Abstract
Extensive studies of the biology of the pigment-producing cell (melanocyte) have resulted in a wealth of knowledge regarding the genetics and developmental mechanisms governing skin and hair pigmentation. The ease of identification of altered pigment phenotypes, particularly in mouse coat color mutants, facilitated early use of the pigmentary system in mammalian genetics and development. In addition to the large collection of developmental genetics data, melanocytes are of interest because their malignancy results in melanoma, a highly aggressive and frequently fatal cancer that is increasing in Caucasian populations worldwide. The genetic programs regulating melanocyte development, function, and malignancy are highly complex and only partially understood. Current research in melanocyte development and pigmentation is revealing new genes important in these processes and additional functions for previously known individual components. A detailed understanding of all the components involved in melanocyte development and function, including interactions with neighboring cells and response to environmental stimuli, will be necessary to fully comprehend this complex system. The inherent characteristics of pigmentation biology as well as the resources available to researchers in the pigment cell community make melanocytes an ideal cell type for analysis using systems biology approaches. In this review, the study of melanocyte development and pigmentation is considered as a candidate for systems biology-based analyses.
Keywords: melanocyte, pigmentation, melanoma, development, genetics
Introduction
Melanocytes are derived from a multipotent stem cell population called the neural crest. Neural crest cells migrate extensively and along specific pathways throughout developing vertebrates while proliferating and differentiating into a variety of cell types, including neurons and schwann cells of the entire peripheral nervous system in addition to melanocytes. Melanocyte precursors, melanoblasts, migrate along a dorsal-lateral pathway beneath the ectoderm, and subsequently colonize skin and hair follicles, where they complete differentiation into mature melanocytes and begin production of melanin pigment, which is transferred to skin keratinocytes and hair. Synthesis of the two forms of melanin, brown/black eumelanin and red/yellow pheomelanin, occurs within specialized organelles known as melanosomes. Pigment production by melanocytes not only produces the extensive and complex coloration patterns seen in mammals, but also provides protection from sun exposure, as melanocytes increase pigment production in skin in response to solar UV radiation (1). Other notable features of melanocytes include complex sub-cellular trafficking of the components needed to assemble functional melanosomes (2), and an intricate network of factors needed for the establishment, maintenance and differentiation of adult melanocyte stem cells within hair follicles (3–5).
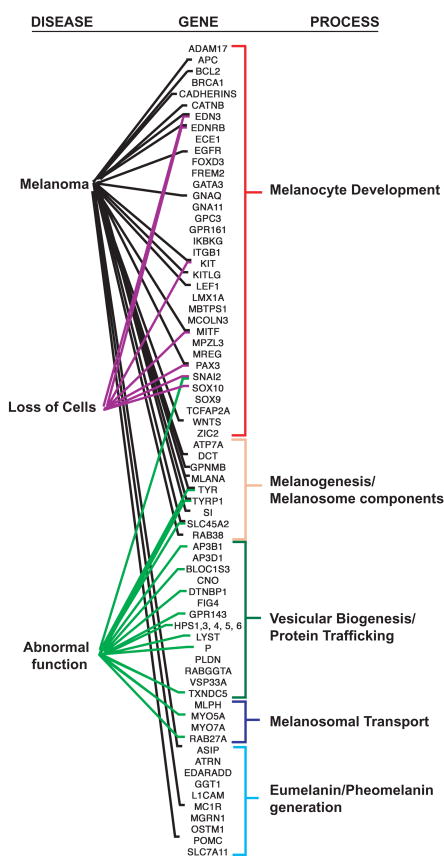
Defects in melanocyte development and function are associated with a variety of human diseases and disorders, including albinism (6), piebaldism (7), Waardenburg syndrome (WS) (8), Tietz syndrome (9), Chediak-Higashi syndrome (CHS) (10), and Hermansky-Pudlack syndrome (HPS) (11). The cancerous growth of melanocytes results in melanoma, an aggressive cancer with a high mortality rate (12). These diseases/disorders are often genetically heterogeneous, and identification of the various genetic insults that cause these abnormalities have revealed many genes and pathways that function in melanocytes (Fig. 1). Analysis and assembly of cellular pathways associated with human disorders has directed much of melanocyte research. However this type of gene-centric, reductionist research approach, which has traditionally been used in biological research, may not capture the complexity of biochemical interactions and pathways regulating neural crest/melanocyte development and pigment production, as these processes exhibit complex spatial, temporal, and environmental variations. Reductionist approaches may also overlook the full spectrum of genes/proteins involved; for example, those with no assayable phenotype when mutated, or those so fundamental that they are lethal before effects can be seen.
Figure 1.
The majority of genes known to affect melanocyte development and function have a known role in only one system or disease. Although a multitude of genes have known functions in individual melanocyte cellular processes, few have been correlated with multiple processes and/or human diseases. Each line between a gene and a cellular process or human disease indicates that this gene has been experimentally demonstrated to have a role in this process or disease. Each line between a gene and disease was validated by the presence of an article relating to both the disease and specific gene, identified by a PubMed search (http://www.ncbi.nlm.nih.gov/sites/entrez?db=PubMed). Loss of cells includes WS, and abnormal function includes albinism, piebaldism, HPS, and CHS. Most functions of the genes in these processes have been discovered on an independent basis, using gene-centric, reductionist research approaches. The absence of multiple roles for many pigmentation genes suggests that a full understanding of all the molecules involved is lacking. In the future, a full understanding of all the roles each of these genes plays will reveal many more functions for each gene, covering this diagram with lines. This figure is not meant to be exhaustive, but to illustrate the current knowledge of gene functions in melanocyte cellular systems.
Systems biology, herein defined as the systematic study of complex interactions in biological systems, endeavors to analyze biological processes from a holistic rather than reductionist perspective, seeking to define and understand systems that are by definition irreducible and unpredictable, where the outcome of the system cannot be fully predicted by independent analysis of the parts. Ultimately this information may propel biological analyses from a descriptive to predictive science, allowing a better understanding of a disease state and highlighting rational avenues for intervention. This field has emerged as large-scale screens and data collection have permitted global examination of the genome/proteome under defined experimental parameters, in combination with novel, computational biology-based modeling programs, which allow manipulation and analysis of these large data sets. Because complex computational model formulation rather than simplified mathematical formulas is the foundation of systems biology (13,14), detailed experimental data are required to lay the groundwork for future analyses at the systems level. The extensive conventional studies of genes/proteins already performed in the field of pigmentation biology will provide a solid framework to direct future analyses and computational modeling that will identify all the genes, proteins, and cofactors that regulate the unique functions attributable to melanocytes, with the ultimate goal of a complete, integrated, and dynamic picture of all communication that occurs within a melanocyte, from detailed biochemistry at the single cell level to cell-cell communication that regulates differentiation and migration. It is this system-wide understanding of all stages of melanocyte biology that will unravel melanocyte developmental programs and pigmentary diseases such as melanoma. In this review, we discuss the advantages and challenges of studying melanocytes using systems biology approaches, as well as current experimental approaches that are collecting the data necessary for future analyses.
Historical perspective
The pigmentary system has been used to model biological and disease systems for more than a century, resulting in a wealth of knowledge regarding the genetics and developmental mechanisms governing pigmentation. Historically, mouse pigmentation mutants were crucial tools for testing Mendelian theories in the early 1900s and were the first loci used to describe genetic linkage analyses in vertebrates (15,16). These studies used readily identifiable variations in pigmentation that were both heritable and easily scored (17). However it was not until the 1990’s that we learned that these traits resulted from alterations in melanocyte development, function, or interaction with neighboring keratinocytes and hairs. In the 1930’s, variations in coat pigmentation patterns of spotted piebald mice were studied as quantitative genetic traits. Crosses of piebald strains with varying patterns predicted the presence of an interactive network of determinants with quantitative effects on the extent and patterns of white spotting (18). These studies utilized holistic approaches and studied the biological system as an entire entity, aiming to understand complex interactions among multiple loci to account for the final pattern observed.
With the advent of linkage mapping and positional cloning, the molecular defects and genes responsible for many of the single gene mutations affecting pigmentation were discovered. Today, more than 100 genes regulating melanocyte development and pigmentation have been identified (19,20). These genes can be categorized by the different functions assigned to them while analyzing observed phenotypic perturbations correlated with the presence of mutant alleles. Many disease-associated genes have also been studied in animal models with similar traits, providing insight into both melanocyte development/function and human diseases (21,22). While these studies have provided a solid base for establishing linear relationships in the melanocytic pathways, our knowledge is often limited to information about a gene’s role in the particular disease or stage of development in which it was discovered. Figure 1 illustrates the known roles for many genes in various melanocyte processes and the focal points from which interaction pathways can be constructed. These processes include melanoblast development and migration, regulated by cascades of transcription factors; formation of the melanosome, regulated by factors that contribute to organelle biogenesis and vesicular transport; melanogenesis, an enzymatic process; melanosome transfer to melanocyte dendrites, regulated by actin transport molecules; and pigment formation in response to environmental stimuli. However, this framework of knowledge lacks a full understanding of all molecules involved and all roles of individual molecules, as reductionist research has by necessity focused on one gene or pathway at a time. It is likely that genes with only one known function in melanocyte systems are in fact working in multiple systems.
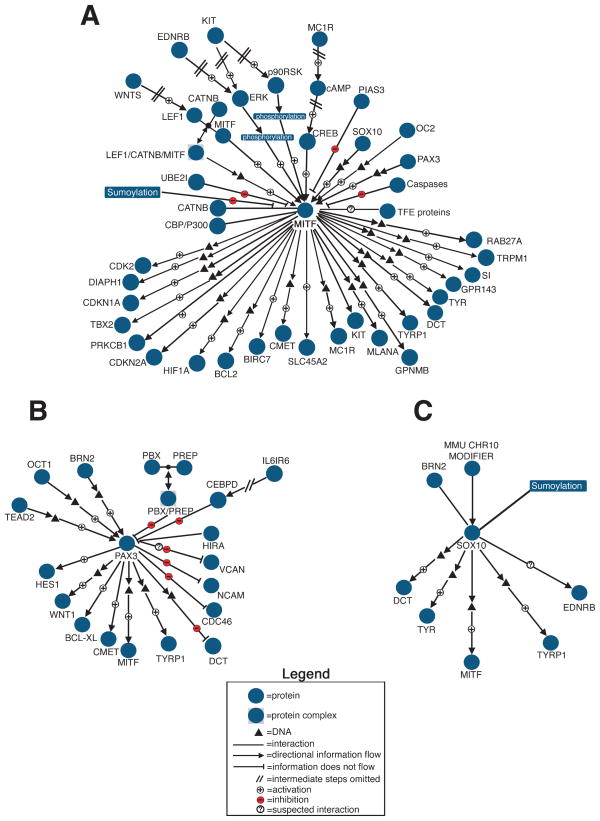
Melanocyte research has often focused on a subset of these genes in which a molecular defect was associated with a mouse model or human disease, providing impetus for a series of studies to understand their mode of action. For example, much has been learned about melanocyte development by studying the function of the three transcription factors MITF, PAX3, and SOX10. The pathways of these genes interact to regulate crucial aspects of melanocyte development and function, and each is mutated in various forms of WS (8). Many of the interactions documented for these genes are shown in Figure 2. Microphthalmia-associated Transcription Factor (MITF) has been termed the “melanocyte master regulator”, because it plays such a central role in melanocyte development and function (Fig. 2a). It is required for melanocyte differentiation and survival, activates transcription of melanogenic enzymes and melanogenic proteins, and is associated with melanoma progression (23–32). MITF also governs numerous other cellular functions in the melanocyte, including environmental response (27,33), cell survival (34,35), cell motility (36), and cell cycle progression (37–39). MITF itself undergoes complex post-transcriptional regulation, including phosphorylation, sumoylation, ubiquitination, and caspase cleavage (40–45). Paired box gene 3 (PAX3) has a broader expression pattern than MITF, regulating neural tube closure, early development of myoblast and neural crest lineages, and the formation of nervous, muscular, cardiovascular and melanocyte systems (Fig. 2b). PAX3 regulation of early neural crest development appears to maintain neural crest stem cell properties via inhibition of apoptosis (46–48), and PAX3 has been proposed to inhibit apoptosis in melanoma (49,50). In melanocytes, PAX3 activates transcription of MITF (51–53) and plays a crucial role in maintaining melanocyte stem cells (3). SRY-box containing gene 10 (SOX10) regulates specification of neural crest-derived melanocytes, neurons, and glia (54) (Fig. 2c). SOX10 strongly activates MITF and regulates expression of melanogenic enzymes (51,52,55–58). Upstream regulation of SOX10, as well as additional downstream targets, interacting factors, and posttranslational modifications are only beginning to be ascertained (59–62). In summary, MITF, PAX3, and SOX10 serve to illustrate that the extensive data currently known on cellular processes governing melanocyte biology will provide an ideal foundation for future systems biology analyses in these cells.
Figure 2.
Navigational maps of the regulation and downstream targets of (A) MITF, (B) PAX3, and (C) SOX10, three key transcription factors that govern melanocyte development. Illustrated are the pathways known to be active in developing neural crest precursors to melanoblasts, melanoblasts themselves, or mature melanocytes of mammalian organisms. The black triangles represent DNA promoters/enhancers, illustrating direct transcription factor binding; the absence of direct binding illustrates indirect regulation. Pathways that appear unique to melanoma, details of post-translational modifications, as well as interactions identified in only non-melanocyte cell types or non-mammalian systems to date are excluded from this diagram.
Alongside traditional gene discovery methods, the techniques of germline mutagenesis and RNA interference (RNAi) are increasing this essential foundation and identifying novel pathways important to melanocyte biology. Chemical mutagenesis screens in model systems such as mice and fish have discovered new molecules that regulate melanocyte development/function (63–69). As mutations in some genes alone may cause embryonic lethality or have melanocyte phenotypes too subtle to measure, screens have been established in mice with a known mutation that disrupts melanoblast homeostasis, thus sensitizing the animals to modest perturbations but resulting in reproducible phenotypes (70–72). While these approaches search for random loci that alter melanocyte function in the context of the whole animal, many aspects of melanocyte biology can be studied in melanocytes isolated away from the organism. Genome wide approaches that knockdown gene expression in melanocyte cell culture have provided useful insights into function and disease, discovering new genes and cellular pathways functioning in melanocytes. For example, a recent genome-wide siRNA screen in human melanocytes identified 92 novel genes that function in melanogenesis, and also revealed a requirement for autophagy in melanin production (73), and an siRNA screen in melanoma identified a candidate gene for metastasis (74).
Both these techniques are powerful tools that can be employed to expand our knowledge of melanocyte biology; however, each technique comes with its own limitations. Using germline mutagenesis to generate multiple functional variants in all genes (ie. loss of function, hypomorphic, and gain of function) would require generation and maintenance of large numbers of animals, which is often cost prohibitive for vertebrate research. Moreover, the phenotypic screening required is labor intensive and requires development of efficient yet sensitive phenotyping methods (75). Screening of genome mutations using cell-based assays avoids some of these cost limitations, but in these assays the cells are usually taken out of context for in vitro phenotype assessment. Moreover, random mutagenesis approaches using cell-based systems often use pooled populations where mutagenic events need to be determined after screening or selection. RNAi screening techniques can overcome this limitation by using an arrayed format of siRNA delivery, but a limited spectrum of gene alterations can be assessed, in most cases hypomorphs. RNAi approaches also require stringent internal control experiments to prevent generation of false positives, necessitating consistent standards of RNAi screening and ongoing development of statistical methodology applicable to RNAi screens (76,77).
Genomic-scale approaches: Moving melanocyte biology towards a systems-level understanding
Given the solid foundation provided by previous melanocyte biology/disease research, the field is poised to take full advantage of the rapid technological advances that are allowing systems biology-focused analysis to be feasible and cost effective. Because it is now possible to analyze melanocytes at various stages of development and disease in both model organisms and humans, the response of migrating melanoblasts to temporal and spatial differences and the alteration of melanoblast patterns based upon their developmental stage and embryonic location can be studied. Global analyses of these various stages will illuminate the detailed molecular programming that regulates development and the misprogramming that occurs in disease. One of the first steps that will lay the groundwork for these stage-specific global analyses is application of genome-wide approaches to identify all the genes and proteins that function in melanocyte systems. Many recent studies have made great strides towards this endeavor, as described below.
Significant effort has been made to catalog the proteins and transcripts of melanocytes, both identifying them and analyzing their changes based upon temporo-spatial or environmental differences. A proteomics analysis of the melanosome provided a catalog of the ≈1500 total proteins present in this organelle, and subdivided these data based upon various stages of melanosome maturation, revealing the similarities and differences of protein expression among melanosome stages (78–80). Gene expression in mature melanocytes has been studied by several groups, including analysis of melanocytes isolated from hair follicles (5,81), fractionated skin layers (82), and melanocytes grown in culture (83,84), thus providing comprehensive information on the mature melanocyte transcriptome. Limitations of proteomics and gene expression profiling techniques involve the inherent problems of variability that occur at multiple levels of the experiment and are unique for each platform used, resulting in false positives or false negatives. For example, hybridization-based gene expression studies are limited to those genes or splice variants represented in the baseline dataset, thus an incomplete representation of the proteome/transcriptome will generate false negatives. Hybridization-based microarray expression analyses are also limited in their ability to discern absolute expression values. However these issues are not present with emerging Next-Generation Sequencing-based technologies, which use counting-based approaches to measure expression values and are unrestricted in sequence selection (85). Regardless of the platform used, RNA/protein source homogeneity is a limitation of these studies, as the experimental results are reflective of a representative average of the pooled population of cells and do not capture individual variations that may occur from variables such as cell cycle or cellular interactions. Also, significant barriers remain in data analysis and in data comparison among platforms and labs (86,87). Nevertheless, these studies represent an excellent starting point for characterization of the proteins and transcripts that make up pigment cells.
The many components involved in transcriptional regulation of melanocytic genes need to be fully elucidated to lay the foundation for a systems biology approach. Transcription start sites for melanocyte-expressed genes have been predicted computationally and many have been validated from alignment of cDNA to genomic sequence (88). A small number of genes have been analyzed to identify consensus sequence elements recognized by a subset of transcription factors, such as MITF (89). These approaches need to be expanded to the entire genome for multiple transcriptional regulators, and also need to include more analysis of genomic elements, modifications and structure, as has been done for a subset of the human genome (90). Comparative analysis of genomic sequence from multiple species has allowed the identification of candidate functional genomic regions based upon their evolutionary conservation, an excellent example being the discovery of regulatory regions for the Sox10 locus (91–93). In the near future, this type of analysis will be expanded using predictive algorithms and Chromatin Immunoprecipitation coupled with Next Generation Sequencing-based technologies (94) to identify relevant transcription factor binding sequences in an unbiased manner. Applying these approaches to melanocytes from various developmental stages and disease states will aid in our interpretation of changes in networks.
The study of differential responses to perturbations in homeostasis can also provide useful insights into how networks respond to defined stimuli. Melanocyte biology has exploited this approach for examining global changes in response to stimulation of signaling pathways as well as environmental insults such as UV exposure. UV radiation results in activation of multiple cellular processes in melanocytes, including the Melanocortin1 receptor pathway, which stimulates cAMP formation and subsequently initiates melanogenesis, stimulates proliferation, inhibits apoptosis, and enhances DNA repair of the direct mutagenic effects of UV radiation on DNA (95). Several studies have made use of microarray analysis of UV-irradiated melanocytes to catalog the resulting changes in mRNA expression (96–98), and applied systems analyses approaches to identify the numerous pathways involved (98). Future studies need to expand these analyses to a more global genomic level and to also measure the effects of additional stimuli.
Applying a systems biology approach to determine how normal melanocyte gene expression is altered during melanoma progression and metastasis with respect to global genomic, RNA, and protein differences will provide a solid foundation to apply target-specific therapeutic approaches to this disease. Such a comprehensive approach to investigate melanoma is required given that melanomas show significant genetic heterogeneity (99–102). Genomic-based technologies have identified large regions of the genome that exhibit DNA copy number changes which correlate with melanoma and genes relevant for melanoma progression (103,104). Numerous gene expression profile studies have been performed using melanoma samples (http://www.ncbi.nlm.nih.gov/geo/), identifying melanoma subtypes based upon global gene expression profiles (99,101,102), providing details of melanoma genetics, such as the discovery of Wnt5a misregulation resulting in increased motility in melanoma (99,105,106), and providing details of basic melanocyte biology, such as the discovery of Rab38 function in melanosomal protein trafficking (107). Numerous independent melanoma expression analysis studies have been performed on comparable microarray platforms (101,102,108–110), allowing for pooling of data that yields increased statistical power in subsequent analysis. This technique facilitated the discovery of novel MITF target genes by identifying transcripts that exhibited a correlation to MITF gene expression (83). These expression analyses have had a tremendous impact on our understanding of melanoma biology, but are only beginning to unravel the complexity of melanoma biology (111). The relative ease with which melanoma cells can be cultured from biopsy samples will provide the biological materials needed for future studies of the network signaling changes associated with various stages of progression of this devastating disease.
The extensive pre-existing knowledge of melanocyte biology has allowed assessment of the consistency and relevance of the genome-wide approaches described above. For example, proteomics analysis of melanosomes detected many of the genes identified by reductionist approaches to be involved in the biology of pigmentation, and gene expression analyses aimed at identifying targets of MITF pinpointed previously identified targets (78–80,83). This correspondence with previously known data validates the ability of comprehensive, genome platform-based techniques to find new molecules and interactions, demonstrating that the integration of reductionist and genome-wide approaches will be useful for both modeling and discovery.
Conclusion
Scientists within the pigment research community are well-integrated (http://www.ifpcs.org/) and readily embrace new technology, two characteristics necessary to advance pigment biology into the realm of systems biology. Pigment biology researchers should keep the goal of a systems biology understanding in mind as they expand our knowledge of the molecules involved in pigmentation, including standardization of techniques to facilitate data comparison, and collection of quantitative data such as protein concentrations or mRNA/protein dynamics. Although key molecules in melanocyte development and function are already known, system-wide approaches will circumvent preconceived hypotheses on the global importance of the few well-studied factors and accelerate our discovery of additional relevant pathways.
Useful resources available for melanocyte research should be fully employed, such as immortalized melanocyte cell lines. These cell lines have been isolated from a variety of animals with different genetic backgrounds and are readily manipulated, allowing introduction of precise developmental or environmental insults and detailed expression analyses. For example, cell lines harboring mutations in pigmentary genes along with Ink4a-Arf deletions allow comparison of defined changes in pigment genes in the context of a uniform, minimally rearranged genomic background (http://www.sgul.ac.uk/depts/anatomy/pages/Dot/Cell%20bank%20holdings.htm). Full-length mouse cDNA libraries constructed from pigmented melanoma and immortalized albino embryonic melanocyte cell lines are a readily available collection of genes expressed in these pigment cells of differing origin (112,113).
Software is available for initial studies annotating and displaying biological systems (for example Cytoscape, http://www.cytoscape.org/features2.php, and Metacore, http://www.genego.com/metacore.php), but subsequent analyses will require more complete computational modeling of biological systems. Much work remains to overcome the computational challenges of biomedical data integration and to develop computational modeling that can demonstrate the complexity of biological systems in a user-friendly output. However, recent advances have begun to allow computational modeling of cellular processes on a large scale, showing promise for these computational tools to be accessible to non-theorists (13). For example, recent mathematical modeling of the dynamics of transcriptional control of gene expression (114) will be relevant to a full understanding of MITF, SOX10, and PAX3 functions.
Additional challenges include the development of experimental approaches to provide precise details of molecular interactions within melanocytes, such as subcellular localization studies, single-cell analyses, kinetic approaches, in vivo analyses, and systematic chemical pertubations. These approaches have the potential to reveal the activities of proteins within individual cells at high resolution, uncovering details of gene/protein expression that could be obscured by averaging data over a population of cells. This was recently shown by retrovirus-mediated fluorescent labeling of proteins within human cells (115), and could be applied to melanocytes, for example to extend the gene expression profiling of individual mouse melanocyte stem cells (5). Cultures of primary neural crest cells also have the potential to allow real-time, single cell studies of melanocyte development.
Single-cell isolation studies necessitate removal from the in vivo environment, thus destroying melanocyte contacts with neighboring keratinocytes or hair cells as well as all extracellular interactions regulating development, growth, or pigment production. These interactions within the correct temporal and spatial context are crucial to a systems biology understanding of melanocyte development and function, and could be addressed by harnessing recent technological advances for in vivo melanocyte studies. For example, innovations in nanoparticle technology allow targeted introduction of small molecules into living systems, coupled with three-dimensional live cell imaging in vivo (116,117), opening up new possibilities in the resolution at which single molecules can be observed, tracked and manipulated to query cellular processes in live cells. These technologies could be tailored to suit the unique biology of the melanocyte, for example by selective labeling of melanosomes, as was recently done for lysosomes of lung cancer cells utilizing pH-activitable fluorophores (118), and even extended to melanocyte visualization within the hair follicle of living organisms, as the hair follicle is ideally suited for in vivo modification with little intervention. The feasibility of in vivo analyses of reporter gene expression with single-cell resolution has recently been demonstrated in C. elegans embryos (119), giving promise that this technology may eventually be extended to vertebrate embryonic development.
In the past, reductionist experiments have been designed to evaluate the results obtained from drastic alterations in a single pathway, which in and of itself creates a bias in the type of data collected. The challenge for the future is to consider ways to query melanocyte biology with respect to the contribution of multiple pathways at varying levels of induction, to gain a more complete understanding of how responses to biological processes are coordinated within the cell and ultimately within the entire organism. Such application of systems biology approaches to investigate melanocyte biology will allow the molecules and the associated quantitative measurements within the networks governing melanogenesis and melanocyte function to be determined. The utilization of this data along with advances in computational modeling will ultimately allow a more detailed understanding of the fundamental systems at work in melanocytes.
Acknowledgments
This research was supported by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health.
Contributor Information
Laura L Baxter, NIH/NHGRI/GDRB.
Stacie K Loftus, NIH/NHGRI/GDRB.
William J Pavan, NIH/NHGRI/GDRB.
References
- 1.Miyamura Y, Coelho SG, Wolber R, Miller SA, Wakamatsu K, Zmudzka BZ, Ito S, Smuda C, Passeron T, Choi W, Batzer J, Yamaguchi Y, Beer JZ, Hearing VJ. Regulation of human skin pigmentation and responses to ultraviolet radiation. Pigment Cell Res. 2007;20(1):2–13. doi: 10.1111/j.1600-0749.2006.00358.x. [DOI] [PubMed] [Google Scholar]
- 2.Hearing VJ. Biogenesis of pigment granules: a sensitive way to regulate melanocyte function. J Dermatol Sci. 2005;37(1):3–14. doi: 10.1016/j.jdermsci.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 3.Lang D, Lu MM, Huang L, Engleka KA, Zhang M, Chu EY, Lipner S, Skoultchi A, Millar SE, Epstein JA. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature. 2005;433(7028):884–887. doi: 10.1038/nature03292. [DOI] [PubMed] [Google Scholar]
- 4.Nishimura EK, Granter SR, Fisher DE. Mechanisms of hair graying: incomplete melanocyte stem cell maintenance in the niche. Science. 2005;307(5710):720–724. doi: 10.1126/science.1099593. [DOI] [PubMed] [Google Scholar]
- 5.Osawa M, Egawa G, Mak SS, Moriyama M, Freter R, Yonetani S, Beermann F, Nishikawa S. Molecular characterization of melanocyte stem cells in their niche. Development. 2005;132(24):5589–5599. doi: 10.1242/dev.02161. [DOI] [PubMed] [Google Scholar]
- 6.Oetting WS, King RA. Molecular basis of albinism: mutations and polymorphisms of pigmentation genes associated with albinism. Hum Mutat. 1999;13(2):99–115. doi: 10.1002/(SICI)1098-1004(1999)13:2<99::AID-HUMU2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 7.Thomas I, Kihiczak GG, Fox MD, Janniger CK, Schwartz RA. Piebaldism: an update. Int J Dermatol. 2004;43(10):716–719. doi: 10.1111/j.1365-4632.2004.02114.x. [DOI] [PubMed] [Google Scholar]
- 8.Tachibana M, Kobayashi Y, Matsushima Y. Mouse models for four types of Waardenburg syndrome. Pigment Cell Res. 2003;16(5):448–454. doi: 10.1034/j.1600-0749.2003.00066.x. [DOI] [PubMed] [Google Scholar]
- 9.Smith SD, Kelley PM, Kenyon JB, Hoover D. Tietz syndrome (hypopigmentation/deafness) caused by mutation of MITF. J Med Genet. 2000;37(6):446–448. doi: 10.1136/jmg.37.6.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shiflett SL, Kaplan J, Ward DM. Chediak-Higashi Syndrome: a rare disorder of lysosomes and lysosome related organelles. Pigment Cell Res. 2002;15(4):251–257. doi: 10.1034/j.1600-0749.2002.02038.x. [DOI] [PubMed] [Google Scholar]
- 11.Wei ML. Hermansky-Pudlak syndrome: a disease of protein trafficking and organelle function. Pigment Cell Res. 2006;19(1):19–42. doi: 10.1111/j.1600-0749.2005.00289.x. [DOI] [PubMed] [Google Scholar]
- 12.Chin L, Garraway LA, Fisher DE. Malignant melanoma: genetics and therapeutics in the genomic era. Genes Dev. 2006;20(16):2149–2182. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- 13.Klauschen F, Angermann BR, Meier-Schellersheim M. Understanding diseases by mouse click: the promise and potential of computational approaches in Systems Biology. Clin Exp Immunol. 2007;149(3):424–429. doi: 10.1111/j.1365-2249.2007.03472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sorger PK. A reductionist’s systems biology: opinion. Curr Opin Cell Biol. 2005;17(1):9–11. doi: 10.1016/j.ceb.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 15.Castle WE, Allen G. The heredity of albinism. Proc Am Acad Arts Sci. 1903;38:603–621. [Google Scholar]
- 16.Haldane JBS, Sprunt AD, Haldane NM. Reduplication in mice. J Genet. 1915;5:133–135. [Google Scholar]
- 17.Silvers WK. The Coat Colors of Mice: A Model for Mammalian Gene Action and Interaction. Springer Verlag; New York: 1979. [Google Scholar]
- 18.Dunn LC, Charles DR. Studies on Spotting Patterns I. Analysis of Quantitative Variations in the Pied Spotting of the House Mouse. Genetics. 1937;22(1):14–42. doi: 10.1093/genetics/22.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennett DC, Lamoreux ML. The color loci of mice--a genetic century. Pigment Cell Res. 2003;16(4):333–344. doi: 10.1034/j.1600-0749.2003.00067.x. [DOI] [PubMed] [Google Scholar]
- 20.Oetting WS, Montoliu L, Bennett DC. Color Genes. Volume 2008: European Society for Pigment Cell Research. http://www.espcr.org/micemut.
- 21.Barsh GS. The genetics of pigmentation: from fancy genes to complex traits. Trends Genet. 1996;12(8):299–305. doi: 10.1016/0168-9525(96)10031-7. [DOI] [PubMed] [Google Scholar]
- 22.Jackson IJ. Homologous pigmentation mutations in human, mouse and other model organisms. Hum Mol Genet. 1997;6(10):1613–1624. doi: 10.1093/hmg/6.10.1613. [DOI] [PubMed] [Google Scholar]
- 23.Baxter LL, Pavan WJ. The oculocutaneous albinism type IV gene Matp is a new marker of pigment cell precursors during mouse embryonic development. Mech Dev. 2002;116(1–2):209–212. doi: 10.1016/s0925-4773(02)00130-2. [DOI] [PubMed] [Google Scholar]
- 24.Baxter LL, Pavan WJ. Pmel17 expression is Mitf-dependent and reveals cranial melanoblast migration during murine development. Gene Expr Patterns. 2003;3(6):703–707. doi: 10.1016/j.modgep.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Bentley NJ, Eisen T, Goding CR. Melanocyte-specific expression of the human tyrosinase promoter: activation by the microphthalmia gene product and role of the initiator. Mol Cell Biol. 1994;14(12):7996–8006. doi: 10.1128/mcb.14.12.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du J, Fisher DE. Identification of Aim-1 as the underwhite mouse mutant and its transcriptional regulation by MITF. J Biol Chem. 2002;277(1):402–406. doi: 10.1074/jbc.M110229200. [DOI] [PubMed] [Google Scholar]
- 27.Aoki H, Moro O. Involvement of microphthalmia-associated transcription factor (MITF) in expression of human melanocortin-1 receptor (MC1R) Life Sci. 2002;71(18):2171–2179. doi: 10.1016/s0024-3205(02)01996-3. [DOI] [PubMed] [Google Scholar]
- 28.Hodgkinson CA, Moore KJ, Nakayama A, Steingrimsson E, Copeland NG, Jenkins NA, Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74(2):395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- 29.Hughes MJ, Lingrel JB, Krakowsky JM, Anderson KP. A helix-loop-helix transcription factor-like gene is located at the mi locus. J Biol Chem. 1993;268(28):20687–20690. [PubMed] [Google Scholar]
- 30.Tachibana M, Takeda K, Nobukuni Y, Urabe K, Long JE, Meyers KA, Aaronson SA, Miki T. Ectopic expression of MITF, a gene for Waardenburg syndrome type 2, converts fibroblasts to cells with melanocyte characteristics. Nat Genet. 1996;14(1):50–54. doi: 10.1038/ng0996-50. [DOI] [PubMed] [Google Scholar]
- 31.Yasumoto K, Yokoyama K, Takahashi K, Tomita Y, Shibahara S. Functional analysis of microphthalmia-associated transcription factor in pigment cell-specific transcription of the human tyrosinase family genes. J Biol Chem. 1997;272(1):503–509. doi: 10.1074/jbc.272.1.503. [DOI] [PubMed] [Google Scholar]
- 32.Yokoyama K, Yasumoto K, Suzuki H, Shibahara S. Cloning of the human DOPAchrome tautomerase/tyrosinase-related protein 2 gene and identification of two regulatory regions required for its pigment cell-specific expression. J Biol Chem. 1994;269(43):27080–27087. [PubMed] [Google Scholar]
- 33.Sato-Jin K, Nishimura EK, Akasaka E, Huber W, Nakano H, Miller A, Du J, Wu M, Hanada K, Sawamura D, Fisher DE, Imokawa G. Epistatic connections between microphthalmia-associated transcription factor and endothelin signaling in Waardenburg syndrome and other pigmentary disorders. FASEB J. 2008;22(4):1155–1168. doi: 10.1096/fj.07-9080com. [DOI] [PubMed] [Google Scholar]
- 34.Buscà R, Berra E, Gaggioli C, Khaled M, Bille K, Marchetti B, Thyss R, Fitsialos G, Larribère L, Bertolotto C, Virolle T, Barbry P, Pouysségur J, Ponzio G, Ballotti R. Hypoxia-inducible factor 1{alpha} is a new target of microphthalmia-associated transcription factor (MITF) in melanoma cells. J Cell Biol. 2005;170(1):49–59. doi: 10.1083/jcb.200501067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGill GG, Horstmann M, Widlund HR, Du J, Motyckova G, Nishimura EK, Lin YL, Ramaswamy S, Avery W, Ding HF, Jordan SA, Jackson IJ, Korsmeyer SJ, Golub TR, Fisher DE. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109(6):707–718. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- 36.McGill GG, Haq R, Nishimura EK, Fisher DE. c-Met expression is regulated by Mitf in the melanocyte lineage. J Biol Chem. 2006;281(15):10365–10373. doi: 10.1074/jbc.M513094200. [DOI] [PubMed] [Google Scholar]
- 37.Carreira S, Goodall J, Aksan I, La Rocca SA, Galibert MD, Denat L, Larue L, Goding CR. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature. 2005;433(7027):764–769. doi: 10.1038/nature03269. [DOI] [PubMed] [Google Scholar]
- 38.Carreira S, Liu B, Goding CR. The gene encoding the T-box factor Tbx2 is a target for the microphthalmia-associated transcription factor in melanocytes. J Biol Chem. 2000;275(29):21920–21927. doi: 10.1074/jbc.M000035200. [DOI] [PubMed] [Google Scholar]
- 39.Du J, Widlund HR, Horstmann MA, Ramaswamy S, Ross K, Huber WE, Nishimura EK, Golub TR, Fisher DE. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell. 2004;6(6):565–576. doi: 10.1016/j.ccr.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 40.Hemesath TJ, Price ER, Takemoto C, Badalian T, Fisher DE. MAP kinase links the transcription factor Microphthalmia to c-Kit signalling in melanocytes. Nature. 1998;391(6664):298–301. doi: 10.1038/34681. [DOI] [PubMed] [Google Scholar]
- 41.Larribere L, Hilmi C, Khaled M, Gaggioli C, Bille K, Auberger P, Ortonne JP, Ballotti R, Bertolotto C. The cleavage of microphthalmia-associated transcription factor, MITF, by caspases plays an essential role in melanocyte and melanoma cell apoptosis. Genes Dev. 2005;19(17):1980–1985. doi: 10.1101/gad.335905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller AJ, Levy C, Davis IJ, Razin E, Fisher DE. Sumoylation of MITF and its related family members TFE3 and TFEB. J Biol Chem. 2005;280(1):146–155. doi: 10.1074/jbc.M411757200. [DOI] [PubMed] [Google Scholar]
- 43.Murakami H, Arnheiter H. Sumoylation modulates transcriptional activity of MITF in a promoter-specific manner. Pigment Cell Res. 2005;18(4):265–277. doi: 10.1111/j.1600-0749.2005.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu M, Hemesath TJ, Takemoto CM, Horstmann MA, Wells AG, Price ER, Fisher DZ, Fisher DE. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev. 2000;14(3):301–312. [PMC free article] [PubMed] [Google Scholar]
- 45.Xu W, Gong L, Haddad MM, Bischof O, Campisi J, Yeh ET, Medrano EE. Regulation of microphthalmia-associated transcription factor MITF protein levels by association with the ubiquitin-conjugating enzyme hUBC9. Exp Cell Res. 2000;255(2):135–143. doi: 10.1006/excr.2000.4803. [DOI] [PubMed] [Google Scholar]
- 46.Margue CM, Bernasconi M, Barr FG, Schafer BW. Transcriptional modulation of the anti-apoptotic protein BCL-XL by the paired box transcription factors PAX3 and PAX3/FKHR. Oncogene. 2000;19(25):2921–2929. doi: 10.1038/sj.onc.1203607. [DOI] [PubMed] [Google Scholar]
- 47.Pani L, Horal M, Loeken MR. Rescue of neural tube defects in Pax-3-deficient embryos by p53 loss of function: implications for Pax-3- dependent development and tumorigenesis. Genes Dev. 2002;16(6):676–680. doi: 10.1101/gad.969302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Underwood TJ, Amin J, Lillycrop KA, Blaydes JP. Dissection of the functional interaction between p53 and the embryonic proto-oncoprotein PAX3. FEBS Lett. 2007;581(30):5831–5835. doi: 10.1016/j.febslet.2007.11.056. [DOI] [PubMed] [Google Scholar]
- 49.He SJ, Stevens G, Braithwaite AW, Eccles MR. Transfection of melanoma cells with antisense PAX3 oligonucleotides additively complements cisplatin-induced cytotoxicity. Mol Cancer Ther. 2005;4(6):996–1003. doi: 10.1158/1535-7163.MCT-04-0252. [DOI] [PubMed] [Google Scholar]
- 50.Muratovska A, Zhou C, He S, Goodyer P, Eccles MR. Paired-Box genes are frequently expressed in cancer and often required for cancer cell survival. Oncogene. 2003;22(39):7989–7997. doi: 10.1038/sj.onc.1206766. [DOI] [PubMed] [Google Scholar]
- 51.Bondurand N, Pingault V, Goerich DE, Lemort N, Sock E, Le Caignec C, Wegner M, Goossens M. Interaction among SOX10, PAX3 and MITF, three genes altered in Waardenburg syndrome. Hum Mol Genet. 2000;9(13):1907–1917. doi: 10.1093/hmg/9.13.1907. [DOI] [PubMed] [Google Scholar]
- 52.Potterf SB, Furumura M, Dunn KJ, Arnheiter H, Pavan WJ. Transcription factor hierarchy in Waardenburg syndrome: regulation of MITF expression by SOX10 and PAX3. Hum Genet. 2000;107(1):1–6. doi: 10.1007/s004390000328. [DOI] [PubMed] [Google Scholar]
- 53.Watanabe A, Takeda K, Ploplis B, Tachibana M. Epistatic relationship between Waardenburg syndrome genes MITF and PAX3. Nat Genet. 1998;18(3):283–286. doi: 10.1038/ng0398-283. [DOI] [PubMed] [Google Scholar]
- 54.Mollaaghababa R, Pavan WJ. The importance of having your SOX on: role of SOX10 in the development of neural crest-derived melanocytes and glia. Oncogene. 2003;22(20):3024–3034. doi: 10.1038/sj.onc.1206442. [DOI] [PubMed] [Google Scholar]
- 55.Hou L, Arnheiter H, Pavan WJ. Interspecies difference in the regulation of melanocyte development by SOX10 and MITF. Proc Natl Acad Sci U S A. 2006;103(24):9081–9085. doi: 10.1073/pnas.0603114103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murisier F, Guichard S, Beermann F. A conserved transcriptional enhancer that specifies Tyrp1 expression to melanocytes. Dev Biol. 2006;298(2):644–655. doi: 10.1016/j.ydbio.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 57.Murisier F, Guichard S, Beermann F. The tyrosinase enhancer is activated by Sox10 and Mitf in mouse melanocytes. Pigment Cell Res. 2007;20(3):173–184. doi: 10.1111/j.1600-0749.2007.00368.x. [DOI] [PubMed] [Google Scholar]
- 58.Potterf SB, Mollaaghababa R, Hou L, Southard-Smith EM, Hornyak TJ, Arnheiter H, Pavan WJ. Analysis of SOX10 function in neural crest-derived melanocyte development: SOX10-dependent transcriptional control of dopachrome tautomerase. Dev Biol. 2001;237(2):245–257. doi: 10.1006/dbio.2001.0372. [DOI] [PubMed] [Google Scholar]
- 59.Cook AL, Smith AG, Smit DJ, Leonard JH, Sturm RA. Co-expression of SOX9 and SOX10 during melanocytic differentiation in vitro. Exp Cell Res. 2005;308(1):222–235. doi: 10.1016/j.yexcr.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 60.Hakami RM, Hou L, Baxter LL, Loftus SK, Southard-Smith EM, Incao A, Cheng J, Pavan WJ. Genetic evidence does not support direct regulation of EDNRB by SOX10 in migratory neural crest and the melanocyte lineage. Mech Dev. 2006;123(2):124–134. doi: 10.1016/j.mod.2005.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stanchina L, Baral V, Robert F, Pingault V, Lemort N, Pachnis V, Goossens M, Bondurand N. Interactions between Sox10, Edn3 and Ednrb during enteric nervous system and melanocyte development. Dev Biol. 2006;295(1):232–249. doi: 10.1016/j.ydbio.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 62.Yokoyama S, Takeda K, Shibahara S. SOX10, in combination with Sp1, regulates the endothelin receptor type B gene in human melanocyte lineage cells. FEBS J. 2006;273(8):1805–1820. doi: 10.1111/j.1742-4658.2006.05200.x. [DOI] [PubMed] [Google Scholar]
- 63.Fitch KR, McGowan KA, Van Raamsdonk CD, Fuchs H, Lee D, Puech A, Hérault Y, Threadgill DW, Hrabé de Angelis M, Barsh GS. Genetics of dark skin in mice. Genes Dev. 2003;17(2):214–228. doi: 10.1101/gad.1023703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kelsh RN, Brand M, Jiang YJ, Heisenberg CP, Lin S, Haffter P, Odenthal J, Mullins MC, van Eeden FJ, Furutani-Seiki M, Granato M, Hammerschmidt M, Kane DA, Warga RM, Beuchle D, Vogelsang L, Nüsslein-Volhard C. Zebrafish pigmentation mutations and the processes of neural crest development. Development. 1996;123:369–389. doi: 10.1242/dev.123.1.369. [DOI] [PubMed] [Google Scholar]
- 65.McGowan KA, Aradhya S, Fuchs H, de Angelis MH, Barsh GS. A mouse keratin 1 mutation causes dark skin and epidermolytic hyperkeratosis. J Invest Dermatol. 2006;126(5):1013–1016. doi: 10.1038/sj.jid.5700241. [DOI] [PubMed] [Google Scholar]
- 66.McGowan KA, Li JZ, Park CY, Beaudry V, Tabor HK, Sabnis AJ, Zhang W, Fuchs H, de Angelis MH, Myers RM, Attardi LD, Barsh GS. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40(8):963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rawls JF, Frieda MR, McAdow AR, Gross JP, Clayton CM, Heyen CK, Johnson SL. Coupled mutagenesis screens and genetic mapping in zebrafish. Genetics. 2003;163(3):997–1009. doi: 10.1093/genetics/163.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van Raamsdonk CD, Fitch KR, Fuchs H, de Angelis MH, Barsh GS. Effects of G-protein mutations on skin color. Nat Genet. 2004;36(9):961–968. doi: 10.1038/ng1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bogani D, Warr N, Elms P, Davies J, Tymowska-Lalanne Z, Goldsworthy M, Cox RD, Keays DA, Flint J, Wilson V, Nolan P, Arkell R. New semidominant mutations that affect mouse development. Genesis. 2004;40(2):109–117. doi: 10.1002/gene.20071. [DOI] [PubMed] [Google Scholar]
- 70.Buac K, Watkins-Chow DE, Loftus SK, Larson DM, Incao A, Gibney G, Pavan WJ. A Sox10 expression screen identifies an amino acid essential for Erbb3 function. PLoS Genet. 2008;4(9):e1000177. doi: 10.1371/journal.pgen.1000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matera I, Cockroft JL, Moran JL, Beier DR, Goldowitz D, Pavan WJ. A mouse model of Waardenburg syndrome type IV resulting from an ENU-induced mutation in endothelin 3. Pigment Cell Res. 2007;20(3):210–215. doi: 10.1111/j.1600-0749.2007.00371.x. [DOI] [PubMed] [Google Scholar]
- 72.Matera I, Watkins-Chow DE, Loftus SK, Hou L, Incao A, Silver DL, Rivas C, Elliott EC, Baxter LL, Pavan WJ. A sensitized mutagenesis screen identifies Gli3 as a modifier of Sox10 neurocristopathy. Hum Mol Genet. 2008;17(14):2118–2131. doi: 10.1093/hmg/ddn110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ganesan AK, Ho H, Bodemann B, Petersen S, Aruri J, Koshy S, Richardson Z, Le LQ, Krasieva T, Roth MG, Farmer P, White MA. Genome-Wide siRNA-Based Functional Genomics of Pigmentation Identifies Novel Genes and Pathways That Impact Melanogenesis in Human Cells. PLoS Genet. 2008;4(12):e1000298. doi: 10.1371/journal.pgen.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gobeil S, Zhu X, Doillon CJ, Green MR. A genome-wide shRNA screen identifies GAS1 as a novel melanoma metastasis suppressor gene. Genes Dev. 2008;22(21):2932–2940. doi: 10.1101/gad.1714608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brown SD, Hardisty RE. Mutagenesis strategies for identifying novel loci associated with disease phenotypes. Semin Cell Dev Biol. 2003;14(1):19–24. doi: 10.1016/s1084-9521(02)00168-4. [DOI] [PubMed] [Google Scholar]
- 76.Echeverri CJ, Beachy PA, Baum B, Boutros M, Buchholz F, Chanda SK, Downward J, Ellenberg J, Fraser AG, Hacohen N, Hahn WC, Jackson AL, Kiger A, Linsley PS, Lum L, Ma Y, Mathey-Prévôt B, Root DE, Sabatini DM, Taipale J, Perrimon N, Bernards R. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Methods. 2006;3(10):777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- 77.Haney SA. Increasing the robustness and validity of RNAi screens. Pharmacogenomics. 2007;8(8):1037–1049. doi: 10.2217/14622416.8.8.1037. [DOI] [PubMed] [Google Scholar]
- 78.Chi A, Valencia JC, Hu ZZ, Watabe H, Yamaguchi H, Mangini NJ, Huang H, Canfield VA, Cheng KC, Yang F, Abe R, Yamagishi S, Shabanowitz J, Hearing VJ, Wu C, Appella E, Hunt DF. Proteomic and bioinformatic characterization of the biogenesis and function of melanosomes. J Proteome Res. 2006;5(11):3135–3144. doi: 10.1021/pr060363j. [DOI] [PubMed] [Google Scholar]
- 79.Hu ZZ, Valencia J, Huang H, Chi A, Shabanowitz J, Hearing VJ, Appella E, Wu C. Comparative Bioinformatics Analyses and Profiling of Lysosome-Related Organelle Proteomes. International journal of mass spectrometry. 2007;259(1–3):147–160. doi: 10.1016/j.ijms.2006.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kushimoto T, Basrur V, Valencia J, Matsunaga J, Vieira WD, Ferrans VJ, Muller J, Appella E, Hearing VJ. A model for melanosome biogenesis based on the purification and analysis of early melanosomes. Proc Natl Acad Sci USA. 2001;98(19):10698–10703. doi: 10.1073/pnas.191184798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rendl M, Lewis L, Fuchs E. Molecular dissection of mesenchymal-epithelial interactions in the hair follicle. PLoS Biol. 2005;3(11):e331. doi: 10.1371/journal.pbio.0030331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.April CS, Barsh GS. Skin layer-specific transcriptional profiles in normal and recessive yellow (Mc1re/Mc1re) mice. Pigment Cell Res. 2006;19(3):194–205. doi: 10.1111/j.1600-0749.2006.00305.x. [DOI] [PubMed] [Google Scholar]
- 83.Hoek KS, Schlegel NC, Eichhoff OM, Widmer DS, Praetorius C, Einarsson SO, Valgeirsdottir S, Bergsteinsdottir K, Schepsky A, Dummer R, Steingrimsson E. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res. 2008 doi: 10.1111/j.1755-148X.2008.00505.x. in press. [DOI] [PubMed] [Google Scholar]
- 84.Loftus SK, Antonellis A, Matera I, Renaud G, Baxter LL, Reid D, Wolfsberg TG, Chen Y, Wang C, Prasad MK, Bessling SL, McCallion AS, Green ED, Bennett DC, Pavan WJ. Gpnmb is a Melanoblast-Expressed, MITF-Dependent Gene. Pigment Cell Melanoma Res. 2008 doi: 10.1111/j.1755-148X.2008.00518.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mardis E. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24(3):133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 86.Cordero F, Botta M, Calogero RA. Microarray data analysis and mining approaches. Briefings in functional genomics & proteomics. 2007;6(4):265–281. doi: 10.1093/bfgp/elm034. [DOI] [PubMed] [Google Scholar]
- 87.Wilkes T, Laux H, Foy CA. Microarray data quality - review of current developments. OMICS. 2007;11(1):1–13. doi: 10.1089/omi.2006.0001. [DOI] [PubMed] [Google Scholar]
- 88.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schönbach C, Sekiguchi K, Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y Consortium, F Group RGERGaGSGGNPC. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 89.Goding CR. Mitf from neural crest to melanoma: signal transduction and transcription in the melanocyte lineage. Genes Dev. 2000;14(14):1712–1728. [PubMed] [Google Scholar]
- 90.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, Drenkow J, Bell I, Zhao X, Srinivasan KG, Sung WK, Ooi HS, Chiu KP, Foissac S, Alioto T, Brent M, Pachter L, Tress ML, Valencia A, Choo SW, Choo CY, Ucla C, Manzano C, Wyss C, Cheung E, Clark TG, Brown JB, Ganesh M, Patel S, Tammana H, Chrast J, Henrichsen CN, Kai C, Kawai J, Nagalakshmi U, Wu J, Lian Z, Lian J, Newburger P, Zhang X, Bickel P, Mattick JS, Carninci P, Hayashizaki Y, Weissman S, Hubbard T, Myers RM, Rogers J, Stadler PF, Lowe TM, Wei CL, Ruan Y, Struhl K, Gerstein M, Antonarakis SE, Fu Y, Green ED, Karaoz U, Siepel A, Taylor J, Liefer LA, Wetterstrand KA, Good PJ, Feingold EA, Guyer MS, Cooper GM, Asimenos G, Dewey CN, Hou M, Nikolaev S, Montoya-Burgos JI, Loytynoja A, Whelan S, Pardi F, Massingham T, Huang H, Zhang NR, Holmes I, Mullikin JC, Ureta-Vidal A, Paten B, Seringhaus M, Church D, Rosenbloom K, Kent WJ, Stone EA, Batzoglou S, Goldman N, Hardison RC, Haussler D, Miller W, Sidow A, Trinklein ND, Zhang ZD, Barrera L, Stuart R, King DC, Ameur A, Enroth S, Bieda MC, Kim J, Bhinge AA, Jiang N, Liu J, Yao F, Vega VB, Lee CW, Ng P, Yang A, Moqtaderi Z, Zhu Z, Xu X, Squazzo S, Oberley MJ, Inman D, Singer MA, Richmond TA, Munn KJ, Rada-Iglesias A, Wallerman O, Komorowski J, Fowler JC, Couttet P, Bruce AW, Dovey OM, Ellis PD, Langford CF, Nix DA, Euskirchen G, Hartman S, Urban AE, Kraus P, Van Calcar S, Heintzman N, Kim TH, Wang K, Qu C, Hon G, Luna R, Glass CK, Rosenfeld MG, Aldred SF, Cooper SJ, Halees A, Lin JM, Shulha HP, Xu M, Haidar JN, Yu Y, Iyer VR, Green RD, Wadelius C, Farnham PJ, Ren B, Harte RA, Hinrichs AS, Trumbower H, Clawson H, Hillman-Jackson J, Zweig AS, Smith K, Thakkapallayil A, Barber G, Kuhn RM, Karolchik D, Armengol L, Bird CP, de Bakker PI, Kern AD, Lopez-Bigas N, Martin JD, Stranger BE, Woodroffe A, Davydov E, Dimas A, Eyras E, Hallgrimsdottir IB, Huppert J, Zody MC, Abecasis GR, Estivill X, Bouffard GG, Guan X, Hansen NF, Idol JR, Maduro VV, Maskeri B, McDowell JC, Park M, Thomas PJ, Young AC, Blakesley RW, Muzny DM, Sodergren E, Wheeler DA, Worley KC, Jiang H, Weinstock GM, Gibbs RA, Graves T, Fulton R, Mardis ER, Wilson RK, Clamp M, Cuff J, Gnerre S, Jaffe DB, Chang JL, Lindblad-Toh K, Lander ES, Koriabine M, Nefedov M, Osoegawa K, Yoshinaga Y, Zhu B, de Jong PJ. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Antonellis A, Bennett WR, Menheniott TR, Prasad AB, Lee-Lin SQ, Program NCS, Green ED, Paisley D, Kelsh RN, Pavan WJ, Ward A. Deletion of long-range sequences at Sox10 compromises developmental expression in a mouse model of Waardenburg-Shah (WS4) syndrome. Hum Mol Genet. 2006;15(2):259–271. doi: 10.1093/hmg/ddi442. [DOI] [PubMed] [Google Scholar]
- 92.Antonellis A, Huynh JL, Lee-Lin SQ, Vinton RM, Renaud G, Loftus SK, Elliot G, Wolfsberg TG, Green ED, McCallion AS, Pavan WJ. Identification of neural crest and glial enhancers at the mouse Sox10 locus through transgenesis in zebrafish. PLoS Genet. 2008;4(9):e1000174. doi: 10.1371/journal.pgen.1000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Werner T, Hammer A, Wahlbuhl M, Bösl MR, Wegner M. Multiple conserved regulatory elements with overlapping functions determine Sox10 expression in mouse embryogenesis. Nucleic Acids Res. 2007;35(19):6526–6538. doi: 10.1093/nar/gkm727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 95.Abdel-Malek ZA, Knittel J, Kadekaro AL, Swope VB, Starner R. The melanocortin 1 receptor and the UV response of human melanocytes--a shift in paradigm. Photochem Photobiol. 2008;84(2):501–508. doi: 10.1111/j.1751-1097.2008.00294.x. [DOI] [PubMed] [Google Scholar]
- 96.Jean S, Bideau C, Bellon L, Halimi G, De Méo M, Orsière T, Dumenil G, Bergé-Lefranc JL, Botta A. The expression of genes induced in melanocytes by exposure to 365-nm UVA: study by cDNA arrays and real-time quantitative RT-PCR. Biochim Biophys Acta. 2001;1522(2):89–96. doi: 10.1016/s0167-4781(01)00326-8. [DOI] [PubMed] [Google Scholar]
- 97.Valéry C, Grob JJ, Verrando P. Identification by cDNA microarray technology of genes modulated by artificial ultraviolet radiation in normal human melanocytes: relation to melanocarcinogenesis. J Invest Dermatol. 2001;117(6):1471–1482. doi: 10.1046/j.0022-202x.2001.01607.x. [DOI] [PubMed] [Google Scholar]
- 98.Yang G, Zhang G, Pittelkow MR, Ramoni M, Tsao H. Expression profiling of UVB response in melanocytes identifies a set of p53-target genes. J Invest Dermatol. 2006;126(11):2490–2506. doi: 10.1038/sj.jid.5700470. [DOI] [PubMed] [Google Scholar]
- 99.Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A, Sampas N, Dougherty E, Wang E, Marincola F, Gooden C, Lueders J, Glatfelter A, Pollock P, Carpten J, Gillanders E, Leja D, Dietrich K, Beaudry C, Berens M, Alberts D, Sondak V. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406(6795):536–540. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- 100.Fecher LA, Cummings SD, Keefe MJ, Alani RM. Toward a molecular classification of melanoma. J Clin Oncol. 2007;25(12):1606–1620. doi: 10.1200/JCO.2006.06.0442. [DOI] [PubMed] [Google Scholar]
- 101.Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R, Weber BL, Nathanson KL, Phillips DJ, Herlyn M, Schadendorf D, Dummer R. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006;19(4):290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- 102.Ryu B, Kim DS, Deluca AM, Alani RM. Comprehensive expression profiling of tumor cell lines identifies molecular signatures of melanoma progression. PLoS ONE. 2007;2(7):e594. doi: 10.1371/journal.pone.0000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bauer J, Bastian BC. Distinguishing melanocytic nevi from melanoma by DNA copy number changes: comparative genomic hybridization as a research and diagnostic tool. Dermatologic therapy. 2006;19(1):40–49. doi: 10.1111/j.1529-8019.2005.00055.x. [DOI] [PubMed] [Google Scholar]
- 104.Lin WM, Baker AC, Beroukhim R, Winckler W, Feng W, Marmion JM, Laine E, Greulich H, Tseng H, Gates C, Hodi FS, Dranoff G, Sellers WR, Thomas RK, Meyerson M, Golub TR, Dummer R, Herlyn M, Getz G, Garraway LA. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 2008;68(3):664–673. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dissanayake SK, Wade M, Johnson CE, O’Connell MP, Leotlela PD, French AD, Shah KV, Hewitt KJ, Rosenthal DT, Indig FE, Jiang Y, Nickoloff BJ, Taub DD, Trent JM, Moon RT, Bittner M, Weeraratna AT. The Wnt5A/protein kinase C pathway mediates motility in melanoma cells via the inhibition of metastasis suppressors and initiation of an epithelial to mesenchymal transition. J Biol Chem. 2007;282(23):17259–17271. doi: 10.1074/jbc.M700075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Weeraratna AT, Jiang Y, Hostetter G, Rosenblatt K, Duray P, Bittner M, Trent JM. Wnt5a signaling directly affects cell motility and invasion of metastatic melanoma. Cancer Cell. 2002;1(3):279–288. doi: 10.1016/s1535-6108(02)00045-4. [DOI] [PubMed] [Google Scholar]
- 107.Loftus SK, Larson DM, Baxter LL, Antonellis A, Chen Y, Wu X, Jiang Y, Bittner M, Hammer JA, 3rd, Pavan WJ. Mutation of melanosome protein RAB38 in chocolate mice. Proc Natl Acad Sci U S A. 2002;99(7):4471–4476. doi: 10.1073/pnas.072087599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Johansson P, Pavey S, Hayward N. Confirmation of a BRAF mutation-associated gene expression signature in melanoma. Pigment Cell Res. 2007;20(3):216–221. doi: 10.1111/j.1600-0749.2007.00375.x. [DOI] [PubMed] [Google Scholar]
- 109.Magnoni C, Tenedini E, Ferrari F, Benassi L, Bernardi C, Gualdi G, Bertazzoni G, Roncaglia E, Fantoni L, Manfredini R, Bicciato S, Ferrari S, Giannetti A, Tagliafico E. Transcriptional profiles in melanocytes from clinically unaffected skin distinguish the neoplastic growth pattern in patients with melanoma. Br J Dermatol. 2007;156(1):62–71. doi: 10.1111/j.1365-2133.2006.07564.x. [DOI] [PubMed] [Google Scholar]
- 110.Wagner KW, Punnoose EA, Januario T, Lawrence DA, Pitti RM, Lancaster K, Lee D, von Goetz M, Yee SF, Totpal K, Huw L, Katta V, Cavet G, Hymowitz SG, Amler L, Ashkenazi A. Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat Med. 2007;13(9):1070–1077. doi: 10.1038/nm1627. [DOI] [PubMed] [Google Scholar]
- 111.Ren S, Liu S, Howell P, Xi Y, Enkemann SA, Ju J, Riker AI. The impact of genomics in understanding human melanoma progression and metastasis. Cancer control : journal of the Moffitt Cancer Center. 2008;15(3):202–215. doi: 10.1177/107327480801500303. [DOI] [PubMed] [Google Scholar]
- 112.Baxter LL, Hsu BJ, Umayam L, Wolfsberg TG, Larson DM, Frith MC, Kawai J, Hayashizaki Y, Carninci P, Pavan WJ. Informatic and genomic analysis of melanocyte cDNA libraries as a resource for the study of melanocyte development and function. Pigment Cell Res. 2007;20(3):201–209. doi: 10.1111/j.1600-0749.2007.00372.x. [DOI] [PubMed] [Google Scholar]
- 113.Carninci P, Waki K, Shiraki T, Konno H, Shibata K, Itoh M, Aizawa K, Arakawa T, Ishii Y, Sasaki D, Bono H, Kondo S, Sugahara Y, Saito R, Osato N, Fukuda S, Sato K, Watahiki A, Hirozane-Kishikawa T, Nakamura M, Shibata Y, Yasunishi A, Kikuchi N, Yoshiki A, Kusakabe M, Gustincich S, Beisel K, Pavan W, Aidinis V, Nakagawara A, Held WA, Iwata H, Kono T, Nakauchi H, Lyons P, Wells C, Hume DA, Fagiolini M, Hensch TK, Brinkmeier M, Camper S, Hirota J, Mombaerts P, Muramatsu M, Okazaki Y, Kawai J, Hayashizaki Y. Targeting a complex transcriptome: the construction of the mouse full-length cDNA encyclopedia. Genome Res. 2003;13(6B):1273–1289. doi: 10.1101/gr.1119703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ben-Tabou de-Leon S, Davidson EH. Modeling the dynamics of transcriptional gene regulatory networks for animal development. Dev Biol. 2008 doi: 10.1016/j.ydbio.2008.10.043. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cohen AA, Geva-Zatorsky N, Eden E, Frenkel-Morgenstern M, Issaeva I, Sigal A, Milo R, Cohen-Saidon C, Liron Y, Kam Z, Cohen L, Danon T, Perzov N, Alon U. Dynamic Proteomics of Individual Cancer Cells in Response to a Drug. Science. 2008;322(5907):1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- 116.Ohyabu Y, Kaul Z, Yoshioka T, Inoue K, Sakai S, Mishima H, Uemura T, Wadhwa R, Kaul SC. Stable and Non-Disruptive In Vitro/In Vivo Labeling of Mesenchymal Stem Cells by Internalizing Quantum Dots. Hum Gene Ther. 2008 doi: 10.1089/hum.2008.100. in press. [DOI] [PubMed] [Google Scholar]
- 117.Ram S, Prabhat P, Chao J, Ward ES, Ober RJ. High accuracy 3D quantum dot tracking with multifocal plane microscopy for the study of fast intracellular dynamics in live cells. Biophys J. 2008 doi: 10.1529/biophysj.108.140392. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Urano Y, Asanuma D, Hama Y, Koyama Y, Barrett T, Kamiya M, Nagano T, Watanabe T, Hasegawa A, Choyke PL, Kobayashi H. Selective molecular imaging of viable cancer cells with pH-activatable fluorescence probes. Nat Med. 2008 doi: 10.1038/nm.1854. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Murray JI, Bao Z, Boyle TJ, Boeck ME, Mericle BL, Nicholas TJ, Zhao Z, Sandel MJ, Waterston RH. Automated analysis of embryonic gene expression with cellular resolution in C. elegans. Nat Methods. 2008;5(8):703–709. doi: 10.1038/nmeth.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]