Abstract
The spinal cord is a unique vertebrate feature that originates, together with the hindbrain, from the caudal neural plate. Whereas the hindbrain subdivides into rhombomeres, the spinal cord remains unsegmented. We have identified Cdx transcription factors as key determinants of the spinal cord region in zebrafish. Loss of Cdx1a and Cdx4 functions causes posterior expansion of the hindbrain at the expense of the unsegmented spinal cord. By contrast, cdx4 overexpression in the hindbrain impairs rhombomere segmentation and patterning and induces the expression of spinal cord-specific genes. Using cell transplantation, we demonstrate that Cdx factors function directly within the neural ectoderm to specify spinal cord. Overexpression of 5′ Hox genes fails to rescue hindbrain and spinal cord defects associated with cdx1a/cdx4 loss-of-function, suggesting a Hox-independent mechanism of spinal cord specification. In the absence of Cdx function, the caudal neural plate retains hindbrain characteristics and remains responsive to surrounding signals, particularly retinoic acid, in a manner similar to the native hindbrain. We propose that by preventing the posterior-most region of the neural plate from following a hindbrain developmental program, Cdx factors help determine the size of the prospective hindbrain and spinal cord territories.
Keywords: Cdx, Caudal, Hox, Retinoic acid, Segmentation, Rhombomeres, Hindbrain, Spinal cord, Central nervous system, Chordates, Vertebrates, Evolution
INTRODUCTION
One of the most prominent characteristics that distinguishes the rostral and caudal regions of the vertebrate central nervous system (CNS) is its segmental nature. Both forebrain and hindbrain have been shown to be organized into boundary-restricted, clonally related groups of cells known as neuromeres (Figdor and Stern, 1993; Fraser et al., 1990; Orr, 1887). The forebrain neuromeres (or prosomeres) and the more familiar hindbrain rhombomeres are important for proper neuronal organization and structural patterning of the adult brain and brain stem (reviewed by Borday et al., 2004; Kiecker and Lumsden, 2005; Puelles and Rubenstein, 2003). In the hindbrain, extensive genetic and molecular data have shown the existence of a regulatory network of transcription and secreted factors that specify the identity and order of rhombomere formation (Waskiewicz et al., 2002) (reviewed by Moens and Prince, 2002). In sharp contrast, the spinal cord remains unsegmented, although an anterior-posterior iterative arrangement of various neuronal populations can be seen to different degrees in Amphioxus, zebrafish and in certain tetrapods (Bone, 1960; Fetcho, 1987; Forehand and Farel, 1982; Myers, 1985; Roberts and Clarke, 1982; Stern et al., 1991). However, these iterated arrangements are not the consequence of cryptic segmentation, as demonstrated by the extensive mixing of cells in clonal analysis experiments (Morin-Kensicki and Eisen, 1997; Stern et al., 1991), but are the result of spinal cord neurons responding to paraxial mesoderm-derived signals (Detwiler, 1934; Eisen and Pike, 1991; Ensini et al., 1998; Keynes and Stern, 1984; Lewis and Eisen 2004). Despite the striking difference in segmentation, the hindbrain and spinal cord share a number of characteristics: both derive from the caudal neural plate (Brown and Storey, 2000; Muhr et al., 1997; Schoenwolf, 1992), are patterned along their anterior-posterior axis by Hox transcription factors (Deschamps and van Nes, 2005; Krumlauf et al., 1993; Lumsden and Krumlauf, 1996) and have a common evolutionary origin (Ghysen, 2003; Hirth et al., 2003). Therefore, we set out to investigate the mechanisms that direct the caudal neural plate to develop as hindbrain or as spinal cord.
Cdx (Caudal) transcription factors have been implicated in the development of trunk and tail structures across all major animal groups by controlling the sequential addition and identity of body segments (Copf et al., 2004). Within vertebrates, the three family members Cdx1, Cdx2 and Cdx4 are expressed in nested domains in the trunk and tail of the embryo (Davidson et al., 2003; Frumkin et al., 1993; Gamer and Wright, 1993; Joly et al., 1992; Lohnes, 2003; Marom et al., 1997; Meyer and Gruss, 1993; Pillemer et al., 1998; Reece-Hoyes et al., 2002). Most of our understanding of Cdx function is restricted to their role in paraxial mesoderm in mouse, where they have been shown to integrate FGF, retinoic acid and Wnt signals into coherent Hox gene expression (reviewed by Deschamps and van Nes, 2005; Lohnes, 2003). This role seems to be conserved in zebrafish and Xenopus (Davidson et al., 2003; Davidson and Zon, 2006; Isaacs et al., 1998; Pownall et al., 1998; Pownall et al., 1996; Shimizu et al., 2006; Shimizu et al., 2005). The function of Cdx genes in CNS development, however, is poorly understood, despite the fact that expression of Cdx genes in the caudal neural plate is highly conserved across vertebrates (Ehrman and Yutzey, 2001; Frumkin et al., 1993; Joly et al., 1992; Marom et al., 1997; Meyer and Gruss, 1993; Nordstrom et al., 2006; Pillemer et al., 1998; Reece-Hoyes et al., 2002).
Using a variety of morphological, cellular and molecular criteria we present evidence that spinal cord specification in zebrafish is dependent on the partially redundant functions of Cdx1a and Cdx4. In agreement with a previous study (Shimizu et al., 2006), we show that zebrafish embryos lacking full Cdx1a and Cdx4 functions develop an expanded hindbrain. In addition, we show that this expanded hindbrain is organized into segmental units arranged in a mirror-image duplicated pattern of ectopic rhombomeres within the trunk region of the embryo. We also show that Cdx factors can induce the development of spinal cord cell types and posterior Hox gene expression, when misexpressed in rostral regions of the CNS. We propose that Cdx transcription factors normally function to prevent rhombomere formation in the caudal neural plate and that by preventing the posterior-most region of the neural plate from following a segmented developmental program, Cdx transcription factors help determine the size of the prospective hindbrain and spinal cord territories. We hypothesize that this newly proposed function of Cdx transcription factors allowed the development of the dorsal, hollow and unsegmented caudal neural tube that is characteristic of the vertebrate lineage.
MATERIALS AND METHODS
Fish care, microinjection, cell transplantation and pharmacological treatments
Zebrafish (Danio rerio) were raised and handled following standard techniques (Westerfield, 1994). Embryos from wild-type AB stock, kgghi2188A (Golling et al., 2002), Tg[isl1:GFP] (Higashijima et al., 2000) and Tg[βactin:GFP] (Gillette-Ferguson et al., 2003) were obtained from natural spawning, grown at 28°C and staged as described (Kimmel et al., 1995). Injections were carried out at the one-cell stage. Antisense cdx1a (5 ng) (Shimizu et al., 2005) and cdx4 (20 ng) (Davidson et al., 2003) morpholino oligonucleotides (Gene Tools LLC) were injected alone or in combination. For mRNA overexpression, 25 pg of hoxc6a, hoxb8a or hoxa9a (Prince et al., 1998a), or 40 pg of gap43-RFP (cell membrane marker, gift of E. Amaya, University of Cambridge, Cambridge, UK) capped sense mRNA (SP6 mMessage mMachine Kit, Ambion) was injected using a standard injection protocol (Bruce et al., 2001). Retinoic acid signaling was blocked by incubating embryos in 1 µM BMS493 in the dark.
Cell transplantation was performed as previously described (Ho and Kane, 1990). Donor embryos were injected at the one-cell stage with 40-kDa lysine-fixable fluorescein (Invitrogen). Donor cells were collected at sphere stage and approximately 20 cells were transplanted 2- to 5 cell-diameters away from the margin of a stage-matched unlabeled host.
Transgenic constructs, genotyping and heat shock
To generate a heat-inducible cdx4 expression construct, full-length zebrafish cdx4 was PCR amplified using the following primers: mCdx5′-forward, 5′-CGATTCCGGGATCCACCGGTCGCCACCATGTATGGATCGTGTTTGCTCGAAAAAGAGGCAAGCATGTATCACCAA- 3′ (BamHI and AgeI sites underlined, translation initiation site in bold); and 3′MCScdx-reverse, 5′-TTGTCTAGAAAGCTTGGTACCGATCGATAGTTTGTA - ATCCTTTTTGACCAC-3′ (XbaI site underlined). The forward primer changes the 5′ end of the gene to that of mouse Cdx4, rendering it unrecognizable to the zebrafish cdx4 morpholino. BamHI/XbaI-digested PCR product was cloned in-frame into pcDNA3.1(−)/Myc-His B (Invitrogen). The modified mouse 5′-zebrafish cdx4-myc-his (m5′zcdx4) gene was digested with SphI, blunt ended, digested with AgeI and gel purified. Separately, the zebrafish hsp70 promoter (Halloran et al., 2000) and the pBS-ISce-II KS vector (Thermes et al., 2002) were digested with SacI/AgeI and SacI/EcoRV, respectively, and purified. A double ligation was then set up using the purified fragments to generate phsp70:m5′zcdx4-ISce. All constructs were confirmed by sequencing.
Stable transgenic fish were generated by injecting 1 µg phsp70:m5′zcdx4-ISce plasmid and 1 unit of ISce-I meganuclease in 1× ISce-I buffer (New England Biolabs) into fertilized eggs during the first 15 minutes of development (Thermes et al., 2002). Embryos were grown to adulthood, pairwise crossed and their embryos genotyped using the 3phsp70-forward (5′-GTATTACTTTGTTAACGTGGC-3′) and BGHrev (5′-TGAAAGBCACAGTCGAGG-3′) primers (IDT). As positive PCR controls, Wnt5a-12 (5′-CAGTTCTCACGTCTGCTACTTGCA-3′) and Wnt-21 (5′-ACTTCCGGCGTGTTGGAGAATTC-3′) primers were included in all reactions. Founder fish whose progeny produced diagnostic bands were out-crossed to wild-type fish. F1 fish were genotyped and individuals carrying the transgene were used as the Tg[phsp70:cdx4] line founders. A standard heat-shock protocol was used to induce transient cdx4 expression (Halloran et al., 2000).
Whole-mount in situ hybridization and immunocytochemistry
Detection of cdx1a (Shimizu et al., 2005); cdx4 (Joly et al., 1992); cyp26a1 (Kudoh et al., 2002); epha4a (Xu et al., 1994); foxb1.2 (mar) (Odenthal and Nusslein-Volhard, 1988); islet1 (Inoue et al., 1994); krx20 (Oxtoby and Jowett, 1993); hoxa2b, hoxb1a, hoxb3a, hoxb8a, hoxc4a, hoxd3a and hoxd4a (Prince et al., 1998a; Prince et al., 1998b); myod (Weinberg et al., 1996); olig2 (Park et al., 2002); radical fringe (rfng) (Cheng et al., 2004); raldh2 (also known as aldh1a2 – ZFIN) (Begemann et al., 2001); retinoic acid receptor alpha (RAR′; also known as raraa – ZFIN) (Hale et al., 2006); and valentino (val) (Moens et al., 1998) expression by in situ hybridization was carried out as previously described (Bruce et al., 2001), using NBT/BCIP or Fast Red as the enzyme substrate.
Antibody labeling was performed as previously described (Svoboda et al., 2001). Monoclonal mouse anti-acetylated Tubulin (Sigma-Aldrich), mouse anti-myosin HC (A4.1025, Developmental Studies Hybridoma Bank, IA, USA), mouse anti-neurofilament 160k (RMO44, Zymed, CA, USA) and polyclonal rabbit anti-GFP conjugated to FITC (Invitrogen) were used at 1:500, 1:100, 1:5000 and 1:1000, respectively. Goat anti-mouse [Alexa Fluor 647 (Invitrogen) and/or FITC (Jackson ImmunoResearch)] or goat anti-rabbit (Alexa Fluor 488, Invitrogen) secondary antibodies were used at 1:2000.
Image processing
Deyolked embryos were manually sectioned using a scalpel. Specimens were photographed with a Nikon D1 digital camera mounted on a Leica MZFL III or Zeiss Axioskop microscope. For confocal microscopy, single optical sections and image stacks were obtained using a Zeiss laser-scanning confocal imaging system (LSM 510). Three-dimensional reconstructions were produced with the Zeiss LSM 510 software and ImageJ 1.32 (NIH). Figure panels were constructed using Photoshop 7.0 (Adobe).
RESULTS
Cdx4 is required for proper anterior-posterior position of the hindbrain-spinal cord transition
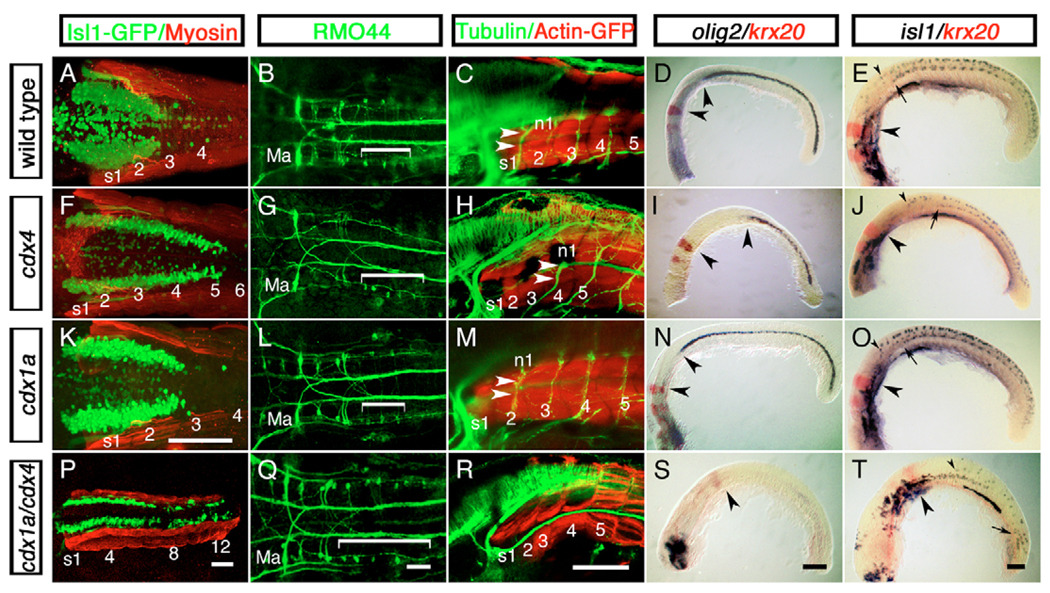
In zebrafish, the Cdx genes cdx1a and cdx4 are expressed in the posterior portion of the embryo (see Fig. S1 in the supplementary material) (Davidson et al., 2003; Davidson and Zon, 2006; Joly et al., 1992; Shimizu et al., 2005). We tested the function of Cdx4 in zebrafish CNS development by examining the distribution of hindbrain and spinal neuronal populations in wild-type and cdx4 morpholino-injected embryos (Fig. 1A–J). These embryos, referred to hereafter as cdx4-deficient embryos, are phenotypically indistinguishable from cdx4 loss-of-function mutants kgghi22188A and kggtv205 (Davidson et al., 2003; Golling et al., 2002). We used the vagal motor neurons and the T reticular interneurons as landmarks for the posterior hindbrain. These neurons are located in the spinobulbar junction, and they have a characteristic organization in vertebrates that is evolutionarily conserved (Fetcho, 1987; Kimmel et al., 1985; Wake, 1993). We found that in cdx4-deficient embryos, the size of the vagus expanded posteriorly by two somites as compared with wild-type siblings (Fig. 1A,F). Similarly, the RMO44-immunopositive T reticular interneurons also expanded posteriorly in these embryos (Fig. 1B,G). Other hindbrain-specific populations, such as the branchiomotor and reticulospinal neurons of rhombomeres (r) 1–6, appeared unaffected (Fig. 1B,G and data not shown). We also examined three distinct spinal cord populations in cdx4-deficient embryos: motor neurons and their exit roots, oligodendrocytes and Rohon-Beard sensory neurons. In wild-type embryos, the axons of the first spinal motor neurons exit at the level of somite 2 (Fig. 1C). By contrast, these axons exited at the level of somite 4 in cdx4-deficient embryos (Fig. 1H), correlating with the two-somite posterior expansion of the hindbrain neuronal populations (Fig. 1F,G). The distribution of olig2-expressing primary motor neurons and oligodendrocytes and isl1-positive Rohon-Beard sensory neurons also shifted posteriorly in cdx4-deficient embryos (Fig. 1D,E,I,J), indicating a generalized posterior shift of the spinal cord territory. Together, these data suggest that Cdx4 is necessary in zebrafish for the proper determination of the axial location of the transition from hindbrain to spinal cord.
Fig. 1. Hindbrain expansion and spinal cord reduction or loss in zebrafish embryos lacking Cdx function.
Distribution of hindbrain and spinal cord cell populations in wild-type embryos (A–E) and in cdx4-(F–J), cdx1a-(K–O) and cdx1a/cdx4-(P–T) deficient embryos. (A,F,K,P) Distribution of isl1:GFP-positive vagal motor neurons (green) in relation to adjacent somites (s, red) at 50 hours post-fertilization (hpf). (B,G,L,Q) Distribution of RMO44-immunopositive reticulospinal neurons at 50 hpf. The region occupied by T reticular interneurons is indicated with a bracket. r2 Mauthner neurons are also indicated (Ma). (C,H,M,R) Distribution of spinal motor neurons visualized with an anti-acetylated Tubulin antibody (n, green) in relation to adjacent somites (s, red) at 72 hpf. Axons of the first spinal motor neuron pool are indicated with arrowheads. (D,I,N,S) Distribution of olig2-expressing spinal cord oligodendrocytes (purple) with respect to krx20-expressing r3 and r5 (red) at the 20-somite stage (19 hpf). Distances between r5 and the rostral-most olig2-positive cells are indicated by arrowheads. Forebrain was removed for mounting purposes except in S, where it was retained to show that the in situ hybridization worked in these embryos. (E,J,O,T) Distribution of spinal cord isl1-positive (purple) motor neurons and Rohon-Beard sensory neurons as compared with krx20 expression in r3 and r5 (red) at 20 hpf. The position of r5 (large arrowheads), the most-rostral spinal motor neurons (arrows) and Rohon-Beard sensory neurons (small arrowhead) are indicated. For each condition, a minimum of 40 embryos from five independent experiments were analyzed. A,B,F,G,K,L,Q, dorsal view; K, oblique view; remaining embryos, lateral view. Scale bars: 100 µm for each column, except in K, for A,F,K.
Cdx1a and Cdx4 act redundantly in the specification of the prospective spinal cord territory
Cdx transcription factors have been shown to act redundantly in the patterning of the mouse paraxial mesoderm (reviewed by Deschamps and van Nes, 2005; Lohnes, 2003), raising the possibility that cdx1a and cdx4 might cooperate in determining the hindbrain-spinal cord transition in zebrafish. To test this hypothesis, we injected wild-type and cdx4-deficient embryos with a cdx1a morpholino (Shimizu et al., 2005) and examined the distribution of cell populations described above (Fig. 1K–T). Whereas wild-type embryos injected with a cdx1a morpholino showed no CNS defects (Fig. 1A–E,K–O), reduction of cdx1a function in a cdx4-deficient background led to an almost complete loss of spinal cord identities: spinal motor neurons and oligodendrocytes were absent, and the number of isl1-positive Rohon-Beard sensory neurons was greatly reduced (Fig. 1R–T). Conversely, there was a posterior expansion of r7 and r8 in these embryos, as shown by the trunk and tail distribution of isl1-GFP–positive branchiomotor neurons (Fig. 1P). Most of these branchiomotor neurons had axonal projections and morphologies characteristic of vagal cells, although separate and distinct neuronal clusters were also observed near the tail of these embryos (Fig. 1P). A posterior expansion of T reticular interneurons was also observed in these embryos (Fig. 1Q). Together, these data suggest that Cdx1a and Cdx4 function redundantly in the specification of spinal cord-specific neuronal identities, and in their absence most of the caudal neural plate takes on an expanded hindbrain fate.
The hindbrain patterning and segmentation program is improperly induced in the CNS of cdx1a/cdx4-deficient embryos
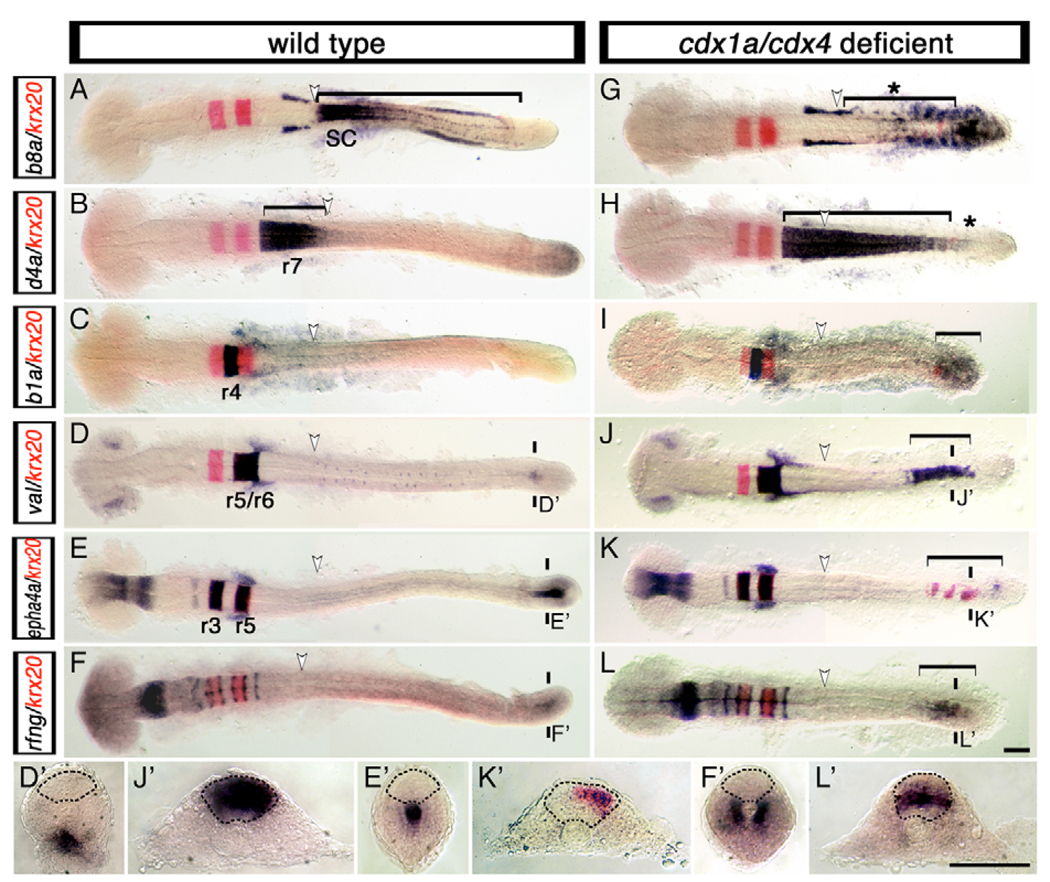
We next examined the expression of Hox patterning genes in the CNS of cdx1a/cdx4-deficient embryos (i.e. in embryos deficient for both genes). In these embryos, the r7 and anterior spinal cord marker hoxd4a was found to be expressed throughout the posterior CNS, whereas expression of the spinal cord markers hoxb6a, hoxb8a and hoxb10a was notably absent (Fig. 2A,B,G,H and data not shown). Within the hindbrain, cdx1a/cdx4-deficient embryos showed no changes in the anterior limit of hoxa2b (r2), hoxb1a (r4), hoxb3a (r5) and hoxd4a expression as compared with wild-type embryos (Fig. 2B,C,H,I and data not shown) suggesting that only spinal cord fates fail to be specified in these embryos. Together with our morphological data, these results show that the native hindbrain region in embryos lacking Cdx1a and Cdx4 activities has a normally ordered and nested set of rhombomeric identities and that the increase in hindbrain size seen in these embryos is mostly owing to an expansion of r7 and r8.
Fig. 2. Loss of Cdx function activates the expression of hindbrain genes in the posterior CNS.
Expression of hindbrain markers (purple) in wild-type (A–F,D′–F′) and cdx1a/cdx4-deficient (G–L,J′–L′) zebrafish embryos counterstained for the r3 and r5 marker krx20 (red signal). (A,G) Spinal cord hoxb8a expression (bracket in A) is lost in cdx1a/cdx4-deficient embryos (bracket with asterisk in G). (B,H) r7/8 hoxd4a expression (bracket in B) is expanded caudally in cdx1a/cdx4-deficient embryos (bracket in H) except for the most caudal tip of the CNS (asterisk in H). (C,I) In addition to its normal domain of expression in r4, hoxb1a expression can also be seen in the posterior CNS of cdx1a/cdx4-deficient embryos (bracket in I). (D,J) In the hindbrain, val is expressed in r5 and r6 of wild-type (D) and cdx1a/cdx4-deficient (J) embryos. In the tail region, val is also expressed in the posterior CNS of cdx1a/cdx4-deficient (bracket in J,J′) but not wild-type (D′) embryos. (E,K) Overlapping expression of epha4a (purple) and krx20 (red) are visualized in r3 and r5 of wild-type (E) and cdx1a/cdx4-deficient (K) embryos. In the tail region, epha4a and krx20 are expressed in the posterior CNS of cdx1a/cdx4-deficient (bracket in K,K′) but not wild-type (E′) embryos. (F,L) In the hindbrain, radical fringe (rfng) is expressed in seven stripes at the rhombomere boundaries in wild-type (F) and cdx1a/cdx4-deficient embryos (L). In the tail region, rfng is also expressed in the posterior CNS of cdx1a/cdx4-deficient (bracket in L,L′) but not wild-type (F′) embryos. For each condition, a minimum of 44 embryos from at least three independent experiments were analyzed, with more than 82% of embryos displaying the phenotype shown. Representative 20-somite, stage-matched, whole-mounted embryos are shown in dorsal view, anterior to the left. The position of somite 3, the hindbrain-spinal cord transition in wild-type embryos, is indicated with a white arrowhead. The planes of section are indicated with two short vertical bars. Sections are dorsal to the top, with the neural rod delineated by the dashed line. Scale bars: 100 µm.
We also noticed that in cdx1a/cdx4-deficient embryos, the expanded hoxd4a-positive r7/8 region did not extend along the entire length of the posterior CNS (Fig. 2H, asterisk). Instead, ectopic expression of more-anterior rhombomere-specific genes such as the r4-specific marker hoxb1a was seen within the posterior CNS (Fig. 2I, bracket). This phenomenon was not restricted to Hox genes. The gene valentino (val; also known as mafb and kr), which is normally expressed in the eyes, hindbrain r5 and r6 and in the tail mesenchyme (Moens et al., 1998), was also found to be expressed in the posterior CNS of these embryos (Fig. 2D,D′,J,J′). Similarly, krx20 (egr2b – ZFIN) expression, normally confined to r3 and r5 (Oxtoby and Jowett, 1993), was now present in the posterior CNS (Fig. 2G–I,K,K′). This shows that in cdx1a/cdx4-deficient embryos, several members of the regulatory network controlling rhombomere patterning are ectopically expressed outside of their native hindbrain region.
The regulatory network controlling hindbrain patterning also controls its segmentation into rhombomeres (Moens and Prince, 2002; Waskiewicz et al., 2002). Therefore, we examined the expression of epha4a, radical fringe (rfng) and foxb1.2 (also known as mar) in cdx1a/cdx4-deficient embryos, as these genes have been shown to be involved in rhombomere cell-sorting and boundary formation (Cheng et al., 2004; Cooke et al., 2001; Cooke et al., 2005; Odenthal and Nusslein-Volhard, 1988). In wild-type embryos, epha4a is expressed within odd-numbered rhombomeres as well as in the forebrain, midbrain and tail notochord, whereas expression of rfng and foxb1.2 in the CNS is restricted to the boundaries between rhombomeres (Fig. 2E–F′). In cdx1a/cdx4-deficient embryos, these genes were ectopically expressed in small discontinuous domains in the posterior CNS (Fig. 2K–L′), showing the induction of hindbrain segmentation genes beyond their normal domain of expression. Together, these data suggest that in the absence of Cdx1a and Cdx4 activities, the caudal neural plate not only fails to acquire spinal cord characteristics, but it also becomes competent to initiate the molecular program leading to the formation of supernumerary hindbrain segments and boundaries.
Development and mirror-image patterning of supernumerary rhombomeres in the CNS of cdx1a/cdx4-deficient embryos
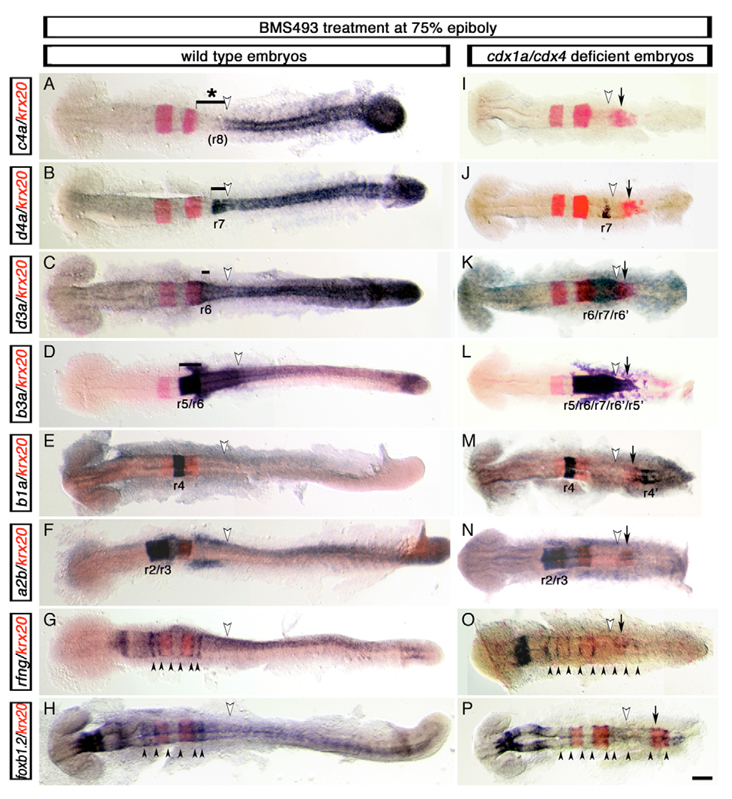
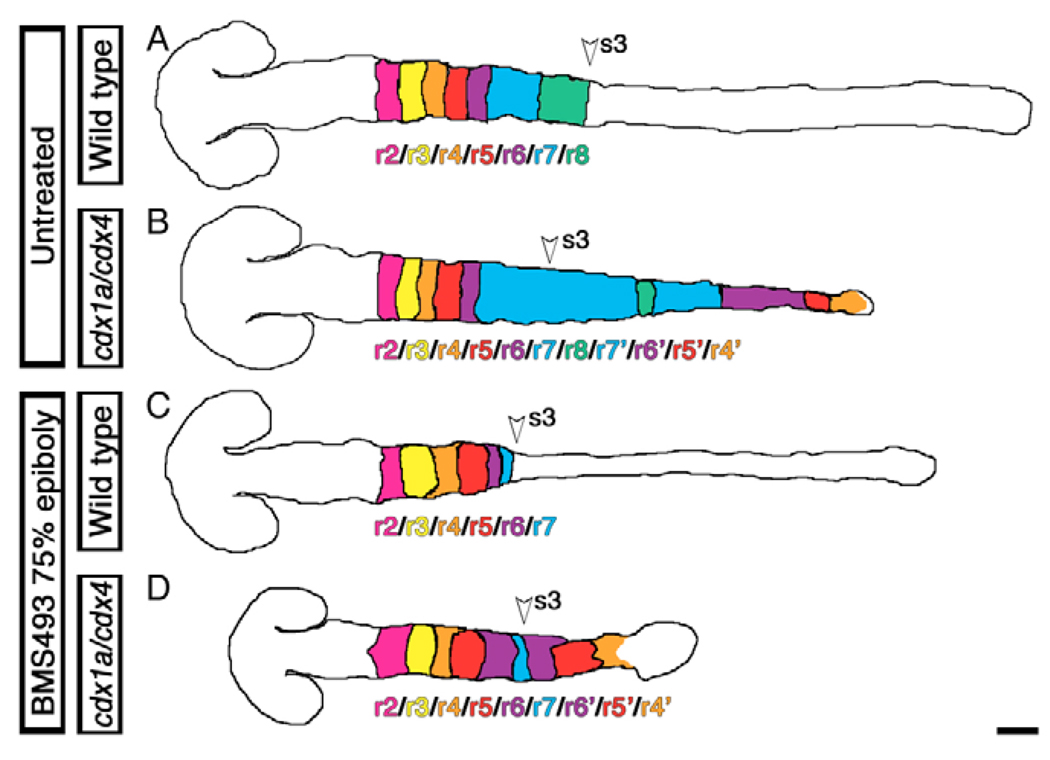
Despite the fact that several hindbrain- and boundary-specific genes were ectopically expressed in the posterior CNS of cdx1a/cdx4-deficient embryos, we noted the absence of definitively sized additional rhombomeres in this region. We hypothesized that the expansion of the native r7/8 region in cdx1a/cdx4-deficient embryos could be inhibiting the formation of correctly sized rhombomeres within the transformed caudal neural plate. In order to experimentally reduce the size of the r7/8 region, we took advantage of the observation that retinoic acid (RA) signaling inhibition results in the loss of r6–8 markers in wild-type (Begemann et al., 2004; Maves and Kimmel, 2005) and cdx1a/cdx4-deficient embryos (Shimizu et al., 2006). We treated embryos with the pan-RA receptor inhibitor BMS493 at midgastrulation to reduce the size of r7/8 without affecting the development of more-rostral rhombomeres (Begemann et al., 2004; Maves and Kimmel, 2005). As predicted, these treatments led to the loss of the r8 marker hoxc4a and an altered expression limit of the r7/8 marker hoxd4a (Fig. 3A,B,I,J). We then examined the expression of more-anterior Hox genes including hoxa2b (r2/r3), hoxb1a (r4), hoxb3a (r5/r6) and hoxd3a (r6) in comparison with the r3 and r5 marker krx20 (Fig. 3C–F,K–N). Excluding hoxa2b, whose expression domain was confined to r2/3 as in wild-type embryos (Fig. 3N) (Prince et al., 1998b), ectopic expression of these markers was observed in rhombomere-sized domains in the posterior CNS (Fig. 3K–M). These embryos expressed krx20 in three definitive stripes (Fig. 3I–N), contrasting with the more loosely organized expression seen in the posterior of cdx1a/cdx4-deficient embryos without BMS493 treatment (Fig. 2). Within the second and third krx20 stripes, we observed broad hoxb3a expression (r5/r6 marker, Fig. 3L) and, nested within it, smaller hoxd3a (r6/7 marker, Fig. 3K) and hoxd4a (r7 marker, Fig. 3J) expression domains. The r4 marker hoxb1a was expressed in two rhombomere-like domains, between the first and second stripe of krx20 expression and posterior to the third krx20-positive domain (Fig. 3M). In addition, we examined the expression of the rhombomere boundary markers rfng and foxb1.2 (Cheng et al., 2004; Odenthal and Nusslein-Volhard, 1988) and found that cdx1a/cdx4/RA-deficient embryos had nine evenly spaced, boundary-like stripes (Fig. 3O,P), instead of the six seen in wild-type embryos (Fig. 2F and data not shown). Together, these results show that upon RA-pathway inhibition, cdx1a/cdx4-deficient embryos can develop three supernumerary rhombomeres in the posterior CNS in addition to the normal seven. These supernumerary rhombomeres express Hox identity genes in a reverse anterior-posterior orientation in what seems a mirror-image duplication of the hindbrain, as follows: r2, r3, r4, r5, r6, r7, r6, r5, r4 (Fig. 3I–L, summarized in Fig. 4).
Fig. 3. Development of supernumerary rhombomeres and their mirror-image patterning in cdx1a/cdx4-deficient zebrafish embryos with compromised RA signaling.
Changes in rfng, foxb1.2 (mar), hox (purple) and krx20 (red) gene expression visualized in wild-type (A–H) and cdx1a/cdx4-deficient (I–P) embryos treated with the retinoic acid (RA) receptor inhibitor BMS493 (BMS) at mid-gastrulation (75% epiboly, 8 hpf). (A,I) hoxc4a expression in r8 is lost in wild-type (bracket with asterisk in A) and cdx1a/cdx4-deficient (I) embryos with compromised RA signaling. (B,J) RA inhibition reduced expression of the r7/8 marker hoxd4a in wild-type (bracket in B) and cdx1a/cdx4-deficient (J) embryos. (C,K) hoxd3a expression in the hindbrain posterior to r5 is reduced in wild-type embryos (bracket in C) and is limited to a central domain of the CNS in cdx1a/cdx4-deficient embryos (K). (D,L) hoxb3a, which is normally expressed posterior to the r4/r5 boundary (bracket in D), is expressed in a central domain in the CNS of cdx1a/cdx4-deficient embryos that includes the krx20 r5 and ectopic expression domains (L). (E,M) hoxb1a is expressed in r4 of wild-type (E) and cdx1a/cdx4-deficient (M) embryos. The latter also shows an additional hoxb1a-positive domain of expression in the posterior CNS. (F,N) hoxa2b is strongly expressed in r2 and r3 and weakly in r4 in wild-type (F) and cdx1a/cdx4-deficient (N) BMS-treated embryos. (G,O) rfng is expressed in six and nine boundary-like stripes (arrowheads) in wild-type (G) and cdx1a/cdx4-deficient (N) BMS-treated embryos, respectively. (H,P) foxb1.2 is expressed in nine boundary-like stripes (arrowheads) in cdx1a/cdx4-deficient BMS-treated embryos (P), compared with the six stripes seen in their wild-type siblings (H). For each condition, a minimum of 44 embryos from at least three independent experiments was analyzed at the equivalent of the 20-somite stage. More than 82% of embryos displayed the phenotype shown. Representative embryos were dorsal flat-mounted, anterior to the left. Ectopic rhombomere-like krx20 domain of expression is labeled with an arrow. Position of somite 3, the hindbrain-spinal cord transition in wild-type embryos, is indicated with a white arrowhead. Supernumerary rhombomeres are labeled r′. Scale bar: 100 µm.
Fig. 4. Summary of CNS transformations caused by the loss of Cdx function.
Summary diagrams showing the hindbrain and spinal cord transformation seen in wild-type (A,C) and cdx1a/cdx4-deficient (B,D) zebrafish embryos with (A,B) or without (C,D) an intact retinoic acid signaling pathway. Diagrams of 20-somite, stage-matched embryos are based on Fig 2 and Fig 3. Rhombomeres are color coded according to their identity; the location of the third somite, the normal position of the hindbrain-spinal cord transition in wild-type embryos, is indicated by an arrowhead. Dorsal views, anterior to the left. Scale bar: 100 µm.
The formation of supernumerary segments in the posterior CNS region of cdx1a/cdx4/RA-deficient embryos is likely to be owing to the loss of Cdx1a and Cdx4 activity rather than RA signaling, as inhibition of the RA pathway in wild-type embryos results in the loss of posterior rhombomeres (Begemann et al., 2004; Begemann and Meyer, 2001; Begemann et al., 2001). In support of this, close reexamination of hindbrain-specific gene expression in cdx1a/cdx4-deficient embryos not treated with BMS493 showed that these embryos had signs of incipient mirror-image duplication in the CNS, despite having normal expression levels of genes involved in the synthesis, reception and degradation of RA (see Fig. S2 in the supplementary material). For example, the expanded hoxd4apositive r7/8 territory was flanked by the r5/6 marker val, which in turn was bordered by the r4-marker hoxb1a (Fig. 2H–J, summarized in Fig. 4). Together, these results suggest that the absence of Cdx1a and Cdx4 functions allows the posterior CNS to adopt a segmented pattern of development with the potential to develop supernumerary, hindbrain rhombomere-like fates.
Cdx function is required in the CNS to prevent hindbrain expansion
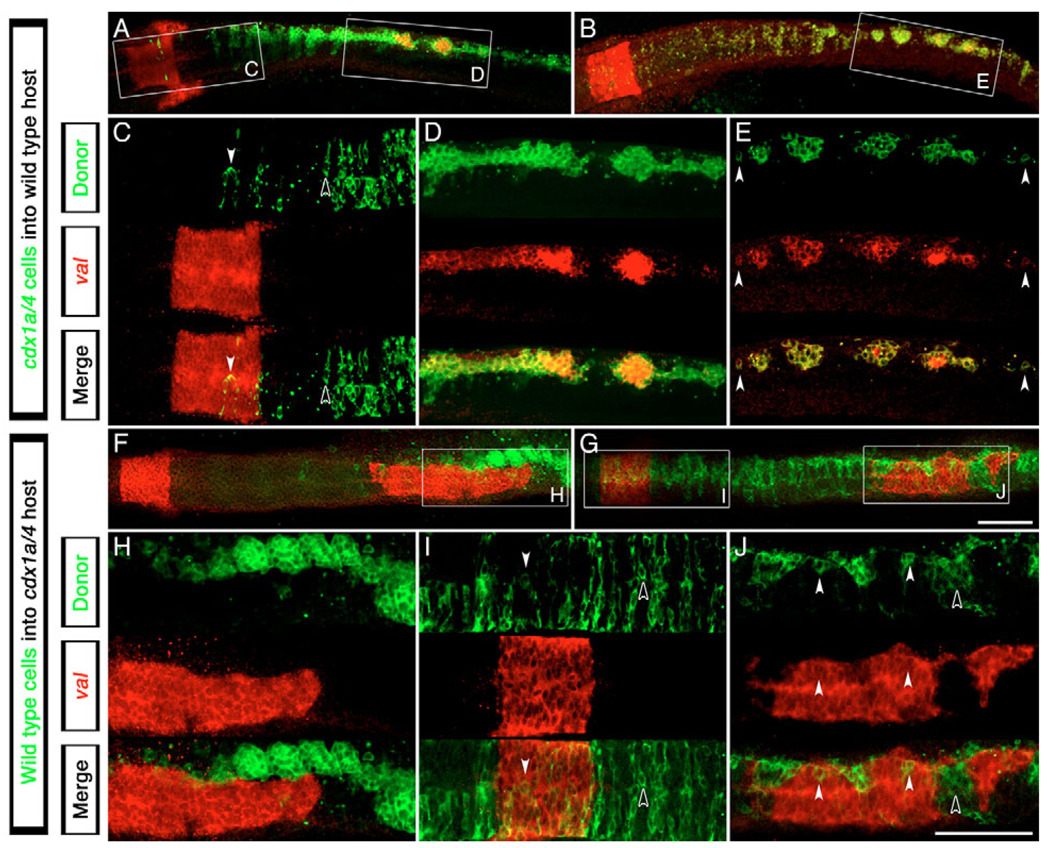
It has been suggested that spinal cord fate may depend on paraxial mesoderm-derived signals (Ensini et al., 1998; Muhr et al., 1999; Muhr et al., 1997; Nordstrom et al., 2002; Nordstrom et al., 2006), raising the question of whether Cdx function is required in the CNS, paraxial mesoderm, or in both tissues, to specify spinal cord. We transplanted cdx1a/cdx4-deficient, fluorescein-labeled cells into the CNS of wild-type host embryos and analyzed the expression of the r5/6 marker val (n=8; Fig. 5A–E), as this gene is ectopically expressed in the posterior CNS of cdx1a/cdx4-deficient but not wild-type embryos (Fig. 2D,J). As shown in Fig. 5, transplanted cdx1a/cdx4-deficient cells were typically found populating the entire length of the CNS by the 20-somite stage. In rostral regions, transplanted cdx1a/cdx4-deficient as well as wild-type host cells located in the native r5/6 territory expressed val (Fig. 5C). In the posterior CNS, only the transplanted cdx1a/cdx4-deficient cells, but not surrounding wild-type cells, expressed val, even when in isolation (n=8; Fig. 5D,E, arrowheads). In reciprocal experiments, transplantation of wild-type cells into the paraxial mesoderm of a cdx1a/cdx4-deficient host failed to prevent val expression within the posterior CNS (n=2; Fig. 5F–H). Taken together, these transplant experiments indicate that Cdx function is required autonomously within neural ectoderm for correct spinal cord fate specification.
Fig. 5. Autonomous requirement of Cdx factors in the zebrafish CNS for hindbrain and spinal cord specification.
Expression analysis of the r5/6 marker val (red staining) in clones of cdx1a/cdx4-deficient cells transplanted into wild-type hosts (A–E) or wild-type cells transplanted into cdx1a/cdx4-deficient host embryos (F–J) (transplanted cells in green). (A,B) cdx1a/cdx4-deficient cells can incorporate into the wild-type host CNS at all axial levels. The boxed regions are shown at higher magnification in C–E. (C) cdx1a/cdx4-deficient cells are evenly distributed in hindbrain and spinal cord regions of the CNS, only expressing val when located in the r5/6 territory (white arrowhead compared with black arrowhead). (D,E) cdx1a/cdx4-deficient cells located in the caudal spinal cord tend to form clusters of cells that express val (n=8). Surrounding wild-type cells do not express this marker. Isolated cells also express this gene (arrowheads). (F,H) cdx1a/cdx4-deficient embryos show ectopic val expression in the posterior CNS despite the presence of wild-type cells in the paraxial mesoderm (n=2). (G) Incorporation of wild-type cells throughout the CNS of cdx1a/cdx4-deficient hosts. The boxed regions are shown at higher magnification in I and J. (I) Uniform distribution of wild-type cells in the hindbrain and surrounding regions of cdx1a/cdx4-deficient host embryos. Cells located within the r5/6 region express the marker val (white arrowheads compared with black arrowheads). (J) In the posterior CNS, most wild-type cells segregate in clusters that fail to express val (black arrowhead). When in isolation, wild-type cells express val (white arrowheads, n=5). Confocal 3 µm sections of dorsal flat-mounted embryos, anterior to the left. Scale bars: in G, 100 µm for A,B,F,G; in J, 100 µm for C–E,H–J.
We also transplanted wild-type cells into the CNS of cdx1a/cdx4-deficient embryos (n=5; Fig. 5G,I,J). Wild-type cells were able to contribute to the entire length of the CNS. In rostral regions, wild-type and cdx1a/cdx4-deficient cells were evenly distributed within the tissue, with transplanted cells expressing val only when located in the native r5/6 territory (Fig. 5I). Interestingly, in posterior regions, wild-type cells were found segregating from host cdx1a/cdx4-deficient cells and did not express val (black arrowhead). However, in the posterior CNS, isolated wild-type cells surrounded by cdx1a/cdx4-deficient cells were occasionally seen expressing val (Fig. 5J, white arrowheads). Together, these results suggest that the expression of the hindbrain-specific val gene can be controlled by the level of Cdx function within the CNS.
cdx4 overexpression in the hindbrain induces spinal cord development
Our experiments indicate that Cdx activity is required for the caudal neural plate to develop as spinal cord instead of as segmented hindbrain. This hypothesis predicts that cdx4 overexpression in the hindbrain should: (1) interfere with the segmentation of this region; (2) change hindbrain neuronal identities; and (3) induce spinal cord neuronal markers. cdx4 overexpression by mRNA injection at the one-cell stage causes severe gastrulation defects (Davidson et al., 2003) (data not shown). To overcome this limitation, we generated a transgenic fish line, Tg[hsp70:cdx4], carrying a 5′-end modified zebrafish cdx4 gene under the control of the heat-inducible hsp70 promoter, which enables the rapid and ubiquitous induction of transgene expression at any point during development by incubating the embryos for 1 hour at 37°C (Halloran et al., 2000) (see Fig. S3A,B in the supplementary material and data not shown).
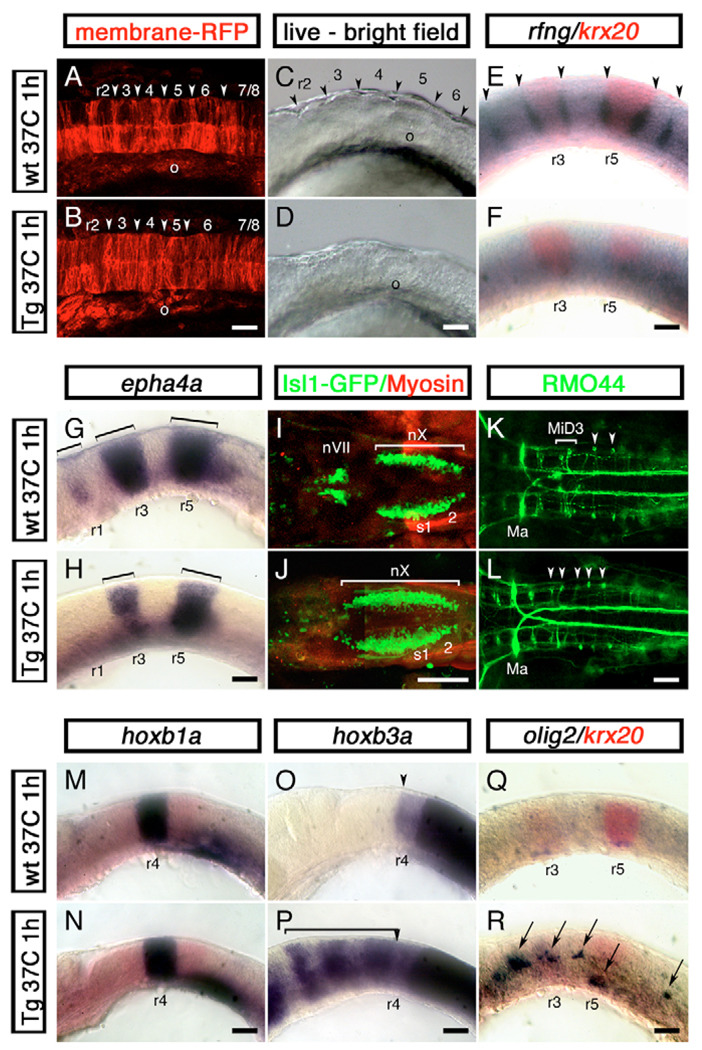
To study the effects of cdx4 overexpression in rhombomere formation, one-cell stage Tg[hsp70:cdx4] embryos were injected with gap43-RFP mRNA, a membrane-tagged red fluorescent protein, to follow the formation of rhombomere boundaries after heat shocking the embryos at the three-somite stage. This labeling method has been used to reveal the rhombomere boundaries before they become morphologically distinct (Moens et al., 1998). In transgenic heat-shocked and control embryos, boundary formation initiated at the six-somite stage (data not shown) and visible boundaries were apparent by the 14-somite stage, although less well defined in embryos carrying the cdx4 transgene (Fig. 6A,B). By the 20-somite stage, however, the characteristic rhombomere bulges seen in wild-type embryos were not present in their transgenic siblings (Fig. 6C,D). At this stage, heat-shocked embryos also showed loss of rfng and foxb1.2 expression at the rhombomere boundaries, and downregulation of the cell adhesion moleculeencoding epha4a gene in r1, r3 and r5 (Fig. 6E–H). Less severe defects were obtained when the transgene was induced at other developmental stages [from 75% epiboly to the ten-somite stage, 8–14 hours post-fertilization (hpf), data not shown]. These results show that Cdx4 interferes with rhombomere cell sorting and boundary formation.
Fig. 6. Cdx4 overexpression in the zebrafish hindbrain disrupts rhombomere formation and promotes spinal cord development.
Wild-type embryos (A,C,E,G,I,K,M,O,Q) and their siblings carrying one copy of a phsp70:cdx4 transgene (B,D,F,H,J,L,N,P,R) were heat shocked for 1 hour at 37°C at the three-somite stage and then grown at 28°C for a total of 16 (14 somites, A,B), 19 (20 somites, C-H,M-R) or 50 (I–L) hours. (A,B) Confocal images of the hindbrain region of 14-somite stage embryos injected with membrane-anchored RFP (gap43-RFP) mRNA to reveal rhombomere furrow formation (arrowhead). In all 12 embryos examined, cdx4 overexpression impaired furrow formation. (C,D) Distinctive rhombomere bulges (arrowheads in C) fail to appear in cdx4-overexpressing embryos (D) at the 20-somite stage (n=15). (E,F) Embryos overexpressing cdx4 fail to express the rhombomere boundary marker rfng (purple, arrowheads). krx20 expression was used to visualize r3 and r5 (red). (G,H) Loss of epha4a expression in r1 and reduction in r3 and r5 (brackets) in embryos overexpressing cdx4. (I,J) cdx4 overexpression results in the rostral expansion of vagal (nX) motor neurons and the loss of facial (nVII) and trigeminal (nV, not shown) motor neurons in isl1-GFP embryos. (K,L) In wild-type embryos, the RMO44-positive MiD3 reticulospinal neurons are found forming a cluster in r6 (brackets, K). This cluster is replaced by individual r7/8-like, T reticular interneurons in cdx4-overexpressing embryos (arrowheads, L). On a few occasions, MiD2 (r5) and Mauthner (r2) neurons (Ma) were also lost in cdx4-overexpressing embryos (data not shown). (M,N) Expression of the r4 marker hoxb1a is not affected in embryos overexpressing cdx4. (O,P) hoxb3a, a gene that is normally transcribed in rhombomeres posterior to r4 (O, arrowhead), is expressed throughout the rostral hindbrain in cdx4-overexpressing embryos (bracket in P). (Q,R) cdx4 overexpression induces ectopic hindbrain expression of the spinal motor neuron and oligodendrocyte marker olig2 (purple, arrows in R). For the 20-somite stage embryos (C–H,M–R), a minimum of 36 embryos from at least three independent experiments were analyzed. All embryos mounted anterior to the left, lateral views except for A,B,I–L, which are dorsal. o, otic vesicle; r, rhombomere; s, somite. Scale bars: 100 µm.
We then examined the effects of cdx4 overexpression on hindbrain neuronal populations and patterning. cdx4 overexpression caused the anterior expansion of r7/8 neuronal populations such as the vagal motor neurons (nX) and the T reticular interneurons (Fig. 6I–L). Furthermore, rostral neuronal populations, including the trigeminal (nV in r2) and facial (nVII in r4) motor neurons as well as the MiD2 (r5), MiD3 (r6) and in some cases Mauthner (r4) reticulospinal neurons, were lost in these embryos (Fig. 6I–L and data not shown). The posteriorization of hindbrain neuronal populations was also reflected at the level of Hox gene expression. Although cdx4 overexpression had no effect on Hox genes normally expressed in r2–4 (hoxa2b in r2/r3, hoxb1a in r4, hoxb2a in r3/r4; Fig. 6M,N and data not shown), it induced generalized hoxb3a, hoxb5a, hoxb8a and hoxb10a hindbrain expression (Fig. 6O,P and data not shown). Despite the variability in response of different rhombomeres to cdx4 overexpression, these results corroborate that Cdx4 has the ability to posteriorize the hindbrain.
Although these results suggest that Cdx4 can interfere with normal hindbrain development, they do not address whether Cdx4 is sufficient to initiate the development of spinal cord neuronal populations. We examined the expression of olig2, a marker for spinal cord primary motor neurons and oligodendrocytes (Park et al., 2002), after cdx4 overexpression. We found that this gene was ectopically expressed in the hindbrain of heat-shocked transgenic but not wild-type embryos (Fig. 6Q,R), suggesting that Cdx4 can divert the development of hindbrain cells to a spinal cord fate. Taken together, these results suggest that Cdx factors normally promote spinal cord development by inducing and later patterning neuronal cell types specific to this region, and by interfering with molecular pathways leading to hindbrain patterning and segmentation.
Hox-independent specification of spinal cord fates by Cdx factors
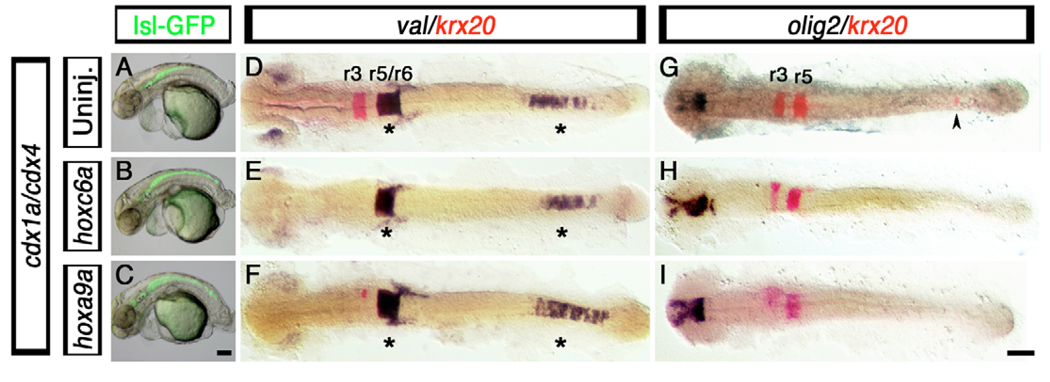
Hindbrain expansion in cdx1a/cdx4-deficient embryos was accompanied by the loss of posterior Hox gene expression (Fig. 2). Since posterior Hox genes are under direct Cdx regulation (Charite et al., 1998) and their activity is required to override anterior Hox function in caudal regions of the embryo (Duboule, 1991), it was important to know whether this hindbrain expansion was due to the lack of posterior Hox activity or more directly to the loss of Cdx function itself. We overexpressed the posterior Hox genes hoxc6a, hoxb8a and hoxa9a by injecting 25 pg of the respective mRNAs into cdx1a/cdx4-deficient isl1-GFP transgenic embryos, and examined the distribution of GFP-positive branchiomotor neurons at 50 hpf (Fig. 7A–C). Previously, it was shown that posterior Hox gene overexpression rescues the loss of the red blood cells observed in cdx1a/cdx4-deficient embryos (Davidson and Zon, 2006). Although we were able to observe the rescue of the red blood cell marker gata1 in our injected embryos (see Fig. S4 in the supplementary material), we never observed the rescue of the CNS defects seen in the cdx1a/cdx4-deficient embryos. For example, branchiomotor neurons were still present along the trunk of cdx1a/cdx4-deficient embryos overexpressing these posterior Hox genes (Fig. 7A–C). This suggests that Cdx factors have a Hox-independent role in spinal cord specification.
Fig. 7. Hindbrain and spinal cord defects associated with loss of Cdx function are not rescued by posterior Hox gene overexpression.
(A–I) Branchial motor neuron distribution (A–C, GFP-positive cells), hindbrain markers krx20 (D–I, red) and val (D–F, purple), and spinal cord oligodendtrocyte marker olig2 (G–I, purple staining), in cdx1a/cdx4-deficient zebrafish embryos injected with 25 pg of hoxc6a and hoxa9a mRNA. (A–C) At 50 hpf, control and hoxc6a and hoxa9a mRNA-injected cdx1a/cdx4-deficient, isl1:GFP transgenic embryos show GFP-positive branchiomotor neurons throughout the posterior CNS. (D–I) At the 20-somite stage (19 hpf), hoxc6a and hoxa9a overexpression in cdx1a/cdx4-deficient embryos results in reduced krx20 expression in r3 and r5 and its loss in the posterior CNS (E,F,H,I, red; caudal expression indicated with an arrowhead), as compared with uninjected controls (D,G and Fig. 2). (D–F) val expression is maintained in r5/6 and posterior CNS of cdx1a/cdx4-deficient embryos overexpressing posterior Hox genes (see Fig. 2 for wild-type control). (G,I) Posterior Hox gene overexpression does not rescue spinal cord olig2 expression in cdx1a/cdx4-deficient embryos (see Fig. 1 for wild-type control). Embryos shown in lateral (A–C) or dorsal (D–I) views, anterior to the left. Asterisk indicates val expression in hindbrain and posterior CNS. Arrowhead indicates ectopic krx20 expression in the posterior CNS. A minimum of 15 embryos in three independent experiments were analyzed, with more than 90% of embryos displaying the phenotypes shown. Scale bars: 100 µm.
We also examined the effect that posterior Hox gene overexpression had on the mirror-image patterning of the CNS of cdx1a/cdx4-deficient embryos (Fig. 7D–I). In the native hindbrain, r3 and r5 krx20 expression was reduced or absent, whereas the r5/r6 val expression domain was mostly unaffected (Fig. 4D–I), consistent with the ability of middle and posterior Hox genes to transform anterior rhombomeres into posterior ones (Bruce et al., 2001). We also found, as previously reported (Shimizu et al., 2006), that posterior Hox gene overexpression prevents ectopic krx20 expression in the posterior CNS of cdx1a/cdx4-deficient embryos (Fig. 7G–I, arrowhead). However, when we additionally examined markers of posterior hindbrain identities, such as val, we observed that the expression of these genes in cdx1a/cdx4-deficient embryos was still present in both the native hindbrain and posterior CNS after Hox gene overexpression (Fig. 7D–F, asterisks). This result shows that hindbrain patterning genes can be differentially affected by Hox gene overexpression in the absence of Cdx activity.
To test if posterior Hox gene overexpression could rescue spinal cord identities in cdx1a/cdx4-deficient embryos, we analyzed the expression of the oligodendrocyte marker olig2 (Fig. 7G–I). None of the tested Hox genes rescued the loss of olig2 expression in cdx1a/cdx4-deficient embryos (Fig. 7G–I and data not shown). This result, together with our hindbrain marker analysis, suggests that both hindbrain size and spinal cord specification might be independent of Hox activities.
DISCUSSION
Our analysis of cdx1a/cdx4-deficient zebrafish embryos has shown that Cdx function is required within the neural tissue for spinal cord specification and patterning: loss of Cdx function causes the unsegmented spinal cord to become segmented and take on hindbrain features. In reciprocal gain-of-function experiments we find that cdx4 overexpression is sufficient to cause the segmented hindbrain to lose aspects of its segmental character and take on features of the spinal cord. We conclude that Cdx factors specify vertebrate spinal cord cell fates and, by regulating posterior Hox gene expression, additionally influence anterior-posterior patterning.
Cdx promotes spinal cord development
In zebrafish, at the beginning of gastrulation, the hindbrain and spinal cord precursor cells are broadly distributed along the margin of the epiblast and are not yet committed to their fate (Woo and Fraser, 1995; Woo and Fraser, 1998). Commitment occurs towards the end of the gastrulation period, when hindbrain and spinal cord cells occupy the anterior and posterior halves of the caudal neural plate, respectively (Woo and Fraser, 1995). This segregation and commitment of prospective hindbrain and spinal cord cells correlates with the restriction of cdx4 transcripts to the posterior third of the neuroectoderm from an initial broad, ventral-to-dorsal gradient of expression at the margin of the epiblast (see Fig. S1 in the supplementary material) (Davidson et al., 2003; Davidson and Zon, 2006; Shimizu et al., 2005). In addition to the posterior CNS expression domain, cdx4 and cdx1a are expressed in a lateral-posterior domain of the tailbud that contains the spinal cord precursor cells, among other lineages (Kanki and Ho, 1997). This nested expression explains the partially redundant function of cdx1a and cdx4 in spinal cord specification and patterning; only the loss of both genes causes severe tail truncations, absence of spinal cord and, as previously shown, lack of hematopoietic stem cells (Davidson and Zon, 2006). This partially redundant function of Cdx1a and Cdx4 in the development of the spinal cord is not unlike the situation described in the paraxial mesoderm of mouse (Chawengsaksophak et al., 2004; van den Akker et al., 2002; van Nes et al., 2006).
Notably, the failure of these Cdx-deficient embryos to develop spinal cord does not appear to be caused simply by a tail truncation, but also involves the posterior expansion of the hindbrain territory. The use of isl1-GFP transgenic animals as well as various hindbrain-specific markers has shown that whereas the native anterior hindbrain regions appear unaffected, the native posterior hindbrain region, especially the r7/8 region, has greatly expanded its domain to take up the majority of the former spinal cord region in these embryos. Posterior to the expanded r7/8 territory, the remaining CNS expresses ectopic hindbrain-specific markers, including anterior Hox genes, at the expense of spinal cord-specific markers. In addition, our morphological studies have shown that spinal cord-specific characteristics, such as the formation of spinal nerve roots, are lost from the nervous systems of cdx1a/cdx4-deficient embryos. Together, our results suggest that Cdx factors are necessary for the specification and development of the spinal cord region.
The loss of Cdx functions in the developing nervous system leads to the formation of a larger than normal hindbrain region in which both expanded and ectopic rhombomeric identities can be found within the former spinal cord territory. We have also observed that in cdx1a/cdx4-deficient embryos, the hindbrain boundary markers rfng and foxb1.2 are ectopically expressed within the CNS in the tailbud region, and that this incipient segmentation resolves into recognizable rhombomere-like structures upon partial inhibition of RA signaling. The involvement of Cdx factors in not only spinal cord specification and anterior-posterior patterning, but perhaps also inhibition of segment formation, places these factors at an important regulatory crossroad.
Cdx repression of hindbrain development
A formal possibility is that Cdx factors might allow spinal cord specification by repressing hindbrain-specific characteristics within the posterior CNS; such a function would be consistent with the expression pattern of cdx4 within the spinal cord region of the zebrafish. The creation of a heat-inducible cdx4 transgenic line has enabled the overexpression of a Cdx factor at the end of the hindbrain determination period. We have shown that cdx4 overexpression affects correct hindbrain formation. For example, the loss of rfng and foxb1.2 expression within the hindbrain suggests that a relatively late step in the hindbrain segmentation cascade, namely the formation of segmentation boundaries, can be disrupted by cdx4 overexpression. Therefore, this type of experiment suggests that by interfering with the hindbrain segmentation program, Cdx factors might be able to direct the caudal neural plate cells to a spinal cord fate.
By contrast, the analysis of more-upstream hindbrain segmentation pathway components such as krx20, epha4a and hoxb3a, gave variable results in our overexpression assays, depending on which region of the hindbrain was analyzed. This variation can be attributed to the heterochrony of the region, as different rhombomeres form at different times during development and express different sets of genes (Moens and Prince, 2002). In these experiments, we confined our analyses to cases in which cdx4 overexpression was accomplished by heat-shock treatment administered at the three-somite stage, the developmental stage at which cdx4 overexpression caused the most severe hindbrain abnormalities. At this time, genes involved in hindbrain patterning and rhombomere boundary formation such as hoxb1a, val and krx20, are already expressed in the hindbrain (Moens et al., 1998; Prince et al., 1998b; Waskiewicz et al., 2002). Under these experimental conditions, the hindbrain region expresses what appears to be a mixed hindbrain/spinal cord identity with some aspects of hindbrain fate, such as the formation of vagal motor neurons, now overlapping with aspects of spinal cord fate, such as the ectopic expression of the spinal motor neuron marker olig2. It is likely that cdx4 overexpression at different times of development will give different outcomes, a possibility we are currently testing. Despite this caveat, we have shown that the overexpression of cdx4 is able to interfere with both the segmentation and specification of individual rhombomeric identities in the zebrafish hindbrain.
Specification and patterning of the spinal cord territory by Cdx
Although our work, like that of many others (reviewed by Deschamps and van Nes, 2005; Lohnes, 2003), shows that Cdx genes have roles in the establishment of Hox gene expression limits, we further propose that the initial function of Cdx in establishing the spinal cord field might be independent of a role in Hox gene regulation. We suggest that Cdx factors initially function to establish the prospective spinal cord territory by preventing the posterior-most region of the caudal neural plate from adopting a segmental developmental program (hindbrain fate) and by inducing or promoting the expression of spinal cord-specific gene expression. Consistent with this hypothesis are our data showing that the overexpression of 5′ Hox genes fails to rescue the loss of the spinal cord markers seen in cdx1a/cdx4-deficient zebrafish embryos and only causes the posterioization of the expanded hindbrain. If Cdx functioned solely through the control of Hox gene expression, then the general overexpression of a posterior Hox gene would be predicted to prevent the expansion seen in cdx1a/cdx4-deficient embryos. Since this was not the case, we propose that separate hindbrain and spinal cord territories must be established prior to becoming receptive to Hox gene functions. In cdx1a/cdx4-deficient embryos, posterior hindbrain identities are still present when 5′ Hox genes are overexpressed. Our conclusion differs from that of Shimizu et al. (Shimizu et al., 2006), who interpreted their overexpression studies as showing that Hox genes could prevent the ectopic expression of posterior hindbrain fates in cdx1a/cdx4-deficient embryos. We note that Shimizu et al. (Shimizu et al., 2006) only utilized the r5 marker krx20; however, when we additionally evaluated the r5/6 marker val it was clear that ectopic hindbrain fates were still present in the Hox-overexpressing embryos (Fig. 7). Therefore, our data suggest that in vertebrates, Cdx might have homeotic functions independent of those of Hox factors, similar to the function of the caudal gene in the Drosophila adult (Moreno and Morata, 1999). This homeotic function may act both prior to and independent of any downstream control of Hox genes, similar to the ability of Drosophila Caudal to repress Abd-B transcription and induce Distal-less, brachyenteron and even skipped gene expression during analia development (Moreno and Morata, 1999). Further work will be required to characterize the Hox-independent function of Cdx during spinal cord development.
Another function of Cdx factors within the nervous system is to allow the hindbrain and spinal cord regions to differentially respond to gradients of FGF, RA and Wnt signals in the embryo. This is illustrated by the striking mirror image expression of ectopic hindbrain patterning genes in cdx1a/cdx4-deficient embryos and by the failure of 5′ Hox gene overexpression to prevent this phenotype. As the caudal neural plate fails to be specified as spinal cord in cdx1a/cdx4-deficient embryos, it retains hindbrain characteristics and remains responsive to surrounding signals, particularly FGF and RA, in a manner similar to the native hindbrain region. For example, in wild-type embryos, r4-derived FGF signals are responsible for inducing and patterning the r5 and r6 regions (Maves et al., 2002). During normal development, the native hindbrain territory is located far removed from the tailbud region, which is another source of FGF signals (Draper et al., 2003; Griffin et al., 1995). However, in cdx1a/cdx4-deficient embryos, the expanded hindbrain now comes into close contact with the tailbud. As shown by Shimizu et al. (Shimizu et al., 2006), tailbud-derived FGF signals are able to mimic the FGF-dependent, r5- and r6-inducing activity of r4. This signaling activity, coupled with paraxial mesoderm-derived RA signals, is responsible for the induction of ectopic r4, r5, r6 and r7/8 identities in the trunk region of the embryo (summarized in Fig. 4). Because the gradients of FGF from the tailbud region of the embryo are reversed relative to the FGF gradients found in the native hindbrain region, the pattern of ectopic rhombomeric identities is likewise reversed within the trunk region. Therefore, the mirror-image pattern of rhombomeric identities seen in the trunk region of cdx1a/cdx4-deficient embryos could be produced by the normal responses of hindbrain tissues to the same types of signals that they would be exposed to in the hindbrain’s native location. This is further supported by transplantation experiments, in which individual cdx1a/cdx4-deficient cells in the spinal cord region of a wild-type host responded to FGF and RA factors as if they were located in the hindbrain. Therefore, we propose that the functions of the cdx1a and cdx4 genes in the nervous system of the zebrafish are to inhibit the hindbrain developmental program by preventing the tissue from inappropriately segmenting and taking on inappropriate anterior-posterior identities.
Evolutionary implications of Cdx function in spinal cord development
Based on patterns of expression and functional similarities across species, we propose that the control of hindbrain and spinal cord development by Cdx transcription factors might be common to all vertebrates. Remarkably, the rostral limit of the most anteriorly expressed Cdx gene coincides with the position of the hindbrain-spinal cord transition in zebrafish, Xenopus, chick and mouse (Cambronero and Puelles, 2000; Frumkin et al., 1993; Lohnes, 2003; Marom et al., 1997; Meyer and Gruss, 1993; Pillemer et al., 1998; Reece-Hoyes et al., 2002) (this work); Cdx factors thus define the prospective spinal cord territory in the caudal neural plate and, by exclusion, the region that will give rise to hindbrain. Careful reexamination of Cdx function in chordate neural tube patterning might prove useful in addressing the underlying developmental mechanisms and evolutionary origin of the vertebrate spinal cord.
Supplementary Material
Acknowledgments
We thank past and present members of the V.E.P. and R.K.H. laboratories for helpful comments and suggestions throughout this work; R. Bielang and J. Chetta for technical assistance and fish care; D.-G. Ahn, A. Bruce, A. Foley, C. Huang, M. Kinkel and R. Muller for critically reading the manuscript; and members of the zebrafish community for probes and reagents, in particular N. Hopkins for the kgghi2811A fish line, E. Amaya for gap43-RFP and D. Stanier for the gift of the cdx4 morpholino. The A4.1025 antibody developed by Helen M. Blau was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242, USA. I.S. was supported by The Helen Hay Whitney Foundation and D.T. by an NSF Graduate Research Fellowship. This study was also supported by grants NIH NS043977 to M.H., NIH DK064973-01 and Juvenile Diabetes Research Foundation Grant 1-2003-257 to V.E.P., and NIH GM67714, NIH DK68286 and MOD FY01-623 to R.K.H.
Footnotes
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/134/11/2147/DC1
References
- Begemann G, Meyer A. Hindbrain patterning revisited: timing and effects of retinoic acid signalling. BioEssays. 2001;23:981–986. doi: 10.1002/bies.1142. [DOI] [PubMed] [Google Scholar]
- Begemann G, Schilling TF, Rauch GJ, Geisler R, Ingham PW. The zebrafish neckless mutation reveals a requirement for raldh2 in mesodermal signals that pattern the hindbrain. Development. 2001;128:3081–3094. doi: 10.1242/dev.128.16.3081. [DOI] [PubMed] [Google Scholar]
- Begemann G, Marx M, Mebus K, Meyer A, Bastmeyer M. Beyond the neckless phenotype: influence of reduced retinoic acid signaling on motor neuron development in the zebrafish hindbrain. Dev. Biol. 2004;271:119–129. doi: 10.1016/j.ydbio.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Bone Q. The central nervous system in Amphioxus. J. Comp. Neurol. 1960;115:27–64. [Google Scholar]
- Borday C, Wrobel L, Fortin G, Champagnat J, Thaeron-Antono C, Thoby-Brisson M. Developmental gene control of brainstem function: views from the embryo. Prog. Biophys. Mol. Biol. 2004;84:89–106. doi: 10.1016/j.pbiomolbio.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Brown JM, Storey KG. A region of the vertebrate neural plate in which neighbouring cells can adopt neural or epidermal fates. Curr. Biol. 2000;10:869–872. doi: 10.1016/s0960-9822(00)00601-1. [DOI] [PubMed] [Google Scholar]
- Bruce AEE, Oates AC, Prince VE, Ho RK. Additional hox clusters in the zebrafish: divergent expression patterns belie equivalent activities of duplicate hoxB5 genes. Evol. Dev. 2001;3:127–144. doi: 10.1046/j.1525-142x.2001.003003127.x. [DOI] [PubMed] [Google Scholar]
- Cambronero F, Puelles L. Rostrocaudal nuclear relationships in the avian medulla oblongata: Fate map with quail chick chimeras. J. Comp. Neurobiol. 2000;427:522–545. doi: 10.1002/1096-9861(20001127)427:4<522::aid-cne3>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Charite J, de Graaff W, Consten D, Reijnen MJ, Korving J, Deschamps J. Transducing positional information to the Hox genes: critical interaction of Cdx gene products with position-sensitive regulatory elements. Development. 1998;125:4349–4358. doi: 10.1242/dev.125.22.4349. [DOI] [PubMed] [Google Scholar]
- Chawengsaksophak K, de Graaff W, Rossant J, Deschamps J, Beck F. Cdx2 is essential for axial elongation in mouse development. Proc. Natl. Acad. Sci. USA. 2004;101:7641–7645. doi: 10.1073/pnas.0401654101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YC, Amoyel M, Qiu X, Jiang YJ, Xu Q, Wilkinson DG. Notch activation regulates the segregation and differentiation of rhombomere boundary cells in the zebrafish hindbrain. Dev. Cell. 2004;6:539–550. doi: 10.1016/s1534-5807(04)00097-8. [DOI] [PubMed] [Google Scholar]
- Cooke J, Moens C, Roth L, Durbin L, Shiomi K, Brennan C, Kimmel C, Wilson S, Holder N. Eph signalling functions downstream of Val to regulate cell sorting and boundary formation in the caudal hindbrain. Development. 2001;128:571–580. doi: 10.1242/dev.128.4.571. [DOI] [PubMed] [Google Scholar]
- Cooke JE, Kemp HA, Moens CB. EphA4 is required for cell adhesion and rhombomere-boundary formation in the zebrafish. Curr. Biol. 2005;15:536–542. doi: 10.1016/j.cub.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Copf T, Schroder R, Averof M. Ancestral role of caudal genes in axis elongation and segmentation. Proc. Natl. Acad. Sci. USA. 2004;101:17711–17715. doi: 10.1073/pnas.0407327102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AJ, Zon LI. The caudal-related homeobox genes cdx1a and cdx4 act redundantly to regulate hox expression and the formation of putative hematopoietic stem cells during zebrafish embryogenesis. Dev. Biol. 2006;292:506–518. doi: 10.1016/j.ydbio.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Davidson AJ, Ernst P, Wang Y, Dekens MP, Kingsley PD, Palis J, Korsmeyer SJ, Daley GQ, Zon LI. cdx4 mutants fail to specify blood progenitors and can be rescued by multiple hox genes. Nature. 2003;425:300–306. doi: 10.1038/nature01973. [DOI] [PubMed] [Google Scholar]
- Deschamps J, van Nes J. Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development. 2005;132:2931–2942. doi: 10.1242/dev.01897. [DOI] [PubMed] [Google Scholar]
- Detwiler SR. An experimental study of spinal nerve segmentation in Amblystoma with reference to the plurisegmental contribution to the brachial plexus. J. Exp. Zool. 1934;67:395–441. [Google Scholar]
- Draper BW, Stock DW, Kimmel CB. Zebrafish Fgf24 functions with Fgf8 to promote posterior mesodermal development. Development. 2003;130:4639–4654. doi: 10.1242/dev.00671. [DOI] [PubMed] [Google Scholar]
- Duboule D. Patterning in the vertebrate limb. Curr. Opin. Genet. Dev. 1991;1:211–216. doi: 10.1016/s0959-437x(05)80072-3. [DOI] [PubMed] [Google Scholar]
- Ehrman LA, Yutzey KE. Anterior expression of the caudal homologue cCdx-B activates a posterior genetic program in avian embryos. Dev. Dyn. 2001;221:412–421. doi: 10.1002/dvdy.1151. [DOI] [PubMed] [Google Scholar]
- Eisen JS, Pike SH. The spt-1 mutation alters segmental arrangement and axonal development of identified neurons in the spinal cord of the embryonic zebrafish. Neuron. 1991;6:767–776. doi: 10.1016/0896-6273(91)90173-w. [DOI] [PubMed] [Google Scholar]
- Ensini M, Tsuchida TN, Belting HG, Jessell TM. The control of rostrocaudal pattern in the developing spinal cord: specification of motor neuron subtype identity is initiated by signals from paraxial mesoderm. Development. 1998;125:969–982. doi: 10.1242/dev.125.6.969. [DOI] [PubMed] [Google Scholar]
- Fetcho JR. A review of the organization and evolution of motoneurons innervating the axial musculature of vertebrates. Brain Res. Rev. 1987;12:243–280. doi: 10.1016/0165-0173(87)90001-4. [DOI] [PubMed] [Google Scholar]
- Figdor MC, Stern CD. Segmental organization of embryonic diencephalon. Nature. 1993;363:630–634. doi: 10.1038/363630a0. [DOI] [PubMed] [Google Scholar]
- Forehand CJ, Farel PB. Spinal cord development in anuran larvae: 1. Primary and secondary neurons. J. Comp. Neurol. 1982;209:386–394. doi: 10.1002/cne.902090408. [DOI] [PubMed] [Google Scholar]
- Fraser S, Keynes R, Lumsden A. Segmentation in the chick embryo hindbrain is defined by cell lineage restrictions. Nature. 1990;344:431–435. doi: 10.1038/344431a0. [DOI] [PubMed] [Google Scholar]
- Frumkin A, Haffner R, Shapira E, Tarcic N, Gruenbaum Y, Fainsod A. The chicken CdxA homeobox gene and axial positioning during gastrulation. Development. 1993;118:553–562. doi: 10.1242/dev.118.2.553. [DOI] [PubMed] [Google Scholar]
- Gamer LW, Wright CV. Murine Cdx-4 bears striking similarities to the Drosophila caudal gene in its homeodomain sequence and early expression pattern. Mech. Dev. 1993;43:71–81. doi: 10.1016/0925-4773(93)90024-r. [DOI] [PubMed] [Google Scholar]
- Ghysen A. The origin and evolution of the nervous system. Int. J. Dev. Biol. 2003;47:555–562. [PubMed] [Google Scholar]
- Gillette-Ferguson I, Ferguson DG, Poss KD, Moorman SJ. Changes in gravitational force induce alterations in gene expression that can be monitored in the live, developing zebrafish heart. Adv. Space Res. 2003;32:1641–1646. doi: 10.1016/S0273-1177(03)90405-4. [DOI] [PubMed] [Google Scholar]
- Golling G, Amsterdam A, Sun Z, Antonelli M, Maldonado E, Chen W, Burgess S, Haldi M, Artzt K, Farrington S, et al. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat. Genet. 2002;31:135–140. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
- Griffin K, Patient R, Holder N. Analysis of FGF function in normal and no tail zebrafish embryos reveals separate mechanisms for formation of the trunk and the tail. Development. 1995;121:2983–2994. doi: 10.1242/dev.121.9.2983. [DOI] [PubMed] [Google Scholar]
- Hale LA, Tallafuss A, Yan YL, Dudley L, Eisen JS, Postlethwait JH. Characterization of the retinoic acid receptor genes raraa, rarab and rarg during zebrafish development. Gene Expr. Patterns. 2006;6:546–555. doi: 10.1016/j.modgep.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Halloran MC, Sato-Maeda M, Warren JT, Su F, Lele Z, Krone PH, Kuwada JY, Shoji W. Laser-induced gene expression in specific cells of transgenic zebrafish. Development. 2000;127:1953–1960. doi: 10.1242/dev.127.9.1953. [DOI] [PubMed] [Google Scholar]
- Higashijima S, Hotta Y, Okamoto H. Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. J. Neurosci. 2000;20:206–218. doi: 10.1523/JNEUROSCI.20-01-00206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirth F, Kammermeier L, Frei E, Walldorf U, Noll M, Reichert H. An urbilaterian origin of the tripartite brain: developmental genetic insights from Drosophila. Development. 2003;130:2365–2373. doi: 10.1242/dev.00438. [DOI] [PubMed] [Google Scholar]
- Ho RK, Kane DA. Cell-autonomous action of zebrafish spt-1 mutation in specific mesodermal precursors. Nature. 1990;348:728–730. doi: 10.1038/348728a0. [DOI] [PubMed] [Google Scholar]
- Inoue A, Takahashi M, Hatta K, Hotta Y, Okamoto H. Developmental regulation of islet-1 mRNA expression during neuronal differentiation in embryonic zebrafish. Dev. Dyn. 1994;199:1–11. doi: 10.1002/aja.1001990102. [DOI] [PubMed] [Google Scholar]
- Isaacs HV, Pownall ME, Slack JM. Regulation of hox gene expression and posterior development by the Xenopus caudal homologue Xcad3. EMBO J. 1998;17:3413–3427. doi: 10.1093/emboj/17.12.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly JS, Maury M, Joly C, Duprey P, Boulekbache H, Condamine H. Expression of a zebrafish caudal homeobox gene correlates with the establishment of posterior cell lineages at gastrulation. Differentiation. 1992;50:75–87. doi: 10.1111/j.1432-0436.1992.tb00488.x. [DOI] [PubMed] [Google Scholar]
- Kanki JP, Ho RK. The development of the posterior body in zebrafish. Development. 1997;124:881–893. doi: 10.1242/dev.124.4.881. [DOI] [PubMed] [Google Scholar]
- Keynes RJ, Stern CD. Segmentation in the vertebrate nervous system. Nature. 1984;310:786–789. doi: 10.1038/310786a0. [DOI] [PubMed] [Google Scholar]
- Kiecker C, Lumsden A. Compartments and their boundaries in vertebrate brain development. Nat. Rev. Neurosci. 2005;6:553–564. doi: 10.1038/nrn1702. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Metcalfe WK, Schabtach E. T reticular interneurons: a class of serially repeating cells in the zebrafish hindbrain. J. Comp. Neurol. 1985;233:365–376. doi: 10.1002/cne.902330306. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Krumlauf R, Marshall H, Struder M, Nonchev S, Sham MH, Lumsden A. Hox homeobox genes and regionalisation of the nervous system. J. Neurobiol. 1993;24:1328–1340. doi: 10.1002/neu.480241006. [DOI] [PubMed] [Google Scholar]
- Kudoh T, Wilson SW, Dawid IB. Distinct roles for Fgf, Wnt and retinoic acid in posteriorizing the neural ectoderm. Development. 2002;129:4335–4346. doi: 10.1242/dev.129.18.4335. [DOI] [PubMed] [Google Scholar]
- Lewis KE, Eisen JS. Paraxial mesoderm specifies zebrafish primary motoneuron subtype identity. Development. 2004;131:891–902. doi: 10.1242/dev.00981. [DOI] [PubMed] [Google Scholar]
- Lohnes D. The Cdx1 homeodomain protein: an integrator of posterior signaling in the mouse. BioEssays. 2003;25:971–980. doi: 10.1002/bies.10340. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Krumlauf R. Patterning the vertebrate neuraxis. Science. 1996;1109:1109–1115. doi: 10.1126/science.274.5290.1109. [DOI] [PubMed] [Google Scholar]
- Marom K, Shapira E, Fainsod A. The chicken caudal genes establish an anterior-posterior gradient by partially overlapping temporal and spatial patterns of expression. Mech. Dev. 1997;64:41–52. doi: 10.1016/s0925-4773(97)00043-9. [DOI] [PubMed] [Google Scholar]
- Maves L, Kimmel CB. Dynamic and sequential patterning of the zebrafish posterior hindbrain by retinoic acid. Dev. Biol. 2005;285:593–605. doi: 10.1016/j.ydbio.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Maves L, Jackman W, Kimmel CB. FGF3 and FGF8 mediate a rhombomere 4 signaling activity in the zebrafish hindbrain. Development. 2002;129:3825–3837. doi: 10.1242/dev.129.16.3825. [DOI] [PubMed] [Google Scholar]
- Meyer BI, Gruss P. Mouse Cdx-1 expression during gastrulation. Development. 1993;117:191–203. doi: 10.1242/dev.117.1.191. [DOI] [PubMed] [Google Scholar]
- Moens CB, Prince VE. Constructing the hindbrain: insights from the zebrafish. Dev. Dyn. 2002;224:1–17. doi: 10.1002/dvdy.10086. [DOI] [PubMed] [Google Scholar]
- Moens C, Cordes SP, Giorgianni MW, Barsh GS, Kimmel C. Equivalence in the genetic control of hindbrain segmentation in fish and mouse. Development. 1998;125:381–391. doi: 10.1242/dev.125.3.381. [DOI] [PubMed] [Google Scholar]
- Moreno E, Morata G. Caudal is the Hox gene that specifies the most posterior Drosophile segment. Nature. 1999;400:873–877. doi: 10.1038/23709. [DOI] [PubMed] [Google Scholar]
- Morin-Kensicki EM, Eisen JS. Sclerotome development and peripheral nervous system segmentation in embryonic zebrafish. Development. 1997;124:159–167. doi: 10.1242/dev.124.1.159. [DOI] [PubMed] [Google Scholar]
- Muhr J, Jessell TM, Edlund T. Assignment of early caudal identity to neural plate cells by a signal from caudal paraxial mesoderm. Neuron. 1997;19:487–502. doi: 10.1016/s0896-6273(00)80366-9. [DOI] [PubMed] [Google Scholar]
- Muhr J, Graziano E, Wilson S, Jessell TM, Edlund T. Convergent inductive signals specify midbrain, hindbrain, and spinal cord identity in gastrula stage chick embryos. Neuron. 1999;23:689–702. doi: 10.1016/s0896-6273(01)80028-3. [DOI] [PubMed] [Google Scholar]
- Myers PZ. Spinal motoneurons of the larval zebrafish. J. Comp. Neurol. 1985;263:555–561. doi: 10.1002/cne.902360411. [DOI] [PubMed] [Google Scholar]
- Nordstrom U, Jessell TM, Edlund T. Progressive induction of caudal neural character by graded Wnt signaling. Nat. Neurosci. 2002;5:525–532. doi: 10.1038/nn0602-854. [DOI] [PubMed] [Google Scholar]
- Nordstrom U, Maier E, Jessell TM, Edlund T. An early role for Wnt signaling in specifying neural patterns of Cdx and Hox gene expression and motor neuron subtype identity. PLoS Biol. 2006;4:1438–1452. doi: 10.1371/journal.pbio.0040252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenthal J, Nusslein-Volhard C. Fork head domain genes in zebrafish. Dev. Genes Evol. 1988;208:245–258. doi: 10.1007/s004270050179. [DOI] [PubMed] [Google Scholar]
- Orr H. Contribution to the embryology of the lizard. J. Morphol. 1887;1:311–372. [Google Scholar]
- Oxtoby E, Jowett T. Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res. 1993;21:1087–1095. doi: 10.1093/nar/21.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HC, Mehta A, Richardson JS, Appel B. olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Dev. Biol. 2002;248:356–368. doi: 10.1006/dbio.2002.0738. [DOI] [PubMed] [Google Scholar]
- Pillemer G, Epstein M, Blumberg B, Yisraeli JK, De Robertis EM, Steinbeisser H, Fainsod A. Nested expression and sequential downregulation of the Xenopus caudal genes along the anterior-posterior axis. Mech. Dev. 1998;71:193–196. doi: 10.1016/s0925-4773(97)00193-7. [DOI] [PubMed] [Google Scholar]
- Pownall ME, Tucker AS, Slack JM, Isaacs HV. eFGF, Xcad3 and Hox genes form a molecular pathway that establishes the anteroposterior axis in Xenopus. Development. 1996;122:3881–3892. doi: 10.1242/dev.122.12.3881. [DOI] [PubMed] [Google Scholar]
- Pownall ME, Isaacs HV, Slack JM. Two phases of Hox gene regulation during early Xenopus development. Curr. Biol. 1998;8:673–676. doi: 10.1016/s0960-9822(98)70257-x. [DOI] [PubMed] [Google Scholar]
- Prince VE, Joly L, Ekker M, Ho RK. Zebrafish hox genes: genomic organization and modified colinear expression patterns in the trunk. Development. 1998a;125:407–420. doi: 10.1242/dev.125.3.407. [DOI] [PubMed] [Google Scholar]
- Prince VE, Moens CB, Kimmel CB, Ho RK. Zebrafish hox genes: expression in the hindbrain region of wild-type and mutants of the segmentation gene, valentino. Development. 1998b;125:393–406. doi: 10.1242/dev.125.3.393. [DOI] [PubMed] [Google Scholar]
- Puelles L, Rubenstein JL. Forebrain gene expression domains and the evolving prosomeric model. Trends Neurosci. 2003;26:469–476. doi: 10.1016/S0166-2236(03)00234-0. [DOI] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Keenan ID, Isaacs HV. Cloning and expression of Cdx family from the frog Xenopus tropicalis. Dev. Dyn. 2002;223:134–140. doi: 10.1002/dvdy.1234. [DOI] [PubMed] [Google Scholar]
- Roberts A, Clarke JDW. The neuroanatomy of an amphibian embryo spinal cord. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1982;296:195–212. doi: 10.1098/rstb.1982.0002. [DOI] [PubMed] [Google Scholar]
- Schoenwolf GC. Morphological and mapping studies of the paranodal and postnodal levels of the neural plate during chick neurulation. Anat. Rec. 1992;233:281–290. doi: 10.1002/ar.1092330211. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Bae YK, Muraoka O, Hibi M. Interaction of Wnt and caudal-related genes in zebrafish posterior body formation. Dev. Biol. 2005;279:125–141. doi: 10.1016/j.ydbio.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Bae YK, Hibi M. Cdx-Hox code controls competence for responding to Fgfs and retinoic acid in zebrafish neural tissue. Development. 2006;133:4709–4719. doi: 10.1242/dev.02660. [DOI] [PubMed] [Google Scholar]
- Stern CD, Jaques KF, Lim TM, Fraser SE, Keynes RJ. Segmental lineage restrictions in the chick embryo spinal cord depend on the adjacent somites. Development. 1991;113:239–244. doi: 10.1242/dev.113.1.239. [DOI] [PubMed] [Google Scholar]
- Svoboda KR, Linares AE, Ribera AB. Activity regulates programmed cell death of zebrafish Rohon-Beard neurons. Development. 2001;128:3511–3520. doi: 10.1242/dev.128.18.3511. [DOI] [PubMed] [Google Scholar]
- Thermes V, Grabher C, Ristoratore F, Bourrat F, Choulika A, Wittbrodt J, Joly JS. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech. Dev. 2002;118:91–98. doi: 10.1016/s0925-4773(02)00218-6. [DOI] [PubMed] [Google Scholar]
- van den Akker E, Forlani S, Chawengsaksophak K, de Graaff W, Beck F, Meyer BI, Deschamps J. Cdx1 and Cdx2 have overlapping functions in anteroposterior patterning and posterior axis elongation. Development. 2002;129:2181–2193. doi: 10.1242/dev.129.9.2181. [DOI] [PubMed] [Google Scholar]
- van Nes J, de Graaff W, Lebrin F, Gerhard M, Beck F, Deschamps J. The Cdx4 mutation affects axial development and reveals an essential role of Cdx genes in the ontogenesis of the placental labyrinth in mice. Development. 2006;133:419–428. doi: 10.1242/dev.02216. [DOI] [PubMed] [Google Scholar]
- Wake DB. Brainstem organization and branchiomeric nerves. Acta Anat. 1993;148:124–131. doi: 10.1159/000147531. [DOI] [PubMed] [Google Scholar]
- Waskiewicz AJ, Rikhof HA, Moens CB. Eliminating zebrafish pbx proteins reveals a hindbrain ground state. Dev. Cell. 2002;3:723–733. doi: 10.1016/s1534-5807(02)00319-2. [DOI] [PubMed] [Google Scholar]
- Weinberg ES, Allende ML, Kelly CS, Abdelhamid A, Murakami T, Andermann P, Doerre OG, Grunwald DJ, Riggleman B. Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development. 1996;122:271–280. doi: 10.1242/dev.122.1.271. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book: A Guide for the Laboratory use of Zebrafish (Danio rerio) Oregon: University of Oregon Press; 1994. [Google Scholar]
- Woo K, Fraser S. Order and coherence in the fate map of the zebrafish nervous system. Development. 1995;121:2595–2609. doi: 10.1242/dev.121.8.2595. [DOI] [PubMed] [Google Scholar]
- Woo K, Fraser S. Specification of the hindbrain fate in the zebrafish. Dev. Biol. 1998;197:283–296. doi: 10.1006/dbio.1998.8870. [DOI] [PubMed] [Google Scholar]
- Xu Q, Holder N, Patient R, Wilson SW. Spatially regulated expression of three receptor tyrosine kinase genes during gastrulation in the zebrafish. Development. 1994;120:287–299. doi: 10.1242/dev.120.2.287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.