation, but not the thymus. J. Exp. Med. 2005;201:127–137. doi: 10.1084/jem.20041201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benson MJ, Pino-Lagos K, Rosemblatt M, Noelle RJ. All-trans retinoic acid mediates enhanced T reg cell growth, differentiation, and gut homing in the face of high levels of co-stimulation. J. Exp. Med. 2007;204:1765–1774. doi: 10.1084/jem.20070719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, Belkaid Y. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J. Exp. Med. 2007;204:1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-{beta} and retinoic acid dependent mechanism. J. Exp. Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haribhai D, Lin W, Relland LM, Truong N, Williams CB, Chatila TA. Regulatory T cells dynamically control the primary immune response to foreign antigen. J. Immunol. 2007;178:2961–2972. doi: 10.4049/jimmunol.178.5.2961. [DOI] [PubMed] [Google Scholar]
- 16.Wong J, Mathis D, Benoist C. TCR-based lineage tracing: no evidence for conversion of conventional into regulatory T cells in response to a natural self-antigen in pancreatic islets. J. Exp. Med. 2007 doi: 10.1084/jem.20070822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marson A, Kretschmer K, Frampton GM, Jacobsen ES, Polansky JK, Macisaac KD, Levine SS, Fraenkel E, von BH, Young RA. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445:931–935. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445:936–940. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- 19.Wan YY, Flavell RA. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature. 2007;445:766–770. doi: 10.1038/nature05479. [DOI] [PubMed] [Google Scholar]
- 20.Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, Schlawe K, Chang HD, Bopp T, Schmitt E, Klein-Hessling S, Serfling E, Hamann A, Huehn J. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS. Biol. 2007;5:e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baron U, Floess S, Wieczorek G, Baumann K, Grutzkau A, Dong J, Thiel A, Boeld TJ, Hoffmann P, Edinger M, Turbachova I, Hamann A, Olek S, Huehn J. DNA demethylation in the human Foxp3 locus discriminates regulatory T cells from activated Foxp3+ conventional T cells. Eur. J. Immunol. 2007 doi: 10.1002/eji.200737594. [DOI] [PubMed] [Google Scholar]
- 22.Sauer S, Bruno L, Hertweck A, Finlay D, Leleu M, Spivakov M, Knight ZA, Cobb BS, Cantrell D, O'Connor E, Shokat KM, Fisher AG, Merkenschlager M. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc. Natl. Acad. Sci. USA. 2008;105:7797–7802. doi: 10.1073/pnas.0800928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pacholczyk R, Ignatowicz H, Kraj P, Ignatowicz L. Origin and T cell receptor diversity of Foxp3+CD4+CD25+ T cells. Immunity. 2006;25:249–259. doi: 10.1016/j.immuni.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 24.Malek TR, Bayer AL. Tolerance, not immunity, crucially depends on IL-2. Nat. Rev. Immunol. 2004;4:665–674. doi: 10.1038/nri1435. [DOI] [PubMed] [Google Scholar]
- 25.Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T cell lineage specification by the forkhead transcription factor Foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 26.Balandina A, Lecart S, Dartevelle P, Saoudi A, Berrih-Aknin S. Functional defect of regulatory CD4+CD25+ T cells in the thymus of patients with autoimmune myasthenia gravis. Blood. 2005;105:735–741. doi: 10.1182/blood-2003-11-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huan J, Culbertson N, Spencer L, Bartholomew R, Burrows GG, Chou YK, Bourdette D, Ziegler SF, Offner H, Vandenbark AA. Decreased Foxp3 levels in multiple sclerosis patients. J. Neurosci. Res. 2005;81:45–52. doi: 10.1002/jnr.20522. [DOI] [PubMed] [Google Scholar]
- 28.Pesu M, Watford WT, Wei L, Xu L, Fuss I, Strober W, Andersson J, Shevach EM, Quezado M, Bouladoux N, Roebroek A, Belkaid Y, Creemers J, O'Shea JJ. T-cell-expressed proprotein convertase furin is essential for maintenance of peripheral immune tolerance. Nature. 2008;455:246–250. doi: 10.1038/nature07210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsieh CS, Zheng Y, Liang Y, Fontenot JD, Rudensky AY. An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nat. Immunol. 2006;7:401–410. doi: 10.1038/ni1318. [DOI] [PubMed] [Google Scholar]
- 30.Wong J, Obst R, Correia-Neves M, Losyev G, Mathis D, Benoist C. Adaptation of TCR repertoires to self-peptides in regulatory and nonregulatory CD4+ T cells. J. Immunol. 2007;178:7032–7041. doi: 10.4049/jimmunol.178.11.7032. [DOI] [PubMed] [Google Scholar]
- 31.Hsieh CS, Liang Y, Tyznik AJ, Self SG, Liggitt D, Rudensky AY. Recognition of the peripheral self by naturally arising CD25+ CD4+ T cell receptors. Immunity. 2004;21:267–277. doi: 10.1016/j.immuni.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 32.Kraj P, Pacholczyk R, Ignatowicz L. alpha/beta TCRs differ in the degree of their specificity for the positively selecting MHC/peptide ligand. J. Immunol. 2001;166:2251–2259. doi: 10.4049/jimmunol.166.4.2251. [DOI] [PubMed] [Google Scholar]
- 33.Malherbe L, Hausl C, Teyton L, Heyzer-Williams MG. Clonal selection of helper T cells is determined by an affinity threshold with no further skewing of TCR binding properties. Immunity. 2004;21:669–679. doi: 10.1016/j.immuni.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 34.Marie JC, Letterio JJ, Gavin M, Rudensky AY. TGF-{beta}1 maintains suppressor function and Foxp3 expression in CD4+CD25+ regulatory T cells. J. Exp. Med. 2005;201:1061–1067. doi: 10.1084/jem.20042276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hill JA, Feuerer M, Tash K, Haxhinasto S, Perez J, Melamed R, Mathis D, Benoist C. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27:786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 36.Lochner M, Peduto L, Cherrier M, Sawa S, Langa F, Varona R, Riethmacher D, Si-Tahar M, Di Santo JP, Eberl G. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORgamma t+ T cells. J. Exp. Med. 2008;205:1381–1393. doi: 10.1084/jem.20080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lathrop SK, Santacruz NA, Pham D, Luo J, Hsieh CS. Antigen-specific peripheral shaping of the natural regulatory T cell population. J. Exp. Med. 2008;205:3105–3117. doi: 10.1084/jem.20081359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Viret C, Wong FS, Janeway CA. Designing and maintaining the mature TCR repertoire: the continuum of self-peptide:self-MHC complex recognition. Immunity. 1999;10:559–568. doi: 10.1016/s1074-7613(00)80055-2. [DOI] [PubMed] [Google Scholar]
- 39.Ernst B, Lee DS, Chang JM, Sprent J, Surh CD. The peptide ligands mediating positive selection in the thymus control T cell survival and homeostatic proliferation in the periphery. Immunity. 1999;11:173–181. doi: 10.1016/s1074-7613(00)80092-8. [DOI] [PubMed] [Google Scholar]
- 40.Saruta M, Yu QT, Fleshner PR, Mantel PY, Schmidt-Weber CB, Banham AH, Papadakis KA. Characterization of Foxp3+CD4+ regulatory T cells in Crohn's disease. Clin. Immunol. 2007;125:281–290. doi: 10.1016/j.clim.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 41.Ruprecht CR, Gattorno M, Ferlito F, Gregorio A, Martini A, Lanzavecchia A, Sallusto F. Coexpression of CD25 and CD27 identifies Foxp3+ regulatory T cells in inflamed synovia. J. Exp. Med. 2005;201:1793–1803. doi: 10.1084/jem.20050085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suffia IJ, Reckling SK, Piccirillo CA, Goldszmid RS, Belkaid Y. Infected site-restricted Foxp3+ natural regulatory T cells are specific for microbial antigens. J. Exp. Med. 2006;203:777–788. doi: 10.1084/jem.20052056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Curotto de Lafaille MA, Kutchukhidze N, Shen S, Ding Y, Yee H, Lafaille JJ. Adaptive Foxp3+ regulatory T cell-dependent and -independent control of allergic inflammation. Immunity. 2008;29:114–126. doi: 10.1016/j.immuni.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 44.Mucida D, Kutchukhidze N, Erazo A, Russo M, Lafaille JJ, Curotto de Lafaille MA. Oral tolerance in the absence of naturally occurring Tregs. J. Clin. Invest. 2005;115:1923–1933. doi: 10.1172/JCI24487. [DOI] [PMC free article] [PubMed] [Google Scholar]