Abstract
Using microarray analysis, in situ hybridization and immunocytochemistry, we found that the transcription factor TBX3 is produced in three discrete neuronal populations of the adult mouse brain, the arcuate nucleus (including in NPY but not dopaminergic neurons), the histaminergic tuberomammillary nucleus and in cholinergic neurons of the solitary tract nucleus. The immunoreactive protein had a nuclear location in these neurons, consistent with its function as a transcription factor. Although the function of tbx3 in these neurons is unknown, a review of the literature strongly suggests that these neuronal populations may be abnormal in Ulnar-Mammary syndrome patients with tbx3 mutations, explaining previously overlooked phenotypes in this syndrome, such as obesity, sexual dysfunction if not sleep abnormalities.
Keywords: tbx3, tuberomammillary, arcuate, solitary tract nucleus, Ulnar-Mammary syndrome, NPY
1. Introduction
The T-box family of genes encodes transcription factors that are defined by the presence of a conserved sequence, the so-called T-box, which encodes the T-domain, a domain involved in DNA-binding (Kispert et al., 1995). Present in all metazoans, T-box genes are involved in early embryonic cell fate decisions, regulation of the development of extraembryonic structures, embryonic patterning, and organogenesis, most notably Tbx1 subfamily members in the developing heart. Seventeen T-box genes have been identified in humans so far. Mutations in these genes cause a wide ranging array of phenotypes and a number of human disorders are caused by mutated T-box genes. Examples of this are Ulnar-Mammary syndrome (UMS) caused by a mutation in tbx3 and ACTH deficiency caused by a mutated tbx19 (Packham and Brook, 2003).
T-box proteins typically act as transcriptional activators but can also repress transcription, as demonstrated for TBX2 and TBX3, which have been reported to act both as activators and/or repressors, depending on promoter context (Naiche et al., 2005 for references). The protein product of tbx3 typically functions as a transcriptional repressor during normal limb bud development (Carlson et al., 2001). Tbx3 haploinsuficiency causes autosomal dominant UMS in humans. In UMS, the ulnar ray of the limb is affected with phenotypes ranging from simple hypoplasia of the fifth digit to complete absence of the forearm and hand. Patients with UMS also have abnormal development of breasts, teeth and genitalia as well as growth retardation and obesity and they typically have delayed onset of puberty (Bamshad et al., 1997; Schinzel et al., 1987). Cardiac development defects can also be present (Meneghini et al., 2006). Northern blot studies have indicated expression of tbx3 in various adult human tissues, but not brain (Bamshad et al., 1999).
During a microarray experiment comparing gene expression in hypocretin versus histaminergic cell-containing regions of the posterior hypothalamus in mice, we found an up to fifty-fold higher expression of tbx3 in the histamine cell tuberomammillary nucleus (TMN). To pursue this surprising finding, we used in situ hybridization (ISH) and immunocytochemistry (ICC) to study tbx3 expression and the TBX3 protein in the posterior hypothalamus. We report that tbx3 is expressed in the arcuate nucleus (ARC), the histaminergic tuberomammillary nucleus (TMN) of the posterior hypothalamus, and a group of cholinergic neurons in the nucleus of the solitary tract (NST) of the dorsal medulla. This expression pattern may explain previously reported phenotypic features in UMS patients, most notably obesity.
2. Results
Microarray gene expression comparisons are reported in table 1. The data are given as the relative expression level in the lateral hypothalamus with the posterior hypothalamus as baseline. As can be seen from the table, hcrt and Hdc were the most highly differentially expressed in the two regions, indicating excellent enrichment. Melanin concentrating hormone (MCH) was also highly enriched in the hypocretin region, as expected. Of the genes that are expressed at higher levels in the hypocretin region, several myelin-related genes were present; this likely reflects the higher amount of white matter in this region compared to the TM region (reflecting the fornix as a white matter track). Likewise, the posterior hypothalamic sample included the arcuate nucleus and the meninges, and another gene from the list that was highly expressed in this region was the transporter protein Xtrp3s1, which was expressed in the meninges (data not shown). Most genes in the list were widely expressed in the brain and got a prominent position in the list because they had a comparatively low expression in either of the compared regions. All genes were studied with ISH, and apart from HDC and tbx3, peripherin (Prph) was found to be expressed in the TM neurons and this result is reported elsewhere (Eriksson et al., 2008). Of the eight genes that had a rank order higher than 15 at both ZT 10 and 22, four genes, HDC, Prph, Xtrp3s1 and tbx3, were confirmed with ISH to have a selective expression in the dissected region.
Table 1.
Gene list of transcripts expressed preferentially in hypocretin-containing versus histamine-containing cell regions of the posterior hypothalamus (for dissection, see Fig. 1A). Expression values in the posterior TMN hypothalamus were used as baseline for the comparison and the 30 genes with the highest and lowest expression, given as log 2 values, at ZT 10 in the lateral hypothalamus are shown. A similar comparison was preformed at ZT 22 and if the genes ranked among the 70 highest or lowest the rank order and log2 difference is shown to the right.
| Genes upregulated in the histamine-containing cell region | ||||
|---|---|---|---|---|
| ZT 10 | ZT 22 | |||
| Rank | Log 2 | Gene Name | Rank | Log 2 |
| 1 | −7.1 | Histidine decarboxylase (Hdc) | 4 | −4.6 |
| 2 | −5.9 | Peripherin (Prph) | 1 | −5.9 |
| 3 | −5.7 | Osteoglycin (Ogn) | 36 | −3 |
| 4 | −5.7 | Wnt inhibitory factor 1 (Wif1) | 23 | −3.5 |
| 5 | −4.9 | Vestigial like 3 (Vgll) | ||
| 6 | −4.7 | Death associated protein-like 1 (Dapl11 | 3 | −4 |
| 7 | −4.6 | X transporter protein. 3 similar 1 (Xtrp3s1) | 8 | −4.3 |
| 8 | −4.5 | Amylase 2. Pancreatic (Amy2) | 7 | −4.4 |
| 9 | −4.5 | Annexin A2 (Anxa2) | 11 | −4.2 |
| 10 | −4.5 | Fibroblast growth factor bind. prot. 1 (Fgfbp1) | 14 | −4 |
| 11 | −4.4 | T-box 3 (tbx3) | 2 | −5.8 |
| 12 | −4.4 | AE binding protein 1 (Aebp1) | 20 | −3.8 |
| 13 | −4.2 | T-cell specific GTPase (Tgtp) | ||
| 14 | −4.1 | Nuclear protein 1 (Nupr) | ||
| 15 | −4.1 | Fibromodulin (Fmod) | 16 | −3.9 |
| 16 | −4 | Retinol binding protein 4 (Rbp4) | ||
| 17 | −4 | AHNAK nucleoprotein (Ahnak) | 24 | −3.5 |
| 18 | −4 | Myocilin (Myoc) | 17 | −3.9 |
| 19 | −3.8 | Paired-like homeodomain transcr. fact. 2 (Pitx2) | 65 | −2.1 |
| 20 | −3.5 | Cholecystokinin (Cck) | 18 | −3.8 |
| 21 | −3.5 | Insulin-like growth factor bind. prot. 6 (Igfbp6) | 48 | −2.6 |
| 22 | −3.5 | Cellular retinoic acid binding protein 1 (Crabp1) | 6 | −4.5 |
| 23 | −3.4 | Tektin 1 (Tekt1) | ||
| 24 | −3.3 | Inter-alpha trypsin inhibitor. heavy ch. 2 (Itih2) | 5 | −4.5 |
| 25 | −3.3 | Natriuretic peptide receptor 3 (Npr3) | 9 | −4.3 |
| 26 | −3.3 | Bcl2-associated athanogene 3 (Bag3) | ||
| 27 | −3.2 | Gene model 70 (Gm70) | ||
| 28 | −3.1 | Annexin A1 (Anxa1) | ||
| 29 | −3.1 | Special AT-rich sequence binding protein 1 (Satb1) | ||
| 30 | −3 | Angiomotin-like 1 (Amotl1) | ||
| Genes upregulated in the hypocretin-containing cell region | ||||
| ZT 10 | ZT 22 | |||
| Rank | Log 2 | Gene Name | Rank | Log 2 |
| 1 | 6.5 | Hypocretin (Hcrt) | 1 | 7.1 |
| 2 | 4.3 | Hypothetical protein LOC100041002 | ||
| 3 | 4 | RIKEN cDNA 2300009A05 gene | ||
| 4 | 2.5 | MCH precursor (Pmch) | ||
| 5 | 2.5 | Myelin-assoc. oligodendroc. basic prot. (Mobp) | 6 | 2.9 |
| 6 | 2.2 | Lipase. hormone sensitive (lipe) | ||
| 7 | 2 | Neurotensin (Nts) | 18 | 2.4 |
| 8 | 1.9 | Endoth. sphingolip. G-prot.-coupl. rec. 8 (EDG8) | 56 | 1.6 |
| 9 | 1.9 | Neurofilament. medium polypeptide (Nefm) | 25 | 2.1 |
| 10 | 1.8 | Ras-GTPase-activat. SH3-dom. bind. Prot.(G3bp) | 4 | 3.3 |
| 11 | 1.8 | Myelin-associated glycoprotein (Mag) | 28 | 2 |
| 12 | 1.8 | Breast carcinoma amplified sequence 1 (Bcas1) | 36 | 1.9 |
| 13 | 1.8 | Procollagen C-endopeptidase enhancer 2 (Pcolce2 | ) | |
| 14 | 1.7 | Ecotropic viral integration site 2a (Evi2a) | 14 | 2.4 |
| 15 | 1.7 | Myelin proteolipid protein 1 (Plp1) | 19 | 2.3 |
| 16 | 1.7 | K+ voltage gated channel shaker related 1 (Kcna1) | ||
| 17 | 1.7 | Anillin, actin binding protein (Anln) | 11 | 2.5 |
| 18 | 1.7 | R-spondin homolog (Rspo1) | 62 | 1.5 |
| 19 | 1.6 | Myelin and lymphocyte protein (Mal) | 22 | 2.1 |
| 20 | 1.6 | Checkpoint kinase 1 (Chek1) | ||
| 21 | 1.6 | RIKEN cDNA 2900052N01 gene | ||
| 22 | 1.6 | Nuclear receptor subfam. 2. gr. F. mem. 2. (Nr2f2) | 41 | 1.8 |
| 23 | 1.6 | RIKEN cDNA A330104H05 gene | ||
| 24 | 1.6 | RIKEN cDNA 9530066K23 gene | ||
| 25 | 1.5 | Tweety homolog 2 (Ttyh2) | 66 | 1.5 |
| 26 | 1.5 | Vesicle-associated membrane protein 1 (Vamp1) | 2 | 5 |
| 27 | 1.5 | Ectonucleotide pyrophosphatase 2 (Enpp2) | 60 | 1.5 |
| 28 | 1.5 | RIKEN cDNA A330104H05 gene | ||
| 29 | 1.5 | Chimerin 2 (Chn2) | ||
| 30 | 1.5 | K+ voltage gated channel. Shawrel. 1(Kcnc1) | 58 | 1.6 |
In situ hybridization with a 35S-labelled RNA probe and film autoradiography revealed expression of tbx3 in the arcuate and tuberomammillary (TM) nuclei (Fig. 1B). Immunostaining revealed that the TBX3 protein located in cell nuclei in these regions (Fig. 1C). Cytoplasmic TBX3 staining was never seen in any part of the brain.
Figure 1. Tissue dissection and T-box3 mRNA and protein in the hypothalamus.
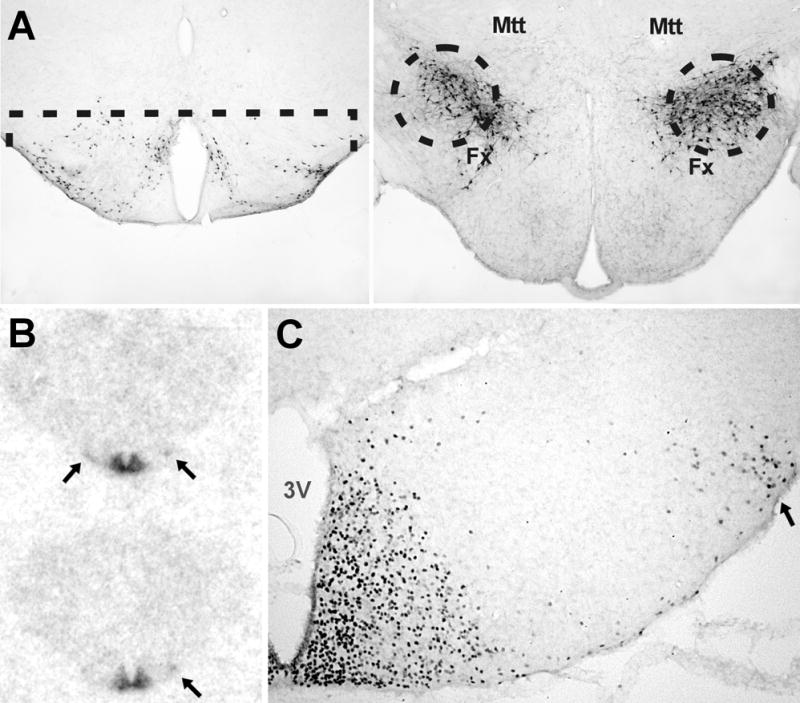
A. The mouse brain regions dissected out for the microarray studies are here indicated on histological sections. The dissection is done on 500 μm-thick vibratome slices under a dissection microscope. The TM region, which is shown here by histamine immunostaining, is dissected by a horizontal cut at the level of the top of the 3rd ventricle. The hcrt region, visualized here by hcrt immunostaining, is punched out with a 0.7 mm punch tube with the position of the fornix (Fx) and mammillothalamic tract (Mtt) as guides for the exact position. B. Autoradiogram of adult mouse brain hybridized with a 35S-labelled RNA probe. The signal in the arcuate nucleus is very obvious, and weaker signal in the TM nucleus is indicated with arrows. C. Immunostaining for TBX3 protein in the posterior hypothalamus. A large number of nuclei are stained in the arcuate nucleus. Stained nuclei are also seen in the TM nucleus (arrow) and dispersed neurons. 3V, third ventricle.
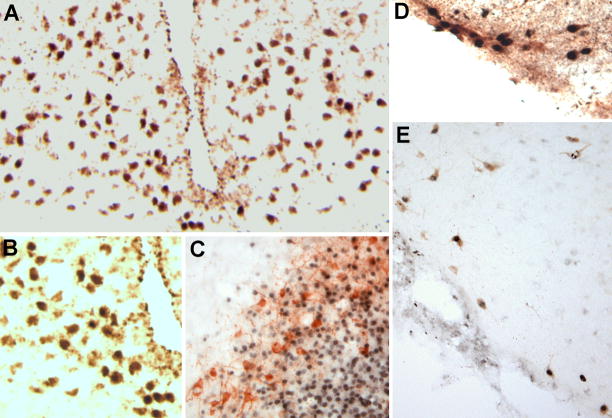
Staining of the arcuate nucleus with an antibody against AgRP revealed that a large portion of the neurons with TBX3-containing nuclei are NPY/AgRP neurons (Fig. 2A,B). We also used an antibody against tyrosine hydroxylase to study if the dopaminergic neurons of the arcuate nucleus are TBX3 positive, and found that none of these neurons contained TBX3 in their nuclei (Fig. 2C).
Figure 2. Neurochemical identification of the tbx3 containing neurons.
A. Double staining for TBX3 and AgRP in the arcuate nucleus. Almost all of the stained nuclei are located to AgRP immunoreactive neurons, and this was seen throughout the nucleus. In B this double staining is shown at a larger magnification. C. Staining for tyrosine hydroxylase reveals that the arcuate dopaminergic neurons are devoid of TBX3 stained nuclei. D. Double staining for TBX3 and HDC. All HDC immunoreactive neurons in the lateral subdivision of the TM nucleus have TBX3 in their nuclei. E. Virtually all of the more diffusely spread histaminergic neurons also had TBX3-positive nuclei, whereas very few nuclei that didn’t appear to be located in histaminergic neurons were seen in this part of the brain.
To find out if the TBX3-positive neurons of the TMN were histaminergic neurons, the region was also double stained using an antiserum recognizing HDC. We found that virtually all HDC positive neurons had TBX3 immunoreactive nuclei. This was the case in both the main, laterally located, concentration of histaminergic neurons (Fig 2D) and for the more diffusely dispersed histaminergic neurons (Fig. 2E).
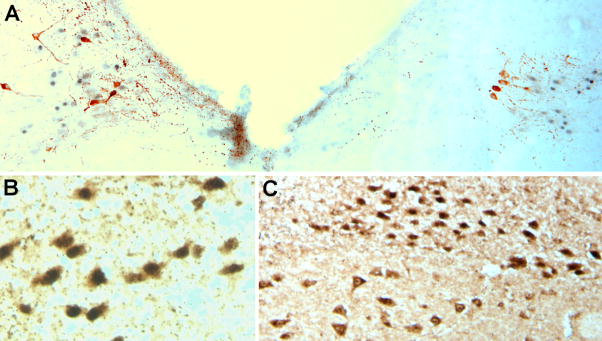
Further mapping of TBX3 immunoreactivity throughout the entire brain indicated staining in a single additional location. We found two bilaterally symmetrical groups of stained nuclei in the dorsal medulla. As these nuclei appeared to be close to the adrenergic C2 region, we stained for phenylethanolamine N-methyltransferase (PNMT), and found out that although the nuclei were very close to the C2 adrenaline producing neuronal groups, they were not identical (Fig. 3A). Staining with an antiserum against choline acetyltransferase (ChAT) revealed that the TBX3 positive neurons are cholinergic, indicating that these TBX3 positive neurons are the cholinergic neurons of the nucleus of the solitary tract (Fig 3B,C).
Figure 3. T-box3 in the dorsal medulla.
A. Overview of the dorsal medulla. In this semicoronal section the rostrocaudal level of the left and right side is slightly different. Black/gray nuclei indicating TBX3 are seen and PNMT IR adrenaline producing neurons of the C2 region are brown. The neurons are located close to the stained nuclei but adrenaline neurons with tbx3 stained nuclei were never seen. B. Double staining for TBX3 and ChAT. All TBX3 immunoreactive nuclei are located to ChAT positive, cholinergic neurons of the NTS. In C double stained neurons are seen in the upper part of the picture, and more ventrally cholinergic neurons in the dorsal motor nucleus of the vagus that lack TBX3 stained nuclei are seen.
3. Discussion
Here we report for the first time that tbx3 is discretely expressed in the brain of adult mice within the posterior hypothalamus and the brainstem. This surprising finding was based on microarray, ISH and ICC data. The nuclear localization of the immunoreactive signal also support these data, as TBX3 is a DNA binding protein with known nuclear localization,
Whereas T-box genes are known to be actively transcribed during development, there are only few reports on the expression of these genes in the adult central nervous system. The T-box gene tbx21 is expressed in projection neurons of the olfactory bulb of the adult mouse (Faedo et al., 2002). Similarly, the TBX1 protein is present in the cytoplasm of neurons in the cerebral cortex, hippocampus and cerebellar Purkinje cells of the rat (Hong and Hsueh, 2007). Another example is the expression of tbx19 in corticotroph and melanotroph cells of the pituitary. T-BOX19 is indeed needed for the expression of POMC in these neurons, where it activates the POMC promoter synergistically with another transcription factor, Pitx1. TBX19 is however absent from all other tissues, including arcuate POMC neurons (Lamolet et al., 2001). A mutation in tbx19 in humans and the resulting lack of pituitary POMC results in adrenal insufficiency (Peckham and Brook, 2003).
The phenotype of UMS patients almost always includes hypogenitalism, delayed puberty and diminished sexual activity. Delayed puberty and hypogenitalism suggests impaired hypothalamic-pituitary-gonadal axis function. Decreased TBX3 activity in the arcuate nucleus could be involved in mediating this phenotype, as gonadotropin releasing hormone (GnRH) neurons are located in this structure, and GnRH is necessary for the production and release of gonadotropin from the pituitaty. This nucleus also contains neurons producing kisspeptin, a peptide known to stimulate gonadotropin release through GnRH neuron excitation (Dungan et al., 2006). Gonadotropins are necessary for the induction of puberty and gonadotropin deficiency has been suggested as the cause for hypogenitalism in UMS (Sasaki et al., 2002). It is thus possible if not likely that haploinsuficiency of tbx3 in the arcuate nucleus affects the expression of GnHR, kisspeptin or both, mediating sexual dysfunction in UMS.
Another common feature of the UMS syndrome is obesity. A possible candidate for this phenotype may be TBX3 containing arcuate NPY neurons, although lesion of these neurons is known to reduce appetite (Luquet et al., 2005). Another candidate may be POMC neurons, which appeared to be tbx3 negative, although this was not conclusively verified histochemically. Finally, tbx3 is expressed in TM histaminergic and NST cholinergic neurons. Decreased histaminergic tone has been implicated in obesity through pharmacological studies (Haas and Panula, 2003). Further, the NST receives gustatory and visceral afferents and is thought to contribute to feeding anticipatory activity and regulation of meal size (Schwartz, 2006 for references). Interestingly, the NST is also central to baroreflex regulation, whereas the TM nucleus supports motivational waking, most notably in the presence of a novel environment (Takahashi et al, 2006; Zlomuzica et al., 2008). Whether or not sleep, cognitive or blood pressure abnormalities are present in UMS patients may be worth investigating.
Our findings are notable as they open the possibility to further study cell fate in these functionally important neuronal populations. Further, it is possible that tbx3 acts in synergy with other transcription factors in these discrete locations, as has been described for tbx19 in the pituitary and for multiple T-box genes in the heart, regulating different gene sets in different nuclei. It would also be interesting to study whether tbx3 is necessary to the maintenance of the respective neurotransmitter phenotypes in these neurons.
Further progress in studying the role of TBX3 in the brain will likely have to involve functional analysis studies in tbx3-mutated mice, and additional anatomical studies in human brains. Interestingly, a mouse model expressing a mutated tbx3 gene has been produced (Davenport et al., 2003), but in this model, haploinsufficieny in various tissues is not always sufficient to induce a phenotype comparable to UMS. T-box3 knockout homozygotes, although stillborn, have a deficiency of mammary gland induction, and exhibit both forelimb and hindlimb abnormalities, confirming a role for tbx3 in these functions. In contrast to this, and unlike the human situation, heterozygote animals have a mild overall phenotype and no mammary/limb development abnormalities. In the heart of tbx3 heterozygotes, however, arrhythmias and functional ectopic pacemakers are present (Hoogaars et al., 2007), suggesting sensitivity to haploinsufficiency in this tissue in mice. Whether or not the populations of neurons described in this publication develop normally in homozygous stillborn and heterozygote tbx3 animals warrants further investigations, together with a closer look at obesity, sleep and hormonal status in these animals.
In conclusion, we found that TBX3 is produced in three discrete neuronal populations of the adult mouse brain, the arcuate, TM and solitary tract nuclei. Although the function of TBX3 in these neurons is unknown, a review of the literature strongly suggests that these neuronal populations may be abnormal in UMS patients, explaining previously overlooked phenotypes, such as obesity and sexual dysfunction.
4. Experimental Procedure
Array studies
For gene expression arrays studies, the hypocretin-containing (perifornical and dorsomedial hypothalamus) and histamine-containing (TMN and surrounding posterior hypothalamic areas) cell areas of the posterior hypothalamus were dissected as displayed in Fig. 1A. Male C57BL/6J mice (12 weeks) were decapitated at ZT 10 and 22, the brain removed and cooled in ice-cold saline. The hypothalamus was dissected and cut on a vibratome starting at the posterior tip of the mammillary bodies; a section of 200 μm was removed to reach the posterior end of the TMN region and a 500 μm coronal section was cut, from which the TMN region was dissected with a horizontal cut just dorsal to the third ventricle. A 400 μm section was next discarded and the next 500 μm section next was used to punch (0.7 mm, using a Palkovits-type puncher) the hypocretin neuron region. The entire procedure was done in iced cold buffer and all appropriate landmarks to establish this procedure, including immunostaining for histamine and hypocretin, had been repeatedly verified in advance. The tissue was rapidly frozen on dry ice and then stored at −80°C. Twenty mice were used to gather sufficient RNA amounts for each array hybridization.
Total RNA was isolated with the RNeasy kit according to protocols provided by the manufacturer (Qiagen, Valencia, CA). After controlling quality and amount of RNA (2–2.5 μg) needed for hybridization, double stranded cDNA and biotinylated cRNA was synthesized without any amplification step according to protocols provided by Affymetrix (Santa Clara, CA) and purified with Affymetrix genechip Sample Cleanup Module. After fragmentation, the biotinylated cRNA was hybridized to Affymetrix U74 v.2 microarrays and analyzed with the MicroArray Suite ver. 5.0 software (Affymetrix).
In situ Hybridization
For all neuronatomical studies, animals were perfused transcardially with 4% paraformaldehyde in 100 mM phosphate buffer (pH 7.3) and postfixed overnight. The in situ hybridization was done in the same way as in our recently published paper (Eriksson et al., 2008), where we describe the procedure in detail. Briefly, the brain tissue was hybridized with a 35S-tagged RNA probe and exposed to X-ray films. The probe was prepared with an EST clone as template (IMAGE clone ID 4166742, [Genbank:BF300742], which corresponds to approximately bases 2200–2900 of both transcript variants 1 and 2 of the tbx3 mRNA.
Immunocytochemistry
Immunostaining was performed on free-floating sections. Antibody incubations and washes were done at room temperature in PBS with 0.25% Triton X-100, and the tissue was washed for at least 20 min between the steps. All antibody solutions contained 2% donkey serum. The tissue was first treated with 1% H2O2 for 30 min to quench endogenous peroxidase, blocked with 5% normal donkey serum for 2–4 hours and incubated overnight with primary antiserum.
Double stainings were done in sequence. The first staining was done using a goat-anti-TBX3 serum (1:100, Santa Cruz Biotechnology, Santa Cruz, CA) over night, biotinylated donkey-anti-goat (1:1000 for 90 min; Jackson Immunoresearch, West Grove, PA), avidin-biotinylated horseradish peroxidase complex (1:1000 for 90 min; Vector Laboratories, Burlingame, CA) and then the immunoreactivity was visualized in 0.05% nickel ammonium sulfate, 0.05% DAB and 0.015% H2O2 to get a black reaction product. After the TBX3 staining, tissue was treated with 1% H2O2 for 45 min and the avidin-biotin blocking kit (Vector) to prevent interference with the second staining. The second stainings were done in the same way as the first TBX3 staining, but the nickel salt was omitted to produce a brown staining.
Primary antisera used in the second staining step were guinea pig-anti-agouti-related protein (AgRP; Chemicon, Temecula, CA), rabbit-anti-tyrosine hydroxylase (Chemicon), guinea pig-anti-histidine decarboxylase (HDC) serum (American Research Products, Belmont, MA), rabbit-anti-phenylethanolamine N-methyltransferase (PNMT; Chemicon) and rabbit-anti-choline acetyl transferase (ChAT; Chemicon). The antisera were diluted 1:1000-1:30,000 depending on the desired staining intensity and the second reagents were biotinylated donkey antisera with the appropriate species reactivity (all from Jackson Immunoresearch).
Acknowledgments
We wish to thank Jennifer Kouragian for her dedicated assistance in processing this manuscript.
This work was supported by the Howard Hughes Medical Institute and National Institutes of Health (MH073435). Dr. Eriksson passed away December 29th 2008.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bamshad M, Le T, Watkins WS, Dixon ME, Kramer BE, Roeder AD, Carey JC, Root S, Schinzel A, Van Maldergem L, Gardner RJ, Lin RC, Seidman CE, Seidman JG, Wallerstein R, Moran E, Sutphen R, Campbell CE, Jorde LB. The spectrum of mutations in TBX3: Genotype/Phenotype relationship in ulnar-mammary syndrome. Am J Hum Genet. 1999;64:1550–62. doi: 10.1086/302417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad M, Lin RC, Law DJ, Watkins WC, Krakowiak PA, Moore ME, Franceschini P, Lala R, Holmes LB, Gebuhr TC, Bruneau BG, Schinzel A, Seidman JG, Seidman CE, Jorde LB. Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome. Nat Genet. 1997;16:311–5. doi: 10.1038/ng0797-311. [DOI] [PubMed] [Google Scholar]
- Carlson H, Ota S, Campbell CE, Hurlin PJ. A dominant repression domain in Tbx3 mediates transcriptional repression and cell immortalization: relevance to mutations in Tbx3 that cause ulnar-mammary syndrome. Hum Mol Genet. 2001;10:2403–13. doi: 10.1093/hmg/10.21.2403. [DOI] [PubMed] [Google Scholar]
- Davenport TG, Jerome-Majewska LA, Papaioannou VE. Mammary gland, limb and yolk sac defects in mice lacking Tbx3, the gene mutated in human ulnar mammary syndrome. Development. 2003;130:2263–73. doi: 10.1242/dev.00431. [DOI] [PubMed] [Google Scholar]
- Dungan HM, Clifton DK, Steiner RA. Minireview: kisspeptin neurons as central processors in the regulation of gonadotropin-releasing hormone secretion. Endocrinology. 2006;147:1154–8. doi: 10.1210/en.2005-1282. [DOI] [PubMed] [Google Scholar]
- Eriksson KS, Zhang S, Lin L, Lariviere RC, Julien JP, Mignot E. The type III neurofilament peripherin is expressed in the tuberomammillary neurons of the mouse. BMC Neurosci. 2008;9:26. doi: 10.1186/1471-2202-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faedo A, Ficara F, Ghiani M, Aiuti A, Rubenstein JL, Bulfone A. Developmental expression of the T-box transcription factor T-bet/Tbx21 during mouse embryogenesis. Mech Dev. 2002;116:157–60. doi: 10.1016/s0925-4773(02)00114-4. [DOI] [PubMed] [Google Scholar]
- Haas H, Panula P. The role of histamine and the tuberomamillary nucleus in the nervous system. Nat Rev Neurosci. 2003;4:121–30. doi: 10.1038/nrn1034. [DOI] [PubMed] [Google Scholar]
- Hong CJ, Hsueh YP. Cytoplasmic distribution of T-box transcription factor Tbr-1 in adult rodent brain. J Chem Neuroanat. 2007;33:124–30. doi: 10.1016/j.jchemneu.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Hoogaars WM, Engel A, Brons JF, Verkerk AO, de Lange FJ, Wong LY, Bakker ML, Clout DE, Wakker V, Barnett P, Ravesloot JH, Moorman AF, Verheijck EE, Christoffels VM. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007;21:1098–112. doi: 10.1101/gad.416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispert A, Koschorz B, Herrmann BG. The T protein encoded by Brachyury is a tissue-specific transcription factor. EMBO J. 1995;14:4763–72. doi: 10.1002/j.1460-2075.1995.tb00158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamolet B, Pulichino AM, Lamonerie T, Gauthier Y, Brue T, Enjalbert A, Drouin J. A pituitary cell-restricted T box factor, Tpit, activates POMC transcription in cooperation with Pitx homeoproteins. Cell. 2001;104:849–59. doi: 10.1016/s0092-8674(01)00282-3. [DOI] [PubMed] [Google Scholar]
- Luquet S, Perez FA, Hnasko TS, Palmiter RD. NPY/AgRP neurons are essential for feeding in adult mice but can be ablated in neonates. Science. 2005;310:683–5. doi: 10.1126/science.1115524. [DOI] [PubMed] [Google Scholar]
- Meneghini V, Odent S, Platonova N, Egeo A, Merlo GR. Novel TBX3 mutation data in families with ulnar-mammary syndrome indicate a genotype-phenotype relationship: mutations that do not disrupt the T-domain are associated with less severe limb defects. Eur J Med Genet. 2006;49:151–8. doi: 10.1016/j.ejmg.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Naiche LA, Harrelson Z, Kelly RG, Papaioannou VE. T-box genes in vertebrate development. Annu Rev Genet. 2005;39:219–39. doi: 10.1146/annurev.genet.39.073003.105925. [DOI] [PubMed] [Google Scholar]
- Packham EA, Brook JD. T-box genes in human disorders. Hum Mol Genet. 2003;12(Spec No 1):R37–44. doi: 10.1093/hmg/ddg077. [DOI] [PubMed] [Google Scholar]
- Sasaki G, Ogata T, Ishii T, Hasegawa T, Sato S, Matsuo N. Novel mutation of TBX3 in a Japanese family with ulnar-mammary syndrome: implication for impaired sex development. Am J Med Genet. 2002;110:365–9. doi: 10.1002/ajmg.10447. [DOI] [PubMed] [Google Scholar]
- Schinzel A, Illig R, Prader A. The ulnar-mammary syndrome: an autosomal dominant pleiotropic gene. Clin Genet. 1987;32:160–8. doi: 10.1111/j.1399-0004.1987.tb03347.x. [DOI] [PubMed] [Google Scholar]
- Schwartz GJ. Integrative capacity of the caudal brainstem in the control of food intake. Philos Trans R Soc Lond B Biol Sci. 2006;361:1275–80. doi: 10.1098/rstb.2006.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Lin JS, Sakai K. Neuronal activity of histaminergic tuberomammillary neurons during wake-sleep states in the mouse. J Neurosci. 2006;26:10292–8. doi: 10.1523/JNEUROSCI.2341-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlomuzica A, Viggiano D, De Souza Silva MA, Ishizuka T, Gironi Carnevale UA, Ruocco LA, Watanabe T, Sadile AG, Huston JP, Dere E. The histamine H1-receptor mediates the motivational effects of novelty. Eur J Neurosci. 2008;27:1461–74. doi: 10.1111/j.1460-9568.2008.06115.x. [DOI] [PubMed] [Google Scholar]