Abstract
During active muscle contraction, tension is generated through many simultaneous, independent interactions between the molecular motor myosin and the actin filaments. The ensemble of myosin motors displays heterogeneous conformations reflecting different kinetic steps of the ATPase pathway. We used electron tomography of actively contracting insect flight muscle fast-frozen, freeze-substituted, Araldite embedded, thin sectioned and stained, to obtain 3-D snapshots of the multiplicity of actin-attached myosin structures. We describe procedures for alignment of the repeating lattice of sub-volumes (38.7 nm cross-bridge repeats bounded by troponin) and multivariate data analysis to identify self-similar repeats for computing class averages. Improvements in alignment and classification of repeat sub-volumes reveals (for the first time in active muscle images) the helix of actin subunits in the thin filament and the troponin density with sufficient clarity that a quasiatomic model of the thin filament can be built into the class averages independent of the myosin cross-bridges. We show how quasiatomic model building can identify both strong and weak myosin attachments to actin. We evaluate the accuracy of image classification to enumerate the different types of actin-myosin attachments.
Keywords: electron tomography, muscle contraction, image processing, image classification, atomic models
INTRODUCTION
The study of muscle contraction has been dominated by methods that yield a single averaged signal from the ensemble of actin-myosin interactions. X-ray diffraction, fluorescent and paramagnetic probes, fiber mechanics and protein biochemistry produce information on the average state of the ensemble within the sample. Spatial averaging methods previously used for 3-D imaging of muscle (Taylor et al., 1984; Taylor et al., 1989b; Taylor et al., 1989a; Taylor et al., 1993; Winkler et al., 1996) have the same limitation. Single molecule methods, which must by necessity be applied to samples outside of the well ordered filament lattice of muscle, lose information on any effects imposed by organization of the lattice.
Electron microscopy is uniquely suited for visualizing the individual molecules in an ensemble in situ and can do so in 3-D using the technique of electron tomography (ET) (Lucic et al., 2005). While ET can produce a 3-D image of even a single copy of a structure, it does so with extremely low stochastic signal-to-noise ratio (SNR) and thus details are usually hidden by the noise. In the muscle lattice, any structures (cross-bridges) that do not occur multiple times constitute a source of structural noise that tends to camouflage patterns of acto-myosin interactions. To achieve higher resolution and detail, averaging methods are necessary both to improve the stochastic SNR and to extract patterns in the data hidden by the structural noise. Ideally, averaging is done over a population of identical conformations of interest, all well aligned so that differences between class members arise due to stochastic noise. The critical step here is identifying patterns of actin-myosin interactions that may be randomly distributed within an ordered lattice of filaments.
Insect flight muscle (IFM) shows the best ordered sarcomeres of any known striated muscle. X-ray diffraction of IFM yields detailed patterns of intense reflections in rigor (Holmes et al., 1980), relaxed (Reedy et al., 1965; Miller and Tregear, 1972; Reedy et al., 1983; AL-Khayat et al., 2003), and even contracting muscle in vivo (Dickinson et al., 2005; Bekyarova et al., 2008). The rich X-ray diagram of relaxed IFM reflects the ordered lattice of actin and myosin filaments with their ordered helical arrangement of subunits, but with little interaction between each. In rigor IFM, the myosin heads form regularly angled “chevrons” every 38.7 nm along the thin filament (Reedy and Reedy, 1985). Despite the well ordered filaments and lattice, there are periodicity differences between the actin and myosin filaments that cause variation in cross-bridge structure even in homogeneous enzymatic states, such as nucleotide-free rigor.
Although IFM actin and myosin filaments share a common 38.7 nm helical repeat, the axial repeat of 5.52 nm between actin subunits and 14.5 nm between “crowns” of 4 myosin heads on the thick filament do not match. Actin filaments have a helical symmetry of 28 subunits in 13 turns that gives rise to a helical repeat of 77.3 nm and a half period crossover repeat of 38.7 nm. The shortest axial repeat is 116 nm equaling 8 × 14.5 nm thick filament repeats and 3 × 38.7 nm actin crossover repeats. The shortest true repeat period over which myosin origins and actin targets come into identical axial and azimuthal register is 232 nm. Consequently, there is a continuous vernier between myosin head origins and actin binding sites along the 116 and 232 nm long periods, so that no single cross-bridge conformation, like that observed in nucleotide-free myosin sub-fragment 1 (S1) bound to actin filaments in vitro (acto-S1), can describe the structure of cross-bridges in situ.
ET of myosin cross-bridges in rigor IFM has characterized the structural variability due to lattice constraints of myosin heads (Schmitz et al., 1996; Winkler and Taylor, 1999; Chen et al., 2002), due to mechanical stretch (Liu et al., 2004), and due to swelling the rigor lattice to show the length and orientation of the S2 domain (Liu et al., 2006). Despite the comparatively high regularity of the rigor lattice, each of these studies has required development of improved methods for analysis of structure variability in order to reveal more molecular details.
ET has also produced 3-D images of nucleotide-bearing cross-bridges in IFM, including the variable conformations of cross-bridges in AMPPNP (Schmitz et al., 1996), and in a weakly-bound equilibrium state produced by AMPPNP and ethylene glycol (Schmitz et al., 1997). However, these studies did not produce high-SNR averages to reveal details of the actin-attached cross-bridges.
Unlike rigor muscle, where nucleotide-free myosin heads are attached to actin in a single kinetic state, in contracting muscle, myosin heads can attach to actin in any of several steps of the kinetic cycle, some of which involve weak actin-myosin attachments and others that are strong attachments (Geeves and Conibear, 1995). Characterization of these attachments requires knowledge of the position of two key parts of the myosin head, its motor domain (MD), which contains the actin binding and catalytic activities, and the lever arm, which binds an essential light chain (ELC) and a regulatory light chain (RLC). Axial movements of the lever arm generate filament sliding. Strong actin-myosin binding involves stereospecific interactions of the MD with actin, but strongly bound myosin heads may have lever arm orientations that vary over a ~90° range (Taylor et al., 1999). Weak actin-myosin attachments involve non-specific, mostly ionic interactions between the MD and actin. Weak actin-myosin attachments are a particular challenge to imaging, because their low actin affinity and heterogeneous structure make them difficult to trap in sufficient numbers and homogeneity to be amenable to visualization by any of the averaging techniques of cryoelectron microscopy such as helical or single particle reconstruction. The tomograms of fast-frozen contracting muscle have numerous factors that produce heterogeneity in the myosin-actin attachments, which must be sorted out if high quality averages are to be obtained without obliterating the meaningful conformations.
Electron micrographs of quick-frozen contracting muscle commonly reveal cross-bridges of highly variable form (Hirose et al., 1994; Lenart et al., 1996). In the only previous application of ET to quick-frozen, actively contracting muscle in a state dubbed high static tension or HST, tilt angles of both the MD and the lever arm of individual cross-bridges were analyzed and a hypothetical sequence of S1 head conformations corresponding to a power stroke of a single motor was modelled (Taylor et al., 1999). Those data were collected using manual selection of tilt angles, irradiated continuously as the tilt series was collected and recorded on film. Tilt angles were estimated from the goniometer vernier and refined during the image alignment process. “Column averaging” along each filament was used to improve the SNR of the cross-bridges. These are 1-D averages along axial columns of density that repeat every 116 nm. These column averages showed active cross-bridges bound to only a restricted number of actin subunits along the thin filament that are termed “actin target zones” and identified both weakly bound and strongly bound cross-bridge attachments. However, column averaging over the 116 nm period cannot resolve individual actin subunits in the thin filaments, an essential criterion if a more precise characterization of the different cross-bridge forms, in particular the weak binding forms, is to be achieved.
Here we report on development of several improvements to the analysis of the 38.7 nm repeats that facilitate a more detailed view of the variety of myosin head forms in fast frozen contracting muscle than has been possible up to now. These improvements include (1) a procedure for aligning individual repeats that resolves the helix of actin subunits within the thin filament, (2) application of multivariate data analysis (MDA) and hierarchical ascendant classification to accurately identify and group self-similar cross-bridge forms regardless of placement within the filament ensemble, (3) approaches to building quasiatomic models to identify weak and strong binding cross-bridge forms and to evaluate their accuracy, and (4) use of MDA and classification to quantify the frequency as well as location of variable actin attachments in situ. The result of these improvements is visualization and quantification of a rich variety of forms, some of which can be readily identified with strong binding myosin heads, some of which may represent weak binding states that precede strong binding, others that may sample collision complexes between the thin filament and myosin or may indicate novel, weak interactions between myosin and actin and possibly troponin (Tn).
MATERIALS AND METHODS
Rapid Freezing and Freeze Substitution
Rapid freezing with simultaneous monitoring of fiber tension was performed on a Heuser Cryopress freezing head (Hirose et al., 1993). Modifications made to the freezing head, and details of the specimen manipulation prior to and subsequent to freezing that are specific for this work have been described (Taylor et al., 2007). Freezing experiments were performed on single fibers of glycerinated Lethocerus dorsal longitudinal muscle activated by raising the [Ca2+] to pCa 4.5 in the presence of Mg ATP. Briefly, after 20–40 sec equilibration in activation buffer the fibres were slam-frozen without cryoprotectant against a liquid He-cooled, Au-coated Cu mirror while monitoring tension up to the moment of impact.
Freeze-slammed fibers were freeze-substituted at −80°C in acetone using a tannic acid-uranyl acetate sequence, dubbed TAURAC (Taylor et al., 2007), and embedded in Araldite-506. Ultrathin (25–30 nm) longitudinal sections were stained by permanganate-lead (Reedy and Reedy, 1985). In cross-section, frozen fibers showed vitrification and native cylindrical morphology to a depth of 3–6 μm. For further details see (Taylor et al., 1999).
Electron Tomography
Dual-axis tilt series of half-sarcomeres covering a maximum range of ±72° were collected on a FEI CM300-FEG at 300 keV using a Gatan model 670 Ultrahigh tilt analytical holder (Gatan, Pleasanton, NJ) and recorded on a TVIPS Tem-Cam F224 2k x 2k CCD camera. Each tilt series was recorded from regions where longitudinal thin sections contained single myosin and actin filament (myac) layers (Reedy, 1968). Tilt angles within each tilt series were calculated according to the Saxton scheme (Saxton et al., 1984). Each tilt series consisted of ~100 images, each recorded with ~200 electrons/pixel (4.2 e−/Å2). One series was collected while tilting along an axis parallel to the filament. The second tilt series was obtained by removing the specimen holder from the microscope, extracting and rotating the specimen grid ~90°, reinserting the holder with its specimen grid into the microscope, and relocating the region recorded in the first tilt series.
The two tilt series were first independently aligned using marker-free alignment and area matching (Winkler and Taylor, 2006). The two resulting tomograms were then merged by patch correlation and volume warp using IMOD (Mastronarde, 1997) to produce the raw, dual-axis tomogram of dimensions 1600 × 1344 × 70. The missing data within the transform of the final tomogram consists of a pyramidal volume. The pixel size, 0.69 nm, was internally calibrated using the axial 116 nm period. We collected two dual-axis tilt series, but only one contained an appreciable number, 515, of well-centered repeat subvolumes. The analysis described here is based on this tomogram.
Subvolume Extraction and Alignment
Repeat subvolumes, hereafter referred to simply as repeats, containing a 38.7 nm axial length of the actin filaments, their bound cross-bridges and adjacent thick filament segments were identified by computing a cross-correlation map between a reference consisting of three successive repeats along the thin filament (116 nm) against the entire raw tomogram. The repeats were centered on the actin target zones, which are seen as segments 11–16 nm long centered between successive Tn densities. Each repeat had dimensions of x=74, y=88, and z=48 voxels, the y axis corresponding to the fiber axis and the x axis to the inter-thick filament axis.
Successful MDA and manageable building of quasiatomic models into the class averages, required that all the 38.7 nm repeat segments be aligned to the same frame of reference. The 28/13 helical structure of the IFM actin filament results in an azimuthal rotation by 180° of successive 38.7 nm segments of each actin filament. Thus, alternate repeat segments along each individual thin filament were rotated azimuthally by 180° before any alignment was begun, thus placing them in identical rotational register with the unrotated repeat segments. Each of the thin filaments within the IFM lattice can assume an azimuthal rotation of either 0° or 180° chosen at random (Holmes et al., 1980). No other relative rotations are possible for the individual thin filaments within a single myac layer. To determine the relative thin filaments rotations, both possible orientations were tested for each repeat. The resulting best orientations were then referred back to the individual filaments to determine the final orientation for that filament. Typically ~80% of the repeats from a single filament would display a consistent rotation which was then imposed on all of its repeat segments.
Repeat alignment was characterized using the mutual correlation function (Van Heel et al., 1992) which reduces the relative weight of low frequency, high amplitude Fourier coefficients and gives a sharper correlation peak. The initial alignment was based on the center of the cross-correlation peak that identified each repeat but taking into account the alternating azimuthal rotation of repeats. This was followed by MDA and classification to compute actin filament class averages. Class averages that showed improved thin filament features were used as multiple references for the next alignment cycle, and the process continued until no further improvement was observed in the correlation coefficient, the degree of translational shift and in the consistency of the azimuthal 0° or 180° rotation. Aside from the 180° rotation to account for the filament symmetry, alignment involved only translational movements.
In this repeat alignment stage, the purpose of MDA and classification is to find references that lead to better alignment and for this only the thin filament features are important. A key aspect of MDA is the construction of the mask, which defines the voxels included in the classification process; voxels within the mask contribute while those outside the mask are ignored. A key part of the alignment procedure is the mask which defines those voxels that will be used for the alignment. Both thin filament alignment and MDA used a cylindrical mask that enclosed the thin filament and a small part of the myosin MD that falls within the cylinder. The MDA mask was a Boolean version of this cylinder so that voxels outside the mask are set to zero and those inside are unchanged. The mask for alignment used this same cylinder but applied a soft apodization edge to minimize Fourier ripple effects on the alignment.
Multivariate Data Analysis
MDA was done using software previously described (Winkler and Taylor, 1999; Winkler, 2007). Grouping in classes was done using hierarchical ascendant classification (van Heel, 1989). Masks were obtained by applying a thresholding tool to a variance map computed from the complete aligned image set. Voxels whose value equalled or exceeded the threshold were included within the mask. Multiple masks were devised from the variance map in order to perform the various focused classifications. These were obtained by dividing up and editing the volume obtained after applying the threshold. Including the mask used for the thin filament alignment, a total of 13 different masks were constructed to identify specific features contained within the repeats. Although it is possible for the masks to have islands of excluded voxels within its outermost boundary, in practice, each mask was uniform throughout with the single boundary being the outer perimeter.
The classification always uses the highest ranked eigenvectors determined by MDA, except for the very first one which is generally equivalent to the global average. For the large classification masks, such as the primary left/right masks, 32 factors are used. For the smaller masks, such as the Tn masks, 16 factors are used.
Model Building
Atomic models were built in a hierarchical fashion with modifications from earlier studies of rigor IFM fibers (Liu et al., 2004; Liu et al., 2006). The F-actin quasiatomic model was constructed of 16 actin subunits built on a 28/13 helical lattice and was taken from PDB 1M8Q. A 16-subunit filament was chosen so that two pairs of Tn atomic models (Pirani et al., 2006) could be mounted onto both ends of the filament segment. A pair of tropomyosin (TM) models that matched the helical symmetry of the IFM actin filament were provided by Dr. Katrina J. V. Poole (Poole et al., 2006). The resolution of the reconstruction was not high enough to reveal the density that might be assigned to TM, so the position of the TM along the thin filament was not altered from the Poole coordinates.
We used two crystal structures for myosin heads to model the in situ cross-bridges. The first was adapted from the chicken skeletal rigor acto/S1 complex (Holmes et al., 2003) (available at ftp://149.217.48.3/pub/holmes) and was used as the starting model for cross-bridges whose lever arm was oriented toward the rigor configuration. For cross-bridges that appeared to be perpendicular to the thin filament or had a lever-arm-up conformation, we used the transition state of scallop myosin S1 (Houdusse et al., 2000) whose MD was pre-aligned to the chicken skeletal rigor MD position as the starting model. The fitting process included rigid body movements of the entire myosin head combined with lever arm modification once the MD was placed into its corresponding density. Adjustments to the lever arm used as pivot points: residue 710 (or 706), 780 (or 775) and 806 (or 801) in the chicken skeletal atomic structure (or scallop transition state structure). Manual fitting was done using the X-ray crystallography model fitting program O (Jones et al., 1991).
Evaluation of Clustering Accuracy
This study uses MDA and classification extensively to identify patterns of cross-bridge structure. We therefore devised the following test to evaluate the accuracy of MDA for identifying similar cross-bridge forms in a highly variable ensemble. A gallery was formed between each class average and the entire class membership of raw repeats, the class average indicating where bridging density was found, and the gallery of class members facilitating comparisons. The presence of cross-bridges placed either M-ward or Z-ward with respect to the target zone actins within the class average was then compared with the presence or absence of this cross-bridge within the raw repeats. Each Z-ward and each M-ward cross-bridge in the raw repeats was counted and the result compared with the prediction from the class average and the number of class members. The comparison did not take into account any variation in orientation of the lever arm among the individual class members relative to the class average value.
RESULTS
Electron Tomograms of HST
Active cross-bridges are more variably angled than rigor “double chevrons”, partly because active heads originate from 14.5 nm periodic shelves, also known as crowns, (Fig. 1A) that are placed variably with respect to the target zones and can approach one target zone from two successive crowns. However, there is an underlying pattern to active cross-bridges which can be seen by sighting across the micrograph from the side at an angle. A distinct banding is present which represents the binding of active myosin heads to the actin target zone (Reedy, 1968). This single banding pattern is distinct from the double bands seen in rigor IFM. The rigor cross-bridges in the actin target zone are “lead” bridges that form the first bands. The second bands in rigor are the “rear” cross-bridges that bind near the Tn complex (Reedy and Reedy, 1985). Although some cross-bridge attachments to Tn are detected here in active contraction, their small numbers and the presence of ATP in the buffer at freezing precludes these being rigor-like rear bridge attachments.
Figure 1.
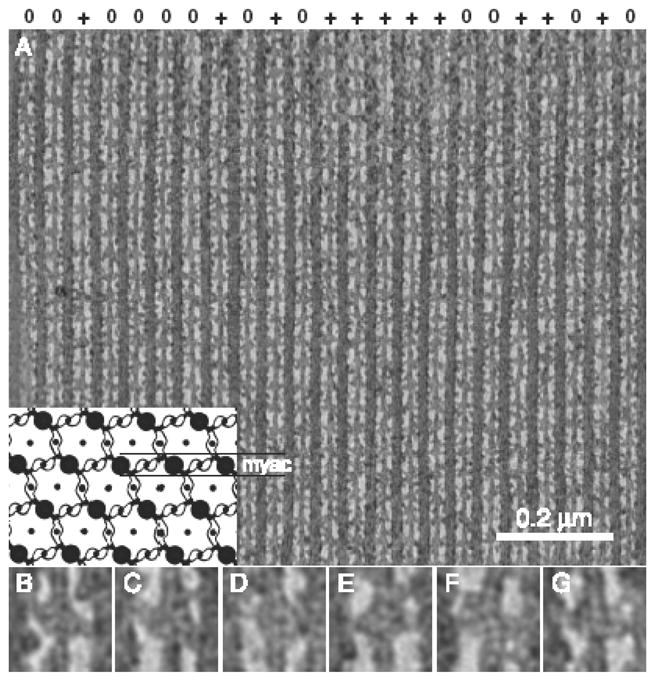
Projection images of the merged dual-axis tomogram of a “myac” layer. (A) Projection of the complete tomogram. Symbols across the top (0,+) indicate whether the thin filament was found to have the same rotation or differs from the others in the population by a 180° rotation about the thin filament axis. Orientation has the Z-line at the bottom, corresponding to the (+) or “barbed” end of the actin filament, and the M-line at the top, corresponding to the (−) or “pointed” end of the actin filament. Insert at the lower left shows a diagram of a cross section of IFM including only one level of cross-bridges. Thick filaments are large black disks, thin filaments are small black disks and connecting cross-bridges are outlined. The myac layer, marked in the diagram by paired horizontal lines, is a longitudinal section consisting of alternating thick and thin filaments. In myac layers, thin filaments only form cross-bridges with the two nearest-neighbor thick filaments; they never form cross-bridges with thick filaments in adjacent myac layers. (B–G) Enlarged images across the bottom illustrate the negative stain effect of the freeze substitution and staining procedure. The black coating on the outside is accumulation of tannic acid and uranyl acetate. Small panels are 44.2 nm wide.
The new tomogram shows a distinct shell of negative staining around the actin filament and the myosin MD (Fig. 1B–G) that can be seen in the raw repeats, but is seen with particular clarity in the global average (the average of all repeats) (Fig. 3B). Because of the negative staining, a single headed cross-bridge appears as a white region of stain exclusion surrounded by dark regions that represent the tannic acid-uranyl acetate coating from the TAURAC freeze substitution. Two headed cross-bridges appear as two white zones and three black zones (Fig. 1B, D). In the raw tomogram, negative staining is pronounced and uniform for the larger structures, such as the myosin MD, the actin filament, and the myosin filament backbone. Smaller structures, such as the lever arm of the myosin heads are variable, sometimes appearing negatively stained (Fig. 1B) and sometimes not (Fig. 1D,G). In class averages, the lever arm never appears negatively stained. The mixture of positive and negative stain in the cross-bridge class averages made it impossible to do quantitative fitting for the myosin heads as practiced earlier (Chen et al., 2002) which required that the density be interpretable everywhere as positive staining. The actin filament, on the other hand, appears everywhere as a negatively stained feature which enabled us to fit quantitatively a quasiatomic model of the thin filament to the density features within the negative stain shell.
Figure 3.

Thin filament detail after alignment and averaging. (A) Global average of originally extracted repeats prior to subvolume alignment. From left to right are a central section of the initial global average, a surface view in the same direction as the original tomogram and a surface view from a direction along the inter-thick-filament axis showing the paired bulges due to troponin. Since this is a stained specimen, regions of protein (stain rich and thus low electron translucency) are dark in this representation (low electron counts) and regions rich in embedding medium, and thus high electron translucency are bright (high electron counts). (B) Global average after subvolume alignment is completed. The 2.75 nm stagger on different sides of the actin filament is now visible as is the different spacing separating troponin densities on opposite sides of the actin filament. The negative stain effect in the specimen is now pronounced. In the surface view, middle, both the troponin densities and the myosin heads have increased definition. The appearance suggests the presence of four myosin heads, two on each actin long pitch helical strand. Viewed along the filament axis, the difference in spacing of the troponin densities on the front and back surfaces (left, red, and right, blue, in this view) is easily visible. (C, D) Images of variance map calculated from all aligned HST repeat volumes. (C) is the central section and (D) is the projection image. Pixels with higher variance are brighter than those with lower variance. Note that electron translucency in the specimen does not necessarily correlate with high variance. Highest variance is localized to the surface of the filaments, low variance is correlated with regions of highest electron translucency (embedding medium) as well as low electron translucency (thick filament backbone). (E) Schematic model of the thin filament illustrating the alternation between wide and narrow spacings. The lettering on the bottom repeat gives the actin protomer name assignments. (F) A mapback of a thin filament viewed along the inter-thick-filament axis. Red lines indicate the centers of the Tn densities.
Alignment of Repeat Subvolumes
The repeat alignment was based on the principle that the thin filament was a feature that could be assumed constant within each repeat and could be used to align all repeats to a common frame of reference. This assumption has consequences for alignment of thick filament features (Liu et al., 2004; Liu et al., 2006) (illustrated schematically in Figure 2). In the filament lattice of IFM, actin filaments individually have a regular helical structure, but are thought to be arranged in the half-sarcomere with a random 180° axial rotation (Holmes et al., 1980). Thick filaments in relaxed muscle are in helical register (Schmitz et al., 1994) but in active muscle they lack even axial registration (Fig. 1A; Fig. 2A (Tregear et al., 2004)). In order to align each repeat to a single reference, successive repeats along individual thin filaments were rotated about the filament axis by 180°, a requirement of the actin filament symmetry. This rotation also brings with it the adjoining thick filament segments within each repeat which become mixed on the left and right hand sides (Fig. 2B). When repeats from adjacent thin filaments are aligned onto the actin filament reference, their accompanying thick filament densities undergo the same transformation (Fig. 2C) even though they do not influence the alignment. Thick filament features become progressively misaligned and mixed between left and right-sides of the thin filament, even while the thin filament alignment improves. The thick filament is featureless in the global average (Fig. 2D). However, as shown below, thick filament features are retained in most class averages demonstrating the linkage between thin filaments, cross-bridges and their thick filament origins.
Figure 2.
Scheme for alignment and averaging of repeat subvolumes. Actin strands are colored blue and red respectively, troponin complexes are yellow for the red strand and green for the blue strand. Shelves of myosin heads (crowns) spaced 14.5 nm apart are black while the thick filament backbones are colored orange, lime or cyan. (A) Repeats consist of actin crossovers centered between troponin densities. (B) When repeats are extracted from the tomogram, the shelves of myosin origins are thus not aligned among the population of repeats. (C) Before starting alignment, alternate repeats on individual actin filament are rotated 180° about the filament axis. This process also switches thick filament segments to the other side of the thin filament. (D) After aligning and averaging all repeats, the 14.5 nm myosin shelves on the thick filaments become blurred.
Each repeat was identified and extracted based on the center of the cross-correlation peak. The order in which repeats were extracted from the tomogram was first along each individual thin filament followed in turn by adjacent filaments across the half-sarcomere from left to right. This order facilitated rotational alignment of repeats on each thin filament since an initial 180° azimuthal rotation could be applied to alternating repeats as dictated by the helical symmetry. Thus, the repeats from each thin filament could be placed in rotational register even when not yet aligned to adjacent filaments. Next, instead of using single reference alignment (Liu et al., 2004) we combined multireference alignment with MDA and classification (Liu et al., 2006), a procedure that prevents reference bias from dominating. Classification produced several class averages with improved definition of the actin subunits; these were selected for multireference alignment. For the final alignment, we returned to a single reference as it was desirable to be able to fit one quasiatomic model to the thin filament that would be used for all subsequent model building. In addition to revealing the actin subunits along the thin filament, the results from the final alignment identified the relative rotation of all the thin filaments across the half-sarcomere. The thin filament orientations showed no apparent pattern across the tomogram (Fig. 1A). This provides the first independent test of the IFM thin filament arrangement proposed by Holmes et al. (1980).
The thin filament occupies literally a central place in the analysis. The use of a single reference for the final alignment and a quasiatomic model for the thin filament that was fit to the global average could potentially generate errors in the subsequent identification of strong and weak binding cross-bridges if there were substantial variance in the azimuthal alignment of the repeats. Because of its importance, we attempted to estimate the angular variance among the repeats. We performed a quantitative alignment using as a reference a density map derived from the thin filament quasiatomic model against which we fitted each of eight thin filament class averages obtained using the thin filament classification mask (see Methods). The search was conducted using an azimuthal angular increment of 2°. The mean value of the rotation was 1° and the standard deviation was 6°. For the displacement, the standard deviation was 0.5 nm (Table 1).
Table 1.
Orientation difference between thin filament class averages.
| Class ID | # of Members | Translation (nm) | Rotation (°) |
|---|---|---|---|
| 0 | 93 | 1.20 | −6.0 |
| 1 | 68 | 1.37 | 6.0 |
| 2 | 49 | 0.69 | −8.0 |
| 3 | 88 | 2.06 | 2.0 |
| 4 | 65 | 1.69 | 2.0 |
| 5 | 72 | 0.69 | 8.0 |
| 6 | 52 | 0.69 | 8.0 |
| 7 | 28 | 0.97 | −10.0 |
| Weighted mean | 1.25 | 1.0 | |
| Standard deviation | 0.5 | 6.2 | |
Before alignment, actin detail is essentially invisible in the initial global average (Fig. 3A), but after alignment the 2.76 nm offset between actin subunits of the two long pitch helical strands becomes clearly visible (Fig. 3B). Less obvious is the improvement in the visualization of Tn because it is located on the front and back surfaces of the thin filament in myac layer views and superposition in projection images produces either a single or a pair of black spots. The IFM filament lattice dictates that all the thin filaments in myac layers have Tn positioned on the front and back surfaces. When viewed from the side, the Tn feature is clearly visible in the global average even before repeat alignment has started (Fig. 3A). IFM Tn has a molecular mass of ~115 kDa (Leonard and Bullard, 2006), which is about as large as the myosin heads of IFM. Since IFM Tn is congruent with the actin crossover spacing, Tn densities on the front (or back) surface of the thin filament would be expected to alternate between a wide axial separation equivalent to 7½ actin subunits (7.5 × 5.5 nm) and a narrow axial separation equivalent to 6½ actin subunits (6.5 × 5.5 nm) and this is exactly what is seen after repeat alignment (Fig. 3B,F). The alternation of wide and narrow Tn separation has not been seen in an IFM 3-D reconstruction until now. In rigor, the attachment of myosin heads in the region of Tn, i.e. the rear bridges of the double chevron (Reedy and Reedy, 1985), obscures the Tn density. Because the actin subunits are now resolved, they can be assigned individual designations (Fig. 3E).
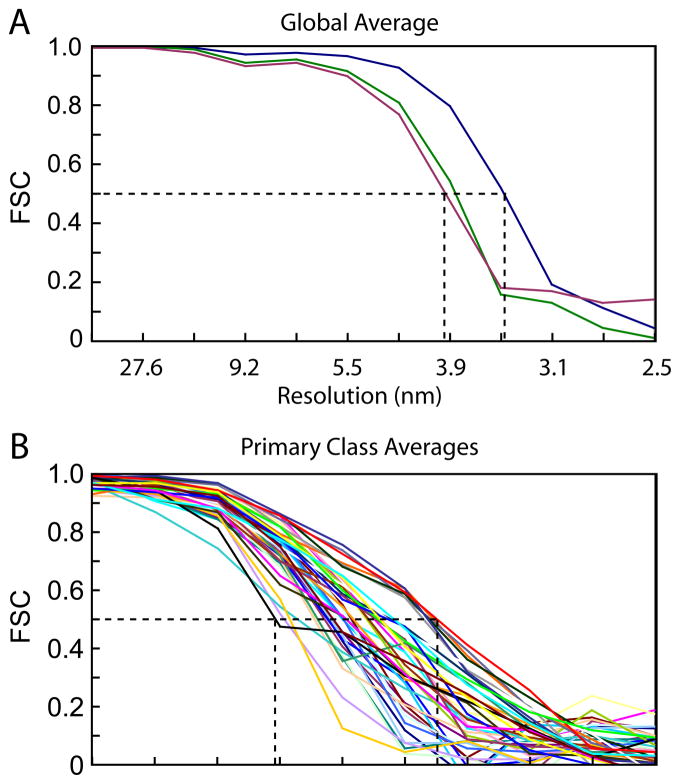
The resolution obtained in our reconstructions was computed using the Fourier Shell Correlation (Harauz and Van Heel, 1986) with the empirical threshold of 0.5 as the criterion. The resolution of the global average was 3.5 nm – 3.9 nm, the former computed for the thin filament region only and the latter including the inter-thick-filament space or the entire repeat (Fig. 4A). Resolution of the 20 primary mask class averages ranges from 5 nm to 9 nm (Fig. 4B) and was assessed on those voxels defined by the MDA masks to which a soft Gaussian boundary of six pixels width was applied to prevent Fourier ripples from sharp edges from defining the resolution. For small volumes such as those obtained from the different classifications, the apodization width may have a significant effect on the FSC leading to artifactual resolution estimates. That the resolution estimated by FSC is at least approximately correct is verified by the appearance of the staggered arrangement of actin subunit densities in the target zone. This would only appear if the resolution was 5 nm or better. All class averages show the actin subunit stagger (Fig. 7), consistent with 5 nm resolution.
Figure 4.
Resolution assessment of isometric IFM cross-bridge repeats. (A) Global average. Resolution is not homogenous through out the entire volume. The blue line is the measurement for the thin filament portion only; the green line is with the inter filament space is added (where the cross-bridges are located) and the red line for when the thick filament portion is further added. (B) FSC plots for the primary mask class averages, range 5~9 nm.
Figure 7.
Reassembled repeats of isometric HST. Orientation of all panels has Z-line at the bottom, M-line at the top. Numbers in the upper right hand corner correspond to raw repeat numbers. Top four rows are 20 reassembled repeats obtained from classification using the primary masks shown in isosurface display. Rows 1 and 2 contain examples of single-headed and double-headed (red circles) cross-bridges. Row 3 contains primarily mask motifs, which consist of two cross-bridge pairs, one pair coming from a thick filament “crown” positioned closer to the M-line side of the 38.7 nm repeat, and the other pair from the adjacent crown closer to the Z-line of the repeat. All cross bridges in this row are single headed. Rows 3 and 4 contain ten reassembled repeats from classifications using the primary and Tn masks. Tn bridges identified by blue ovals. Most repeats have no Tn bridges or have Tn bridges on only one side. In occasional instances, e.g. 366, Tn bridges are found on both sides. Bottom 4 rows are central sections through the same reassembled repeats showing the staggered arrangement of actin protomers and their bound myosin heads along the thin filament.
In the actin target zone, i.e. midway between Tn densities, the actin subunits in the global average appear to extend radially further than predicted from the actin filament structure alone (Fig. 3B). This effect is attributable to density contributed by the attached myosin heads. However, there are no densities bridging thick and thin filaments (Fig. 3B), only density attributable to the MD of the attached target zone cross-bridges, showing that their lever arms are highly variable.
Multivariate Data Analysis
The purpose of MDA and classification is to sort the highly variable cross-bridge forms within the repeats into self-similar groups (see also Methods). The averages computed from these self-similar groups (hereafter referred to as class averages) represent a high-SNR version of its members. The principle of this averaging process is to find the balance between SNR improvement, which is achieved by computing fewer classes with more class members, and the retention of structure variability, which is achieved by computing more classes with fewer class members.
The number of ways that actin bound myosin heads can be distributed within an individual repeat is very large when considered in terms of the probability of binding to a particular actin subunit within the repeat combined with differences in structure of the bound heads. However, the number of possibilities is smaller, and more manageable if considered separately for specific regions of the repeat, such as the left and right hand sides of the thin filament. Thin filament labeling by myosin heads from one side is independent of the labeling from the other side so there is no compelling reason for classifying the density within the entire repeat with a single MDA mask.
The results of MDA are highly dependent on the binary mask that defines the enclosed volume of interest. Voxels outside of the mask are ignored in the analysis and only those within the mask have relevance. For the cross-bridge classifications the region containing the actin filament was specifically excluded because the repeat alignment had minimized its contribution to the inter-repeat variability. We constructed 12 different focused MDA masks to target myosin heads bound to specific regions of the thin filament (Fig. 5). The first set of classifications used the two primary classification masks to target myosin heads separately on the left and right-side of the repeat (Fig. 5A). This left-right classification scheme was modelled on that used to analyze cross-bridges in rigor IFM fibers swollen in low ionic strength buffer (Liu et al., 2006). These two primary masks were created from the variance map of the global average (Fig. 3C, D) so that it concentrated the classification on the most variable regions of the reconstruction, at and between the thick and thin filament surfaces.
Figure 5.
MDA masks. Masks are shown as translucent colored surfaces superimposed on the global average as a solid gray surface to provide spatial reference point. (A) Surface display of left (light purple) and right (magenta) primary masks for primary cross-bridge classifications. The actin target zone is positioned in the middle of the mask. View perspectives from left to right: top view (looking toward Z-line), front view, and tilted view. (B, C) Surface display of masks specific for troponin bridges. (B) Mask for back-left (yellow) and front-right (cyan) masks. (C) Mask for front-left (green) and back-right (orange) masks. (D) Surface display of masks specific for the thick filament surface. (E) Special mask to identify out-of-target-zone cross bridges in four separate regions of the repeat.
The class averages obtained from the two primary masks were not designed to identify Tn bridges and no Tn bridges were found in primary mask class averages. To identify those repeats that contain apparent myosin contacts with the Tn complex, we constructed four “Tn-bridge” masks. Two of these cover the side from which rear bridges of rigor approach (Fig. 5B); the other two are complementary (Fig. 5C). Later to verify the origin of the myosin head on the thick filament we constructed another pair of masks (Fig. 5D). Finally, a set of four specific masks were used to identify cross-bridges bound outside of the target zone (Fig. 5E). Averages from primary and Tn mask classifications would later be reassembled to make composite class averages. Although these masks can partially overlap at the borders, each is unique for the enclosed structure being analyzed.
We computed 20 left-side and 20 right-side class averages using the two primary masks. Each primary mask class average had between 16 and 38 members. The improvement in stochastic SNR can be computed from the number of repeats averaged and is 4–6 fold. However, stochastic noise is not particularly high in these tomograms, compared with those from ice embedded specimens. Features in the individual images of the tilt series are well defined and have high contrast by virtue of the heavy metal staining. The tomogram combines the electron counts for both tilt series, each of which consists of over 100 images with 4.2 e−/Å2 exposure. The main noise contribution arises from structure, either conformational variability or differences in preservation and staining. For either cause, it is desirable to reduce the noise variation to facilitate interpretation and quasiatomic model building.
Repeat Reassembly
Use of so many different classification masks generates several problems for display of the final results. Although classification was focused on specific regions, class averages were always computed over the entire repeat subvolume in order to retain the surrounding context. However, the relatively homogeneous class averages produced by MDA have relevance only within the volume defined by the subregion mask; averaged features outside of the masked area are lacking in detail because they remain heterogeneous. It takes some viewer effort to keep separate the difference in relevance between classified and unclassified regions when viewing the class averages. To circumvent such uneven feature definition in the focused class averages, we developed a “reassembly” procedure, wherein an entire raw repeat is reconstructed from just the classified parts of the class averages to which it contributed. The result is a high-SNR averaged version of the underlying pattern contained in the original raw repeats (by averaging with other repeats that had the same pattern) and removing noise, whether stochastic or structural. A much larger range of variation could be recovered in this manner without sacrificing SNR improvement. However, SNR improvement is variable across the reassembled repeat since it depends on the number of repeats averaged in each class.
The reassembly process is illustrated for a single raw repeat (Fig. 6). Density is present in this repeat in both the target zone and in the region of Tn plus other density that is not easily interpreted, perhaps representing heads not attached to actin. The first step in the reassembly involves computing the six required focused class averages (Fig. 6B, C). Notable is the poor quality and variability of the averaged (and ultimately irrelevant) bridging density recovered within the target zones of the four Tn bridge classes. The target zone bridges are heterogeneous in each Tn bridge class because the membership of each Tn bridge class is different from the membership of the primary classes. The left and right primary class averages are combined but these show no bridging density in the region of Tn (Fig. 6D). The Tn bridge classes are then combined, but only two of these showed cross-bridges in the averages (Fig. 6D). The complete repeat is then combined using the top part containing the primary mask class averages with the lower part containing the Tn bridge class averages (Fig. 6E).
Figure 6.
Repeat reassembly. Different masked regions of each raw repeat are subjected to separate MDA and classification using left and right-side primary masks and four Tn region masks. Each classified region is represented by a corresponding class average. The original repeat is represented by a reassembled repeat from all these class averages. From left to right: column A: a raw repeat; column B: Six different MDA masks, which are the same as depicted in Figure 5, are applied to this repeat for separate MDA and classification; column C: class averages computed for each of the different masked regions of the raw repeat; column D: partially reassembled repeat, the top image combines the class averages from primary left- and right-side classifications and the bottom one combines the class averages from different Tn bridge mask classifications; column E: the whole repeat is reassembled by combining the highlighted portions in column D.
Although the process is described as straightforward, the actual combination is done across seams that do not arbitrarily cut through cross-bridges. The seam is therefore not identical for all reassembled repeats and must be decided by manual intervention. Fortunately, there were comparatively few Tn bridges which are the ones most affected. When recombined, seams are sometimes seen at the class average boundaries where it cuts across thick and thin filaments. The seam is usually eliminated by applying a low pass filter to the reassembled repeat to the same limiting resolution that was applied for the entire tomogram.
In principle, the 20 primary mask class averages from each side could be reassembled in all possible combinations to give a total of 400 different reassemblies. However, recombination is not done for all possible combinations, but only for those actually observed. For example, for primary mask class averages, the reassembly of any raw repeat was formed by combining the left-side average of the class to which it belonged with the right-side average of the class to which it belonged. If a repeat also contributed to a Tn bridge class, then this class average was included in the reassembled repeat. Usually a small number of reassembled repeats can show nearly all the different primary and Tn bridge class averages (Fig. 7) but not in all possible combinations.
The reassembled class averages displayed a wide range of appearances. Somewhat surprisingly, there were more two-headed cross-bridges than expected (red circles in Figure 7). Usually only zero or one Tn bridge was found in most repeats, but occasionally two were found (repeat 366). Mask motifs (predominately in the third row) were also quite variable in appearance compared to the structures observed previously using column averages (Taylor et al., 1999). The majority of cross-bridges were single-headed attachments, and that includes all Tn bridges and most mask motifs.
Building Quasiatomic Models
The complexity of the repeat reassembly made quasiatomic model building potentially complex. However, the high similarity among the thin filament class averages mentioned above supports the suggestion that a single thin filament quasiatomic model constructed to the global average would be sufficient for the fitting of the myosin cross-bridges into the different class averages. Every reassembled repeat showed the actin helix and the Tn density within which the thin filament quasiatomic model generally was positioned neatly and without need for modification.
The model fitting for the cross-bridge densities was started by placing the two myosin head structures, one representing a post power stroke orientation and the other a plausible prepower stroke orientation but with its MD aligned to the rigor acto-S1 structure, on any actin subunit proximal to bridging density. Using two starting atomic structures, one with the lever arm up and the other with lever arm down, for fitting strongly bound myosin heads, minimized the amount of axial lever arm changes needed to fit the density.
One important question had to be answered before the myosin head could be rebuilt to fit the outline of the cross-bridge density; is the myosin head found on that actin subunit binding strongly or weakly? The most widely accepted measure of strong actin binding is whether the actin binding cleft on the MD is open or closed (Holmes and Geeves, 2000). The resolution in the rebuilt class average is insufficient to visualize the actin binding cleft, so another characteristic was necessary. The question of strong or weak binding was answered in the following way. If the starting model’s MD fit the cross-bridge density well without modification, it was kept in this strong binding configuration (Fig. 8A). The cross-bridge was then considered to be strongly bound to actin. An acceptable fit was determined by whether the contour of the density followed the outline of the MD at a particular contour threshold, not by whether the MD fell entirely within the envelope at that threshold. No single contour was a good fit for all cross-bridges so the quality of the fit was evaluated at different thresholds.
Figure 8.
Fitting of weak and strong binding myosin heads. The color convention is that strong binding cross-bridges are red and weak binding cross-bridges are magenta. The two actin strands are green and light blue. Left image is an axial view and right image is an azimuthal view looking towards the Z-disk. (A, B) Identification of a strong binding cross-bridge. (A) The MD of the initial rigor myosin head on a target zone actin fits without modification and the lever arm is approximately in the correct axial position. (B) Model after repositioning the lever arm into the density. The spur of density at the top of the cross-bridge in the azimuthal view is probably an averaged remnant of disordered free myosin heads. (C–F) Fitting of a weak binding myosin head into an M-ward member of a mask motif. (C) Strong binding starting model of rigor myosin. (D) Strong binding starting model of scallop transition state, which has a closer match to the lever arm, but the MD is far off. (E) Rigid body movement of the entire scallop transition structure to position the MD within the density envelope. (F) Readjustment of the lever arm in the azimuthal direction completes the fitting. (G) Color rendered fit of a repeat containing two weak binding myosin heads and one strong binding myosin head. The TM strand is colored yellow, Tn orange, ELC blue, RLC cyan. Target zone actins are in a darker shade of their actin strand color for easier identification.
Which of the two initial models would be used to fit the lever arm depended on the relative orientation of the bridging density. If oriented toward the rigor position, the lever arm was fit starting from the rigor initial structure. If oriented 90° or above, then the rebuilding started with the scallop transition state structure. Lever arm adjustments in the azimuthal direction were sometimes quite large (Fig. 8A,B).
If the MD had to be moved to place it in the density, the cross-bridge was considered to be weak binding (Fig. 8C,D). All weak binding cross-bridges were fit starting from the scallop transition state structure. Some of these movements were of necessity quite large, e.g. Fig. 8D,E, sometimes more than 6 nm. The structure illustrated in Figure 8C–F is part of a “mask motif” which consists of myosin heads originating from two different 14.5 nm shelves along the thick filament but terminating on a single target zone. For fitting the weak binding cross-bridges, we used a set of very restrictive rules because the shape of the density was insufficiently defined to support unconstrained model fitting. The whole S1 model was moved first as a single rigid body, with special attention paid to avoid the TM (assumed to be in the “closed” position) and to minimize the possible lever arm adjustment performed later, especially the azimuthal bending; no rocking of the MD model along the axial direction appeared to be needed. However, we can’t exclude the possibility that some rocking occurs, but the fitting could be satisfactorily performed without it. An empty shell in the density map due to negative staining of the myosin head usually indicated the position of the MD. We then adjusted its lever arm position if necessary by rotating around three pivot points (Fig. 8E,F).
Not all weak binding myosin-actin attachments were as obvious as the one illustrated in Figure 8. Sometimes only part of the myosin head extended outside of the cross-bridge envelope (Figure 9). Again, when the MD fell outside the bridging density, the starting model used to fit the density was always the scallop transition state structure (Fig. 9A,B). The MD required largely rotation within the azimuthal plane to place the portion extending outside of the envelope back inside (Fig. 9C). Finally, the lever arm is moved to obtain the final fit (Fig. 9D).
Figure 9.
Fitting of a weak cross-bridge from isometric HST. The two strands of F-actin are colored in green and light blue. (A) Starting atomic structure from rigor acto/S1 complex colored red. (B) The initial scallop transition state structure colored magenta. Comparison of the two starting models shows that the lever arm of the transition state is a closer fit, but neither MD falls within the envelope and a lever arm movement to fit would be extreme. (C) The myosin head is repositioned in a single rigid body movement, leaving a small lever arm movement to obtain a fit. Note that the best contour threshold for surface display is not generally the same one used for the fitting which sometimes leaves part of the myosin head outside of the envelope.
We built quasiatomic models into all 20 left-side and all 20 right-side class averages for a total of 40 models encompassing bridges that bind in the target zone or just M-ward of it. These could then be combined as required by the class memberships to produce a quasiatomic model for all reassembled class averages. The result (Fig. 8G) provides a complex atomic interpretation of the many cross-bridge forms observed in contracting muscle. Because we have used the TM atomic structure without modification, there are instances where steric clashes are present for strong binding bridges. Since we do not resolve TM in the class averages, even in the global average, there is little to guide a modification to eliminate the steric clash. Current models for thin filament regulation include the concept that strong binding myosin heads move TM further from the blocked to the open position to give complete activation (Geeves and Conibear, 1995). Thus, the presence of a steric clash where strong binding heads are found is expected.
The class averages produced in this work do not combine a large number of images so the SNR improvement is not large, certainly not large compared to that obtained in single particle image reconstructions. The low amount of averaging is limiting and may affect the quality of the quasiatomic models. This effect is likely to be largest for the modifications to the lever arm. To evaluate this, we computed class averages using the two thick filament surface masks (Fig. 5D) and compared them with two primary class averages, each of which contained 16 members. These thick filament surface class averages focus on the region of the repeats containing the lever arms and produced class averages revealing variable axial and azimuthal lever arm angles. Each of these thick filament surface classes have members from other primary classes and the membership of each primary mask class average is distributed over several thick filament surface class averages. The thick filament surface classes allowed for a set of alterative positions of the lever arm angle and the position of its C-terminus (Table 2). The resulting analysis, weighted according to the number of class members indicated that for the axial angle, the standard deviation could be as little as 6.4° or as high as 16.6°, but overall was 12.6°. For the azimuth, the standard deviation could be as low as 7°, as high as 10.7° but overall was 9.2°. Discarding one outlier (first member of the second group) that is more than 3 standard deviations from the rest, results in a standard deviation overall of 10° for the axial angle and 8° for the azimuth. These numbers should be compared to the overall range of angles which is greater than 90° for the axial angle and ~90° for the azimuthal angle.
Table 2.
Evaluation of fitting uncertainty for lever arm angles.
| Example 1 | ||||
|---|---|---|---|---|
| Class ID | # of members | Axial angle (°) | Axial coord (nm) | Azimuthal angle (°) |
| 0 | 2 | 87.1 | 0.90 | 79.9 |
| 7 | 1 | 93.8 | 2.09 | 112.2 |
| 8 | 2 | 89.2 | 1.30 | 89.4 |
| 10 | 1 | 83.4 | 0.44 | 88.6 |
| 13 | 1 | 109.3 | 4.98 | 93.5 |
| 17 | 9 | 98.8 | 3.09 | 86.3 |
| mean | 95.5 | 2.48 | 88.1 | |
| σ | 6.4 | 1.16 | 7.0 | |
| Example 2 | ||||
|---|---|---|---|---|
| Class ID | # of members | Axial angle (°) | Axial coord (nm) | Azimuthal (°) |
| 0 | 1 | 87.1 | 0.90 | 79.9 |
| 2 | 1 | 141.2 | 9.44 | 113.4 |
| 3 | 5 | 142.5 | 9.61 | 113.2 |
| 4 | 2 | 137.9 | 9.05 | 112.0 |
| 5 | 3 | 120.4 | 6.88 | 92.3 |
| 11 | 2 | 141.8 | 9.47 | 114.6 |
| 14 | 2 | 107.7 | 4.63 | 100.9 |
| mean | 129.8 | 7.83 | 105.7 | |
| σ | 16.6 | 2.47 | 10.7 | |
| overall σ | 12.60 | 1.93 | 9.02 | |
The more important quantity for motor action is the axial displacement of the lever arm C-terminus since this is directly related to the amount of filament sliding. For Example 1, the standard deviation is 1.2 nm and for Example 2, it is 2.5 nm with an overall value of 1.9 nm. Again, the single outlier (one of 16 members) artificially distorts the result and eliminating it gives a standard deviation of 1.8 nm for Example 2 and an overall value of 1.5 nm. The latter value should be evaluated in light of the resolution of the reconstructions, which is generally about 5 nm.
Quantification of Cross bridge Forms
Application of MDA and classification to the grouping of self-similar cross-bridge forms also provides a mechanism for enumerating the frequency of these forms. If a class average displays a cross-bridge on a particular actin subunit, the frequency with which that cross-bridge occurs in the ensemble can be assumed equal to the number of class members. This process can be continued to enumerate the frequency with which each of the 14 actin subunits in the repeats binds a cross-bridge. Thus, statistics can be gathered at the same time that SNR improvements are obtained for the individual structures. The accuracy of this process, however, is not immediately obvious. MDA and classification themselves are not exact, especially for such heterogeneous data.
We refer to cross-bridges as being either Z-ward or M-ward if they were members of mask motifs, i.e. myosin heads originating from successive 14.5 nm shelves and binding to a single target zone. Tn bridges were also counted. If a single cross-bridge was present, it was considered to be a Z-ward cross-bridge. All primary mask class averages had Z-ward cross-bridges and none contained Tn bridges. M-ward bridges were variable and present in only 11 out of 20 classes.
To obtain some estimate of this accuracy, we examined galleries of the class membership associated with 20 primary mask class averages and manually counted similarly placed densities (Fig. 10). At the top (Fig. 10A–D) outlined in green is an M-ward cross-bridge in azimuthal view. The outline is drawn from the class average (Fig. 10D1) and is placed identically over each of the other 15 panels that show the class membership. While there is density within each green outline there is also variability, possibly due to differences in combination of staining or tannic acid build up. For the M-ward cross-bridge in the axial view, density is present within the green outline in 14 of the 15 class members and appears to be missing in G3. However, in the corresponding azimuthal view (C3), the myosin head is simply placed more toward the front of the outline. For the Z-ward cross-bridge outlined in cyan in rows E–H, one of the heads appears to be missing, but like the missing M-ward cross-bridge, the position of the Z-ward cross-bridge was not centered within this plane. This is one of the reasons why the bridge analysis was done using the 3-D images and not projections, the other being that in projection, one cannot be sure of an actin contact.
Figure 10.
Comparison of a class averages computed using the right-side primary classification mask with the class members. Top rows (A–D) show a section through a non-target zone cross-bridge in a weak binding configuration similar to that shown in Figure 8G. Features on the left-side of the thin filament in the averages (H1 and D1) are not significant. The class average is shown in the lower left hand corner (D1) and the rest of the panels are the class members. The red oval outlines the actin filament position while green outlines the averaged cross bridge and is placed identically in all the panels, each of which has bridging density within the outline. Bottom rows (E–H) shows an axial view of the same class average and 15 raw repeats with the average in the lower left hand corner (H1). Outlined in green is the position of the cross-bridge shown in rows A–D, while outlined in cyan is a Z-ward target zone cross-bridge in rows E–H.
This process was carried out for a complete set of left-side primary mask class averages (Table 3). The RMS deviation of the difference between the predicted number of Z-ward cross-bridges and the actual number of Z-ward cross-bridges in the individual classes was 15.7%. The manual counting over all the Z-ward bridges produced a total for Z-ward bridges that was 91.7% of the total predicted from the class membership. The predicted number of bridges must exceed the number obtained by manual counting because all class averages contained Z-ward bridges and thus individual repeats can contain only true positives or false positives (the averaged bridge is missing in the class member).
Table 3.
Crossbridge Prediction Precision
| Class # | # Class Members | Z-ward bridges by MDA | Z-ward bridge counts | Z-ward Difference | M-ward Class Average | M-ward bridges by MDA | M-ward bridge counts | M-ward Difference | Tn bridge counts |
|---|---|---|---|---|---|---|---|---|---|
| 00 | 37 | 37 | 37 | 0 | 0 | 0 | 5 | −5 | 6 |
| 01 | 27 | 27 | 26 | 1 | 1 | 27 | 21 | 6 | 9 |
| 02 | 33 | 33 | 33 | 0 | 1 | 33 | 27 | 6 | 4 |
| 03 | 23 | 23 | 19 | 4 | 1 | 23 | 17 | 6 | 10 |
| 04 | 22 | 22 | 20 | 2 | 0 | 0 | 2 | −2 | 12 |
| 05 | 22 | 22 | 21 | 1 | 1 | 22 | 19 | 3 | 4 |
| 06 | 34 | 34 | 30 | 4 | 1 | 34 | 27 | 7 | 13 |
| 07 | 28 | 28 | 28 | 0 | 0 | 0 | 5 | −5 | 8 |
| 08 | 32 | 32 | 29 | 3 | 1 | 32 | 30 | 2 | 10 |
| 09 | 33 | 33 | 33 | 0 | 1 | 33 | 24 | 9 | 5 |
| 10 | 16 | 16 | 16 | 0 | 0 | 0 | 1 | −1 | 3 |
| 11 | 38 | 38 | 38 | 0 | 0 | 0 | 19 | −19 | 4 |
| 12 | 24 | 24 | 22 | 2 | 0 | 0 | 5 | −5 | 7 |
| 13 | 24 | 24 | 24 | 0 | 1 | 24 | 23 | 1 | 7 |
| 14 | 18 | 18 | 18 | 0 | 0 | 0 | 2 | −2 | 7 |
| 15 | 18 | 18 | 18 | 0 | 0 | 0 | 0 | 0 | 12 |
| 16 | 30 | 30 | 16 | 14 | 1 | 30 | 30 | 0 | 8 |
| 17 | 16 | 16 | 8 | 8 | 1 | 16 | 15 | 1 | 4 |
| 18 | 22 | 22 | 22 | 0 | 0 | 0 | 11 | −11 | 6 |
| 19 | 18 | 18 | 14 | 4 | 1 | 18 | 14 | 4 | 2 |
| Totals | 515 | 515 | 472 | 43 | 11 | 292 | 297 | −5 | 141 |
| %(Counts/Predicted) | 91.7 | 101.7 | |||||||
| % RMS deviation | 15.7 |
43.5 22.0§ |
|||||||
| % of all bridges | 52 | 33 | 15 | ||||||
RMS deviation for only those classes that show M-ward bridges
For M-ward bridges, the numbers are less impressive. M-ward bridges occur much less frequently than Z-ward bridges (about 60% as frequent) and are more heterogeneous. Although they are identified using the primary mask, in many class averages, their density often falls below a useful contour threshold. Only 11 out of 20 class averages showed M-ward bridges but the class members in 19 of 20 classes contained M-ward cross-bridges by manual counting. Thus, for less frequent cross-bridge forms both false positives and false negatives are present, and actual counts can be either less than or greater than predicted counts. There is some compensation between false positives and false negatives because the total number of M-ward cross-bridges actually counted is only 1.7% more than the number predicted based on class membership. The RMS deviation between manual counts and predicted counts for all 20 class averages is 43.5%. For just the 11 classes whose average contain M-ward bridges, the RMS deviation between manual and predicted counts was 22.0%.
The primary MDA mask was not designed to identify Tn bridges, explaining the lack of Tn bridges in the class averages. Tn bridges turned out to be relatively significant according to manual counts, forming 15% of the total number of cross-bridges counted. By designing a set of masks specifically for identifying Tn bridges among the ensemble of aligned repeats (Fig. 5B,C), we could form class averages of this comparatively small but not insignificant population of cross-bridges.
DISCUSSION
ET has the potential to make a strong contribution to the visualization of dynamic structural changes, displayed in an ensemble of molecules distributed over multiple enzymatic states. This is particularly true of the actions of motor molecules, of which force generation in muscle is one such example. Another includes the action of dynein motors in flagella and cilia (Nicastro et al., 2005). The advantage of ET for such studies is the ability to produce 3-D images of structures and ensembles of structures that are unique. A myac layer of Lethocerus IFM, is effectively a unique ensemble because no two sections would reveal the same pattern of actin attached and unattached myosin heads. However, they might contain the same distribution of myosin head structures, just differently arranged within the filament lattice.
The main difficulty with these unique 3-D images is their low SNR. The noise can be stochastic arising from the limited electron counts, or can be structural in that there are many objects in the image that are of little interest to the structure under investigation. If the specimen is unstained and frozen hydrated, much of the noise will be stochastic due to the limitations of radiation damage and the low contrast. With plastic embedded and stained specimens, such as the IFM specimens described here, stochastic noise is much lower because higher electron exposures can be tolerated, but variations in preservation, staining and structural state produce increased “structural” noise. Regardless of the source, interpretation of the reconstruction in terms of molecular conformation requires that averages be produced that are high-SNR representations of an ensemble of ideally identical states. If these states are distributed randomly throughout the specimen, the challenge is to know how many states are present and to identify them so that high quality averages that reinforce details are produced. It is to the problem of identifying self-similar forms in a heterogeneous distribution that the present work is directed.
New instruments and automated tilt series data collection (Koster et al., 1992) have made routine even the collection of tilt series from frozen hydrated biological material. Although our specimens are more radiation resistant than ice embedded specimens, the reduced electron exposure to the specimen achieved with automated tracking and image recording is beneficial. Once sarcomeres are visualized and their power spectra checked to confirm good preservation, the most well-centered myac layer is selected and made eucentric. All other manipulations are made just outside the area of interest, and subsequent specimen exposure is limited to that needed to record the image of interest, i.e. about 1 sec/exposure. The computerized goniometer provides accurate tilt angles, particularly the increments between angles.
Although the sections used in the present study were cut from the same specimen blocks used for Taylor et al. (1999), the improvement in detail available from these tomograms is striking. All data analysis procedures described here were also tried on the 1999 data, but without improvement, particularly in the ability to resolve the actin subunits on the thin filament. This indicates that the new tomograms are of significantly higher quality, most likely due to the automated data collection and reduced electron exposure. Other contributing factors are the recording of images on CCD camera, the larger number of tilt angles present in the tilt series (which leads to much finer sampling of views and fewer back projection artifacts), the combination of dual-axis tilt series to compute a single tomogram with less missing data, more accurate and constant defocus, and better software for tilt series image alignment (Winkler and Taylor, 2006).
The general improvement provided by this procedure over column averaging (Taylor et al., 1999) can be seen in the actin detail (Figure 3). Column averaging was restricted to the axial 116 nm period common to both actin and myosin, of which there are about 8 repeats per 1 μm long thin filament (Taylor et al., 1999). Layer lines from orders of the axial 116 nm period were not visible beyond 12.9 nm which severely restricted the resolution. Averaging over the 116 nm period blurs the actin subunit arrangement because an odd number of crossover periods are included in each long period. The alternative of column averaging over the axial 232 nm period, would overcome the blurring of actin subunits, but only provides 4-fold averaging and a 2-fold improvement in stochastic SNR. By aligning all 515 repeat subvolumes to a single actin filament, we obtain a SNR improvement of (515)1/2 or 22.7 fold for the actin filament in the global average. This improvement enables fitting of an actin filament quasiatomic model to the thin filament density, independent of other features in the sub-volume repeat. The exact disposition of actin subunits in the quasiatomic models is crucial for the identification of different kinds of myosin attachment, either strong or weak.
Identifying and Quantifying Structural States
A paracrystalline specimen, such as striated muscle provides a unique opportunity to develop the means for identifying self-similar structures among a heterogenous ensemble, because some parts of the structure can be considered invariant at the present resolution (although possibly not at higher resolution). Invariant common structures can be used to minimize misalignment as a source of variation. In the muscle lattice, both the actin filament and the thick filament backbone can be considered large invariant structures, but they are not equally “shapely” to the extent that the thick filament backbone lacks easy to resolve features from which to determine the axial displacements. Alignment on the actin filament eliminates repeat misalignment as one variable in the structure and at the same time provides a large structure with multiple features (actin subunits and Tn) to help define the alignment. The thin filament segment used for alignment of each repeat contains actin, TM and Tn, with a total mass of at least 1.5 MDa (20 × 42 kDa + 20 × 10 kDa + 4 × 115 kDa + unquantifiable small fractions of target zone HC) making it much larger than any of the features that would be identified through repeat classification. This is important because the variability that we are investigating is in comparatively small structures, such as the myosin lever arm (MW ~42 kDa) or even the entire myosin head (MW ~130 kDa) which by themselves do not provide enough shape for accurate alignment, but can be seen in the raw images and identified using the procedures described here.
The goal of recovering the most structure variability in the reconstruction is complicated by the issue of structure noise. Structure noise is usually attributed to differences in preservation, but in the present case can mean much more. In the context of MDA, one of any structure constitutes noise as the procedure is designed to identify patterns in the data. Counts of number of myosin heads interacting with actin in isometric HST based on the methods reported here is 53%. This means that 47% of the heads are not interacting with actin and these are presumably distributed throughout the reconstruction in a manner that is largely random and lacking in any pattern with the exception of those heads aligned along the 14.5 nm crowns. Myosin heads unattached to actin constitute a major source of structural noise in the reconstruction.
An important question for extracting different conformations is the number of conformations. The question cannot be answered precisely because the structures are not known, so the practical solution is to plan for as many as possible. However, improving SNR while retaining structural variations that may be significant are conflicting goals; high SNR requires averaging as many repeats as possible, whereas recovering structural variability requires computing the largest number of classes. The solution is to align on a common structure provided by the thin filament and then classify on separate regions that surround it. The approach is not novel, having been used successfully in single particle averaging of myosin and dynein in projection (Burgess et al., 2004). The separate left-side and right-side classification scheme was used by us previously to identify different structures in rigor IFM swollen in low ionic strength buffer (Liu et al., 2006). The same procedure applied here for active muscle fibers recovered considerably more variability in the region of the target zone than was encountered in swollen rigor fibers. However, when comparing class averages with the original raw repeats (Table 3), we could recognize many instances of bridging densities connecting the thick filament to Tn that were not being recovered in the class averages. Thus, the procedure was missing what could be an important feature of contracting IFM.
To obtain a more accurate picture of the myosin heads that were interacting with the thin filament, we expanded the classification in three ways (Figure 5). First we designed four additional masks, and thus classification steps, that focused on the Tn region of the thin filament. Second, to verify the origin of the myosin heads on the thick filament backbone observed using the primary mask class averages, an additional classification was done to enhance this feature on the thick filament surface. Third, we computed a set of four classifications to enumerate myosin heads attached to actin outside the target zones but not bound to Tn. The reassembly of the different class averages into high SNR versions of each repeat recovered a wide range of myosin attachments to the thin filament.
Weak versus Strong Actin Binding
An important issue in muscle contraction is the structure of weak binding cross-bridges that are thought to constitute myosin’s initial interaction with actin. The resolution of the present work is more than double the previous attempt so that an actin filament atomic model can be fit to the global average. This is a major improvement over the earlier work (Taylor et al., 1999), which required that column averages have a cross-bridge form that resembled a strong binding conformation before an actin filament could be fit to the map. Now the actin filament can be fit independent of the myosin heads, providing an objective basis by which weak and strong actin-myosin attachments can be distinguished by the quality of the initial fit of a strongly bound myosin MD to the density.
This criterion could categorize some potentially strong attachments to actin as weak if the actin filament changed structure in some way. One possibility is that the regular helical structure of the actin filament changed, an idea that was suggested by us based on our first reconstruction of rigor IFM (Taylor et al., 1984) and is consistent with the observation of random variable twist in actin filaments (Egelman et al., 1982; Egelman and DeRosier, 1992). However, rigor leads to far more strong binding attachments to actin, some of which deform the myosin heads considerably, at least when compared to crystal structures and acto-S1 (Chen et al., 2002), supporting the idea that they can deform the actin filament as well. Far fewer strong binding attachments occur in contracting muscle and these are concentrated on a smaller segment of the actin filament. If they deform the actin filament, it is possible that the deformations occur at the level of actin subunits, as opposed to helix perturbations, which would make them more difficult to detect at our resolution.
Very little is known of the structure of weak actin-myosin attachments. These are thought to include all kinetic states of the myosin ATPase which have ATP or ADP·Pi in the active site. A weak binding state has been visualized in 3-D only once (Volkmann et al., 2005) with a non-muscle myosin, for which the actin affinity of myosin with ATP in the active site was unusually strong (Tauhata et al., 2001). The procedures developed here detect weak actin-myosin attachments virtually all along the thin filament, but the most significant for muscle contraction may be those that occur in the target zone as these can potentially convert to strong binding states, whereas those outside the target zone do not and may be merely collision complexes.
Stain Distribution
The raw repeats, and especially the class averages, show a strong buildup of stain on the surfaces of the actin filaments, the myosin heads and the thick filament backbone. This gives the reconstruction a strong appearance of negative staining. This stain layer is most likely due to the buildup of tannic acid and uranyl acetate. The TAURAC sequence for freeze substitution does not utilize any osmium or glutaraldehyde, substances known to be damaging to actin filaments in fixed and embedded specimens (Begg et al., 1978; Maupin-Szamier and Pollard, 1978; Lehrer, 1981). This stain coating, which we presume covers all surfaces relatively uniformly, acts as both the preservative and contrasting agent and has an odd consequence for the tomogram and the class averages.
The hollow appearance of the thin filament in the region of the target zone when viewed as an isodensity surface (Fig. 3A,B; Fig. 6) is suggestive of similar appearances obtained using spatial averaging methods which are approaches that treat all repeats as if they were identical (Taylor et al., 1984; Taylor et al., 1989b; Taylor et al., 1989a). At that time this was thought to be due to incomplete data, such as the missing pyramid that characterizes this work (Taylor et al., 1986) or even physical separation of the two long pitch strands of actin subunits, as similar images were also observed in negative stain projections of actin filaments (Bremer et al., 1991). The axial resolution of the spatial averages was ~12.9 nm, insufficient to resolve actin subunits. The present work resolves the actin subunits and the images do not show a gap between the two actin strands when viewed as continuous density. Moreover, the atomic models we built do not require any changes in the radius of actin subunits to obtain a fit so any “strand separation” must be below our resolution. Rather, the evidence seems as originally thought to support anisotropic resolution (the resolution is lowest in the direction perpendicular to the section plane) as the main contributor. Negative staining will be least visible (most blurred) for surfaces oriented parallel to the myac layer section plane and thus most difficult to show in an isosurface display.
The main drawback to the negative staining is its non-uniformity. In the averages, large structures appear negatively stained, whereas small features, such as the myosin lever arm, usually appear as if they were positively stained. All structures are probably both positive and negative stained. The stain density on the front and back surfaces of the thin filament appears weak which makes it difficult to display the reconstruction within a completely enclosed isodensity surface. Consequently the front surface of the thin filament is often absent. Not all TAURAC preserved IFM shows this effect. Repeat averages of freeze substituted stretched rigor fibers (Liu et al., 2004) and chemically fixed swollen rigor fibers (Liu et al., 2006) did not, either because they had more tannic acid buildup, or more likely because the data are more homogeneous in rigor compared to HST.
The negative staining effect has its major consequence on quantitative model building. Most quantitative model fitting software currently requires that the density be interpretable everywhere as protein (Chen et al., 2002; Tama et al., 2004). This is possible for true negative or positive staining, but in the present case, the negative stain shell results in a pair of contour surfaces defining large objects like the MD and actin subunits and only a single contour for other objects like the lever arm. At the moment, there is no adequate treatment of this particular condition, for which the solution is either a manual fit (as used here) or separate fittings for large and small objects.
Quasiatomic Model Precision
The goal of this work is an interpretation of the tomogram in terms of the structures of the molecules contributing to muscle contraction, which is best realized by fitting the atomic structures into their corresponding density. It is generally thought that for a 2 nm resolution reconstruction, that an atomic structure can be docked with a precision of at least 5 times the resolution, or within 0.4 nm (Baker and Johnson, 1996; Rossmann, 2000; Saibil, 2000). The precision is quite likely to depend on both the resolution (since at lower resolution there is less shape definition), and the preservation method, the above estimates being largely determined for frozen-hydrated specimens. We have done several tests to evaluate our fitting precision for different features in our reconstructions. Not surprisingly, we found the precision is quite variable depending on the size of the structure being docked and the resolution of the class average.
The largest structure that was fit was the thin filament, which had a mass of 1.5 MDa. By fitting a density map derived from the thin filament atomic model we evaluated the precision of the docking. The critical parameter is the accuracy of the azimuthal orientation which affects the decision for whether to assign the cross-bridge as a weak or a strong myosin-actin attachment. The precision for fitting the quasiatomic model of the thin filament, originally fit into the global average, into eight thin filament class averages with ~64 members each was ±6°. At a radius of 4 nm, which defines the outer diameter of the actin filament, a ±6° fitting error translates to ±0.41 nm, which is better than 10x the resolution of the global average. The more important uncertainty is at the outer limit of the MD when docked onto the thin filament (radius 7.5 nm), and this value, ±0.78 nm or 6.4 times the resolution (~5 nm), is more in line with expectations.
For smaller structures, such as the lever arm, we made a separate estimate utilizing an alternative determination of the position of the C-terminus from the thick filament surface classifications. The lever arm is the smallest structure that we actually fit into the density, and its changes in orientation express a key parameter for understanding muscle contraction. While the thin filament quasiatomic model is fit to the global average of 515 repeats, the class averages into which the lever arms fit contained fewer class members (between 16 and 38), and hence have lower SNR. The resolution was nevertheless similar because actin subunits were resolved in all the class averages. Our estimate of the precision of lever arm fitting was 1.5 nm, or about 3x the resolution. To put this into perspective, the range of axial motion of the lever arm observed for isometric HST is 13 nm (Taylor et al., 1999), about 8x the fitting precision.
The results obtained in the present work, are within expectations based on previous estimates that have been derived from frozen hydrated specimens. The fitting precision varies depending on the size of the object being fit, 1.5 MDa for the thin filament to 42 kDa for the lever arm. The precision also depends on the SNR, being highest for the thin filament class averages, which have the largest number of class members and lowest for the lever arm, which was fit into class averages which have fewer class members.
Accuracy of Quantifying Cross bridge Forms
To understand the relationship between cross-bridge form and tension development, requires an enumeration of the different structures. Such data would also be essential for extending the EM results to a model explaining the X-ray diffraction of muscle fibers (Bekyarova et al., 2008), and even from live insects (Dickinson et al., 2005). The relative importance of the different cross-bridge forms will depend on their frequency; structures that are rare, will not generally be observed by X-ray diffraction, where the observed intensity will depend on the square of the relative frequency of a particular structure.
Counting cross-bridges manually, as was done here and previously (Tregear et al., 2004) is exceedingly tedious and is prone to some inaccuracies in that many variations in structure cannot usually be distinguished in the raw tomograms. An alternative is to use the results from MDA and classification to perform the counting, a process that is objective. However, it is not known how accurately this can be done. MDA and classification are not exact unless the structures being compared are well differentiated. In the present case, we are arguably “binning” a continuum of structures. We therefore compared cross-bridge counts obtained manually from class averages, with predictions from the class membership. The precision of the results obtained by MDA and classification using the primary classification mask (Fig. 4A) depends on the frequency and homogeneity of the cross-bridges.
The numbers of cross-bridges predicted from the class membership can be higher or lower than actual counts obtained manually. Overcounting, when the prediction from the class membership exceeds the actual manual count, can occur when a class member lacks a cross-bridge present in the class average (false positive). Undercounting, when a class member has a cross-bridge that is not present in the class average (false negative), can occur when this cross-bridge is present infrequently. That is to say it is unique, but this uniqueness may not be detectible by simple visual inspection. Unique structures constitute noise in MDA.
The cross-bridges enumerated from the class membership will always be more frequent than if they were counted manually due to a small rate of false positives. The enumeration can be quite accurate for cross-bridge types that occur frequently, such as the Z-ward cross-bridges within the target zone (Table 3). Total Z-ward bridge count over all classes was 91.7% of that predicted by the class membership and the RMS deviation over all 20 classes 15.7%. The error is more than compensated by the fact that the cross-bridges identified by MDA and classification can be assigned to individual actin protomers and a given structure, something that is almost impossible to do manually from raw repeats.
On the other hand, cross-bridges that occur infrequently within the class membership will not form a class and thus will not be counted at all. M-ward cross-bridges are much less frequent as well as more variable than Z-ward cross-bridges so that eleven class averages have M-ward cross-bridges. Manual counts for these M-ward bridges were 84.6% of the counts from the class members and the RMS deviation was 22%. However, manual counts showed that 19 classes contained at least one M-ward cross-bridge. If the 9 classes that lacked any M-ward bridges is included, the RMS deviation rises to 43.5%, but the total number of M-ward bridges over all classes counted by MDA is 101.7% of the totals predicted from the class membership. In this case, the greater accuracy for the totals compared with Z-ward bridges is due to compensation by false positives and false negatives. The more important quantities are the total counts and the RMS deviation for the 11 classes containing M-ward cross-bridges. These are the cross-bridges that will be used to infer mechanism. Although manual counting was effective at identifying the presence of the M-ward cross-bridge, it was ineffective at evaluating their homogeneity. MDA will treat heterogeneous structures as noise.
In isometric HST, the M-ward cross-bridges were predominately on actin subunits that are out of the target zone on the M-ward side. Target zone cross-bridges are the most frequent cross-bridges and are expected to dominate any MDA classification to the detriment of less frequent forms outside of the target zone. The best way to improve the counting accuracy for the bridge types that were infrequent was a more focused mask and this was the reasoning behind the masks that identified Tn bridges (Fig. 4B,C) and out-of-target-zone bridges (Fig. 4E). The four out-of-target-zone masks each cover two actin subunits on a single long pitch strand, which in combination with the two actin subunits of the target zone covers six of seven actin subunits; the Tn bridge masks cover the seventh. In this manner we have been able to obtain myosin head counts on each actin subunit.
The frequency of cross-bridges in the vicinity of Tn was a surprise. By manual counts, these occur at the rate of ~15% of all cross-bridges. Tn bridges occur in the same vicinity as the rear bridges of rigor IFM. That these are not rigor-like rear cross-bridges is supported by the fact that, though numerous, they are still far less frequent than in rigor. In addition, the solvent contains ATP which would preclude these being rigor-like cross-bridges, at least at these frequencies. Finally, on addition of AMPPNP, a non-hydrolyzable analog of ATP to rigor IFM, rear bridges detach and are not seen at all in averaged images (Winkler et al., 1996) and with very much less frequency in electron tomograms (Schmitz et al., 1996).
At least some of the Tn bridges could be precursors to rigor rear bridges which are bound to actin near the location of Tn. Quasiatomic model building of rigor rear bridges requires large rearrangements of the light chain lever arm to place them within the density envelope (Chen et al., 2002). The presence of Tn bridges in contracting muscle indicate, as would be expected, that myosin heads are reaching the actin targets of rigor rear bridges, even if their weak actin affinity prevents them from attaching to the myosin binding site on actin.
Acknowledgments
This work was supported by NIH grants GM30598 (KAT) and AR14317 (MKR). For catching and shipping the live Lethocerus waterbugs from which glycerinated IFM was prepared, we thank Torben Poulsen (Heidelberg, Germany & Chiang Mai, Thailand), Gene Scott (Clewiston, FL, USA), and Claudius and Aneta Samuels (Black River, Jamaica). The I-HST tomogram, aligned subvolumes, reassembled repeats and other metadata have been deposited in the European Bioinformatics Institute under accession code EMD-1561. The quasiatomic models have been deposited in the Protein Data Bank under accession codes 2w4a, 2w49, 2w4t.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- AL-Khayat HA, Hudson L, Reedy MK, Irving TC, Squire JM. Myosin head configuration in relaxed insect flight muscle: X-ray modelled resting crossbridges in a pre-powerstroke state are poised for actin binding. Biophys J. 2003;85:1063–1079. doi: 10.1016/S0006-3495(03)74545-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Johnson JE. Low resolution meets high: towards a resolution continuum from cells to atoms. Curr Opin Struct Biol. 1996;6:585–594. doi: 10.1016/s0959-440x(96)80023-6. [DOI] [PubMed] [Google Scholar]
- Begg DA, Rodewald R, Rebhun LI. The visualization of actin filament polarity in thin sections. Evidence for the uniform polarity of membrane-associated filaments. J Cell Biol. 1978;79:846–852. doi: 10.1083/jcb.79.3.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekyarova TI, Reedy MC, Baumann BA, Tregear RT, Ward A, Krzic U, Prince KM, Perz-Edwards RJ, Reconditi M, Gore D, et al. Reverse actin sliding triggers strong myosin binding that moves tropomyosin. Proc Natl Acad Sci U S A. 2008;105:10372–10377. doi: 10.1073/pnas.0709877105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer A, Millonig RC, Sutterlin R, Engel A, Pollard TD, Aebi U. The structural basis for the intrinsic disorder of the actin filament: the “lateral slipping” model. J Cell Biol. 1991;115:689–703. doi: 10.1083/jcb.115.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SA, Walker ML, Thirumurugan K, Trinick J, Knight PJ. Use of negative stain and single-particle image processing to explore dynamic properties of flexible macromolecules. J Struct Biol. 2004;147:247–258. doi: 10.1016/j.jsb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Chen LF, Winkler H, Reedy MK, Reedy MC, Taylor KA. Molecular modeling of averaged rigor crossbridges from tomograms of insect flight muscle. J Struct Biol. 2002;138:92–104. doi: 10.1016/s1047-8477(02)00013-8. [DOI] [PubMed] [Google Scholar]
- Dickinson M, Farman G, Frye M, Bekyarova T, Gore D, Maughan D, Irving T. Molecular dynamics of cyclically contracting insect flight muscle in vivo. Nature. 2005;433:330–334. doi: 10.1038/nature03230. [DOI] [PubMed] [Google Scholar]
- Egelman EH, DeRosier DJ. Image analysis shows that variations in actin crossover spacings are random, not compensatory. Biophys J. 1992;63:1299–1305. doi: 10.1016/S0006-3495(92)81716-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH, Francis N, DeRosier DJ. F-actin is a helix with a random variable twist. Nature. 1982;298:131–135. doi: 10.1038/298131a0. [DOI] [PubMed] [Google Scholar]
- Geeves MA, Conibear PB. The role of three-state docking of myosin S1 with actin in force generation. Biophys J. 1995;68:194S–199S. discussion 199S–201S. [PMC free article] [PubMed] [Google Scholar]
- Harauz G, Van Heel M. Exact filters for general geometry three-dimensional reconstruction. Optik. 1986;73:146–156. [Google Scholar]
- Hirose K, Franzini-Armstrong C, Goldman YE, Murray JM. Structural changes in muscle crossbridges accompanying force generation. J Cell Biol. 1994;127:763–778. doi: 10.1083/jcb.127.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose K, Lenart TD, Murray JM, Franzini-Armstrong C, Goldman YE. Flash and smash: Rapid freezing of muscle fibers activated by photolysis of caged ATP. Biophys J. 1993;65:397–408. doi: 10.1016/S0006-3495(93)81061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KC, Angert I, Kull FJ, Jahn W, Schroder RR. Electron cryomicroscopy shows how strong binding of myosin to actin releases nucleotide. Nature. 2003;425:423–427. doi: 10.1038/nature02005. [DOI] [PubMed] [Google Scholar]
- Holmes KC, Geeves MA. The structural basis of muscle contraction. Philos Trans R Soc Lond B Biol Sci. 2000;355:419–431. doi: 10.1098/rstb.2000.0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KC, Tregear RT, Barrington Leigh J. Interpretation of the low angle X-ray diffraction from insect muscle in rigor. Proc Roy Soc (London) - Series B: Biological. 1980;207:13–33. [Google Scholar]
- Houdusse A, Szent-Gyorgyi AG, Cohen C. Three conformational states of scallop myosin S1. Proc Natl Acad Sci U S A. 2000;97:11238–11243. doi: 10.1073/pnas.200376897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallog sect A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Koster AJ, Chen H, Sedat JW, Agard DA. Automated microscopy for electron tomography. Ultramicroscopy. 1992;46:207–227. doi: 10.1016/0304-3991(92)90016-d. [DOI] [PubMed] [Google Scholar]
- Lehrer SS. Damage to actin filaments by glutaraldehyde: protection by tropomyosin. J Cell Biol. 1981;90:459–466. doi: 10.1083/jcb.90.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenart TD, Murray JM, Franzini-Armstrong C, Goldman YE. Structure and periodicities of crossbridges in relaxation and during contraction initiated by photolysis of caged calcium. Biophys J. 1996;71:2289–2306. doi: 10.1016/S0006-3495(96)79464-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard K, Bullard B. The thin filament in insect flight muscle. In: Vigoreaux JO, editor. Nature’s Versatile Engine: Insect Flight Muscle Inside and Out. Georgetown, TX: Landes BioScience; 2006. pp. 141–149. [Google Scholar]
- Liu J, Reedy MC, Goldman YE, Franzini-Armstrong C, Sasaki H, Tregear RT, Lucaveche C, Winkler H, Baumann BAJ, Squire JM, et al. Electron tomography of fast frozen, stretched rigor fibers reveals elastic distortions in the myosin crossbridges. J Struct Biol. 2004;147:268–282. doi: 10.1016/j.jsb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Liu J, Wu S, Reedy MC, Winkler H, Lucaveche C, Cheng Y, Reedy MK, Taylor KA. Electron tomography of swollen rigor fibers of insect flight muscle reveals a short and variably angled S2 domain. J Mol Biol. 2006;362:844–860. doi: 10.1016/j.jmb.2006.07.084. [DOI] [PubMed] [Google Scholar]
- Lucic V, Forster F, Baumeister W. Structural studies by electron tomography: from cells to molecules. Annu Rev Biochem. 2005;74:833–865. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- Maupin-Szamier P, Pollard TD. Actin filament destruction by osmium tetroxide. J Cell Biol. 1978;77:837–852. doi: 10.1083/jcb.77.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A, Tregear RT. The structure of insect fibrillar flight muscle in the presence and absence of ATP. J Mol Biol. 1972;70:85–104. doi: 10.1016/0022-2836(72)90165-9. [DOI] [PubMed] [Google Scholar]
- Nicastro D, McIntosh JR, Baumeister W. 3D structure of eukaryotic flagella in a quiescent state revealed by cryo-electron tomography. Proc Natl Acad Sci U S A. 2005;102:15889–15894. doi: 10.1073/pnas.0508274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirani A, Vinogradova MV, Curmi PM, King WA, Fletterick RJ, Craig R, Tobacman LS, Xu C, Hatch V, Lehman W. An atomic model of the thin filament in the relaxed and Ca2+-activated states. J Mol Biol. 2006;357:707–717. doi: 10.1016/j.jmb.2005.12.050. [DOI] [PubMed] [Google Scholar]
- Poole KJ, Lorenz M, Evans G, Rosenbaum G, Pirani A, Craig R, Tobacman LS, Lehman W, Holmes KC. A comparison of muscle thin filament models obtained from electron microscopy reconstructions and low-angle X-ray fibre diagrams from non-overlap muscle. J Struct Biol. 2006;155:273–284. doi: 10.1016/j.jsb.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Reedy MK. Ultrastructure of insect flight muscle. I. Screw sense and structural grouping in the rigor cross-bridge lattice. J Mol Biol. 1968;31:155–176. doi: 10.1016/0022-2836(68)90437-3. [DOI] [PubMed] [Google Scholar]
- Reedy MK, Goody RS, Hofmann W, Rosenbaum G. Co-ordinated electron microscopy and X-ray studies of glycerinated insect flight muscle. I. X-ray diffraction monitoring during preparation for electron microscopy of muscle fibers fixed in rigor, in ATP and in AMPPNP. J Muscle Res Cell Motil. 1983;4:25–53. doi: 10.1007/BF00711957. [DOI] [PubMed] [Google Scholar]
- Reedy MK, Holmes KC, Tregear RT. Induced changes in orientation of the crossbridges of glycerinated insect flight muscle. Nature. 1965;207:1276–1280. doi: 10.1038/2071276a0. [DOI] [PubMed] [Google Scholar]
- Reedy MK, Reedy MC. Rigor cross-bridge structure in tilted single filament layers and flared-X formations from insect flight muscle. J Mol Biol. 1985;185:145–176. doi: 10.1016/0022-2836(85)90188-3. [DOI] [PubMed] [Google Scholar]
- Rossmann MG. Fitting atomic models into electron-microscopy maps. Acta Crystallogr D Biol Crystallogr. 2000;56:1341–1349. doi: 10.1107/s0907444900009562. [DOI] [PubMed] [Google Scholar]
- Saibil HR. Conformational changes studied by cryo-electron microscopy. Nat Struct Biol. 2000;7:711–714. doi: 10.1038/78923. [DOI] [PubMed] [Google Scholar]
- Saxton WO, Baumeister W, Hahn M. Three-dimensional reconstruction of imperfect two-dimensional crystals. Ultramicroscopy. 1984;13:57–70. doi: 10.1016/0304-3991(84)90057-3. [DOI] [PubMed] [Google Scholar]
- Schmitz H, Lucaveche C, Reedy MK, Taylor KA. Oblique section 3-D reconstruction of relaxed insect flight muscle reveals the crossbridge lattice in helical registration. Biophys J. 1994;67:1620–1633. doi: 10.1016/S0006-3495(94)80635-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz H, Reedy MC, Reedy MK, Tregear RT, Taylor KA. Tomographic three-dimensional reconstruction of insect flight muscle partially relaxed by AMPPNP and ethylene glycol. J Cell Biol. 1997;139:695–707. doi: 10.1083/jcb.139.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz H, Reedy MC, Reedy MK, Tregear RT, Winkler H, Taylor KA. Electron tomography of insect flight muscle in rigor and AMPPNP at 23°C. J Mol Biol. 1996;264:279–301. doi: 10.1006/jmbi.1996.0641. [DOI] [PubMed] [Google Scholar]
- Tama F, Miyashita O, Brooks CL., 3rd Normal mode based flexible fitting of high-resolution structure into low-resolution experimental data from cryo-EM. J Struct Biol. 2004;147:315–326. doi: 10.1016/j.jsb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Tauhata SB, dos Santos DV, Taylor EW, Mooseker MS, Larson RE. High affinity binding of brain myosin-Va to F-actin induced by calcium in the presence of ATP. J Biol Chem. 2001;276:39812–39818. doi: 10.1074/jbc.M102583200. [DOI] [PubMed] [Google Scholar]
- Taylor KA, Reedy MC, Cordova L, Reedy MK. Three-dimensional reconstruction of rigor insect flight muscle from tilted thin sections. Nature. 1984;310:285–291. doi: 10.1038/310285a0. [DOI] [PubMed] [Google Scholar]
- Taylor KA, Reedy MC, Cordova L, Reedy MK. Image reconstruction using electron micrographs of insect flight muscle: Use of thick transverse sections to supplement data from tilted thin longitudinal sections. Biophys J. 1986;49:353–364. doi: 10.1016/S0006-3495(86)83648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KA, Reedy MC, Córdova L, Reedy MK. Three-dimensional image reconstruction of insect flight muscle. I The rigor myac layer. J Cell Biol. 1989a;109:1085–1102. doi: 10.1083/jcb.109.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KA, Reedy MC, Córdova L, Reedy MK. Three-dimensional image reconstruction of insect flight muscle. II The rigor actin layer. J Cell Biol. 1989b;109:1103–1123. doi: 10.1083/jcb.109.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KA, Reedy MC, Reedy MK, Crowther RA. Crossbridges in the complete unit cell of rigor insect flight muscle imaged by three-dimensional reconstruction from oblique sections. J Mol Biol. 1993;233:86–108. doi: 10.1006/jmbi.1993.1487. [DOI] [PubMed] [Google Scholar]
- Taylor KA, Schmitz H, Reedy MC, Goldman YE, Franzini-Armstrong C, Sasaki H, Tregear RT, Poole KJV, Lucaveche C, Edwards RJ, et al. Tomographic 3-D reconstruction of quick frozen, Ca2+-activated contracting insect flight muscle. Cell. 1999;99:421–431. doi: 10.1016/s0092-8674(00)81528-7. [DOI] [PubMed] [Google Scholar]
- Taylor KA, Wu S, Reedy MC, Reedy MK. Imaging actomyosin in situ. In: McIntosh JR, editor. Cellular Electron Microscopy. San Diego, CA: Academic Press; 2007. pp. 321–368. [Google Scholar]
- Tregear RT, Reedy MC, Goldman YE, Taylor KA, Winkler H, Franzini-Armstrong C, Sasaki H, Lucaveche C, Reedy MK. Cross-bridge number, position, and angle in target zones of cryofixed isometrically active insect flight muscle. Biophys J. 2004;86:3009–3019. doi: 10.1016/S0006-3495(04)74350-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M. Classification of very large electron microscopical image data sets. Optik. 1989;82:114–126. [Google Scholar]
- Van Heel M, Schatz M, Orlova E. Correlation functions revisited. Ultramicroscopy. 1992;46:307–316. [Google Scholar]
- Volkmann N, Liu H, Hazelwood L, Krementsova EB, Lowey S, Trybus KM, Hanein D. The structural basis of myosin V processive movement as revealed by electron cryomicroscopy. Mol Cell. 2005;19:595–605. doi: 10.1016/j.molcel.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Winkler H. 3D reconstruction and processing of volumetric data in cryo-electron tomography. J Struct Biol. 2007;157:126–137. doi: 10.1016/j.jsb.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Winkler H, Reedy MC, Reedy MK, Tregear R, Taylor KA. Three-dimensional structure of nucleotide-bearing cross-bridges in situ: oblique section reconstruction of insect flight muscle in AMPPNP at 23 degrees C. J Mol Biol. 1996;264:302–322. doi: 10.1006/jmbi.1996.0642. [DOI] [PubMed] [Google Scholar]
- Winkler H, Taylor KA. Multivariate statistical analysis of three-dimensional cross-bridge motifs in insect flight muscle. Ultramicroscopy. 1999;77:141–152. [Google Scholar]
- Winkler H, Taylor KA. Accurate marker-free alignment with simultaneous geometry determination and reconstruction of tilt series in electron tomography. Ultramicroscopy. 2006;106:240–254. doi: 10.1016/j.ultramic.2005.07.007. [DOI] [PubMed] [Google Scholar]