Abstract
This paper describes two SlipChip-based approaches to protein crystallization: a SlipChip-based free interface diffusion (FID) method and a SlipChip-based composite method that simultaneously performs microbatch and FID crystallization methods in a single device. The FID SlipChip was designed to screen multiple reagents, each at multiple diffusion equilibration times, and was validated by screening conditions for crystallization of two proteins, enoyl-CoA hydratase from Mycobacterium tuberculosis and dihydrofolate reductase/thymidylate synthase from Babesia bovis against 48 different reagents at 5 different equilibration times each, consuming 12 μL of each protein for a total of 480 experiments using three SlipChips. The composite SlipChip was designed to screen multiple reagents, each at multiple mixing ratios and multiple equilibration times, and was validated by screening conditions for crystallization of two proteins, enoyl-CoA hydratase from Mycobacterium tuberculosis and dihydrofolate reductase/thymidylate synthase from Babesia bovis. To prevent cross-contamination while keeping the solution in the neck channels for FID stable, the plates of the SlipChip were etched with a pattern of nanowells. This nanopattern was used to increase the contact angle of aqueous solutions on the surface of the silanized glass. The composite SlipChip increased the number of successful crystallization conditions and identified more conditions for crystallization than separate FID and microbatch screenings. Crystallization experiments were scaled up in well plates using conditions identified during the SlipChip screenings, and X-ray diffraction data were obtained to yield the protein structure of dihydrofolate reductase/thymidylate synthase at 1.95 Å resolution. This free-interface diffusion approach provides a convenient and high-throughput method of setting up gradients in microfluidic devices, and may find additional applications in cell-based assays.
Introduction
This paper describes a SlipChip-based approach to simultaneously perform two methods for protein crystallization, microbatch and free interface diffusion (FID), in a single microfluidic device. Currently, there are three challenges to protein crystallization: 1) to crystallize proteins, a large chemical space must be searched to determine the conditions required. The search for the right precipitants and the right concentrations of protein and precipitant is expedited by faster experiments and smaller sample sizes,1–4 and a simple, fast, and controllable system would advance the discovery of new protein structures. 2) A particularly attractive method to crystallize proteins is nanoliter-scale FID because it explores the phase diagram for crystallization as both the concentration of protein and the concentration of precipitant are gradually changed by diffusion, provides a higher transient supersaturation level for crystal nucleation, and eliminates precipitation induced by fast mixing.5,6 Nanoliter-scale FID is consequently efficient for crystallization,7 but currently it is only implemented with valve-based systems.7–10 We emphasize that FID is mechanistically very similar to the well-established counter diffusion methods11 that are typically implemented on microliter scales, including chip-based12 and gel acupuncture-based approaches.13 The use of valves in FID requires external control equipment, and valves are often composed of PDMS. PDMS devices have the additional complication of requiring control of the atmosphere and evaporation.14 Valve-free, equipment-free approaches to implement FID would simply the method and make it more widely available. 3) Different methods of crystallization explore different paths towards the equilibrated condition where crystals of protein form, therefore yield different crystallization results.7,15 These methods can be modified to alter the kinetics of crystallization7,16 and thus explore different routes to form crystals of proteins; however, different methods require different techniques to combine the protein solution and precipitant solution. While it is desirable to use more than one method of crystallization, it is technologically challenging to use two techniques in one experiment.
The SlipChip technology described in this paper addresses these challenges. It has been demonstrated in both pre-loaded17 and user-loaded18 formats, and for simplicity in this paper we illustrate the ideas using the user-loaded format. This paper makes two advances: We developed a FID technique based on SlipChip and also combined FID and microbatch techniques in one “composite” SlipChip.
Results and Discussion
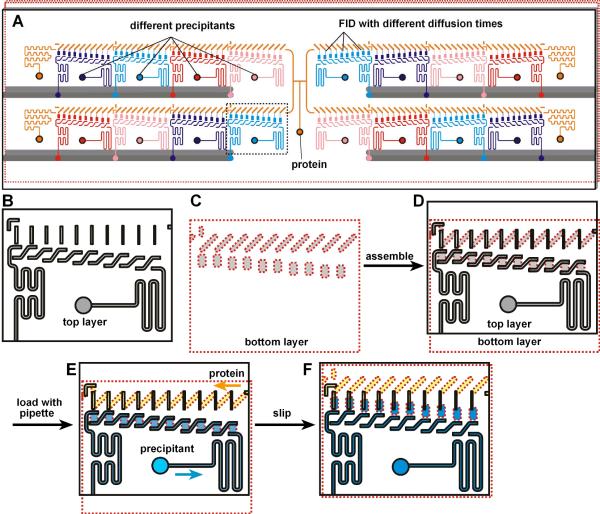
We first designed the SlipChip to incorporate the FID method (Figure 1). The SlipChip was designed to screen a sample against 16 different precipitants at five different equilibration times. Each equilibration time was investigated in duplicate, for a total of 160 experiments in a single SlipChip. The SlipChip could be configured to form 16 separate fluidic paths for the precipitants, each containing 10 wells, and a single fluidic path for the protein sample containing 160 wells (Figure 1A). The general construction of the SlipChip was the same as previously described18 (see Supporting Information). To incorporate the FID method, when the SlipChip was “slipped” to connect the protein wells and the precipitant wells, the microchannels (ducts, 21 μm in depth) that had formed the continuous fluidic path for the protein sample became the neck channel connecting the protein well to the precipitant well (Figure 1D, F, see also supporting movie S1). By gradually increasing the distance between the protein wells and the precipitant wells, the length of the neck was increased from 91 μm to 491 μm; by decreasing the width of the ducts, the width of the neck was decreased from 104 μm to 58 μm (Figure 1F, from left to right). The geometry of the necks, defined as the length of the neck channel divided by the cross-sectional area of the channel, was consequently altered (neck parameters are provided in supporting information Table S1)
Figure 1.
A SlipChip designed to screen a protein against 16 different precipitants using the FID method of crystallization. A) A schematic of the SlipChip. Multiple precipitants (purple, blue, red, and pink), as well as multiple equilibration times for mixing the protein (orange) with each precipitant, can be screened on the same SlipChip. B–F) A zoomed-in schematic of the area outlined in A showing the operation of the SlipChp. B) The top plate (contoured in black) contains ducts for the protein and ducts for the precipitant. The ducts for the protein will become the neck channels that connect the protein wells and the precipitant wells, and these ducts gradually decrease in width from left to right, gradually changing the equilibration time. C) The bottom plate (contoured in red) has wells for the protein and wells for the precipitant. The distance between the wells for the protein and wells for the precipitant is gradually increased from left to right, gradually changing the equilibration time. D) When the two plates are assembled, the fluidic path for the protein and the fluidic path for the precipitants are formed. E) Solutions of protein (yellow) and precipitant (blue) after loading by pipetting. F) After “slipping”, protein and precipitant wells from the bottom plate are bridged by narrow channels in the top plate.
The geometry of the neck controlled the equilibration time (Figure 2, see also supporting movie S1), and we found that the equilibration time increased linearly with the neck geometry (Figure 2E), which was consistent with our expectations and numerical simulations (data not shown). We emphasize that equilibration time occurring in the steady state with fully developed diffusion profiles is different that the time to establish these profiles, the latter time is expected to scale with the square of distance. The FID experiments were set up easily in the SlipChip, requiring no valves and only involving pipetting and slipping. In this approach, the ducts for the protein sample were used to set up the FID experiments, so little sample was wasted. Because the necks were designed to be thin compared to the wells containing precipitant or protein, the change in volume caused by changing the neck geometry was negligible compared to the total volume of the crystallization trial. The volume of the neck constituted only 4–8% of the total volume of the crystallization trial. In these experiments, we focused on how changing the equilibration time affects protein crystallization and not on how changing the volume affects protein crystallization.
Figure 2.
Changing the geometry of the channel changes the equilibration time in the SlipChip. A–C) Microphotographs of food dye diffusing in the FID SlipChip. Each condition represents a different equilibration time, and was done in duplicate. A) Immediately after slipping, T1 = 0 min. B) At time T2 = 24 min. C) At time T3 = 141 min. D) Diffusion profiles were obtained for various neck geometries by using a model fluorescent dye, DTPA. Average intensities in the well for protein were measured by linescan through the wells (See, Figure S3). The diffusion profiles depended on the neck geometry. Conditions correspond to the microphotographs in A–C, time of the microphotographs taken in A–C are marked on the curve with dashed lines. E) The 50% equilibration time and neck geometry are linearly related. 50% equilibration time was defined as the time it took for the average intensity in the protein wells to reach half of the maximum equilibrated intensity; neck geometry was defined by the length of the neck divided by the cross-sectional area of the neck. F) Microphotograph of the FID SlipChip containing the protein (photosynthetic reaction center from Blastochloris viridis) and the precipitant (4 M (NH4 )2 SO4 in 50 mM Na2 HPO4 /NaH2 PO4 buffer, pH 6.0). At the shortest equilibration time, only precipitates were obtained (left zoom-in). As equilibration time increased, fewer, larger crystals were obtained (middle and right zoom-ins).
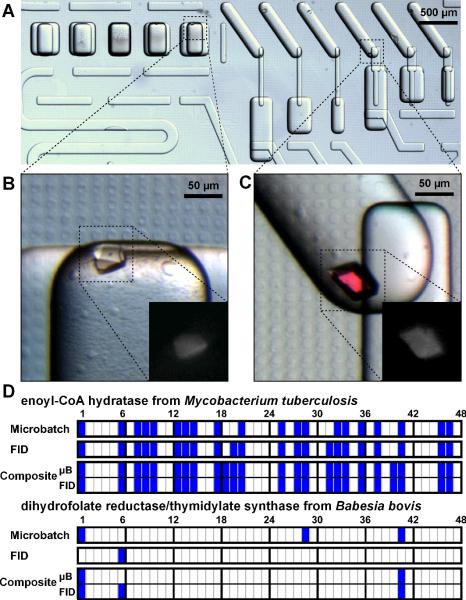
We first tested the effect of equilibration time on the kinetics of crystallization by crystallizing the photosynthetic reaction center from Blastochloris viridis using the FID SlipChip. As expected, we found that as the equilibration time increased, the protein progressed from precipitate to many small crystals to fewer larger crystals (Figure 2F). We then used the FID SlipChip to screen crystallization conditions for two proteins, enoyl-CoA hydratase from Mycobacterium tuberculosis and dihydrofolate reductase/thymidylate synthase from Babesia bovis. Approximately 12 μL of each protein was consumed to screen against a screening kit containing 48 precipitants for a total of 480 experiments (see Supporting Information Table S2). This experiment was performed on three SlipChips, each SlipChip with 16 precipitants and five conditions in duplicate per precipitant, for a total of 160 experiments per chip and consuming 4 μL of protein per chip We also screened both proteins using our previously described user-loaded SlipChip18 using the microbatch method against the same precipitants, and compared the microbatch results to the FID results (Figure 6D).
Figure 6.
Re-screening of crystallization conditions for proteins using the composite SlipChip reproduces results from microbatch and FID methods. (A) A microphotograph of the composite SlipChip used to screen conditions to crystallize dihydrofolate reductase/thymidylate synthase. All wells contain reagent 41 (45% (W/V) PEG-3000, 0.1 M CHES, pH 9.5). (B) Using the microbatch method, crystals formed at a mixing ratio of 1:2 (protein:precipitant). (C) Using the FID method, crystals formed at a mixing ratio of 1:2. Zoom views in B and C show a UV-microscope picture of the protein crystal. (D) A summary of the crystallization results using microbatch, FID, and composite SlipChips. Highlighted cells indicate that crystals formed. Numbers above the cells refer to reagents listed in Table S1. The composite method produced as many or more crystallization hits than either microbatch or FID alone for both enoyl-CoA hydratase and dihydrofolate reductase/thymidylate synthase.
The two proteins studied represent different kinetics of nucleation: enoyl-CoA hydratase nucleates quickly while dihydrofolate reductase/thymidylate synthase nucleates slowly. For enoyl-CoA hydratase, FID minimizes nucleation and yields crystals in conditions where only precipitation is observed in microbatch. Using the FID SlipChip, we were able to obtain crystals of enoyl-CoA hydratase under several conditions (Figure 3). Under conditions that yield crystals in both methods, such as for the photosynthetic reaction center from Blastochloris viridis, FID yields fewer large crystals (Figure 2F) while microbatch yields many small crystals. For dihydrofolate reductase/thymidylate synthase, few crystallization trials were successful in forming crystals. In trials where crystals formed, few crystals were obtained in each trial, indicating that the crystallization of dihydrofolate reductase/thymidylate synthase is nucleation-limited. Only one precipitant condition produced crystals using the FID method, but three precipitant conditions produced crystals in the microbatch method (Figure 6D). These results imply that proteins with different nucleation kinetics will require different crystallization techniques, and using multiple techniques in parallel increases the likelihood of identifying suitable conditions to produce protein crystals.
Figure 3.
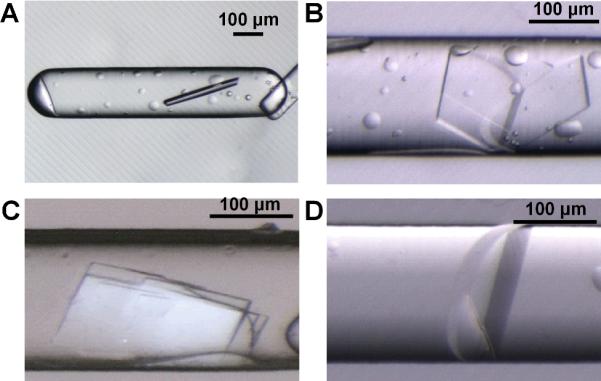
Microphotographs of crystals of enoyl-CoA hydratase from Mycobacterium tuberculosis obtained from SlipChip-based FID. A) A crystal obtained from reagent 15, 20% (w/v) PEG-3000 in imidazole buffer, pH 8.0; B) A crystal obtained from reagent 41, 45% (w/v) PEG-3000 in 0.1 M CHES buffer, pH 9.5; C) A crystal obtained from reagent 8, 2.8 M (NH4)2SO4 in 0.1 M citrate buffer, pH 5.5; D) A crystal obtained from reagent 14, 1.4 M sodium citrate in 0.1M cacodylate buffer, pH 6.5. Reagent numbers correspond to numbering in Table S1.
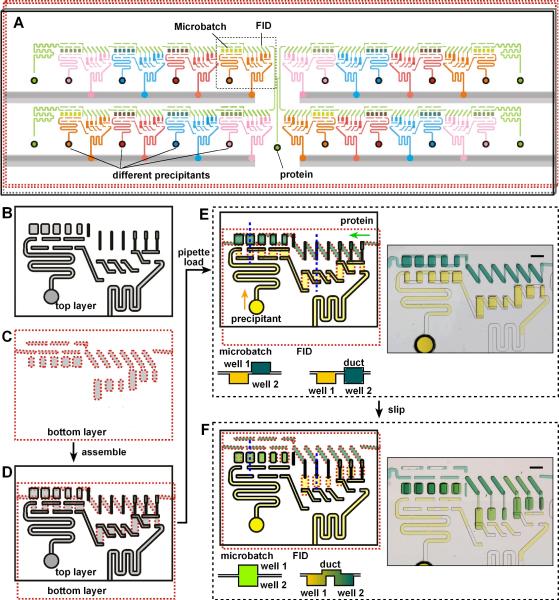
We designed a SlipChp that added another dimension to the screening process: in addition to identifying a precipitant and its concentration for crystallization, the two methods (FID and microbatch) were screened simultaneously (Figure 4). In this SlipChip one could also configure one continuous fluidic path for the protein sample and 16 separate fluidic paths for different precipitants. Wells designed for microbatch experiments and wells designed for FID experiments were in each fluidic path, allowing a single protein to be screened against 16 precipitants each at multiple mixing ratios and equilibration times. We also designed the FID wells to have multiple mixing ratios (1:2, 1:1, and 2:1), for a total of 176 experiments per chip, five microbatch experiments and six FID experiments for each of 16 precipitants.
Figure 4.
Development of a composite SlipChip to combine microbatch and FID methods. (A) A schematic of the composite SlipChip. Multiple precipitants (pink, blue, red, and orange) and multiple volumes and equilibration times for mixing the protein (green) can be screened on the same SlipChip using both microbath and FID methods. B–F) A zoomed-in schematic of the area outlined in A showing the operation of the SlipChip. B) The top plate (contoured in black) contains wells for the protein and ducts for the precipitant (microbatch) and ducts for both the protein and precipitant (FID). C) The bottom plate (contoured in red) has ducts for the protein and wells for the precipitant (microbatch) and wells for both the protein and precipitant (FID). D) When the two plates are assembled, the fluidic path for the protein and the fluidic paths for the precipitants are formed to fill wells for both microbatch and FID methods. E) A schematic of solutions of protein (yellow) and precipitant (blue) filling wells after loading by pipetting. The cross-sectional view (taken along the blue dotted lines in the schematic) shows the relative position of the wells in the microbatch and FID methods before “slipping”. A microphotograph of the food dye experiment demonstrates loading of the wells. F) A schematic of how protein and precipitant wells from one plate can be connected after “slipping”. The cross-sectional view (taken along the dotted blue lines in the schematic) shows how the wells containing protein and wells containing precipitant are connected. In microbatch (left) the two wells are aligned with one another, in FID (right) the two wells are connected by a narrow channel. A microphotogragh of the food dye experiment demonstrates how the protein and precipitant are combined in both microbatch and FID methods.
As we described previously,18 cross-contamination could potentially occur during the slipping step (between Figure 4E and Figure 4F): a thin film of solution can form between the two plates of the SlipChip, connecting the ducts and wells that should be separated. To eliminate this cross-contamination, the contact angle between the solutions and the plates of the SlipChip in the lubricant fluorocarbon must be greater than ~130°, and in other experiments we have spin-coated the plates with thin layers of fluorinated ethylene propylene.18 In the FID method, the solution in the neck channel is not stable at such high contact angles and tends to break up to minimize the surface energy.19 We solved this problem by patterning the surface of the SlipChip to make it more hydrophobic than the surface inside the wells and neck channels. To do so, we introduced an extra step of fine etching before washing off the coating left from the previous etching steps (see Supporting Information). This generated patterns of 10 μm diameter wells that were 250 nm deep (Figure 5). Without nanopatterning, the average contact angle of the 0.1% N,N-Dimethyldodecylamine N-oxide (LDAO) sample solution was only 112.2° (Figure 5C), with nanopatterning, the average contact angle of the same LDAO sample solution was 134.2°. In addition, nanopatterning decreased the surface area of glass that was directly exposed to the solution edge during the slipping step. The small wells trapped lubricating fluid and created a barrier to prevent solution leakage.
Figure 5.
Patterning hydrophobicity on the surface of the SlipChip with nanometer deep micropatterns. A) Microphotograph of the surface of the SlipChip after nanopatterning. No nanopatterning is present on the surface of the wells or microchannels. B) Zoomed-in microphotograph shows the surface patterning of small wells 10 μm in diameter and 250 nm deep. C) Microphotograph of a 4-μLaqueous droplet deposited on a surface silanized without nanopatterning. The droplet contained 0.1% (w/v) LDAO. The contact angle was measured right after the deposition, and was 115.4°. D) Microphotograph of a 4-μL aqueous droplet deposited on a surface silanized with nanopatterning of 250 nm depth. The droplet contained 0.1% (w/v) LDAO. The contact angle was measured right after the deposition, and was 137.9°. E) Contact angle of a 0.1% LDAO solution on silanized glass with different nanopatterns. The nanopatterns were obtained with the same nanopatterning photomask used to construct the SlipChips with nanopatterning (10 μm size cross mesh with 10 μm spacing), but different nanowell depths were obtained by changing the etching time. The contact angle was measured immediately after the droplet was deposited on the glass plate (at 0 min) and again 5 min after the deposition.
The performance of the nanopatterning was affected by the geometry of the nanopattern, including the nanowell size, spacing, and etched depth. We varied these parameters, and measured the contact angle of each nanopatterning (Figure 5E). The contact angle was plotted against the etched depth, but as the etched depth changed the surface area of the nanowells was also changed, because etching of glass is isotropic. Both the depth and the surface area of the nanowells should affect the contact angle, but here we have not done a detailed analysis of the relative importance of these two factors; related effects have been studied previously.20,21 All silanized glass with nanopatterning had a contact angle higher than glass without nanopatterning, and the contact angle increased with the depth of etching. The contact angle was above 130° for those glass plates where the nanopatterning depth was in the range of 196 nm ~3.81 μm. For nanopatterns with depth of 3.81 μm, the maximum contact angle was 153.62° (RSD=1.01%, n=5, measured after 5min of droplet setup). In all experiments, we found that the contact angle decreased with time, as observed by measuring contact angle 5 min later. The amount of the decrease was affected by the nanopattern depth. Nanopatterns with less than 200 nm depth had a faster decrease in contact angle than those nanopaterns that were deeper than 200 nm.
We then tested the composite SlipChip by using it to screen conditions for crystallization of the same two proteins we had already studied using separate FID and microbatch experiments, enoyl-CoA hydratase from Mycobacterium tuberculosis and dihydrofolate reductase/thymidylate synthase from Babesia bovis (Figure 6). The composite approach made the search for relevant crystallization conditions more efficient, as two routes to nucleation and crystal growth were investigated simultaneously, while the same small amount of protein (~ 12 μL) was consumed to screen each protein against the same screening kit. Both microbatch and free-interface diffusion components of the composite SlipChip functioned, and identified crystallization conditions for both proteins (Figure 6D). In the composite SlipChip, the majority of conditions identified by separate microbatch and FID screenings were also identified. For enoyl-CoA hydratase, two new conditions not identified in either of the individual screens were picked up by the hybrid screen. For dihydrofolate reductase/thymidylate synthase, one condition identified in the individual screens was lost. It was surprising to find new conditions using the composite SlipChip, since fewer concentrations were being sampled for each crystallization condition, but we doubt that this is due to any “magic” in the composite approach that leads to more hits or due to accidental slipping after the mixtures are generated. The most likely explanation of these observations is the intrinsic stochasticity of protein crystallization. In addition, the cause of the discrepancy between individual experiments and the composite experiment could be due to the variability among the protein samples used on different days, or slight variability in surface coatings of the SlipChips. We expect that as these SlipChips are used with more protein samples, these effects will become better understood.
Finally, to test whether the conditions identified could be scaled up, we scaled up one of the three conditions for crystallization of dihydrofolate reductase/thymidylate synthase identified in the microbatch SlipChip. The condition chosen was the protein sample at a mixing ratio of 0.33:0.57 with 20 % (w/v) PEG-8000, 0.2 M NaCl and 0.1 M CHES, pH 9.5. We scaled up dihydrofolate reductase/thymidylate synthase instead of enoyl-CoA hydratase because dihydrofolate reductase/thymidylate synthase is more difficult to crystallize, as indicated by fewer recognized hits (Figure 6D). The precipitant, 20 % (w/v) PEG-8000, 0.2 M NaCl and 0.1 M CHES, pH 9.5, produced crystals with best-defined shape at the chosen mixing ratio. The microbatch method is straightforward to translate the crystallization trial from SlipChips to well plates,18 and we successfully obtained crystals from the scale up experiment. We collected a full X-ray diffraction data set and determined the structure at a resolution of 1.95 Å, space group P212121 (Figure 7 and Table S3). The structure has been deposited in the Protein Data Bank (www.pdb.org), PBDid: 3KJR. The same protein was screened in parallel using Seattle Structural Genomics Center for Infectious Disease (SSGCID) and Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) facilities to yield crystals using microfluidic microbatch in a crystal card22 in conditions using 20% (w/v) PEG-8000, 0.1M CHES pH 9.5. These crystals yielded a 2.35 Å structure, space group P1 (PDBid 3I3R). We emphasize that the screens were conducted double-blind, without any information about crystallization conditions shared until after the screens were completed and crystals were obtained--the screening of crystallization of dihydrofolate reductase/thymidylate synthase on the SlipChip and the concomitant sale up experiments were performed without any knowledge of conditions obtained by the screening in facilities SSGCID and ATCG3D. We are encouraged that similar conditions, sharing the same PEG and buffer and different only by the presence of NaCl in the SlipChip screen, were independently discovered to yield structures. It was even more encouraging that we obtained a higher resolution structure, with a different space group. The 1.95 Å structure obtained in this paper, by virtue of a different space group, provided complementary structural information to the 2.35 Å structure (PDBid 3I3R). The interpretation of the structures is beyond the scope of this paper and will be published elsewhere.
Figure 7.

Determination of the crystal structure of dihydrofolate reductase/thymidylate synthase (PDBid: 3KJR). (A) A microphotograph of crystals of dihydrofolate reductase/thymidylate synthase grown in scale up experiments performed in well plates, by using the exact condition recognized in microbatch SlipChip, at a mixing ratio of 0.33:0.67 with 20 % (w/v) PEG-8000, 0.2 M NaCl and 0.1 M CHES, pH 9.5. (B) An X-ray diffraction pattern obtained from crystals in (A) at a region of 1.9 Å resolution. (C) Structure of dihydrofolate reductase/thymidylate synthase (refined to 1.95 Å resolution) obtained from crystals grown in scale-up experiments. The structure is displayed in cartoon generated using Pymol.
Conclusions
This paper demonstrates a SlipChip-based FID approach to crystallize proteins and a composite SlipChip-based approach to use microbatch and FID crystallization techniques simultaneously. The SlipChip provides a simple and relatively easy-to-use method to set up 160 experiments in free interface diffusion and 176 experiments in both microbatch and free interface diffusion, and all experiments can be setup simultaneously with a single slip. For applications where each experiment needs to be controlled individually, valve-based systems can be attractive. For applications such as protein crystallization, where each trial does not need to be controlled individually, the absence of valves dramatically simplifies both the execution of experiments and fabrication of devices. Fabrication of devices is further simplified by using a SlipChip platform, because the SlipChip should be compatible with inexpensive molding technologies and common plastics. More advanced techniques already demonstrated in plug-based crystallization techniques23–25 should be compatible with the SlipChip design. In addition to screening multiple precipitants, mixing ratios, and equilibration times, the composite SlipChip enables the comparison of two different protein crystallization techniques on the nanoliter scale in the same device. By using a single device, the surface chemistries and solutions used are the same, and any advantage of one method over the other can be identified and realized. Microbatch corresponds to rapid mixing through a larger interface, leading to more rapid nucleation. Free interface diffusion corresponds to slower mixing through a smaller interface, corresponding to slower nucleation. Control of the neck geometry enables the exploration of the continuum of methods bridging microbatch and FID methods. Crystallization based on counter diffusion approaches is mechanistically similar to FID methods. Using methodologies similar to the one described here, counter diffusion for crystallization can be implemented on SlipChip on smaller scale and in more multiplexed format than in traditional methods. The composite SlipChip provides a platform on which to test many proteins and the opportunity to learn more about important characteristics of protein crystallization.
After crystallization conditions are identified, high-quality crystals suitable for X-ray diffraction are needed to characterize the crystals and determine protein structures. To produce crystals large enough for X-ray diffraction, typically a minimum trial volume of ~10 nl is required,7,26 and even much smaller crystals can be analyzed using recent advances in synchrotron x-ray science,27so the crystals obtained in the SlipChip should be large enough for structural characterization. There are two options to obtain X-ray diffraction data from crystals grown in a SlipChip: extraction of crystals or in situ diffraction. The SlipChip is not sealed, therefore, our preliminary experiments indicate, the two plates can be separated and crystals extracted as has been done for a well-based chip,28 although this remains to be carefully tested with the SlipChip. Diffraction in situ may prevent damage to the crystals during post-crystallization manipulations and may increase throughput, but also remains to be tested with the SlipChip. To enable X-ray diffraction in situ in the SlipChip, the the SlipChip can be constructed of material that is compatible with in situ diffraction, such as PDMS, PMMA, and cyclo-olefin-copolymers, or the glass can be etched to create wells with thinner walls.23,29,30
If it is found that for whatever reason some crystals grown in a SlipChip cannot give high-quality X-ray diffraction data, the crystallization experiments can be scaled up using the conditions identified by the SlipChip screenings. Microbatch experiments are easily scaled-up in well plates, as we have shown for this paper. Another success has been achieved using the same strategy with ribose-phosphate pyrophosphokinase from Burkholderia pseudomallei. The condition (20% (w/v) PEG-3350, 0.2M magnesium formate, pH 5.9) recognized by conventional vapor diffusion method yielded crystals in space group of I222. The crystal structure was determined at 2.3 Å resolution (PDBid: 3DAH). In parallel using the SlipChip, we recognized a different condition (11% (w/v) PEG-8000, 37 mM sodium citrate, pH 5.5) yielding crystals in space group of P43212. We obtained a data set at 1.83 Å with crystals produced by scaling up, and the structural determination and PDB deposition is in progress. FID experiments are less trivial to scale up because the diffusion profiles and kinetics need to be replicated and thoughtfully controlled on a larger scale. The predictable diffusion profile we determined for FID SlipChip (Figure 2) should enable rational design of scalable SlipChips both down to picoliter-scales and up to microliter-scales. If the predicted diffusion profile applies at all scales, we can screen the crystallization conditions at a very small scale and scale up the hits to the desirable volume. Such work is in progress.
The technology described here has a number of additional applications beyond protein crystallization. For example, the nanometer-scale etching used to create a superhydrophobic surface that we introduce in this paper will impact surface patterning technologies.31,32 In addition, the techniques used for the FID method can be expanded to control equilibration times when combining solutions in other experiments. This control of equilibration can be useful for setting up concentration gradients in a range of applications, e.g. when studying chemotaxis and in other cell-based assays.
Supplementary Material
ACKNOWLEDGMENT
This work was supported in part by NIH Roadmap for Medical Research R01 GM075827, by the NIH Protein Structure Initiative Specialized Centers Grant U54 GM074961 (ATCG3D), and by the NIH Director's Pioneer Award (1DP1OD003584). Use of the Argonne National Laboratory GM/CA beamlines at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract No. DE-AC02-06CH11357. GM/CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). We thank James Norris of the University of Chicago for the generous gift of RC and SSGCID for the samples of glutaryl-CoA dehydrogenase. SSGCID is supported by Federal Contract No.: HHSN272200700057C from NIAID to the Seattle Biomedical Research Institute and its collaborating subcontractors. We thank Bart Staker and Thomas Edwards for buddy-checking the structure of dihydrofolate reductase/thymidylate synthase and for helpful discussions. We thank Rebecca Pompano for helpful discussions and Heidi Park for contributions to writing and editing this manuscript.
Footnotes
Supporting Information Available. Chemicals and materials, experimental procedures, additional figures and tables, and supporting movie. This material is available free of charge via the internet at http://pubs.acs.org. (PDF).
REFERENCES
- (1).Chen DL, Gerdts CJ, Ismagilov RF. J. Amer. Chem. Soc. 2005;127:9672–9673. doi: 10.1021/ja052279v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Li L, Ismagilov RF. Ann. Rev. Biophys. 2009 doi: 10.1146/annurev.biophys.050708.133630. in press. [DOI] [PubMed] [Google Scholar]
- (3).Zheng B, Gerdts CJ, Ismagilov RF. Curr. Opin. Struc. Biol. 2005;15:548–555. doi: 10.1016/j.sbi.2005.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Cohen AE, Ellis PJ, Miller MD, Deacon AM, Phizackerley RP. J. Appl. Crystallogr. 2002;35:720–726. doi: 10.1107/s0021889802016709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Salemme FR. Arch. Biochem. Biophys. 1972;151:533–539. doi: 10.1016/0003-9861(72)90530-9. [DOI] [PubMed] [Google Scholar]
- (6).Chayen NE. Curr. Opin. Struc. Biol. 2004;14:577–583. doi: 10.1016/j.sbi.2004.08.002. [DOI] [PubMed] [Google Scholar]
- (7).Hansen CL, Skordalakes E, Berger JM, Quake SR. P. Natl. Acad. Sci. USA. 2002;99:16531–16536. doi: 10.1073/pnas.262485199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- (9).Hansen CL, Classen S, Berger JM, Quake SR. J. Amer. Chem. Soc. 2006;128:3142–3143. doi: 10.1021/ja0576637. [DOI] [PubMed] [Google Scholar]
- (10).Anderson MJ, Hansen CL, Quake SR. P. Natl. Acad. Sci. USA. 2006;103:16746–16751. doi: 10.1073/pnas.0605293103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Ng JD, Gavira JA, Garcia-Ruiz JM. J. Struct. Biol. 2003;142:218–231. doi: 10.1016/s1047-8477(03)00052-2. [DOI] [PubMed] [Google Scholar]
- (12).Ng JD, Clark PJ, Stevens RC, Kuhn P. Acta Crystallogr. Sect. D-Biol. Crystallogr. 2008;64:189–197. doi: 10.1107/S0907444907060064. [DOI] [PubMed] [Google Scholar]
- (13).Garciaruiz JM, Moreno A. Acta Crystallogr. Sect. D-Biol. Crystallogr. 1994;50:484–490. doi: 10.1107/S0907444993014350. [DOI] [PubMed] [Google Scholar]
- (14).Thorsen T, Maerkl SJ, Quake SR. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- (15).Chayen NE. Acta Crystallogr. Sect. D-Biol. Crystallogr. 1998;54:8–15. doi: 10.1107/s0907444997005374. [DOI] [PubMed] [Google Scholar]
- (16).Chayen NE. J. Appl. Crystallogr. 1997;30:198–202. [Google Scholar]
- (17).Du WB, Li L, Nichols KP, Ismagilov RF. Lab Chip. 2009;9:2286–2292. doi: 10.1039/b908978k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Li L, Du WB, Ismagilov RF. J. Am. Chem. Soc. 2009 doi: 10.1021/ja908558m. (DOI: 10.1021/ja908555n) [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wu L, Li GP, Xu W, Bachman M. Appl. Phys. Lett. 2006;89 [Google Scholar]
- (20).Reyssat M, Yeomans JM, Quere D. EPL (Europhysics Letters) 2008:26006. [Google Scholar]
- (21).Quere D. Ann. Rev. Mater. Res. 2008;38:71–99. [Google Scholar]
- (22).Gerdts CJ, Elliott M, Lovell S, Mixon MB, Napuli AJ, Staker BL, Nollert P, Stewart L. Acta Crystallogr. D. 2008;64:1116–1122. doi: 10.1107/S0907444908028060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Zheng B, Tice JD, Roach LS, Ismagilov RF. Angew. Chem. Intl. Edit. 2004;43:2508–2511. doi: 10.1002/anie.200453974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Gerdts CJ, Tereshko V, Yadav MK, Dementieva I, Collart F, Joachimiak A, Stevens RC, Kuhn P, Kossiakoff A, Ismagilov RF. Angew. Chem. Int. Edit. 2006;45:8156–8160. doi: 10.1002/anie.200602946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Li L, Fu Q, Kors CA, Stewart L, Nollert P, Laible PD, Ismagilov RF. Microfluid. Nanofluid. 2009 doi: 10.1007/s10404-009-0512-8. DOI: 10.1007-/s10404-009-0512-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Li L, Mustafi D, Fu Q, Tereshko V, Chen DLL, Tice JD, Ismagilov RF. P. Natl. Acad. Sci. USA. 2006;103:19243–19248. doi: 10.1073/pnas.0607502103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Sanishvili R, Nagarajan V, Yoder D, Becker M, Xu SL, Corcoran S, Akey DL, Smith JL, Fischetti RF. Acta Crystallogr. Sect. D-Biol. Crystallogr. 2008;64:425–435. doi: 10.1107/S0907444908001741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Zhou X, Lau L, Lam WWL, Au SWN, Zheng B. Anal. Chem. 2007;79:4924–4930. doi: 10.1021/ac070306p. [DOI] [PubMed] [Google Scholar]
- (29).Emamzadah S, Petty TJ, De Almeida V, Nishimura T, Joly J, Ferrer JL, Halazonetis TD. Acta Crystallogr. Sect. D-Biol. Crystallogr. 2009;65:913–920. doi: 10.1107/S0907444909021489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Dhouib K, Malek CK, Pfleging W, Gauthier-Manuel B, Duffait R, Thuillier G, Ferrigno R, Jacquamet L, Ohana J, Ferrer JL, Theobald-Dietrich A, Giege R, Lorber B, Sauter C. Lab Chip. 2009;9:1412–1421. doi: 10.1039/b819362b. [DOI] [PubMed] [Google Scholar]
- (31).Tuteja A, Choi W, Mabry JM, McKinley GH, Cohen RE. Proc. Natl. Acad. Sci. U. S. A. 2008;105:18200–18205. doi: 10.1073/pnas.0804872105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Barbieri L, Wagner E, Hoffmann P. Langmuir. 2007;23:1723–1734. doi: 10.1021/la0617964. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.