Abstract
Reversible phosphorylation modulates nearly every step of glycogenesis and glycogenolysis. Multiple metabolic disorders are the result of defective enzymes that control these phosphorylation events, enzymes that were identified biochemically before the advent of the molecular biology era. Lafora disease is a metabolic disorder resulting in insoluble glucan accumulation in the cytoplasm, and manifests as a debilitating neurodegeneration that ends with the death of the patient. Unlike most metabolic disorders, the link between Lafora disease and metabolism has not been defined in almost 100 years. Recent results from mammalian cells, mouse models, eukaryotic algae, and plants have begun to define the molecular mechanisms that cause Lafora disease. The emerging theme identifies a new phosphorylation substrate in glycogen metabolism, the glucan itself.
Nearly a century of Lafora disease history
In 1911 Dr. Gonzalo Lafora, a student of Dr. Alois Alzheimer, reported autopsy results from patients with “teenage-onset myoclonus epilepsy with dementia” and described “amyloid bodies in the protoplasm of the ganglion cells” [1, 2]. Although amyloid was later shown to be proteinaceous, the term originally referred to any material that stained in a similar manner as starch, a mixture of amylose and amylopectin [3]. The deposit Dr. Lafora described was later shown to be an accumulation of insoluble glucans, i.e. polymers of glucose linked by glycosidic bonds, and named a Lafora body (LB) [4–6]. Like the “amyloid deposits”, the disease Dr. Lafora described now also bears his name, Lafora disease (LD) (OMIM 254780).
LD is an autosomal recessive neurodegenerative disorder resulting in severe epilepsy and death. It is one of five major progressive myoclonus epilepsies (PMEs) [7–10]. Unlike most other forms of epilepsy, LD is only moderately managed by medication for a brief period of time. LD commonly presents as a single seizure in the second decade of the patient’s life; this single event is followed by progressive central nervous system degeneration and ends with the death of the patient within ten years of the first seizure [8, 11–13].
LD is unique among the PMEs because of the patient’s rapid neurological deterioration and the accumulation of cytoplasmic LBs, which contain 80–93% polyglucans [1, 5]. LD is also unique among neurodegenerative diseases in that it involves formation of an inclusion body that is largely non-proteinaceous. Whereas LBs are found in the cytoplasm of cells from most tissues, clinical features of LD are confined to the CNS and non-neurologic symptoms are rare [12]. LD patients exhibit increased neuronal cell death, number of seizures, and LB accumulation as they age; thus it is hypothesized that LBs trigger these symptoms and ultimately the death of the patient [5].
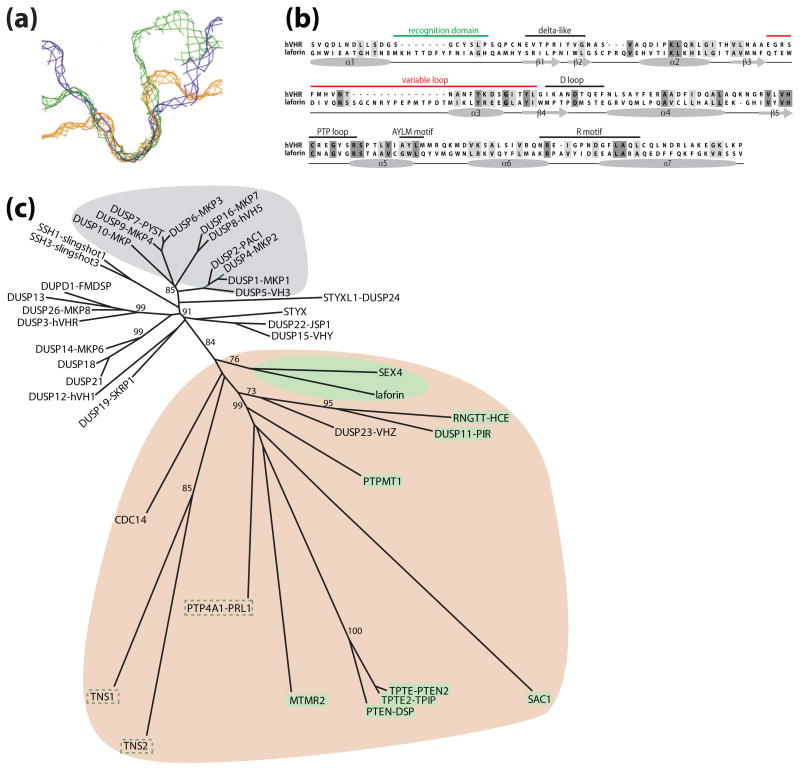
Two groups independently identified EPM2A (epilepsy, progressive myoclonus 2A) as a gene mutated in approximately 48% of LD cases [14, 15]. EPM2A encodes the bimodular protein laforin that contains a canonical dual specificity phosphatase (DSP) active site motif, HCXXGXXRS/T (Cx5R), and a carbohydrate binding module (CBM) (Fig. 1A). Accordingly, recombinant laforin is able to hydrolyze phosphotyrosine and phosphoserine/threonine substrates in vitro [16, 17]. The laforin amino-terminus contains a CBM belonging to family 20 (CBM20), that targets laforin to subcellular sites of glycogen synthesis and promotes the binding of laforin to glycogen both in vitro and in vivo [17]. Intriguingly, out of the 107 human protein tyrosine phosphatases (PTP superfamily), 15 human phosphoprotein phosphatases (PPP family), and 16 human phosphoprotein metallo-dependent phosphatases (PPM family) [18, 19], laforin is the only phosphatase that possesses a CBM of any type. Greater than 70% of proteins that contain a CBM are amylases, glucohydrolases, or cellulases (i.e. enzymes that act on the carbohydrate itself) of plant, fungal, bacterial, or parasitic origins [20–22]. Of the LD cases without mutations in EPM2A, 40% are the result of mutations in EPM2B (epilepsy, progressive myoclonus 2B), and 12% might have mutations in non-coding regions of EPM2A or EPM2B, could be the result of copy number variant, or could be attributed to mutations in an unidentified gene [23].
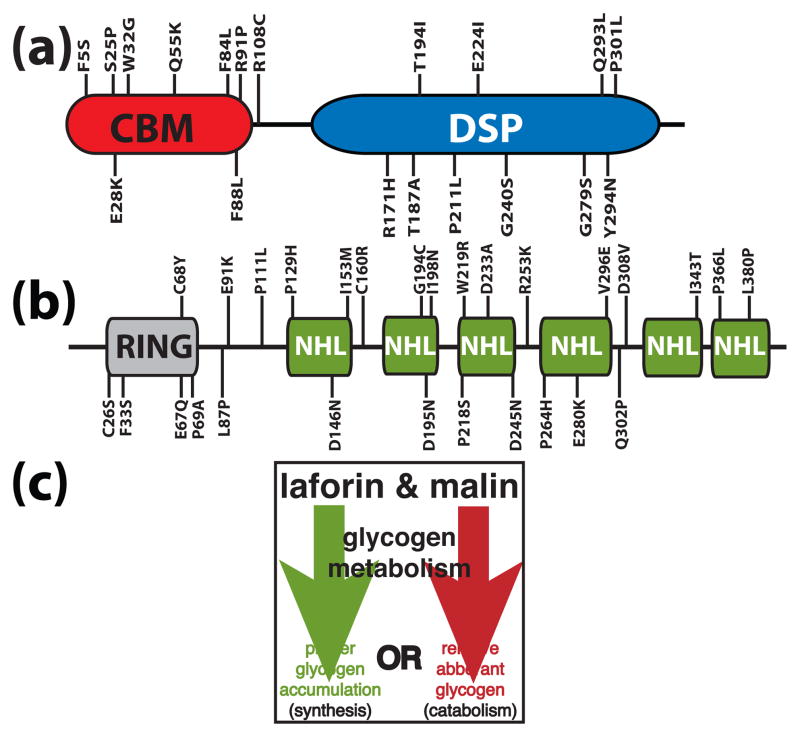
Figure 1.
A schematic of laforin and malin. Amino acid substitutions stemming from Lafora disease missense mutations are shown for laforin and malin. (a) Laforin contains a carbohydrate binding module (CBM) and a dual specificity phosphatase domain (DSP). (b) Malin contains a RING domain followed by six NHL repeats. (c) Malin and laforin are involved in one of the two branches of glycogen metabolism.
EPM2B encodes a 395 amino acid protein called malin [24]. Malin contains a consensus RING domain and six NHL domains (Fig. 1B). RING domains are characteristic of one class of E3 ubiquitin ligases [25]. NHL domains form a six-bladed β-propeller and are involved in protein-protein interactions, similar to WD40 repeats [26, 27]. We demonstrated that malin functions as an E3 ubiquitin ligase in vitro and in vivo [28]. We found that malin directly binds, ubiquitylates, and promotes the degradation of laforin [28]. This result was initially counter-intuitive as mutation of either malin or laforin results in LD. Why then would malin degrade laforin if they both inhibit Lafora disease? More recent data has allowed us to propose a model that better explains this perplexing result, and this work will be discussed in detail below in The Destructive Side of Laforin. In addition, malin regulates glycogen synthesis by ubiquitylating and promoting the degradation of enzymes that orchestrate glycogen synthesis: glycogen debranching enzyme (AGL/GDE), protein targeting to glycogen (PTG), and the muscle isoform of glycogen synthase, which is also expressed in neurons [29–33].
Two transgenic mouse models have been developed for LD. One disrupted Epm2a to generate null mice [34] and one generated transgenic mice by over-expressing inactivated laforin [35] in all tissues. The mouse models mimicked the human disease in that Lafora bodies are present and the mice develop epilepsy, but differ in the respect that the life span of the transgenic mice is not shortened. Neither study determined a molecular role for laforin in LD.
Although the mouse models did not determine the molecular etiology of LD, the data cumulatively placed laforin in the context of being involved in regulating glycogen metabolism. As laforin inhibits the formation of LBs, we and others proposed that laforin functions to either actively promote proper glycogen accumulation or to actively remove aberrant glycogen (Fig. 1C). Lafora initially proposed that the disease was a result of “abnormal metabolism” [1]. However, multiple studies have reported that all known enzymes involved in glycogen metabolism from LD patients posses normal activities [11, 36, 37]. Thus, it seems probable that LD is the result of a defect in a previously uncovered aspect of glycogen metabolism.
What is a Lafora body?
A glucan is one of a variety of complex carbohydrates composed of glucose moieties linked together by glycosidic bonds. One such glucan is glycogen. Glycogen is a branched polymer of glucose produced in the cytoplasm of the majority of archaebacterial, bacterial, fungal, and animal species and is an energy storage molecule. Most non-photosynthetic eukaryotes produce glycogen from UDPglucose, while most bacteria synthesize glycogen from ADPglucose. Glycogen is composed of α-1,4-glycosidic linkages between glucose residues, formed by glycogen synthase, with branches occurring in a continuous pattern every 12–14 residues via α-1,6-glycosidic linkages, formed by the branching enzyme [38]. The branches are referred to as tiers, with a single glycogen molecule being composed of up to twelve tiers [39, 40]. These characteristics make glycogen a water-soluble polymer (Fig 2A and Table 1). Interestingly, two groups reported that glycogen contains small amounts of phosphate, but the purpose of the phosphate or the enzymes responsible for the phosphate were never determined [41–43].
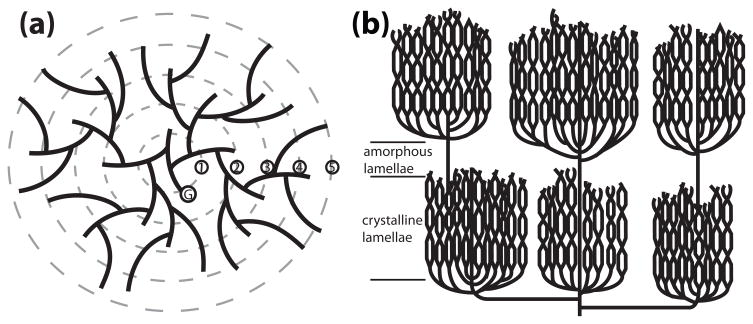
Figure 2.
Models of glycogen and amylopectin. A three dimensional structure of glycogen and starch cannot be determined experimentally due to their polydispersity, but these are the widely accepted models [98–100]. In each model, solid lines represent glucan chains. (a) Glycogen production is initiated when glycogenin (G) covalently attaches glucose to itself at Tyr194 and continues with the autocatalytic addition of about 10 glucosyl residues. This protein–glucosyl complex serves as the starting point that glycogen synthase and branching enzyme utilize to link glucose by α-1,4-glycosidic linkages with branches linked by α-1,6-glycosidic linkages every 12–14 residues. Glycogen synthase and branching enzyme construct up to twelve tiers of branches, five of which are depicted here. These tiers are organized in a continuous manner, rendering glycogen water soluble. (b) Amylopectin is also composed of α-1,4-glycosidic linkages with α-1,6-glycosidic branches, but with branches arranged in clusters at regular intervals. The glucan chains within the clusters interact and this is represented by intersection of the adjacent chains, which form double helices and organize into crystalline lamellae. Between each cluster is a non-branched region that makes up the amorphous lamellae. The decreased branching and the crystalline lamellae render amylopectin, and starch, water insoluble.
Table 1.
Biochemical and physical properties of glucans.
| residues/branch | branching pattern | water soluble | phosphate content | |
|---|---|---|---|---|
| Eukaryotic glycogen | 12–14 | continuous | yes | 0.064–0.25% w/w |
| Bacterial glycogen | 10–15 | continuous | yes | N.D. |
| Amylopectin | 12–25 | discontinuous | no | 0.1–0.5% w/w |
| Floridean starch | 12–20 | discontinuous | no | N.D. |
| Lafora body | 12–30+ | discontinuous | no | 0.35–1.0% w/w |
Starch is the functional equivalent of glycogen for photosynthetic eukaryotes. In green algae and higher plants, starch is produced in a plastid, one type of which is a chloroplast [44]. In contrast to glycogen, starch is an insoluble, semi-crystalline mixture of <10%% amylose and >80%% amylopectin produced in diurnal cycles in Arabidopsis leaves [45]. Amylose is a linear molecule with very few α-1,6-glycosidic linkages. Amylopectin, like glycogen, is composed of α-1,4-glycosidic linkages with α-1,6-glycosidic branches, but with branches arranged in clusters at regular intervals (Fig. 2B and Table 1). Within the clusters, adjacent chains form double helices and the clusters organize into crystalline lamellae. The decreased branching and the crystalline lamellae render amylopectin, and thus starch, water insoluble.
Although glycogen is the normal glucan storage molecule for animals, LBs are accumulations of poorly branched, insoluble glucans and are not defined as glycogen. In fact, studies from the 1960s defined the biochemical composition of LBs and characterized them as more closely resembling plant amylopectin than glycogen [5, 46, 47]. Therefore, a LB is an aberrantly formed glucan. In this sense, LBs are similar to misfolded proteinaceous accumulations seen in multiple neurodegenerative diseases. The biochemical characterization of LBs in the 1960s was largely overlooked, but this work clearly and convincingly demonstrated that LBs are more similar to plant amylopectin than animal glycogen [5, 46, 47]. This work spurred us to examine the literature on the composition of another insoluble glucan called floridean starch.
Floridean starch is synthesized from UDPglucose in the cytoplasm of a group of eukaryotic, non-photosynthetic organisms and photosynthetic red algae, all of which are derivatives of Kingdom Plantae/Archaeplastida [48–52]. Floridean starch was originally isolated from the multicellular red agla Florideophycidae and is composed of amylopectin and amylose [50, 53]. The major difference between floridean starch and starch is that floridean starch is generated in the cytoplasm and starch in plastids [44, 52]. Upon probing the genome of organisms that generate floridean starch, we found that laforin is conserved in a subset of protozoans, Toxoplasma gondii, Eimeria tenella, Tetrahymena thermophila, Paramecium tetraurelia, and Cyanidioschyzon merolae [54]. These organisms synthesize floridean starch and utilize it during a hibernation state of their life cycle. Thus, laforin is conserved in all vertebrates and in a small, defined group of protists. This finding caused us to re-examine the molecular role of laforin in Lafora disease and prompted us to investigate unique possibilities.
The substrate is the key
Although mouse models existed that faithfully mimicked Lafora disease, the molecular etiology of LD remained a mystery largely because the function of laforin was unknown, i.e. the substrate was not identified. Two very plausible hypotheses dominated the LD field.
As glycogen metabolism is driven by the coordinated activity of glycogen synthase (GS) and branching enzyme (BE), one hypothesis stated that LBs formed as a result of misregulation of one of these enzymes [12, 34, 55–57]. This hypothesis postulated that GS and BE were at opposite ends of a fulcrum and that misregulation of either one would lead to an accumulation of a glucan with decreased branching and decreased solubility, biochemical hallmarks of LBs. The best evidence for this model came from the surprising finding that overexpression of glycogen synthase in mouse muscle resulted in aberrant glycogen that resembled a LB [56, 57]. However, Roach and colleagues later definitively demonstrated that both arms of this pathway (GS and BE) are normal in a mouse lacking laforin [58]. In addition, multiple earlier studies examining patient tissue came to similar conclusions [11, 36, 37]. A second hypothesis postulated that laforin was involved in “destroying” LBs possibly by targeting them to lysosomes [12, 34, 35, 58–60]. This model was proposed by Ganesh et al. and Minassian and co-workers and was based on the observation that laforin preferentially binds LBs over glycogen. Later this hypothesis was bolstered by the work described above that demonstrated no changes in glycogen metabolizing enzymes in the LD mouse [23, 58, 59]. These groups put forth that laforin’s function begins after the appearance of “nascent-LBs” and that laforin is involved in monitoring and preventing the accumulation of LBs [23, 59]. Additionally, they speculated that laforin might promote the transport or destruction of “nascent-LBs” before they become detrimental. This hypothesis is supported by recent work demonstrating that deletion or mutation of laforin exacerbates the unfolded protein response due to endoplasm reticulum stress [61, 62].
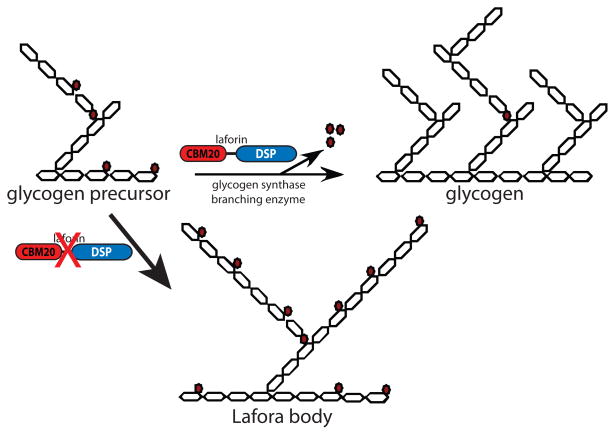
We proposed a third hypothesis when we discovered that laforin possesses the unique ability to dephosphorylate phospho-glucans [63]. We suggested that laforin functions to dephosphorylate glycogen molecules as they are synthesized. We postulated that in the absence of laforin glycogen becomes hyperphosphorylated, phosphate molecules disrupt and decrease normal branching, and a LB forms (Fig. 3). Surprisingly, two groups reported almost forty years ago that LBs from human patients contain increased phosphate and decreased branching compared to glycogen [46, 47]. Although LBs contain increased phosphate, no data exists that places laforin on the anabolism or catabolism side of glycogen metabolism (Fig. 1C). Thus, a similar theme as above, but one placing laforin on the catabolism side of glycogen metabolism, is equally as likely. In this scenario, glycogen metabolism enzymes would release glucose from glycogen and laforin would remove phosphate from glucose as it was exposed. During glycogen metabolism, the outer glucose tiers (Fig. 2A) are released and the inner tiers serve as the foundation for subsequent rounds of glycogen anabolism. One could envision that phosphate groups could block the action of glycogen catabolism enzymes, similar as to that recently described in plants and discussed below (Fig. 6A) [64]. In the absence of laforin, each round of glycogen metabolism would result in a slightly more phosphorylated glucan and would eventually result in a Lafora body. Roach and colleagues have suggested a similar model where laforin acts as part of a “repair or corrective mechanism” and in the absence of laforin glycogen gradually accumulates “structural defects” that eventually develop into LBs [65].
Figure 3.
Model of Lafora body formation caused by loss of laforin. Glucose moieties are depicted as hexagons. Glucose is linked by α-1,4-glycosidic linkages with branches via α-1,6-glycosidic linkages. Glycogen contains small amounts of covalently linked phosphate (0.25% w/w), present as both phosphomonoesters and phosphodiesters [41–43, 65]. Phosphate is represented by red-filled circles, with phosphomonesters adjacent glucose hexagons and phosphodiesters between two glucose hexagons. As nascent glycogen molecules are being synthesized by glycogen synthase and branching enzyme, phosphomonoesters and phosphodiesters accumulate by an unknown mechanism. Laforin removes phosphomonoesters so that glycogen production proceeds normally. In the absence of laforin, phosphomonoesters accumulate and negatively impact glycogen branching and lead to Lafora body (LB) formation. LBs contain increased amounts of phosphate and decreased branching compared to glycogen, and these two characteristics make LBs insoluble.
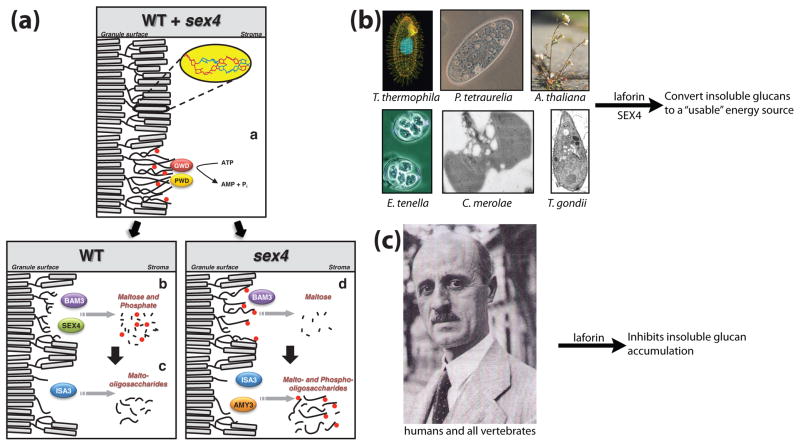
Figure 6.
Models depicting the role of laforin and SEX4 in glucan metabolism. (a) Proposed model of starch breakdown (Copyright American Society of Plant Biologists, www.plantcell.org) [64]. Starch is phosphorylated (red circles) at night by GWD and PWD (i), leading to unwinding of amylopectin double helices. In WT plants, β-amylase isozyme 3 (BAM3) and SEX4 release maltose and phosphate, respectively (ii), and isoamylase 3 (ISA3) hydrolyzes branch points and releases malto-oligosaccharides (iii). In sex4 mutants, phosphate is not hydrolyzed by SEX4, leading to reduced maltose release by BAM3 (iv). Subsequently, α-amylase (AMY3) and ISA3 release both malto- and phospho-oligosaccharides. Following degradation of the outer layer, a new round of degradationcan begin with the phosphorylation of the granule surface by GWD and PWD (i). (b) We propose that in plants and protists, laforin and SEX4 are involved in converting insoluble glucans into usable energy. (c) In humans, all other vertebrates, and at least two invertebrates (Nematostella and Branchiostoma), laforin inhibits insoluble glucan accumulation by dephosphorylating nascent glycogen molecules as proposed in Figure 3. The photograph of Dr. Gonzalo Rodriguez Lafora was and the image of a Tetrahymena was reproduced with permission [104]. All other images were generated by the authors or obtained from non-restricted copyright sources.
In support of the above glucan phosphatase models, Roach and colleagues confirmed the earlier reports of hyperphosphorylated LBs by demonstrating that the mouse lacking laforin has increased glucan phosphate and decreased branching compared to wild-type controls [65, 66]. In addition, they demonstrated that wild-type glycogen contains measurable amounts of phosphate and that laforin releases phosphate from glycogen. Therefore, the above two models are supported by both biochemical and patient data and have been recapitulated in a LD mouse model.
This hypothesis is bolstered by bioinformatic data and structural properties of laforin. The catalytic cleft of most dual specificity phosphatases (DSPs) is shallow and narrow, to accommodate both pSer/pThr and pTyr (Fig 4A). This architecture is typical of the DSPs that dephosphorylate proteinaceous substrates. Alternatively, this region is deep and narrow in protein tyrosine phosphatases so that they only accommodate pTyr and are not capable of dephosphorylating pSer/pThr (Fig. 4A). Phosphoinositol phosphatases exhibit a deep and wide catalytic cleft to accommodate the large phosphoinositol head groups (Fig. 4A) [71, 67–69]. The architecture of this cleft is largely due to the length of the recognition region and variable loop of the phosphatase, with DSPs possessing fewer amino acids in these regions [71, 70]. The recognition domain and variable loop of laforin are both twice as long as that of human VHR, a prototypical proteinaceous DSP (Fig 4B). Thus, laforin is predicted to have a deeper and wider cleft, more similar to phosphoinositol phosphatases (e.g. PTEN and the myotubularins) than to proteinaceous DSPs. This deeper and wider cleft could more easily accommodate phosphorylated glucans. Furthermore, when one generates a phylogeny of all DSPs using only the phosphatase domain there are three distinct clusters (Fig. 4C). The more evolutionarily recent “classical” DSPs cluster together tightly (grey), away from the more ancient and divergent “atypical” DSPs [71]. Within the atypical DSPs is a very divergent group (tan) that includes laforin. Many of the DSPs within this clade dephosphorylate non-proteinaceous substrates (e.g. phosphoinositols, RNA, and glucans; highlighted in green boxes), whereas others have non-defined substrates and/or have activity against non-proteinaceous substrates in vitro (green dashed line). Collectively, these structural qualities suggest that laforin does not dephosphorylate a proteinaceous substrate and support our finding that laforin is indeed a glucan phosphatase. However, the definitive structural data will come from a crystal structure of laforin.
Figure 4.
Structural and bioinformatic properties of laforin. (a) Slices of the active site surface of three classes of PTPs: 1) the deep and wide active site of the phosphoinositol phosphatase MTMR2 in blue, 2) the deep and narrow active site of the pTyr-specific phosphatase PTP1B in green, and 3) the shallow and narrow active site of the dual-specific phosphatase VHR in orange [101]. Used with permission from Current Opinion in Structural Biology. (b) An alignment and secondary structure prediction of human laforin and VHR (hVHR) were generated using PROMALS [102]. The accepted phosphatase motifs are indicated above each segment and the secondary structure is indicated below. Similar amino acids are boxed in light grey and identical amino acids in dark grey. The recognition domain and variable loop are highlighted in green and red, respectively. (c) A phylogeny built using the catalytic domain of the dual specificity phosphatases. The more recently evolved MAPK phosphatases or “classical” DSPs are highlighted with a grey background. The more ancient and divergent “atypical” DSPs fall into two groups. One group is relatively tightly clustered and utilizes proteinaceous substrates and this group has no highlighted background. The second group is more divergent and has a tan background, and includes laforin and SEX4. Most of the DSPs within this clade dephosphorylate non-proteinaceous substrates (e.g. phosphoinositols, RNA, and glucans), highlighted in green boxes. Some of the DSPs in ths clade have undefined in vivo substrates, but they have activity against non-proteinaceous substrates in vitro, highlighted with a green dashed line. The phylogenetic tree was generated from a PROMALS multiple sequence alignment using PROTDIST and FITCH from the PHYLIP 3.65 software package and displayed using HYPERTREE 1.0.0 [102, 103].
Although the above model resolves many questions about laforin and LD, a fourth hypothesis has recently been proposed. A mouse expressing simian virus 40 large tumor antigen was engineered with a transgenic rearranged T-cell receptor (TCR) [72]. These mice are immunocompromised and develop a high rate of lymphoma [73]. Zhang and colleagues later showed that the TCR transgene serendipitously inserted into the laforin gene locus [73]. They presented convincing data that laforin suppresses tumor growth in these immunocompromised mice. In addition, they presented data and stated that laforin dephosphorylates GSK3β, but did not recapitulate this in vitro using recombinant laforin. Instead, they overexpressed laforin in HEK293 cells, immunoprecipitated it, and showed that this mixture of proteins dephosphorylated a 20-mer peptide containing pSer9 of GSK3β. Subsequently, both ourselves as well as Roach and colleagues demonstrated that laforin does not dephosphorylate GSK3β at Ser9 in vitro, nor is there an increase in phosphorylation of GSK3β at Ser9 in multiple tissues from laforin deficient mice [63, 66]. In contrast to these findings, using wild-type and laforin-deficient MEFs, Zheng and Minassian subsequently published that laforin dephosphorylates Ser9 of GSK3β [74]. Surprisingly, they did not examine the state of GSK3β Ser9 in tissue from wild-type versus laforin-deficient mice.
Although there is no consensus concerning the endogenous substrate(s) of laforin, it is striking that no one has reported an increase in tumors in either LD patients or laforin-deficient mice. In addition, Roach and colleagues contributed a convincing correlative study where they took multiple lines of transgenic mice that accumulated more glycogen than normal and found that laforin protein levels increase with glycogen levels, suggesting that more laforin is “needed” as glycogen levels increase to presumably dephosphorylates the excess glycogen [75]. The exact molecular etiology of Lafora disease is still unclear and each of the above models could contribute to the pathology of the disease. However, we feel the data presented above strongly support laforin as a glucan phosphatase and that this role at least partially explains how laforin inhibits LD.
Lessons from plants and protists
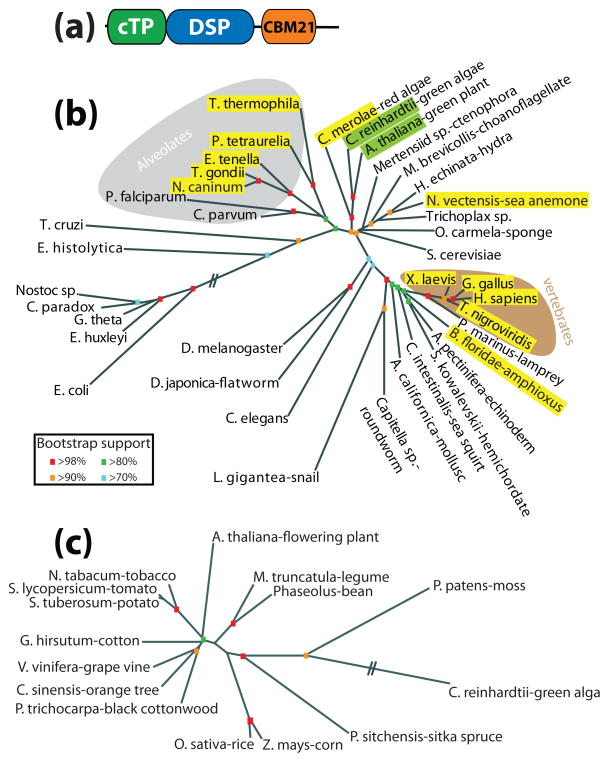
It is rare and very informative when fields as diverse as neuroscience and plant starch metabolism intersect. Niittyla et al. discovered a gene in plants they called starch excess 4 [SC1](SEX4) that contains a DSP followed by a CBM, the same domains as laforin but in the opposite orientation (Fig. 5A) [76]. Strikingly, mutations in SEX4 result in a similar cellular phenotype seen in LD patients, namely an increase in insoluble glucans. We went on to demonstrate that SEX4 has the same biochemical properties as laforin (i.e. it binds glucans, possesses phosphatase activity, and releases phosphate from glucans) and showed that laforin is a functional equivalent of SEX4 by rescuing the plant phenotype with human laforin [54].
Figure 5.
Evolutionary conservation of laforin and SEX4. (a) Schematic of SEX4. SEX4 is composed of a chloroplast targeting peptide (cTP), dual specificity phosphatase domain (DSP), and carbohydrate binding module family 21 (CBM21). (b) Unrooted phylogeny of the small subunit ribosomal RNA (SSU rRNA) from organisms representing many evolutionary niches (modified [78]). Organisms containing laforin are boxed in yellow and those containing SEX4 are boxed in green. Alveolates are shaded with a grey background and vertebrates with a brown background. Bootstrap values are indicated by color coding in the inset. (c) Unrooted phylogeny of all SEX4 orthologs. Bootstrap values are as in b. The phylogenetic trees were generated as in Figure 4. Double hash marks indicate a place where intervening segment was removed due to space limitations.
The Arabidopsis experiments were initiated when we discovered the gene encoding laforin is not confined to vertebrates, as previously thought, but that it is also present in a small set of protists (Fig. 5B) [54]. These protists all produce floridean starch, which is very similar to LBs and plant amylopectin (Table 1) [48, 77]. We found that each protist that contains laforin produces floridean starch. Conversely, protists that do not produce floridean starch or a similar glucan lack laforin. Thus, laforin is absent in the majority of protists, including yeast. Although laforin is conserved in vertebrates and a small subset of non-vertebrate organisms, SEX4 is found in all organisms of green algal descent (Fig. 5C) [78]. The fact that SEX4 is conserved in all members of Archaeplastida/Kingdom Plantae argues for a conserved necessary function from unicellular alga to multicellular plants.
Our understanding of the role of glycogen phosphorylation is still in its infancy, but plant researchers have elucidated a mechanistic cause and effect for starch phosphorylation. Arabidopsis has two kinases that directly phosphorylate starch. Glucan water dikinase (GWD) transfers the β-phosphate of ATP onto the C6 position of glucose in starch. Similarly, phosphoglucan water dikinase (PWD) phosphorylates the C3 position after GWD has phosphorylated the C6 position [79–84]. Mutations in either GWD or PWD result in a similar starch excess accumulation as seen in plants with mutations in SEX4. The emerging theme of starch phosphorylation is described in detail in Figure 6A, but simply stated it appears that glucan phosphorylation solubilizes the outer surface and allows access to the degradation machinery, then phosphate is removed at the C3 and/or C6 position by SEX4 so that another round of degradation can begin [64].
Although much progress has been made regarding glucan phosphorylation in plants, the very existence of phosphate in glycogen remained murky until the 1980s when two groups definitively showed that phosphate is present in both a mono- and diester form [41, 43, 85]. As glycogen, like starch, contains both phosphate and a phosphatase to remove phosphate, one could envision a similar theme for glycogen as described in Figure 6A for starch. Towards this end, we and others have performed bioinformatics searches to identify vertebrate homologs of GWD and/or PWD, but none have been identified to date. However, one group identified an activity in rabbit skeletal muscle that positions glucose 1-phosphate on the C6 of glucose residues in glycogen and likely accounts for the phosphodiester in glycogen [43]. They named this enzme UDPglucose:glycogen glucose 1-phosphotransferase, but it was never purified. They also proposed that phosphomonoester groups in glycogen could arise by removal of glucose moieties originally transferred as glucose 1-phosphate. The phosphatase activity of laforin could be necessary to counter-balance these events.
Collectively, it seems that laforin and SEX4 are involved in degrading insoluble glucans. We propose that in protists and plants, laforin and SEX4 dephosphorylate glucans during catabolism, that this event is downstream of the action of GWD and PWD, and that dephosphorylation is necessary for energy production through release of the stored glucans (Fig. 6B). Similarly, in vertebrates laforin is inhibiting or degrading insoluble glucans before detrimental Lafora bodies form (Fig. 6C). We feel it is probable that laforin dephosphorylates nascent glycogen molecules, as presented in Figure 3, but the details of this model have not been fully elucidated and the exact mechanism in plants and humans likely differ to some degree.
The destructive side of laforin
Although glucan dephosphorylation is likely part of the molecular mechanism driving LD, it is not the entire story. As previously stated, we found that malin directly binds laforin, ubiquitylates it, and triggers the degradation of laforin [28]. Based purely on genetics, this finding is unexpected, as both malin and laforin inhibit LB formation. Why would malin trigger the destruction of laforin?
We and others have noted that laforin binds glucans very efficiently in vitro and once bound might not readily release [17, 60]. If laforin functions to dephosphorylate glycogen as it forms, ubiquitylation could be a method to release laforin from glycogen. Once released, laforin could be de-ubiquitylated and re-cycled, or it could be targeted for degradation.
In addition to ubiquitylating laforin, we and others found that malin has other targets, although this finding is disputed [65]. The targets of malin are all involved in glycogen metabolism, they include protein targeting to glycogen (PTG), glycogen synthase (GS), and glycogen debranching enzyme (AGL/GDE) [29–32]. Surprisingly, laforin is needed for malin to ubiquitylate and trigger degradation of PTG and GS [29, 30, 32]. Therefore, laforin acts as a scaffold to bring malin to additional substrates and malin may ubiquitinate multiple proteins concentrated in this area. Thus, the laforin–malin complex might act as a controlled “garbage” disposal to ubiquitylate and degrade many proteins involved in glycogen metabolism. This is a mechanism that might be shared with other E3 ubiquitin ligases given their propensity to ubiquitinate multiple substrates.
One signal that regulates these events was recently described by Sanz and colleagues when they found that AMP-activated protein kinase (AMPK) phosphorylates PTG [33, 86]. AMPK is a heterotrimeric protein that senses and responds to perturbations in both the cellular AMP:ATP ratio as well as glycogen stores, and thus is considered a key regulator of energy metabolism [87, 88]. Sanz also demonstrated that phosphorylation of PTG by AMPK increases the malin-laforin-dependent degradation of PTG [33]. Not only did they provide biochemical data from tissue culture models, but they also confirmed these findings by using LD patient data significantly strengthening this previously disputed result. Therefore, the signals that regulate these degradation events and the timing of the events are currently being elucidated.
Why do only neurons die?
Whereas glycogen is generated in virtually all liver and skeletal muscle cells, glycogen is only generated in astrocytes and not in neurons in the mature brain [89]. Paradoxically, whereas neurons do not produce or store glycogen, neurons of LD patients accumulate LBs and are the only cells in LD patients that are reported to exhibit a cellular phenotype [34]. Recent work by Guinovart and co-workers solved the perplexing problem of how neurons were capable of generating LBs without generating glycogen. They demonstrated that neurons express low levels of muscle glycogen synthase (MGS) and that MGS in neurons is kept in an inactive, hyperphosphorylated state [30]. In addition, they demonstrated that a malin–laforin complex utilizes ubiquitylation to ensure low levels of MGS in neurons and they elegantly showed that when MGS is dephosphorylated by protein phosphatase 1 (PP1) an aberrant poorly branched polyglucan forms, eventually leading to LBs in LD patients. This work greatly expanded our knowledge of neuronal metabolism, but it did not define why only neurons undergo cell death in LD patients. The molecular mechanism that triggers neuronal apoptosis in LD patients is unknown, but we present four hypotheses that are not mutually exclusive. Given that LD symptoms take 15 years to manifest it may be that multiple mechanisms contribute to the pathogenesis.
Glial cells outnumber neurons approximately 10:1 and provide them with most energy needs. Neurons store minimal to no glycogen and have increased energy needs due to numerous ion channels, and inherently live in an “energy crisis.” Thus, glial glycogen is thought of as a safety net that ensures neurons maintain an energy source during periods of intense activation (reviewed in [89, 90]). The degree of branching and the length of glucose chains in glycogen are optimized for maximal glucose storage in the smallest volume and maximal energy release [39, 91]. As neurons have very limited energy stores and are both dependent on glial glycogen and utilize large amounts of energy, they are hypersensitive to energy perturbations. LBs likely accumulate a significant amount of “trapped” and “unusable” energy. If this trapped energy causes a temporary disruption in energy release, then cells that lack their own energy stores, e.g. neurons, would be the first to undergo apoptosis. In this scenario, neurons are the “canary in the coalmine” and are responding to decreased energy availability. Because neuronal apoptosis leads to an early death of the patient then no other cells have a chance to undergo apoptosis or cell death.
A second possibility is that LBs present a major trafficking problem. Whereas a neuron is 4–100 μ in diameter, neuronal LBs range from 3 to 40 μ [13]. Therefore, LBs might form a blockade in the cytoplasm. In addition, the distance neurons transport intracellular cargos is considerably further than other cells. Thus, a trafficking defect could first present in them and result in neuronal apoptosis.
Third, neuronal death in LD patients could be the only cellular phenotype because of the age of neurons. Neurons abolish mitotic division and have a drastically increased lifespan compared to most other cell types. Neurons in LD patients might be the only cells undergoing apoptosis because of their advanced “age”, and other cell types do not live long enough to experience the detrimental affects. This hypothesis would explain why neurons in murine models that exhibit LBs do not undergo wide-spread apoptosis and why LD mouse and dog models do not die at a young age [92–94]. The lifespan of murine and canine neurons is not as long as human neurons and would not be long enough for LBs to cause massive apoptosis, which takes 15+ years in humans.
Lastly, it is possible that LBs are not the cause of LD, but rather a cellular defense mechanism to sequester and dispose of aberrantly folded glucans. This hypothesis has gained support among researchers studying the multiple neurodegenerative diseases involving proteinaceous accumulations. Corroborating this hypothesis in LD is the fact that not all neurons that undergo cell death in the laforin deficient mouse model have visible LBs. This result could mean that LBs are the end result of multiple aberrant steps of glycogen synthesis, and that the earlier, non-visible products are the pathogenic cause of neuronal apoptosis. Alternatively, LBs might not be the pathogenic cause of LD. Ganesh and colleagues have suggested that a defect in autophagy or the ubiquitin proteasome system is a driving force in LD and LD pathology [95, 96]. However, their studies examining autophagy in cell models utilize overexpressed proteins and treatment with proteasomal inhibitors, thus the data regarding this hypothesis are not yet entirely convincing. Nonetheless, it is striking that like the proteinopathies, mutations in an E3 ubiquitin ligase, malin, result in LD.
Concluding remarks and future perspectives
Collectively, biochemistry, mouse models, cell biology, and LD patient data suggest two essential roles for laforin, 1) dephosphorylation of glycogen, or nascent glucans, to inhibit excess glycogen phosphorylation and LB formation, and 2) recruitment of malin to the site of glycogen synthesis so that malin can ubiquitinate PTG, GS, AGL, laforin, and possibly other proteins to inhibit LB formation. Thus, laforin performs two essential functions and malin one in maintaining proper glycogen metabolism. This idea of laforin having two roles is also supported by LD patient data. LD patients with mutations in the malin gene live 25% longer than patients with laforin mutations [97]. Thus, these data suggest that the function of laforin could be downstream of malin, or that laforin plays a disproportionate role in glycogen metabolism.
As many pathways are regulated by ubiquitylation, it is not overtly surprising that ubiquitination also regulates glycogen metabolism. However, we now must identify and define both the auxiliary proteins mediating these events and the signals that regulate these proteins at the cellular, tissue, and organismal levels. As discussed above, an emerging regulator of these events is AMPK; however, the extent to which AMPK orchestrates this regulation is still being determined.
A surprising discovery from the Lafora disease field is the identification of a glucan phosphatase activity that is conserved from plants to humans. Although the picture is becoming increasingly clear as to how plants utilize glucan phosphorylation and dephosphorylation to store and release energy, respectively, it is not clear how or why vertebrate glucans become phosphorylated. Is there a glucan kinase in vertebrates, similar to GWD and PWD in plants? Multiple groups have performed bioinformatic searches and have yet to identify a similar kinase in vertebrates. Alternatively, the actions of a phospho-glucotransferase could result in glycogen phosphorylation, but this enzyme has not been identified. Is glycogen phosphorylation the result of an evolutionary remnant, i.e. a mistake, or does it have an undefined purpose? Lastly, as yeast, flies, and worms all lack laforin and malin, how do they deal with insoluble glucan accumulations and can we identify an alternative pathway in these model organisms? The answers to these questions will further define Lafora disease at a molecular level, likely uncover potential therapies, will further identify similarities and differences between glycogen and starch metabolism, and may provide mechanisms to modulate glucan (i.e. energy) production in a variety of organisms.
Acknowledgments
We apologize to colleagues whose work we were not able to cite due to space constraints. The authors would like to acknowledge the support of NIH grants NS061803, RR0202171, and University of Kentucky College of Medicine startup funds (to M.S.G.); and NIH grant DK0118849, the Walther Cancer Institute, and the Howard Hughes Medical Institute (to J.E.D.).
Glossary
- CBM
a carbohydrate binding module. CBMs are defined by their tertiary fold, which allows them to bind to one or many types of carbohydrates. They are classified into one of fifty-three families based on amino acid similarity, substrate binding preferences, polypeptide folds, and evolutionary relationships
- CX5R
the catalytic active site motif of the protein tyrosine phosphatase (PTP) superfamily
- DSP
the dual specificity phosphatases are a heterogeneous group of phosphatases that are more evolutionarily diverse than the classical PTPs. They dephosphorylate pTyr, pSer, pThr, and non-proteinaceous substrates (e.g. phosphoinositols, RNA, and glucans)
- Glucan
polymer of glucose linked by glycosidic bonds, e.g. starch, glycogen, amylopectin, cellulose, Lafora body
- GWD
α-glucan, water dikinase. A plant kinase that transfers the β-phosphate of ATP to the C6 position of glucose in starch
- Lafora body
insoluble glucan that closely resembles plant amylopectin and accumulates in the cytoplasm of most cells in LD patients
- Protist
a diverse group of eukaryotic organisms with a unicellular level of organization
- PTP
the protein tyrosine phosphatase superfamily, which is encoded by the largest family of phosphatase genes. PTPs are defined by the active-site motif CX5R in which the cysteine functions as a nucleophile and is essential for activity. They are divided into the classical PTPs that dephosphorylate pTyr, and the DSPs that dephosphorylate pTyr, pSer, pThr, and non-proteinaceous substrates
- PWD
phosphoglucan, water dikinase. A plant kinase that transfers the β-phosphate of ATP to the C3 position of glucose in starch subsequent to phosphorylation of the C6 position
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lafora G, Glick G. Beitrag zur histopathologie der myoklonischen epilepsie. Z Ges Neurol Psychiatr. 1911;6:1–14. [Google Scholar]
- 2.Lafora GR. Uber des Vorkommen amyloider KJrperchen im innern der Ganglienzellen. Virchows Arch f Path Anat. 1911;205:295. [Google Scholar]
- 3.Virchow RLK. Die Cellularpathologie inihrer Begründung auf physiologische and pathologische Gewebelehre. Berlin: Hirschwald; 1858. [Google Scholar]
- 4.Yokoi S, Austin J, Witmer F. Isolation and characterization of Lafora bodies in two cases of myoclonus epilepsy. Journal of Neuropathology and Experimental Neurology. 1967;26(1):125–127. [PubMed] [Google Scholar]
- 5.Yokoi S, Austin J, Witmer F, Sakai M. Studies in myoclonus epilepsy (Lafora body form). I. Isolation and preliminary characterization of Lafora bodies in two cases. Arch Neurol. 1968;19(1):15–33. doi: 10.1001/archneur.1968.00480010033002. [DOI] [PubMed] [Google Scholar]
- 6.Harriman DG, Millar JH, Stevenson AC. Progressive familial myoclonic epilepsy in three families: its clinical features and pathological basis. Brain. 1955;78(3):325–349. doi: 10.1093/brain/78.3.325. [DOI] [PubMed] [Google Scholar]
- 7.Hodskins MB, et al. Anatomico-clinical observations on myclonus in epileptics and on related symptom complexes. American Journal of Psychiatry. 1930;86:827–848. [Google Scholar]
- 8.Berkovic SF, So NK, Andermann F. Progressive myoclonus epilepsies: clinical and neurophysiological diagnosis. J Clin Neurophysiol. 1991;8(3):261–274. [PubMed] [Google Scholar]
- 9.Harriman DGHaM, JHD Progressive familial myclonic epilepsy in three families: Its clinical features and patholgical basis. Brain. 1955;78:325–349. doi: 10.1093/brain/78.3.325. [DOI] [PubMed] [Google Scholar]
- 10.Berkovic SF, Andermann F, Carpenter S, Wolfe LS. Progressive myoclonus epilepsies: specific causes and diagnosis. N Engl J Med. 1986;315(5):296–305. doi: 10.1056/NEJM198607313150506. [DOI] [PubMed] [Google Scholar]
- 11.Janeway R, Ravens JR, Pearce LA, Odor DL, Suzuki K. Progressive myoclonus epilepsy with Lafora inclusion bodies. I. Clinical, genetic, histopathologic, and biochemical aspects. Arch Neurol. 1967;16(6):565–582. doi: 10.1001/archneur.1967.00470240003001. [DOI] [PubMed] [Google Scholar]
- 12.Minassian BA. Lafora’s disease: towards a clinical, pathologic, and molecular synthesis. Pediatr Neurol. 2001;25(1):21–29. doi: 10.1016/s0887-8994(00)00276-9. [DOI] [PubMed] [Google Scholar]
- 13.Van Heycop Ten Ham MW. Lafora disease, a form of progressive myoclonus epilepsy. In: Vinken PJaB GW, editor. Handbook of Clinical neurology. Vol. 15. Holland, Amsterdam: North Holland Publishing Company; 1975. pp. 382–422. [Google Scholar]
- 14.Minassian BA, Lee JR, Herbrick JA, Huizenga J, Soder S, Mungall AJ, Dunham I, Gardner R, Fong CY, Carpenter S, et al. Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy. Nat Genet. 1998;20(2):171–174. doi: 10.1038/2470. [DOI] [PubMed] [Google Scholar]
- 15.Serratosa JM, Gomez-Garre P, Gallardo ME, Anta B, de Bernabe DB, Lindhout D, Augustijn PB, Tassinari CA, Malafosse RM, Topcu M, et al. A novel protein tyrosine phosphatase gene is mutated in progressive myoclonus epilepsy of the Lafora type (EPM2) Hum Mol Genet. 1999;8(2):345–352. doi: 10.1093/hmg/8.2.345. [DOI] [PubMed] [Google Scholar]
- 16.Ganesh S, Agarwala KL, Ueda K, Akagi T, Shoda K, Usui T, Hashikawa T, Osada H, Delgado-Escueta AV, Yamakawa K. Laforin, defective in the progressive myoclonus epilepsy of Lafora type, is a dual-specificity phosphatase associated with polyribosomes. Hum Mol Genet. 2000;9(15):2251–2261. doi: 10.1093/oxfordjournals.hmg.a018916. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Stuckey JA, Wishart MJ, Dixon JE. A unique carbohydrate binding domain targets the lafora disease phosphatase to glycogen. J Biol Chem. 2002;277(4):2377–2380. doi: 10.1074/jbc.C100686200. [DOI] [PubMed] [Google Scholar]
- 18.Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein Tyrosine Phosphatases in the Human Genome. Cell. 2004;117(6):699. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 19.Almo SC, et al. Structural genomics of protein phosphatases. J Struct Funct Genomics. 2007;8:121–140. doi: 10.1007/s10969-007-9036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boraston AB, Bolam DN, Gilbert HJ, Daview GJ. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. The Biochemical Journal. 2004;382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coutinho PM, Henrissat B. Carbohydrate-active enzymes: an integrated database approach. In: Gilbert GDHJ, Henrissat B, Svensson B, editors. Recent Advances in Carbohydrate Bioengineering. Cambridge: The Royal Society of Chemistry; 1999. pp. 3–12. [Google Scholar]
- 22.Rodriguez-Sanoja R, Oviedo N, Sanchez S. Microbial starch-binding domain. Curr Opin Microbiol. 2005;8(3):260–267. doi: 10.1016/j.mib.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 23.Chan EM, Omer S, Ahmed M, Bridges LR, Bennett C, Scherer SW, Minassian BA. Progressive myoclonus epilepsy with polyglucosans (Lafora disease): evidence for a third locus. Neurology. 2004;63(3):565–567. doi: 10.1212/01.wnl.0000133215.65836.03. [DOI] [PubMed] [Google Scholar]
- 24.Chan EM, Young EJ, Ianzano L, Munteanu I, Zhao X, Christopoulos CC, Avanzini G, Elia M, Ackerley CA, Jovic NJ, et al. Mutations in NHLRC1 cause progressive myoclonus epilepsy. Nat Genet. 2003;35(2):125–127. doi: 10.1038/ng1238. [DOI] [PubMed] [Google Scholar]
- 25.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 26.Edwards TA, Wilkinson BD, Wharton RP, Aggarwal AK. Model of the brain tumor-Pumilio translation repressor complex. Genes Dev. 2003;17(20):2508–2513. doi: 10.1101/gad.1119403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slack FJ, Ruvkun G. A novel repeat domain that is often associated with RING finger and B-box motifs. Trends Biochem Sci. 1998;23(12):474–475. doi: 10.1016/s0968-0004(98)01299-7. [DOI] [PubMed] [Google Scholar]
- 28.Gentry MS, Worby CA, Dixon JE. Insights into Lafora disease: malin is an E3 ubiquitin ligase that ubiquitinates and promotes the degradation of laforin. Proc Natl Acad Sci U S A. 2005;102(24):8501–8506. doi: 10.1073/pnas.0503285102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Solaz-Fuster MD, Gimeno-Alcaniz JV, Ros S, Fernandez-Sanchez ME, Garcia-Fojeda B, Garcia OC, Vilchez D, Dominguez J, Garcia-Rocha M, Sanchez-Piris M, et al. Regulation of glycogen synthesis by the laforin-malin complex is modulated by the AMP-activated protein kinase pathway. Hum Mol Genet. 2007 doi: 10.1093/hmg/ddm339. [DOI] [PubMed] [Google Scholar]
- 30.Vilchez D, Ros S, Cifuentes D, Pujadas L, Valles J, Garcia-Fojeda B, Criado-Garcia O, Fernandez-Sanchez E, Medrano-Fernandez I, Dominguez J, et al. Mechanism suppressing glycogen synthesis in neurons and its demise in progressive myoclonus epilepsy. Nat Neurosci. 2007;10(11):1407–1413. doi: 10.1038/nn1998. [DOI] [PubMed] [Google Scholar]
- 31.Cheng A, Zhang M, Gentry MS, Worby CA, Dixon JE, Saltiel AR. A role for AGL ubiquitination in the glycogen storage disorders of Lafora and Cori’s disease. Genes Dev. 2007;21(19):2399–2409. doi: 10.1101/gad.1553207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Worby CA, Gentry MS, Dixon JE. Malin decreases glycogen accumulation by promoting the degradation of protein targeting to glycogen (PTG) J Biol Chem. 2008;283(7):4069–4076. doi: 10.1074/jbc.M708712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vernia S, Solaz-Fuster MC, Gimeno-Alcaniz JV, Rubio T, Garcia-Haro L, Foretz M, Rodriguez de Cordoba S, Sanz P. AMP-activated protein kinase phosphorylates R5/PTG, the glycogen targeting subunt of the R5/PTG-PP1 holoenzyme and accelerates its downregulation by the laforin-malin complex. J Biol Chem. 2009 doi: 10.1074/jbc.M808492200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ganesh S, Delgado-Escueta AV, Sakamoto T, Avila MR, Machado-Salas J, Hoshii Y, Akagi T, Gomi H, Suzuki T, Amano K, et al. Targeted disruption of the Epm2a gene causes formation of Lafora inclusion bodies, neurodegeneration, ataxia, myoclonus epilepsy and impaired behavioral response in mice. Hum Mol Genet. 2002;11(11):1251–1262. doi: 10.1093/hmg/11.11.1251. [DOI] [PubMed] [Google Scholar]
- 35.Chan EM, Ackerley CA, Lohi H, Ianzano L, Cortez MA, Shannon P, Scherer SW, Minassian BA. Laforin preferentially binds the neurotoxic starch-like polyglucosans, which form in its absence in progressive myoclonus epilepsy. Hum Mol Genet. 2004;13(11):1117–1129. doi: 10.1093/hmg/ddh130. [DOI] [PubMed] [Google Scholar]
- 36.Coleman DL, Gambetti P, Mauro SD, Blume RE. Muscle in Lafora disease. Arch Neurol. 1974;31(6):396–406. doi: 10.1001/archneur.1974.00490420062007. [DOI] [PubMed] [Google Scholar]
- 37.Gambetti P, Di Mauro S, Hirt L, Blume RP. Myoclonic epilepsy with lafora bodies. Some ultrastructural, histochemical, and biochemical aspects. Arch Neurol. 1971;25 (6):483–493. doi: 10.1001/archneur.1971.00490060017002. [DOI] [PubMed] [Google Scholar]
- 38.Roach PJ, Skurat AV, Harris RA. Regulation of glycogen metabolism. In: Jefferson LS, Cherrington AD, editors. The Endocrine Pancreas and Regulation of Metabolism. New York, NY: Oxford University Press, Inc; 2001. pp. 609–647. [Google Scholar]
- 39.Melendez R, Melendez-Hevia E, Cascante M. How did glycogen structure evolve to satisfy the requirement for rapid mobilization of glucose? A problem of physical constraints in structure building. J Mol Evol. 1997;45(4):446–455. doi: 10.1007/pl00006249. [DOI] [PubMed] [Google Scholar]
- 40.Shearer J, Graham TE. Novel aspects of skeletal muscle glycogen and its regulation during rest and exercise. Exerc Sport Sci Rev. 2004;32(3):120–126. doi: 10.1097/00003677-200407000-00008. [DOI] [PubMed] [Google Scholar]
- 41.Fontana JD. The presence of phosphate in glycogen. FEBS Lett. 1980;109(1):85–92. doi: 10.1016/0014-5793(80)81317-2. [DOI] [PubMed] [Google Scholar]
- 42.Lomako J, Lomako WM, Kirkman BR, Whelan WJ. The role of phosphate in muscle glycogen. Biofactors. 1994;4(3–4):167–171. [PubMed] [Google Scholar]
- 43.Lomako J, Lomako WM, Whelan WJ, Marchase RB. Glycogen contains phosphodiester groups that can be introduced by UDPglucose: glycogen glucose 1-phosphotransferase. FEBS Lett. 1993;329(3):263–267. doi: 10.1016/0014-5793(93)80234-l. [DOI] [PubMed] [Google Scholar]
- 44.Ball SG, Morell MK. FROM BACTERIAL GLYCOGEN TO STARCH: Understanding the Biogenesis of the Plant Starch Granule. Annual Review of Plant Biology. 2003;54 (1):207–233. doi: 10.1146/annurev.arplant.54.031902.134927. [DOI] [PubMed] [Google Scholar]
- 45.Zeeman SC, Smith SM, Smith AM. The diurnal metabolism of leaf starch. Biochemical Journal. 2007;401:13–28. doi: 10.1042/BJ20061393. [DOI] [PubMed] [Google Scholar]
- 46.Schnabel R, Seitelberger F. Histophysical and histochemical investigations of myoclonus bodies. Pathol Eur. 1968;3(2):218–226. [PubMed] [Google Scholar]
- 47.Sakai M, Austin J, Witmer F, Trueb L. Studies in myoclonus epilepsy (Lafora body form). II. Polyglucosans in the systemic deposits of myoclonus epilepsy and in corpora amylacea. Neurology. 1970;20(2):160–176. doi: 10.1212/wnl.20.2.160. [DOI] [PubMed] [Google Scholar]
- 48.Coppin A, Varré J, Lienard L, Dauvillée D, Guérardel Y, Soyer-Gobillard M, Buléon A, Ball S, Tomavo Stanislas. Evolution of Plant-Like Crystalline Storage Polysaccharide in the Protozoan Parasite Toxoplasma gondii Argues for a Red Alga Ancestry. Journal of Molecular Evolution. 2005;60(2):257–267. doi: 10.1007/s00239-004-0185-6. [DOI] [PubMed] [Google Scholar]
- 49.Guérardel Y, Leleu D, Coppin A, Liénard L, Slomianny C, Strecker G, Ball S, Tomavo S. Amylopectin biogenesis and characterization in the protozoan parasite Toxoplasma gondii, the intracellular development of which is restricted in the HepG2 cell line. Microbes and Infection. 2005;7(1):41–48. doi: 10.1016/j.micinf.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 50.Meeuse BJD, Andries M, Wood JA. Floridean Starch. J Exp Bot. 1960;11(2):129–140. [Google Scholar]
- 51.Nyvall P, Pelloux P, Davies HV, Pedersen M, Roberto Viola. Purification and characterisation of a novel starch synthase selective for uridine 5′-diphosphate glucose from the red alga Gracilaria tenuistipitata. Planta. 1999;209:143–152. doi: 10.1007/s004250050616. [DOI] [PubMed] [Google Scholar]
- 52.Viola R, Nyvall P, Pedersén M. The unique features of starch metabolism in red algae. Proc R Soc Lond B. 2001;268:1417–1422. doi: 10.1098/rspb.2001.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCracken DA, Cain JR. Amylose in Floridean Starch. New Phytologist. 1981;88 (1):67–71. [Google Scholar]
- 54.Gentry MS, Dowen RH, 3rd, Worby CA, Mattoo S, Ecker JR, Dixon JE. The phosphatase laforin crosses evolutionary boundaries and links carbohydrate metabolism to neuronal disease. J Cell Biol. 2007;178(3):477–488. doi: 10.1083/jcb.200704094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Minassian BA. Progressive myoclonus epilepsy with polyglucosan bodies: Lafora disease. Adv Neurol. 2002;89:199–210. [PubMed] [Google Scholar]
- 56.Pederson BA, Csitkovits AG, Simon R, Schroeder JM, Wang W, Skurat AV, Roach PJ. Overexpression of glycogen synthase in mouse muscle results in less branched glycogen. Biochem Biophys Res Commun. 2003;305(4):826–830. doi: 10.1016/s0006-291x(03)00862-3. [DOI] [PubMed] [Google Scholar]
- 57.Raben N, Danon M, Lu N, Lee E, Shliselfeld L, Skurat AV, Roach PJ, Lawrence JC, Jr, Musumeci O, Shanske S, et al. Surprises of genetic engineering: A possible model of polyglucosan body disease. Neurology. 2001;56(12):1739–1745. doi: 10.1212/wnl.56.12.1739. [DOI] [PubMed] [Google Scholar]
- 58.Wang W, Lohi H, Skurat AV, Depaoli-Roach AA, Minassian BA, Roach PJ. Glycogen metabolism in tissues from a mouse model of Lafora disease. Arch Biochem Biophys. 2007;457(2):264–269. doi: 10.1016/j.abb.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ganesh S, Tsurutani N, Suzuki T, Hoshii Y, Ishihara T, Delgado-Escueta AV, Yamakawa K. The carbohydrate-binding domain of Lafora disease protein targets Lafora polyglucosan bodies. Biochem Biophys Res Commun. 2004;313(4):1101–1109. doi: 10.1016/j.bbrc.2003.12.043. [DOI] [PubMed] [Google Scholar]
- 60.Wang W, Roach PJ. Glycogen and related polysaccharides inhibit the laforin dual-specificity protein phosphatase. Biochem Biophys Res Commun. 2004;325(3):726–730. doi: 10.1016/j.bbrc.2004.10.083. [DOI] [PubMed] [Google Scholar]
- 61.Liu Y, Wang Y, Wu C, Zheng P. Deletions and missense mutations of EPM2A exacerbate unfolded protein response and apoptosis of neuronal cells induced by endoplasm reticulum stress. Hum Mol Genet. 2009;18(14):2622–2631. doi: 10.1093/hmg/ddp196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vernia S, Rubio T, Heredia M, Rodriguez de Cordoba S, Sanz P. Increased endoplasmic reticulum stress and decreased proteasomal function in lafora disease models lacking the phosphatase laforin. PLoS One. 2009;4(6):e5907. doi: 10.1371/journal.pone.0005907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Worby CA, Gentry MS, Dixon JE. Laforin: A dual specificity phosphatase that dephosphorylates complex carbohydrates. J Biol Chem. 2006;281(41):30412–30418. doi: 10.1074/jbc.M606117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kotting O, Santelia D, Edner C, Eicke S, Marthaler T, Gentry MS, Comparot-Moss S, Chen J, Smith AM, Steup M, et al. STARCH-EXCESS4 Is a Laforin-Like Phosphoglucan Phosphatase Required for Starch Degradation in Arabidopsis thaliana. Plant Cell. 2009;21(1):334–346. doi: 10.1105/tpc.108.064360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tagliabracci VS, Girard JM, Segvich D, Meyer C, Turnbull J, Zhao X, Minassian BA, Depaoli-Roach AA, Roach PJ. Abnormal metabolism of glycogen phosphate as a cause for lafora disease. J Biol Chem. 2008 doi: 10.1074/jbc.M807428200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tagliabracci VS, Turnbull J, Wang W, Girard JM, Zhao X, Skurat AV, Delgado-Escueta AV, Minassian BA, Depaoli-Roach AA, Roach PJ. Laforin is a glycogen phosphatase, deficiency of which leads to elevated phosphorylation of glycogen in vivo. Proc Natl Acad Sci U S A. 2007;104(49):19262–19266. doi: 10.1073/pnas.0707952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barford D, Flint AJ, Tonks NK. Crystal structure of human protein tyrosine phosphatase 1B. Science. 1994;263(5152):1397–1404. [PubMed] [Google Scholar]
- 68.Lee J-O, Yang H, Georgescu M-M, Di Cristofano A, Maehama T, Shi Y, Dixon JE, Pandolfi P, Pavletich NP. Crystal Structure of the PTEN Tumor Suppressor: Implications for Its Phosphoinositide Phosphatase Activity and Membrane Association. Cell. 1999;99(3):323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 69.Guan KL, Broyles SS, Dixon JE. A Tyr/Ser protein phosphatase encoded by vaccinia virus. Nature. 1991;350(6316):359–362. doi: 10.1038/350359a0. [DOI] [PubMed] [Google Scholar]
- 70.Yuvaniyama J, Denu JM, Dixon JE, Saper MA. Crystal Structure of the Dual Specificity Protein Phosphatase VHR. Science. 1996;272:1328–1331. doi: 10.1126/science.272.5266.1328. [DOI] [PubMed] [Google Scholar]
- 71.Alonso A, Rojas A, Godzik A, Mustelin T. The dual-specific protein tyrosine phosphatase family. Vol. 5. Berlin: Springer; 2003. [Google Scholar]
- 72.Geiger T, Gooding LR, Flavell RA. T-cell responsiveness to an oncogenic peripheral protein and spontaneous autoimmunity in transgenic mice. Proc Natl Acad Sci U S A. 1992;89(7):2985–2989. doi: 10.1073/pnas.89.7.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Y, Liu Y, Wu C, Zhang H, Zheng X, Zheng Z, Geiger TL, Nuovo GJ, Liu Y, Zheng P. Epm2a suppresses tumor growth in an immunocompromised host by inhibiting Wnt signaling. Cancer Cell. 2006;10(3):179–190. doi: 10.1016/j.ccr.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 74.Liu R, Wang L, Chen C, Liu Y, Zhou P, Wang Y, Wang X, Turnbull J, Minassian BA, Zheng P. Laforin negatively regulates cell cycle progression through GSK-3{beta} dependent mechanisms. Mol Cell Biol. 2008 doi: 10.1128/MCB.01334-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang W, Parker GE, Skurat AV, Raben N, DePaoli-Roach AA, Roach PJ. Relationship between glycogen accumulation and the laforin dual specificity phosphatase. Biochem Biophys Res Commun. 2006;350(3):588–592. doi: 10.1016/j.bbrc.2006.09.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Niittyla T, Comparot-Moss S, Lue W-L, Messerli G, Trevisan M, Seymour MDJ, Gatehouse JA, Villadsen D, Smith SM, Chen J, et al. Similar protein phosphatases control starch metabolism in plants and glycogen metabolism in mammals. J Biol Chem. 2006;281(17):11815–11818. doi: 10.1074/jbc.M600519200. [DOI] [PubMed] [Google Scholar]
- 77.Peat S, Turvey JR, Evans JM. The structure of floridean starch. Part II. Enzymic hydrolysis and other studies. J Chem Soc. 1959:3341–3344. [Google Scholar]
- 78.Gentry MS, Pace RM. Conservation of the glucan phosphatase laforin is linked to rates of molecular evolution and the glycogen metabolism of the organism. BMC Evol Biol. 2009;9(1):138. doi: 10.1186/1471-2148-9-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lorberth R, Ritte G, Willmitzer L, Kossmann J. Inhibition of a starch-granule-bound protein leads to modified starch and repression of cold sweetening. Nat Biotechnol. 1998;16(5):473–477. doi: 10.1038/nbt0598-473. [DOI] [PubMed] [Google Scholar]
- 80.Baunsgaard L, Lutken H, Mikkelsen R, Glaring MA, Pham TT, Blennow A. A novel isoform of glucan, water dikinase phosphorylates pre-phosphorylated alpha-glucans and is involved in starch degradation in Arabidopsis. Plant J. 2005;41 (4):595–605. doi: 10.1111/j.1365-313X.2004.02322.x. [DOI] [PubMed] [Google Scholar]
- 81.Kotting O, Pusch K, Tiessen A, Geigenberger P, Steup M, Ritte G. Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves. The phosphoglucan, water dikinase. Plant Physiol. 2005;137(1):242–252. doi: 10.1104/pp.104.055954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ritte G, Heydenreich M, Mahlow S, Haebel S, Kotting O, Steup M. Phosphorylation of C6- and C3-positions of glucosyl residues in starch is catalysed by distinct dikinases. FEBS Lett. 2006;580(20):4872–4876. doi: 10.1016/j.febslet.2006.07.085. [DOI] [PubMed] [Google Scholar]
- 83.Ritte G, Lloyd JR, Eckermann N, Rottmann A, Kossmann J, Steup M. The starch-related R1 protein is an alpha -glucan, water dikinase. Proc Natl Acad Sci U S A. 2002;99(10):7166–7171. doi: 10.1073/pnas.062053099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Blennow A, Nielsen TH, Baunsgaard L, Mikkelsen R, Engelsen SB. Starch phosphorylation: a new front line in starch research. Trends Plant Sci. 2002;7 (10):445–450. doi: 10.1016/s1360-1385(02)02332-4. [DOI] [PubMed] [Google Scholar]
- 85.Kirkman BR, Whelan WJ. Glucosamine is a normal component of liver glycogen. FEBS Lett. 1986;194(1):6–11. doi: 10.1016/0014-5793(86)80041-2. [DOI] [PubMed] [Google Scholar]
- 86.Solaz-Fuster MC, Gimeno-Alcaniz JV, Ros S, Fernandez-Sanchez ME, Garcia-Fojeda B, Criado Garcia O, Vilchez D, Dominguez J, Garcia-Rocha M, Sanchez-Piris M, et al. Regulation of glycogen synthesis by the laforin-malin complex is modulated by the AMP-activated protein kinase pathway. Hum Mol Genet. 2008;17(5):667–678. doi: 10.1093/hmg/ddm339. [DOI] [PubMed] [Google Scholar]
- 87.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8(10):774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 88.McBride A, Ghilagaber S, Nikolaev A, Hardie DG. The glycogen-binding domain on the AMPK beta subunit allows the kinase to act as a glycogen sensor. Cell Metab. 2009;9(1):23–34. doi: 10.1016/j.cmet.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brown AM, Ransom BR. Astrocyte glycogen and brain energy metabolism. Glia. 2007;55(12):1263–1271. doi: 10.1002/glia.20557. [DOI] [PubMed] [Google Scholar]
- 90.Swanson RA. Physiologic coupling of glial glycogen metabolism to neuronal activity in brain. Can J Physiol Pharmacol. 1992;70 (Suppl):S138–144. doi: 10.1139/y92-255. [DOI] [PubMed] [Google Scholar]
- 91.Melendez-Hevia E, Waddell TG, Shelton ED. Optimization of molecular design in the evolution of metabolism: the glycogen molecule. Biochem J. 1993;295 ( Pt 2):477–483. doi: 10.1042/bj2950477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cavanagh JB. Corpora-amylacea and the family of polyglucosan diseases. Brain Res Brain Res Rev. 1999;29(2–3):265–295. doi: 10.1016/s0165-0173(99)00003-x. [DOI] [PubMed] [Google Scholar]
- 93.Lohi H, Young EJ, Fitzmaurice SN, Rusbridge C, Chan EM, Vervoort M, Turnbull J, Zhao XC, Ianzano L, Paterson AD, et al. Expanded repeat in canine epilepsy. Science. 2005;307(5706):81. doi: 10.1126/science.1102832. [DOI] [PubMed] [Google Scholar]
- 94.Yamanami S, Ishihara T, Takahashi M, Uchino F. Comparative study of intraneuronal polyglucosan bodies in brains from patients with Lafora disease and aged dogs. Acta Pathol Jpn. 1992;42(11):787–792. doi: 10.1111/j.1440-1827.1992.tb01879.x. [DOI] [PubMed] [Google Scholar]
- 95.Garyali P, Siwach P, Singh PK, Puri R, Mittal S, Sengupta S, Parihar R, Ganesh S. The malin-laforin complex suppresses the cellular toxicity of misfolded proteins by promoting their degradation through the ubiquitin-proteasome system. Hum Mol Genet. 2009;18(4):688–700. doi: 10.1093/hmg/ddn398. [DOI] [PubMed] [Google Scholar]
- 96.Mittal S, Dubey D, Yamakawa K, Ganesh S. Lafora disease proteins malin and laforin are recruited to aggresomes in response to proteasomal impairment. Hum Mol Genet. 2007;16(7):753–762. doi: 10.1093/hmg/ddm006. [DOI] [PubMed] [Google Scholar]
- 97.Gomez-Abad C, Gomez-Garre P, Gutierrez-Delicado E, Saygi S, Michelucci R, Tassinari CA, Rodriguez de Cordoba S, Serratosa JM. Lafora disease due to EPM2B mutations: a clinical and genetic study. Neurology. 2005;64(6):982–986. doi: 10.1212/01.WNL.0000154519.10805.F7. [DOI] [PubMed] [Google Scholar]
- 98.Buleon A, Colonna P, Planchot V, Ball S. Starch granules: structure and biosynthesis. Int J Biol Macromol. 1998;23(2):85–112. doi: 10.1016/s0141-8130(98)00040-3. [DOI] [PubMed] [Google Scholar]
- 99.Gunja-Smith Z, Marshall JJ, Mercier C, Smith EE, Whelan WJ. A revision of the Meyer-Bernfeld model of glycogen and amylopectin. FEBS Lett. 1970;12(2):101–104. doi: 10.1016/0014-5793(70)80573-7. [DOI] [PubMed] [Google Scholar]
- 100.Myers AM, Morell MK, James MG, Ball SG. Recent Progress toward Understanding Biosynthesis of the Amylopectin Crystal. Plant Physiol. 2000;122(4):989–998. doi: 10.1104/pp.122.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Begley MJ, Dixon JE. The structure and regulation of myotubularin phosphatases. Current Opinion in Structural Biology. 2005;15(6):614–620. doi: 10.1016/j.sbi.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 102.Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36(7):2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bingham J, Sudarsanam S. Visualizing large hierarchical clusters in hyperbolic space. Bioinformatics. 2000;16(7):660–661. doi: 10.1093/bioinformatics/16.7.660. [DOI] [PubMed] [Google Scholar]
- 104.Robinson R. Ciliate Genome Sequence Reveals Unique Features of a Model Eukaryote. PLoS Biology. 2006;4(9):e304. doi: 10.1371/journal.pbio.0040304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Goldsmith E, Sprang S, Fletterick R. Structure of maltoheptaose by difference Fourier methods and a model for glycogen. J Mol Biol. 1982;156(2):411–427. doi: 10.1016/0022-2836(82)90336-9. [DOI] [PubMed] [Google Scholar]
- 106.Ryman BE, Whelan WJ. New aspects of glycogen metabolism. Adv Enzymol Relat Areas Mol Biol. 1971;34:285–443. doi: 10.1002/9780470122792.ch6. [DOI] [PubMed] [Google Scholar]
- 107.Yu TS, Kofler H, Hausler RE, Hille D, Flugge UI, Zeeman SC, Smith AM, Kossmann J, Lloyd J, Ritte G, et al. The Arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. Plant Cell. 2001;13(8):1907–1918. doi: 10.1105/TPC.010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tabata S, Hizukuri S. Studies on Starch Phosphate. Part 2. Isolation of Glucose 3-Phosphate and Maltose Phosphate by Acid Hydrolysis of Potato Starch. Starch. 1971;23(8):267–272. [Google Scholar]
- 109.Gallant DJ, Bouchet B, Baldwin PM. Microscopy of starch: evidence of a new level of granule organization. Carbohydrate Polymers. 1997;32(3–4):177–191. [Google Scholar]
- 110.Manners DJ, WRight A. Alpha-1,4-Glucosans. Part XIV. The interaction of concanavalin-A with glycogens. J Chem Soc. 1962:4592–4595. [Google Scholar]
- 111.Manners DJ, Melville LW, Tipson RS. Advances in Carbohydrate Chemistry. Vol. 12. Academic Press; 1957. The Molecular Structure of Glycogens; pp. 261–298. [DOI] [PubMed] [Google Scholar]