Abstract
The Salmonella Pathogenicity Island-2 (i.e. SPI-2) encodes a unique type III secretion system that delivers effector proteins from the Salmonella-containing vacuole (SCV) into the host cell. The SPI-2 locus also encodes translocated effectors as well as a two-component system – termed SpiR/SsrB – that is essential for the expression of SPI-2 genes. Transcription of the horizontally acquired SPI-2 genes requires the ancestral nucleoid-associated proteins (i.e. NAPs) IHF and Fis, the regulatory protein SlyA, and the two-component systems PhoP/PhoQ and OmpR/EnvZ, as well as the DNA binding protein HilD encoded in a different pathogenicity island. Some of these positive SPI-2 regulators act to antagonize the robust silencing promoted by the NAPs H-NS, Hha, and YdgT.
Introduction
Pathogenic bacteria synthesize their virulence determinants when they are needed. This requires signal transduction systems that respond to particular host and/or environmental signals by modifying the level and/or activity of the transcription circuits, regulatory RNAs and modulator proteins governing the expression of the virulence determinants. The physiological control of virulence genes is crucial for a pathogen's ability to cause disease as their constitutive expression can strongly attenuate virulence. For example, the two-component regulatory systems CpxR/CpxA and PhoP/PhoQ are required for virulence in several bacterial pathogens including the gastroenteritis-causing Salmonella enterica serovar Typhimurium [1,2]. However, constitutive activation of the CpxA sensor significantly reduces Salmonella's ability to proliferate within mouse organs following oral inoculation [2], and the pho-24 mutant (encoding a variant PhoQ protein often referred to as PhoPC) is as attenuated for mouse virulence as a strain deleted for the phoP or phoQ genes [3].
The Salmonella Pathogenicity Island 2 (SPI-2) is a 25-kb locus of horizontally acquired virulence genes that is required for Salmonella replication inside host cells and systemic infection in mice [4,5]. The SPI-2 locus encodes a type III secretion system responsible for delivering effector proteins to the host cell, effectors translocated by the type III secretion system as well as a two-component regulatory system that controls not only its own expression but also that of the secretion apparatus and secreted effectors [4,5]. The effectors facilitate Salmonella survival and replication in host cells by altering a variety of cell properties including its cytoskeletal structure, signal transduction pathways, and vesicular trafficking (reviewed in [6]). Interestingly, some of the genes coding for the effectors translocated by the SPI-2-encoded type III secretion system and regulated by the SPI-2-encoded two-component system are located outside the SPI-2 [7]. The SPI-2 and associated effector genes are expressed primarily when Salmonella is present in a modified phago-some known as the Salmonella-containing vacuole (SCV) [8].
Like many other horizontally acquired genes, the SPI-2 DNA has a higher A + T content than that of the ancestral Salmonella DNA [4,5]. This AT-rich DNA is bound by the histone-like nucleoid structuring protein (H-NS), which results in transcriptional silencing of the corresponding genes [9•,10•]. The H-NS-promoted silencing could protect Salmonella from the detrimental effects of expressing virulence determinants at inappropriate times and/or locations. Yet, Salmonella does express the SPI-2-encoded (as well as other horizontally acquired) virulence genes indicative that it has evolved means to overcome the silencing effect mediated by H-NS.
In this review we discuss how expression of SPI-2 genes and associated effector genes located outside SPI-2 is controlled. We explore the distinct roles played by positive and negative regulators of SPI-2 expression as well as the distinct tasks performed by horizontally acquired and ancestral regulatory proteins. And we examine the possible effects of regulation of genes in one pathogenicity island being affected by a regulator encoded in a different pathogenicity island.
The SPI-2 encoded SpiR/SsrB two-component regulatory system
SpiR/SsrB is a two-component system encoded in the SPI-2 pathogenicity island. SsrB is the response regulator and SpiR is the predicted integral membrane cognate sensor (even though spiR is often referred to as ssrA, we have retained the original designation [4,5] to avoid confusion with the tag-adding ssrA gene [11].) The SsrB protein binds to the promoters of all SPI-2 functional gene clusters [12•] and is essential for expression of the SPI-2-encoded type III secretion system and its effectors, including those coded for outside the SPI-2 locus [7]. SsrB binds to and regulates transcription from both its own promoter and that corresponding to the upstream spiR gene [13] (Figure 1). This is atypical for two-component systems because sensor and regulator proteins encoded by adjacent genes are usually co-regulated where the same promoter(s) generates a polycistronic mRNA. The use of separate promoters to transcribe the ssrB and spiR genes may depend on the growth condition because others reported the presence of a single promoter upstream of spiR driving transcription of a polycystronic spiR–ssrB message [14•]. The SpiR protein is predicted to have two transmembrane domains, which define a large periplasmic region presumably involved in sensing a signal(s) the identity of which remains unknown.
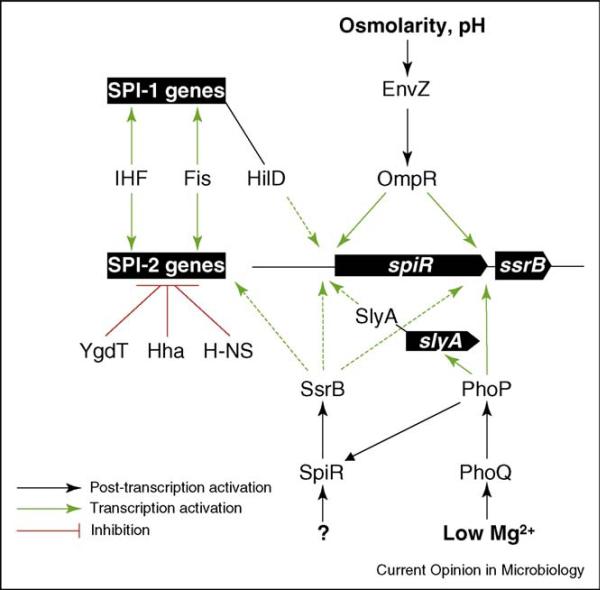
Figure 1.
Schematics of the known SPI-2 regulatory factors. Three different two-component systems, SpiR/SsrB, EnvZ/OmpR and PhoQ/PhoP, are involved in SPI-2 regulation. OmpR activates SPI-2 genes by binding to the promoter and inducing the expression of SpiR and SsrB. PhoP directly binds the ssrB promoter and could induce spiR transcription indirectly by inducing transcription/activation of the transcription factor SlyA. PhoP indirectly enhances SpiR levels. SsrB binds to all SPI-2 promoters including the spiR and its own promoter where it is required for antagonizing the repression activity of H-NS. The SP-1 encoded HilD transcription factor is also involved (under certain conditions) in antagonizing H-NS activity. The NAPs H-NS, Hha, and YdgT function as general negative SPI-2 regulators. Fis and IHF, on the contrary, are NAPs that have a positive effect on SPI-2 as well as SPI-1 gene expression. Transcription inhibition—red lines, Post-transcription activation—black arrows, transcription activation—green arrows, H-NS derepression—dashed green arrows.
Negative and positive regulation of SPI-2 gene expression by nucleoid-associated proteins
Nucleoid-associated proteins as negative regulators of SPI-2 genes
Enteric bacteria harbor nucleoid-associated proteins (or NAPs for short) that function as global regulators by controlling the expression of a large number of genes throughout their genomes [15]. H-NS is a NAP that prevents the uncontrolled expression of horizontally acquired genes. Deletion of the hns gene results in derepression of genes that are silent under laboratory conditions (reviewed in [15]). Thus, inactivation of the hns gene in Salmonella can be quite unhealthy and has to be carried out in strains also harboring mutations in either the phoP or rpoS regulatory genes [9•], suggesting that overexpression and/or super-repression of a PhoP-regulated and/or RpoS-regulated gene(s) results in bacterial lethality.
H-NS binds to AT-rich regions of the genome, such as that corresponding to SPI-2, silencing the expression of the corresponding genes [9•,10•,16]. When the activity of wild-type H-NS is neutralized by expression of the dominant negative H-NSQ92am allele, the level of SPI-2-encoded effectors increases [17]. Likewise, transcription of SPI-2-encoded genes is derepressed in an hns mutant strain experiencing non-SPI-2-inducing conditions [12•]. H-NS exerts its effect directly by binding to the spiR promoter [14•]. Then, what is the mechanism by which H-NS repression of SPI-2 genes is overcome?
As discussed above, the SsrB protein binds to the SPI-2 promoters and is necessary for SPI-2 expression [12•,18], raising the possibility that it functions both by activating gene transcription and by antagonizing H-NS-mediated silencing. Consistent with this notion, transcription of the SPI-2 gene sseA starts within the region footprinted by the SsrB protein in vitro [12•]. Moreover, an ssrB hns double mutant experiencing SPI-2 inducing conditions displays a reduction in transcription of SPI-2-encoded genes relative to an hns mutant; but this reduction is relatively modest when compared to that observed in an hns+ background [12•]. In addition to SsrB, the HilD protein, which is encoded in the SPI-1 pathogenicity island, relieves HNS-promoted repression under particular SPI-2 inducing conditions (i.e. during stationary phase following growth in LB medium) but not under other conditions [14•]. HilD appears to exert its effect directly as gel shift experiments show that it binds to the spiR promoter [14•].
Antagonizing H-NS-promoted silencing is not exclusive to horizontally acquired regulatory determinants because the ancestral SlyA protein binds to the spiR promoter [19] and because genetic studies [13,19] and cDNA microarray analysis [20] implicate SlyA in expression of SPI-2 genes. Furthermore, the intramacrophage activation of the SsrB-activated sifA and sifB promoters [21] is significantly reduced when SlyA is absent (though that corresponding to the SsrB- activated srfH promoter is not [13]). SlyA is required also for expression of the horizontally acquired PhoP-activated pagC and ugtL genes [20], which are neither located within SPI-2 nor dependent on SsrB for expression. Because in vitro transcription of the pagC and ugtL promoters required SlyA only when H-NS was present in the reaction [22], it is possible that SlyA's role as a SPI-2 activator might be limited to overcoming silencing by H-NS as well.
YdgT and Hha are two NAPs that repress transcription of SPI-2 genes. The Salmonella ydgT gene is not expressed at early stages of macrophage infection, a time when SPI-2 genes are being upregulated. Moreover, transcription of most SPI-2 genes was derepressed in a ydgT mutant when Salmonella was grown under SPI-2 inducing conditions, indicating that YdgT functions, genetically speaking, as a SPI-2 repressor [23]. When mice were inoculated with mixtures of wild-type and ydgT mutant Salmonella, the ydgT mutant displayed enhanced early survival advantage in mouse organs; however, during the course of infection the ydgT mutant lost its advantage and was recovered at significantly lower numbers compared with the wild-type strain. At the early stages of infection, the ydgT mutant showed a competitive survival advantage also over the ssrB–ydgT double mutant, indicating that the phenotype displayed by the ydgT depends on the SPI-2 regulator SsrB [23].
Hha is a YdgT homolog that represses SPI-2 gene expression stronger than YdgT, at least under certain conditions [24]. Similar to the behavior of the ydgT mutant, a Salmonella strain defective in the hha gene is significantly attenuated for virulence at late stages of a systemic infection in mice [24]. A strong derepression of the sseA promoter in SPI-2 was reported for an hha mutant but not for the hha-ssrB double mutant when grown in rich medium (e.g. LB at early log phase), which is normally non-conducive to SPI-2 gene transcription. This suggests that Hha is an important SPI-2 silencer when bacteria are not experiencing an intracellular environment [24].
Because YdgT and Hha can form heterodimeric complexes with H-NS [25,26], it has been proposed that YdgT and Hha repress SPI-2 gene expression indirectly, in concert with other NAPs such as H-NS [23,24,27]. However, binding of YdgT and Hha to SPI-2 promoters has not been reported. Furthermore, if YdgT and Hha repress SPI-2 by associating with H-NS, then, the requirement for SsrB should also be reduced when YdgT or Hha are absent as observed when H-NS is removed [12•]. However, the hha ssrB and ydgT ssrB double mutants fail to induce SPI-2 expression [23,24] arguing against the possibility that YdgT and Hha function simply by forming a repressor complex with H-NS. An alternative scenario is that YdgT and Hha do associate with H-NS to prevent SsrB from overcoming H-NS-promoted repression.
Nucleoid-associated proteins as positive regulators of SPI-2 genes
The integration host factor (IHF) and the factor for inversion simulation (Fis) are two NAPs that, in contrast to the repression properties of H-NS, YdgT and Hha, are necessary for full expression of SPI-2 genes. IHF plays an important role in DNA bending and compaction, and is important for the transcriptional regulation of many genes. Genome-wide expression experiments have shown that in the absence of IHF, SPI-2 genes are downregulated when grown in LB mainly in late log phase when expression of IHF usually peaks and SPI-2 genes are induced [28]. Because IHF expression is important for SPI-1 expression at early to late log exponential growth phase, it has been proposed that IHF levels may play a role in coordinating expression of genes located in the SPI-1 and SPI-2 pathogenicity islands [28].
Fis is a direct activator of SPI-2 genes because the Fis protein binds to the promoter regions of the SPI-2 encoded spiR and ssaG genes [29,30], and because a fis mutant displayed decreased expression of SPI-2 genes following growth in LB to stationary phase [30,31]. When grown in LB media, Fis expression is maximal at early log phase and is dramatically reduced by late log phase and stationary phase [29], when expression of SPI-2 genes is induced. Thus, Fis may exert its activating action on SPI-2 genes at a very particular growth condition because optimum SpiR expression occurs at a crucial concentration of Fis and that too much Fis results in diminished SpiR expression [32]. There is a strong correlation between the transcription pattern of fis and the SPI-2-encoded genes when Salmonella is inside macrophages [32]. Fis is known to influence transcription through its ability to preserve intermediate supercoiled forms of DNA from shifting to more relaxed or more negatively supercoiled topology [33]. During the course of macrophage infection the degree of DNA supercoiling of the test plasmid pUC18 was progressively relaxed and SPI-2 genes upregulated. This correlated with Fis activity, suggesting a model in which Fis protein affects SPI-2 genes by controlling SPI-2 promoter topology [32].
Fis may also regulate SPI-2 expression indirectly, by its ability to control expression of other regulatory genes such as phoP and SPI-1 encoded genes. Consistent with this notion, PhoP is an important SPI-2 regulator [7,34–36] and SPI-1 encodes the transcription factor HilD, which has recently been found to be required for SPI-2 activation at late log phase Salmonella grown in LB broth [14•]. Nevertheless, it may be possible to evaluate the relative contributions of the direct and indirect pathways by which Fis controls SPI-2 transcription by exploring the phenotype of strains mutated in the Fis-binding site(s) in SPI-2 promoters.
Regulation of SPI-2 expression by ancestral two-component systems
Ancestral regulatory proteins regulate horizontally acquired genes, perhaps as a means to coordinate the expression of the newly acquired genetic information with that of the rest of the ancestral genome. This is also the case of SPI-2 as the two-component system SpiR/SsrB is regulated by the ancestral two-component systems OmpR/EnvZ and PhoP/PhoQ. OmpR/EnvZ plays an important role in SPI-2 expression when Salmonella is grown under certain laboratory media conducive to SPI-2 expression and also inside host cells [7,13,34,37,38]. The response regulator OmpR binds to the spiR [34] and ssrB [13] promoters leading to the proposal that OmpR regulates SPI-2 genes by promoting the expression of its master regulator: SpiR/SsrB. Even though the OmpR binding sites in the spiR promoter overlap with the SsrB binding sites, OmpR does not seem to antagonize H-NS-promoted silencing as SsrB does because OmpR was required for SPI-2 expression also when H-NS was inactivated by the dominant negative H-NSQ92am protein [14•].
The PhoP/PhoQ system is required for Salmonella virulence [1], controlling a number of pathogenicity determinants including mgtC [39], mig-14 [40], and macAB [41]. Like the SPI-2 genes, the PhoP/PhoQ system is required for Salmonella's ability to survive within macrophages [42] and PhoP-activated genes are typically expressed when Salmonella is inside host cells [8,43,44], suggesting the possibility that the PhoP/PhoQ system regulates expression of SPI-2 genes. Indeed, when Salmonella were grown in media with low Mg2+ [36] and low pH [34], PhoP was required for SPI-2 induction. Moreover, the expression of SsrB-regulated effectors was significantly affected inside macrophages in a phoP mutant [7]. Furthermore, PhoP has been shown to regulate SPI-2 transcription in Salmonella-infected macrophages where PhoP directly binds to the ssrB promoter [35]. In contrast to OmpR, which binds to both the spiR [34] and ssrB [13] promoters, PhoP controls SpiR levels post-transcriptionally in a process that appears to involve the 5′ leader of the spiR mRNA [35]. In addition, because PhoP regulates the expression [45] and/or activation of the SlyA protein [20], and because SlyA is a positive SPI-2 regulator, one might expect PhoP to play a role in SPI-2 expression.
The participation of PhoP/PhoQ in SPI-2 expression has been disputed because, using a plasmid-based fusion of the ssaG and spiR promoters to a promoterless gfp gene, fluorescence was detected inside macrophages also in a phoP mutant [8,34]. Likewise, the intramacrophage promoter activity of the sspH2 gene, coding for an SsrB-regulated effector, was induced 13-fold in a phoP mutant compared to a phoP spiR double mutant, implying that PhoP is dispensable for SPI-2 expression [46]. A comparison of the number of phoP and spiR Salmonella recovered from mice spleen two days after intraperitoneal mixed infections demonstrated that the ratio of the bacterial level of the phoP spiR double mutant to either the phoP or spiR single mutants was similar to the ratio of the corresponding single mutants phoP or spiR, and the wild-type strain. The authors concluded that PhoP-modulated virulence is independent of SPI-2 [47], which is not surprising given that, as discussed above, PhoP has been known to control expression of several virulence determinants unrelated to SPI-2 [39].
How can one reconcile the different findings regarding the role of the PhoP/PhoQ system in SPI-2 expression? One possibility is that the PhoP is necessary to promote SPI-2 expression only under certain circumstances. For example, induction of SPI-2 genes is expected to require factors that can antagonize the strong silencing effects of NAPs, especially when induced after preexisting conditions that promote abundance of such proteins. These factors may not be required if Salmonella senses SPI-2-inducing conditions when coming from a preexisting environment that promoted expression of SPI-2 activators. Thus, even when Salmonella experiences the same signal for SPI-2 induction, the contextual preexisting conditions may determine whether certain SPI-2 regulators, such as PhoP/PhoQ, are required for their induction. In agreement with this idea, HilD is required for SPI-2 expression when induction takes place in LB but not following growth in N minimal media [14•]. Thus, depending on what conditions bacteria experience before entering a host cell, PhoP, HilD, and potentially other proteins, may (or may not) be required for transcription of SPI-2 genes. As discussed above for Fis, the role that PhoP and HilD play in SPI-2 expression during infection of an animal host may be evaluated by inactivating the PhoP or HilD binding sites in the SPI-2-regulated promoters.
Regulation of SPI-2 expression by a horizontally acquired regulator encoded outside SPI-2
Crucial to Salmonella's ability to cause disease is the capacity to coordinate the expression of the numerous virulence factors required in the various tissues experienced during infection. One possible mechanism is the control of genes encoded in one pathogenicity island by a regulatory protein encoded in a different pathogencity island. For example, expression of SPI-5 genes pipB and sopB is distinctly regulated by transcription factors encoded in the SPI-2 and SPI-1 loci, respectively [48]. Interestingly, transcription of SPI-2 genes when Salmonella is grown in LB to stationary phase requires the SPI-1-encoded HilD protein, which promotes expression of SPI-1 genes at early log phase in LB a condition that is normally non-conducive to expression of SPI-2 genes. Accordingly, HilD has been proposed to control the switch from SPI-1 to SPI-2 induction. HilD antagonizes H-NS silencing by binding to the hilA-promoter in SPI-1 and the spiR-promoter in SPI-2. Because binding to the spiR-promoter requires a higher level of HilD protein, this may explain how HilD could differentially regulate SPI-1 and SPI-2 genes. The striking similarity between the growth conditions in which HilD and IHF promote SPI-1 and SPI-2 expression [28] suggests that the coordinate regulation of SPI-2 after SPI-1 by IHF might occur indirectly via HilD. Yet, the role of HilD in SPI-2 expression when Salmonella is inside eukaryotic cells is yet to be reported. This is crucial because deletion of SPI-1 does not affect Salmonella's ability to cause a lethal infection following intraperitoneal inoculation whereas inactivation of SPI-2 does.
Conclusions
The horizontally acquired SPI-2 genes are expressed primarily when Salmonella is inside host cells. This is achieved both by positive and negative regulators (Figure 1), some of which are widespread within enteric bacteria and others specific to the Salmonella genome. The positive regulators of SPI-2 expression exert their action by counteracting the silencing effects of H-NS and other nucleoid-associated proteins, and/or by recruiting RNA polymerase to particular promoters. However, it is not clear how these factors achieve the spatiotemporal control of SPI-2 genes. Whereas laboratory conditions that induce SPI-2 genes are static, the natural environment experienced by Salmonella within the SCV is dynamic as SPI-2 transcription inside mammalian cells peaks 2–4 h after infection and is then significantly reduced [49,50]. Not surprisingly, the importance of some of the SPI-2 regulators is observed specifically under certain growth conditions. Other SPI-2 regulators may be required to optimize expression of SPI-2 genes encoding functionally different products such as effectors, apparatus components, and regulators. Finally, because only one or two SPI-2 apparatuses are expressed per cell [51], additional levels of SPI-2 regulation are yet to be uncovered.
Acknowledgements
Our research on expression of the SPI-2 genes is supported by grants from the National Institutes of Health to EAG who is an investigator of the Howard Hughes Medical Institute.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
•of special interest
•of outstanding interest
- 1.Groisman EA. The pleiotropic two-component regulatory system PhoP–PhoQ. J Bacteriol. 2001;183:1835–1842. doi: 10.1128/JB.183.6.1835-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Humphreys S, Rowley G, Stevenson A, Anjum MF, Woodward MJ, Gilbert S, Kormanec J, Roberts M. Role of the two-component regulator CpxAR in the virulence of Salmonella enterica serotype Typhimurium. Infect Immun. 2004;72:4654–4661. doi: 10.1128/IAI.72.8.4654-4661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller SI, Mekalanos JJ. Constitutive expression of the phoP regulon attenuates Salmonella virulence and survival within macrophages. J Bacteriol. 1990;172:2485–2490. doi: 10.1128/jb.172.5.2485-2490.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ochman H, Soncini FC, Solomon F, Groisman EA. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci U S A. 1996;93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shea JE, Hensel M, Gleeson C, Holden DW. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci U S A. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abrahams GL, Hensel M. Manipulating cellular transport and immune responses: dynamic interactions between intracellular Salmonella enterica and its host cells. Cell Microbiol. 2006;8:728–737. doi: 10.1111/j.1462-5822.2006.00706.x. [DOI] [PubMed] [Google Scholar]
- 7.Worley MJ, Ching KH, Heffron F. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol Microbiol. 2000;36:749–761. doi: 10.1046/j.1365-2958.2000.01902.x. [DOI] [PubMed] [Google Scholar]
- 8.Valdivia RH, Falkow S. Fluorescence-based isolation of bacterial genes expressed within host cells. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- 9•.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [The authors show that H-NS binds and mediates repression of horizontally acquired genes with A + T content that is higher than the overall Salmonella genome. These findings indicate that H-NS provides a mechanism for recognizing and repressing newly acquired DNA, which enables acquisition of exogenous DNA while avoiding detrimental affects of their unregulated expression] [DOI] [PubMed] [Google Scholar]
- 10•.Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;2:e81. doi: 10.1371/journal.ppat.0020081. [The authors show that H-NS binds and mediates repression of horizontally acquired genes with A + T content that is higher than the overall Salmonella genome. These findings indicate that H-NS provides a mechanism for recognizing and repressing newly acquired DNA, which enables acquisition of exogenous DNA while avoiding detrimental affects of their unregulated expression] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karzai AW, Roche ED, Sauer RT. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat Struct Biol. 2000;7:449–455. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- 12•.Walthers D, Carroll RK, Navarre WW, Libby SJ, Fang FC, Kenney LJ. The response regulator SsrB activates expression of diverse Salmonella pathogenicity island 2 promoters and counters silencing by the nucleoid-associated protein H-NS. Mol Microbiol. 2007;65:477–493. doi: 10.1111/j.1365-2958.2007.05800.x. [Using the isolated C-terminus of SsrB in a DNase I protecting footprinting assay the authors map and determine the binding of SsrB to the intergenic promoter regions of the SPI-2 gene clusters (ssaB, sseA, ssaG, and ssaM). The authors demonstrate that the activity of the SPI-2 promoter is SsrB-dependent; however, in the absence of H-NS the SsrB requirement for induction of SPI-2 genes is greatly reduced. These findings suggest a crucial role for SsrB in antagonizing the H-NS-mediated repression of SPI-2 genes] [DOI] [PubMed] [Google Scholar]
- 13.Feng X, Oropeza R, Kenney LJ. Dual regulation by phospho-OmpR of ssrA/B gene expression in Salmonella pathogenicity island 2. Mol Microbiol. 2003;48:1131–1143. doi: 10.1046/j.1365-2958.2003.03502.x. [DOI] [PubMed] [Google Scholar]
- 14•.Bustamante VH, Martinez LC, Santana FJ, Knodler LA, Steele-Mortimer O, Puente JL. HilD-mediated transcriptional crosstalk between SPI-1 and SPI-2. Proc Natl Acad Sci U S A. 2008;105:14591–14596. doi: 10.1073/pnas.0801205105. [By following the kinetics expression of SPI-1 and SPI-2 gene in different Salmonella strains the authors show that the SPI-1 encoded transcription factor HilD that is required for SPI-1 induction when growth enters stationary phase is also required for SPI-2 induction later at the stationary phase. These findings demonstrate a unique case where a transcription factor is required to antagonize H-NS repression of its encoding pathogenicity island and at a different stages of another pathogenicity island, potentially functioning as a transcriptional coordinator of two sets of horizontally acquired genes] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoebel DM, Free A, Dorman CJ. Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology. 2008;154:2533–2545. doi: 10.1099/mic.0.2008/020693-0. [DOI] [PubMed] [Google Scholar]
- 16.Navarre WW, McClelland M, Libby SJ, Fang FC. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 2007;21:1456–1471. doi: 10.1101/gad.1543107. [DOI] [PubMed] [Google Scholar]
- 17.Duong N, Osborne S, Bustamante VH, Tomljenovic AM, Puente JL, Coombes BK. Thermosensing coordinates a cis-regulatory module for transcriptional activation of the intracellular virulence system in Salmonella enterica serovar Typhimurium. J Biol Chem. 2007;282:34077–34084. doi: 10.1074/jbc.M707352200. [DOI] [PubMed] [Google Scholar]
- 18.Feng X, Walthers D, Oropeza R, Kenney LJ. The response regulator SsrB activates transcription and binds to a region overlapping OmpR binding sites at Salmonella pathogenicity island 2. Mol Microbiol. 2004;54:823–835. doi: 10.1111/j.1365-2958.2004.04317.x. [DOI] [PubMed] [Google Scholar]
- 19.Okada N, Oi Y, Takeda-Shitaka M, Kanou K, Umeyama H, Haneda T, Miki T, Hosoya S, Danbara H. Identification of amino acid residues of Salmonella SlyA that are critical for transcriptional regulation. Microbiology. 2007;153:548–560. doi: 10.1099/mic.0.29259-0. [DOI] [PubMed] [Google Scholar]
- 20.Navarre WW, Halsey TA, Walthers D, Frye J, McClelland M, Potter JL, Kenney LJ, Gunn JS, Fang FC, Libby SJ. Co-regulation of Salmonella enterica genes required for virulence and resistance to antimicrobial peptides by SlyA and PhoP/PhoQ. Mol Microbiol. 2005;56:492–508. doi: 10.1111/j.1365-2958.2005.04553.x. [DOI] [PubMed] [Google Scholar]
- 21.Linehan SA, Rytkonen A, Yu XJ, Liu M, Holden DW. SlyA regulates function of Salmonella pathogenicity island 2 (SPI-2) and expression of SPI-2-associated genes. Infect Immun. 2005;73:4354–4362. doi: 10.1128/IAI.73.7.4354-4362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez JC, Latifi T, Groisman EA. Overcoming H-NS-mediated transcriptional silencing of horizontally acquired genes by the PhoP and SlyA proteins in Salmonella enterica. J Biol Chem. 2008;283:10773–10783. doi: 10.1074/jbc.M709843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coombes BK, Wickham ME, Lowden MJ, Brown NF, Finlay BB. Negative regulation of Salmonella pathogenicity island 2 is required for contextual control of virulence during typhoid. Proc Natl Acad Sci U S A. 2005;102:17460–17465. doi: 10.1073/pnas.0505401102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silphaduang U, Mascarenhas M, Karmali M, Coombes BK. Repression of intracellular virulence factors in Salmonella by the Hha and YdgT nucleoid-associated proteins. J Bacteriol. 2007;189:3669–3673. doi: 10.1128/JB.00002-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nieto JM, Madrid C, Miquelay E, Parra JL, Rodriguez S, Juarez A. Evidence for direct protein-protein interaction between members of the enterobacterial Hha/YmoA and H-NS families of proteins. J Bacteriol. 2002;184:629–635. doi: 10.1128/JB.184.3.629-635.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paytubi S, Madrid C, Forns N, Nieto JM, Balsalobre C, Uhlin BE, Juarez A. YdgT, the Hha paralogue in Escherichia coli, forms heteromeric complexes with H-NS and StpA. Mol Microbiol. 2004;54:251–263. doi: 10.1111/j.1365-2958.2004.04268.x. [DOI] [PubMed] [Google Scholar]
- 27.Vivero A, Banos RC, Mariscotti JF, Oliveros JC, Garcia-del Portillo F, Juarez A, Madrid C. Modulation of horizontally acquired genes by the Hha-YdgT proteins in Salmonella enterica serovar Typhimurium. J Bacteriol. 2008;190:1152–1156. doi: 10.1128/JB.01206-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mangan MW, Lucchini S, Danino V, Croinin TO, Hinton JC, Dorman CJ. The integration host factor (IHF) integrates stationary-phase and virulence gene expression in Salmonella enterica serovar Typhimurium. Mol Microbiol. 2006;59:1831–1847. doi: 10.1111/j.1365-2958.2006.05062.x. [DOI] [PubMed] [Google Scholar]
- 29.Kelly A, Goldberg MD, Carroll RK, Danino V, Hinton JC, Dorman CJ. A global role for Fis in the transcriptional control of metabolism and type III secretion in Salmonella enterica serovar Typhimurium. Microbiology. 2004;150:2037–2053. doi: 10.1099/mic.0.27209-0. [DOI] [PubMed] [Google Scholar]
- 30.Lim S, Kim B, Choi HS, Lee Y, Ryu S. Fis is required for proper regulation of ssaG expression in Salmonella enterica serovar Typhimurium. Microb Pathog. 2006;41:33–42. doi: 10.1016/j.micpath.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 31.Yoon H, Lim S, Heu S, Choi S, Ryu S. Proteome analysis of Salmonella enterica serovar Typhimurium fis mutant. FEMS Microbiol Lett. 2003;226:391–396. doi: 10.1016/S0378-1097(03)00641-4. [DOI] [PubMed] [Google Scholar]
- 32.O Cróinin T, Carroll RK, Kelly A, Dorman CJ. Roles for DNA supercoiling and the Fis protein in modulating expression of virulence genes during intracellular growth of Salmonella enterica serovar Typhimurium. Mol Microbiol. 2006;62:869–882. doi: 10.1111/j.1365-2958.2006.05416.x. [DOI] [PubMed] [Google Scholar]
- 33.Travers A, Schneider R, Muskhelishvili G. DNA supercoiling and transcription in Escherichia coli: The FIS connection. Biochimie. 2001;83:213–217. doi: 10.1016/s0300-9084(00)01217-7. [DOI] [PubMed] [Google Scholar]
- 34.Lee AK, Detweiler CS, Falkow S. OmpR regulates the two-component system SsrA–ssrB in Salmonella pathogenicity island 2. J Bacteriol. 2000;182:771–781. doi: 10.1128/jb.182.3.771-781.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bijlsma JJ, Groisman EA. The PhoP/PhoQ system controls the intramacrophage type three secretion system of Salmonella enterica. Mol Microbiol. 2005;57:85–96. doi: 10.1111/j.1365-2958.2005.04668.x. [DOI] [PubMed] [Google Scholar]
- 36.Deiwick J, Nikolaus T, Erdogan S, Hensel M. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol Microbiol. 1999;31:1759–1773. doi: 10.1046/j.1365-2958.1999.01312.x. [DOI] [PubMed] [Google Scholar]
- 37.Garmendia J, Beuzon CR, Ruiz-Albert J, Holden DW. The roles of SsrA–SsrB and OmpR–EnvZ in the regulation of genes encoding the Salmonella typhimurium SPI-2 type III secretion system. Microbiology. 2003;149:2385–2396. doi: 10.1099/mic.0.26397-0. [DOI] [PubMed] [Google Scholar]
- 38.Kim CC, Falkow S. Delineation of upstream signaling events in the salmonella pathogenicity island 2 transcriptional activation pathway. J Bacteriol. 2004;186:4694–4704. doi: 10.1128/JB.186.14.4694-4704.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blanc-Potard AB, Groisman EA. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 1997;16:5376–5385. doi: 10.1093/emboj/16.17.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brodsky IE, Ernst RK, Miller SI, Falkow S. mig-14 is a Salmonella gene that plays a role in bacterial resistance to antimicrobial peptides. J Bacteriol. 2002;184:3203–3213. doi: 10.1128/JB.184.12.3203-3213.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishino K, Latifi T, Groisman EA. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol Microbiol. 2006;59:126–141. doi: 10.1111/j.1365-2958.2005.04940.x. [DOI] [PubMed] [Google Scholar]
- 42.Miller SI, Kukral AM, Mekalanos JJ. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci U S A. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heithoff DM, Conner CP, Hentschel U, Govantes F, Hanna PC, Mahan MJ. Coordinate intracellular expression of Salmonella genes induced during infection. J Bacteriol. 1999;181:799–807. doi: 10.1128/jb.181.3.799-807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mouslim C, Groisman EA. Control of the Salmonella ugd gene by three two-component regulatory systems. Mol Microbiol. 2003;47:335–344. doi: 10.1046/j.1365-2958.2003.03318.x. [DOI] [PubMed] [Google Scholar]
- 45.Cano DA, Martinez-Moya M, Pucciarelli MG, Groisman EA, Casadesus J, Garcia-Del Portillo F. Salmonella enterica serovar Typhimurium response involved in attenuation of pathogen intracellular proliferation. Infect Immun. 2001;69:6463–6474. doi: 10.1128/IAI.69.10.6463-6474.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miao EA, Freeman JA, Miller SI. Transcription of the SsrAB regulon is repressed by alkaline pH and is independent of PhoPQ and magnesium concentration. J Bacteriol. 2002;184:1493–1497. doi: 10.1128/JB.184.5.1493-1497.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beuzon CR, Unsworth KE, Holden DW. In vivo genetic analysis indicates that PhoP–PhoQ and the Salmonella pathogenicity island 2 type III secretion system contribute independently to Salmonella enterica serovar Typhimurium virulence. Infect Immun. 2001;69:7254–7261. doi: 10.1128/IAI.69.12.7254-7261.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knodler LA, Celli J, Hardt WD, Vallance BA, Yip C, Finlay BB. Salmonella effectors within a single pathogenicity island are differentially expressed and translocated by separate type III secretion systems. Mol Microbiol. 2002;43:1089–1103. doi: 10.1046/j.1365-2958.2002.02820.x. [DOI] [PubMed] [Google Scholar]
- 49.Cirillo DM, Valdivia RH, Monack DM, Falkow S. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol. 1998;30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- 50.Hautefort I, Thompson A, Eriksson-Ygberg S, Parker ML, Lucchini S, Danino V, Bongaerts RJ, Ahmad N, Rhen M, Hinton JC. During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell Microbiol. 2008;10:958–984. doi: 10.1111/j.1462-5822.2007.01099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chakravortty D, Rohde M, Jager L, Deiwick J, Hensel M. Formation of a novel surface structure encoded by Salmonella Pathogenicity Island 2. EMBO J. 2005;24:2043–2052. doi: 10.1038/sj.emboj.7600676. [DOI] [PMC free article] [PubMed] [Google Scholar]