Abstract
Human high-density lipoproteins (HDLs) are involved in the transport of cholesterol. The mechanism by which HDL assembles and functions is not well understood owing to a lack of structural information on circulating spherical HDL. Here, we report a series of molecular dynamics simulations that describe the maturation of discoidal HDL into spherical HDL upon incorporation of cholesterol ester as well as the resulting atomic level structure of a mature circulating spherical HDL particle. Sixty cholesterol ester molecules were added in a stepwise fashion to a discoidal HDL particle containing two apolipoproteins wrapped around a 160 dipalmitoylphosphatidylcholine lipid bilayer. The resulting matured particle, captured in a coarse-grained description, was then described in a consistent all-atom representation and analysed in chemical detail. The simulations show that maturation results from the formation of a highly dynamic hydrophobic core comprised of cholesterol ester surrounded by phospholipid and protein; the two apolipoprotein strands remain in a belt-like conformation as seen in the discoidal HDL particle, but with flexible N- and C-terminal helices and a central region stabilized by salt bridges. In the otherwise flexible lipoproteins, a less mobile central region provides an ideal location to bind lecithin cholesterol acyltransferase, the key enzyme that converts cholesterol to cholesterol ester during HDL maturation.
Keywords: high-density lipoprotein, apolipoprotein A-I, coarse-grained modelling, reverse coarse graining, all-atom molecular dynamics
1. Introduction
High-density lipoproteins (HDLs) are assemblies of proteins and lipids involved in the transport of cholesterol and fatty acids through the vasculature in a process known as reverse cholesterol transport (figure 1). HDL is known as ‘good cholesterol’ given the inverse correlation between HDL levels and the incidences of cardiovascular disease. Until recently, raising circulating HDL levels has been thought of as a treatment for arteriosclerosis, while today increasing specific functional forms of HDL, and not just total HDL levels, is considered a more viable treatment (Chhabria et al. 2007; Joy & Hegele 2008). Puzzling in this regard is that HDL particles upon lipidation form a wide range of shapes and sizes, with the ability to transform from lipid-poor globular proteins to discoidal and spherical particles. Apparently, it is not the concentration of the final HDL particle but the subspecies involved in and the efficiency of the maturation process that convey critical health benefits. Indeed, HDL during maturation must bind and activate enzymes taking part in reverse cholesterol transport (Wang & Briggs 2004). The structural variability needed for the overall function makes mature HDL particles difficult to characterize experimentally. Computational modelling, however, is well suited for the characterization of disordered systems as amply proven for lipid bilayers (Heller et al. 1993; Shelley et al. 2001a,b; Ayton & Voth 2002; Lopez et al. 2002; Marrink et al. 2004, 2007; Stevens 2004; Markvoort et al. 2005).
Figure 1.
Initial steps of the reverse cholesterol transport pathway. (a) Lipid-free apo A-I is synthesized in the liver as well as in the intestines and secreted into the blood where it absorbs cholesterol and lipids forming (b) discoidal HDL particles. The maturation of HDL occurs upon the esterification of cholesterol into cholesterol ester by the enzyme LCAT resulting in the formation of (c) spherical HDL particles.
A combination of coarse-grained and all-atom molecular dynamics (MD) simulations was already successful in describing the assembly and disassembly of discoidal HDL, showing excellent agreement with small-angle X-ray scattering (Shih et al. 2005, 2007a,c), chemical cross-linking and mass spectrometry (Bhat et al. 2007) and nuclear magnetic resonance (Li et al. 2006) measurements. Clearly, the main goal of research in HDL particles concerns now the elusive spheroidal type loaded with cholesterol ester. Given the described achievements of molecular modelling, the step towards characterizing spheroidal HDL can now be taken. Owing to the highly dynamic and heterogeneous nature of HDL particles, techniques to capture their mechanism need to cover both large conformational transitions and chemical detail. This is possible on the modelling side through the combination of coarse-grained MD, covering readily microsecond processes, and all-atom MD, exhibiting chemical detail. In the present study, we seek to describe first the maturation of discoidal HDL into spherical HDL through the addition of cholesterol ester molecules using coarse-grained MD. Novel reverse coarse-graining methods (Shih et al. 2007c, 2008a; Freddolino et al. 2008; Thøgersen et al. 2008; Miao & Schulten 2009) are then used to generate a corresponding atomic level description of the spherical HDL particle in order to analyse the detailed structure of mature HDL, in particular lecithin cholesterol acyltransferase (LCAT) binding sites.
Indeed, spherical HDL particles have not been well characterized as reflected by reports of apolipoprotein A-I (apo A-I), lipid and cholesterol ester of wide ranging compositions: (i) 56 lipids, 16 cholesterol ester molecules and 2 apo A-I strands forming particles of 7.4 nm diameter (Sparks et al. 1992); (ii) 132 lipids, 15 cholesterol, 57 cholesterol ester molecules and 3 apo A-I forming particles of 9.3 nm diameter (Jonas et al. 1990); and (iii) reports of larger 12.6 nm diameter particles formed with 232 lipids, 84 cholesterol ester molecules and 4 apo A-I strands (Sparks et al. 1992). While the exact structure of these spherical HDL particles is unclear, even less is known about the orientation and conformation of the apo A-I proteins involved. The conversion of free cholesterol to cholesterol ester by LCAT, an enzyme that binds to and is activated by apo A-I, is a critical step during HDL maturation. The cholesterol ester molecules are believed to partition from the surface of the HDL particle to the centre forming a hydrophobic core surrounded by a continuous lipid monolayer (Davidson & Silva 2005). The apo A-I proteins are thought to float on the surface of this monolayer, offering a scaffold for interactions with enzymes and stabilizing the particle (Mishra et al. 1994). It is generally thought that the apo A-I conformation in spherical HDL differs from that in discoidal HDL, but to what extent is still unknown (Davidson & Silva 2005; Davidson & Thompson 2007). Additionally, it has been demonstrated that the N-terminal domain is not involved in the binding of lipids in discoidal HDL particles (Denisov et al. 2005; Shih et al. 2005, 2007c).
As the spherical HDL particles have not yet been characterized in detail, neither experimentally nor computationally, we felt it prudent to start our simulations from the well-established structure of a discoidal HDL particle consisting of 160 dipalmitoylphosphatidylcholine (DPPC) lipids and 2 truncated Δ (1–54) apo A-I strands (Bayburt et al. 2002; Denisov et al. 2005; Shih et al. 2005, 2006, 2007a,b,c). These HDL particles, made from truncated proteins, offer a sound platform for investigating the maturation process, as the precise stoichiometry of its components and its resulting structure are well characterized (Denisov et al. 2004, 2005; Shih et al. 2005, 2007c). Our study focuses on a crucial step in HDL maturation, namely, cholesterol ester absorption. Changes in protein–lipid stoichiometry should affect maturation dynamics and ought to be investigated. However, for a start, it seems to be prudent to study solely the key cholesterol ester absorption step in HDL maturation for the stoichiometry of the best characterized cholesterol-free HDL particle.
2. Material and methods
We start our simulations from a preformed discoidal HDL particle containing two truncated apo A-I proteins and 160 DPPC lipids; the structure of the particle had been verified using small-angle X-ray scattering (Shih et al. 2005, 2007a). For the MD simulations of the HDL maturation process, we employed our recently developed coarse-grained protein–lipid model for lipoproteins (Shih et al. 2006, 2007a,b,c, 2008a,b; Freddolino et al. 2008) that permits three orders of magnitude faster computation over traditional all-atom MD simulations (Shih et al. 2006). A ‘reverse coarse-graining’ technique developed by us (Shih et al. 2007c) was used to transform the final coarse-grained model of spherical HDL into an atomic level model that permitted us to test the feasibility of the predicted structure using standard all-atom MD with established force fields, as well as to inspect the resulting model for physical and chemical properties. The simulations performed in this study are listed in table 1. Simulation times were long enough to ensure that averaged characteristics such as protein helicity, solvent-accessible hydrophobic surface area and salt-bridge formation are adequately sampled.
Table 1.
List of simulations. (Type: CG denotes coarse grained and AA all-atom equilibrium simulations. CE denotes the number of cholesterol ester molecules absorbed.)
| name | type | initial CE | final CE | time (ns) |
|---|---|---|---|---|
| Sim00 | CG | 0 | 0 | 100 |
| Sim06 | CG | 0 | 6 | 250 |
| Sim12 | CG | 6 | 12 | 250 |
| Sim18 | CG | 12 | 18 | 250 |
| Sim24 | CG | 18 | 24 | 250 |
| Sim30 | CG | 24 | 30 | 1000 |
| Sim36 | CG | 30 | 36 | 250 |
| Sim42 | CG | 36 | 42 | 250 |
| Sim48 | CG | 42 | 48 | 250 |
| Sim54 | CG | 48 | 54 | 250 |
| Sim60 | CG | 54 | 60 | 1000 |
| SimAA | AA | 60 | 60 | 25 |
Transformation of discoidal to spherical HDL was induced by the stepwise addition of cholesterol ester to an initially discoidal HDL particle containing two Δ (1–54) apo A-I proteins surrounding a 160 DPPC lipid bilayer in an anti-parallel belt-like fashion, with the two protein belts overlapping at residue G129 (Shih et al. 2006, 2007b,c). The initial discoidal HDL particle was simulated in a coarse-grained manner for 100 ns (table 1). Six cholesterol ester molecules (oleates) were then added to the surface of the HDL particle, three per side, and allowed to freely diffuse into the particle through 250 ns of simulation. The resulting particle was then used as the starting structure for the addition of another six cholesterol ester molecules. This process was repeated until a total of 60 cholesterol ester molecules had been added (and spontaneously diffused) into a single HDL particle, with 250 or 1000 ns of simulation at each step. At the start of each of the coarse-grained simulations, labelled Sim00-Sim60, the systems were re-solvated with a minimum of 15 Å of coarse-grained water around them and reionized with coarse-grained Na+ and/or Cl− ions. System sizes ranged from 12 500 to 28 500 beads; periodic boundaries were assumed.
The coarse-grained simulations were all performed using a model that the authors and others had developed previously (Shih et al. 2006, 2007a,b,c, 2008b). This model was originally consistent with the Marrink coarse-grained lipid model (Marrink et al. 2004) and implemented using NAMD 2.5 (Kalé et al. 1999; Phillips et al. 2005; Shih et al. 2006). The Marrink model has since been updated (Marrink et al. 2007), and the new force field, named MARTINI, includes improved parameters for lipids and cholesterol; recently, protein parameters were also introduced (Monticelli et al. 2008). In response to the updated MARTINI force field, we have also updated our protein–lipid model such that the lipid/cholesterol model used in the present study is consistent with the MARTINI force field; by reassigning protein backbone and side-chain bead types of our previous protein model to be consistent, the new description assumes the bead types found in the MARTINI model. Protein backbone bead types are defined as neutral with hydrogen bond donating and accepting properties (Nda). Protein side-chain bead types (except for glycine that does not have a side-chain bead) are defined as described in Shih et al. (2008b). All protein parameters remain as described previously (Shih et al. 2006, 2007b). Coarse-grained cholesterol oleate ester (Catte et al. 2008) was constructed by attaching an oleyl lipid tail (from palmitoyloleoylphosphatidylcholine (POPC) lipid) to a cholesterol molecule (figure 2). The ‘constraint’ cholesterol bonds in the MARTINI model were implemented through the use of a strong Kbond of 23.885 kcal mol−1 Å−2.
Figure 2.
All-atom and coarse-grained representation of a cholesterol oleate molecule. The all-atom structure of a cholesterol oleate molecule (shown in licorice representation without hydrogen) is overlaid with the respective coarse-grained beads (transparent sphere representation). The oleyl lipid tail is shown in blue while the cholesterol moiety is shown in green, the two moieties being linked by an ester bond shown in red.
The coarse-grained simulations were performed by initially minimizing the respective starting structures using 1000 steps of conjugate gradient minimization followed by 400 ps of equilibration with the protein backbone positions and the cholesterol oleate beads (figure 2) constrained. The backbone constraints were then removed, and the entire system simulated for 750 ns as an NPT ensemble maintaining particle number (N), pressure (P) and temperature (T). A constant temperature of 310 K was maintained through Langevin dynamics, with a damping coefficient of 5 ps−1; constant pressure was maintained with a Nóse–Hoover piston with a period of 1000 fs and a decay time of 500 fs. A 20 fs integration time step was assumed. Non-bonded interactions were calculated using 1–2 exclusion with shifting starting at 9 Å and a complete cut-off at 12Å.
Following the coarse-grained stepwise addition of 60 cholesterol ester molecules to an HDL particle, the final spherical HDL particle was reverse coarse grained back into an all-atom representation using the CGTools plugin in VMD (http://www.ks.uiuc.edu/Research/vmd/plugins/cgtools/). The reverse coarse graining is performed by mapping the centre of mass of the group of atoms represented by a single CG bead to the bead's location (Shih et al. 2007c). The resulting all-atom system was then solvated, with 15 Å of padding on all sides and 0.5 mol l−1 of Na+ and Cl− ions added, resulting in a total system size of 198 338 atoms. The system was then subjected to three cycles of simulated annealing: (i) with the centres of mass of the atoms represented by a single CG bead restrained to that bead's location (Shih et al. 2007c); (ii) with the protein Cα-atoms and DPPC lipid P-atoms restrained; and (iii) with the protein N-atoms and the DPPC lipid N-atoms restrained. Each cycle of simulated annealing consisted of an initial 100 000 step conjugate gradient minimization followed by heating from 310 to 500 K in steps of 5 K and each step simulated for 10 ps. The systems were then cooled from 500 to 310 K in steps of −5 K, being simulated for 10 ps per step. After the three cycles of simulated annealing, the constraints were removed and 100 000 steps of minimization followed by 100 ps of equilibration were performed at 310 K. The system was subjected to 25 ns of equilibrium simulation in order to generate the all-atom model of a spherical HDL particle.
The all-atom simulations were performed using NAMD 2.6 (Phillips et al. 2005), with CHARMM 22 CMAP (MacKerrel et al. 2004) parameters for the proteins and CHARMM 27 (MacKerell et al. 1998) parameters for the lipid. Cholesterol ester parameters were developed from analogy to cholesterol parameters in Pitman et al. (2004) and from oleyl lipid tail parameters from POPC lipids (MacKerell et al. 1998). A multiple-time-stepping algorithm was used to evaluate electrostatics with bonded interactions computed every 1 fs, short-range non-bonded interactions computed every 2 fs and long-range interactions computed every 4 fs. Long-range electrostatic forces were evaluated using the particle mesh Ewald summation method with a grid spacing of less than 1 Å. Temperature was maintained using Langevin dynamics, and a constant pressure of 1 atm was maintained using a Nóse–Hoover Langevin piston. Periodic boundary conditions were assumed. Non-bonded interactions were calculated using scaled 1–4 exclusion with shifting starting at 8 Å and a complete cut-off at 12 Å.
3. Results and discussion
The conversion from their discoidal to their spherical form during the maturation of circulating HDL particles involves the incorporation of cholesterol ester molecules. The dynamics of this transformation has not been captured until now. Previous experimental and theoretical studies focused on either the cholesterol ester-free discoidal (Phillips et al. 1997; Segrest et al. 1999; Shih et al. 2005; Catte et al. 2006; Wu et al. 2007) or the cholesterol ester-filled spherical structure (Catte et al. 2008; Silva et al. 2008), but not on the pathway between the two.
MD simulations carried out in this study revealed how a discoidal HDL spontaneously forms a spherical particle through the stepwise addition of cholesterol ester. The addition mimics how cholesterol ester molecules are naturally incorporated into HDL when LCAT converts cholesterol into its ester form at the surface of HDL. The simulations permit a gradual absorption of cholesterol ester molecules and show indeed the formation of a spherical HDL particle without direct insertion of the molecules as previously done (Catte et al. 2008). The final spherical HDL particle was assessed for cholesterol ester orientation, protein helicity and intermolecular salt bridging. The results of the simulations are described and discussed in detail below and provide concrete images of HDL particle maturation.
The maturation of HDL into a small spherical particle was achieved through coarse-grained MD. Starting from an initially discoidal HDL, six cholesterol ester molecules (three on the top and three on the bottom) were added to the surface of the particle and the resulting system simulated for 250 ns, which is sufficiently long for the cholesterol ester molecules to be absorbed (requiring on average approx. 25 ns), to partition and to equilibrate in the centre of the HDL particle (see figure S1 in the electronic supplementary material showing RMSD values of Sim06-Sim60). This process was repeated 10 times, until 60 molecules of cholesterol ester were absorbed. The number of cholesterol ester molecules added was chosen according to experimental reports of 9.3 nm diameter spherical HDL containing approximately 60 cholesterol ester molecules (Jonas et al. 1990). Figure 3 shows snapshots after cholesterol ester had been absorbed into the particle in steps of six. One can clearly discern complete absorption at each step and a change in particle shape with the increased number of cholesterol ester molecules, as well as the eventual maturation into a spherical HDL particle. The two apo A-I protein strands remain in a ‘double-belt’ configuration (Segrest et al. 1999) as in the initial discoidal HDL (cf. figure 3a), the proteins dynamically bending into and out of a saddle position, similar to what was reported when lipids are removed from discoidal HDL (Catte et al. 2008).
Figure 3.
Maturation of HDL from discoidal to spherical. Coarse-grained MD simulates the maturation of HDL. Snapshots (a–e) show the structures resulting from the stepwise addition of cholesterol ester and illustrate the resulting shape change. The two apo A-I proteins are shown in blue and red, and the DPPC lipids are shown in light tan. The remaining coloured molecules, which can barely be seen, are the cholesterol ester molecules that are quickly absorbed into the HDL particle forming a hydrophobic core. See figure 4 for a detailed view of the hydrophobic core.
The addition of only a few cholesterol ester molecules (i.e. 6–12) does not cause the HDL particles to become spherical. However, already the first few molecules partition to the centre of the HDL, forming a hydrophobic core (figure 4a–c) as well as occupying space between the lipid tail groups. Initially, the hydrophobic core is planar; with further addition of cholesterol ester molecules (figure 4d–j), the region between the lipid tail groups occupied by the molecules swells, forcing the conversion of the lipid bilayer of the initial discoidal HDL into a lipid monolayer, the HDL particle transitioning from a disc to a sphere. The presence of a hydrophobic core within spherical HDL had been proposed and modelled previously (Catte et al. 2008); in our simulations, the hydrophobic core emerges spontaneously, i.e. it is not assumed a priori.
Figure 4.

Growth of the hydrophobic core during maturation of HDL. Stepwise addition of cholesterol ester molecules results in the transformation of an originally discoidal HDL (top) to a spherical HDL particle (bottom). Initial (left) and final (right) snapshots from simulations Sim06 to Sim60 (a–j) are shown with the HDL particle represented through a transparent surface and cholesterol ester colour coded based on when the molecules were added to the system, from light blue, green, light green, yellow, light orange, dark orange, brown, purple, magenta and pink.
The spontaneously matured HDL particle can be inspected in detail and physically characterized. One finds that the hydrophobic core is fluid in nature. The dynamic movement of the cholesterol ester molecules within the HDL particles of Sim30 and Sim60 is illustrated in movies 1 and 2 provided in the electronic supplementary material. The cholesterol ester molecules forming the hydrophobic core within the HDL particle are highly mobile, penetrating far between the DPPC lipid tails and occasionally interacting with the apo A-I protein strands; the absorbed molecules within the spherical HDL particles preferentially orient with the more rigid cholesterol moiety towards the surface of the particle (approx. 64 ± 5%) and with the more flexible oleyl tail moiety towards the particle centre, regardless of concentration (approx. 65 ± 5%). A similar preferential orientation of the cholesterol ester molecules reported by Catte et al. (2008) in simulations of small spherical HDL consists of 56 POPC, 16 cholesterol ester and 2 truncated apo A-I proteins. It remains unclear whether the dynamic movement of the cholesterol ester molecules is due to the limited space they are allowed to occupy in HDL particles and whether, if given more space, the molecules would form an ordered structure as had been suggested for larger lipoproteins (Laggner et al. 1984).
The absorption of cholesterol ester is accompanied by an increase in the solvent-accessible hydrophobic surface area of the DPPC lipids (as monitored over the last 100 ns of simulation for each absorption step). The increase, from approximately 11 to 17 nm2 (see figure S2 in the electronic supplementary material) in going from discoidal HDL without cholesterol to spherical HDL with 60 cholesterol ester molecules, suggests that during maturation additional apo A-I strands and/or additional lipids can be recruited to cover the exposed hydrophobic surface. Indeed, phospholipid efflux along with cholesterol efflux is known to arise in HDL maturation (Wang & Briggs 2004). Native discoidal and small spherical HDL particles have also been reported to contain a varied number (2, 3 or 4) of protein strands. The particular type of HDL particle that is formed probably depends on the balance of protein, lipid and cholesterol concentrations in the immediate environment. For HDL particles circulating in the bloodstream, the maturation process as seen in our simulations results in an exposed hydrophobic surface that readily attracts cholesterol. The cholesterol is then converted to cholesterol ester by LCAT and absorbed by the HDL particle, partitioning to the growing central hydrophobic core. Prior simulations, as described in Shih et al. (2008b), of discoidal HDL particles containing cholesterol molecules showed that cholesterol, in contrast to cholesterol ester, does not partition to form a central hydrophobic core (see figure S3 and movie 4 in the electronic supplementary material). However, cholesterol transformed into cholesterol ester partitions into the HDL particle inducing it to swell, exposing additional hydrophobic surface area, thus perpetuating a mechanism by which HDL particles can perform their biological function, i.e. accumulating cholesterol.
As the HDL maturation process is too slow to be described by conventional all-atom MD, coarse-grained MD was employed. However, once the HDL particle matured, reverse coarse graining (Shih et al. 2007c) allowed us to regain atomic level resolution, constructing for the final particle an all-atom structure as detailed in movie 3 in the electronic supplementary material and shown in figure 5. During the course of 25 ns equilibration (see the movie of SimAA in the electronic supplementary material), the spherical HDL particle (figure 5a) together with its hydrophobic cholesterol core remained stable (figure 5b). The apolipoprotein helicity decreased from the initial 70 per cent found in discoidal HDL (Jonas et al. 1990; Sparks et al. 1995; Shih et al. 2006) to approximately 53 per cent in spherical HDL (the per cent helicity over the course of SimAA is shown in figure S4 in the electronic supplementary material), consistent with experimental measurement (Sparks et al. 1992). The final spherical HDL particle measured 9.4 nm in diameter, which approximates the size of the spherical rHDL particles reported in Jonas et al. (1990), albeit with only two apo A-I strands in our model as opposed to three seen in the experiment.
Figure 5.
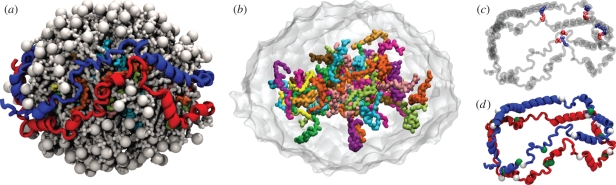
All-atom structure of a spherical HDL particle. The model of spherical HDL was generated using coarse-grained MD with stepwise addition of cholesterol ester into an initially discoidal HDL particle. The final coarse-grained spherical HDL particle was reverse coarse grained into an all-atom structure and 25 ns of equilibration was performed (SimAA). (a) The spherical HDL particle is shown with the two apolipoproteins in blue and red, 160 DPPC lipids in white and the cholesterol ester colour coded based on when the molecules were added in the coarse-grained simulations (figure 4). (b) A hydrophobic core is formed by cholesterol ester (the remainder of the HDL particle is shown as a transparent grey surface). The two apolipoproteins form (c) four stable salt bridges (protein is transparent for clarity) and (d) the proteins form helices that are seen to kink and break at proline and glycine residues (white and green spheres, respectively).
In the coarse-grained simulations, Sim00 to Sim60, performed to generate the spherical HDL structure, the two apo A-I strands are initially located around the circumference of the discoidal particle and move to the surface of the spherical particles upon maturation. After the coarse-grained spherical HDL particle was ‘reverse-coarse grained’ into the associated all-atom structure and equilibrated (with a Cα RMSD of 6.3 Å following the 25 ns of simulation, see figure S5 in the electronic supplementary material), the overall double-belt-like configuration of the two apo A-I strands remained intact; however, with slight separation at several locations (figure 5a). As in previous simulations of discoidal HDL (Shih et al. 2005; Catte et al. 2006), the apo A-I strands in the spherical HDL particle were observed to kink, the kinks being primarily due to the presence of prolines and glycines (figure 5d).
The two protein strands in the spherical HDL particle as seen in SimAA appear to be stabilized by a small number of salt bridges. In the final structure reached, eight salt bridges have formed between the two proteins. This finding is consistent with previous suggestions that salt bridging between apolipoprotein strands plays a role in protein alignment and in stabilizing discoidal and spherical HDL particles (Segrest et al. 1999; Klon et al. 2002a,b; Catte et al. 2008). However, half of the final eight salt bridges are only transient, i.e. they were seen to break and reform during the course of the 25 ns equilibration. Only salt bridges D89/E92-R173, K133/K140-E125, R151-E111 and R173-D89 (the first listed residue refers to the blue coloured apolipoprotein and the second listed residue to the red coloured one as shown in figure 5) remained formed over 25 ns (figure 5c). These salt bridges stabilize the helix pairings: helix 3–helix 7, helix 5–helix 5, helix 6–helix 4 and helix 7–helix 3 (the helices are described as in Klon et al. 2002a). Fewer salt bridges pair the protein strands in the spherical HDL particle than in the discoidal HDL particle, the latter exhibiting approximately 18 salt bridges (as measured from all-atom simulations of discoidal HDL reported in Shih et al. 2005). Apparently, breakage of salt bridges is necessary during maturation of HDL, which is consistent with the observations that oxidation of HDL particles leads to enhanced conversion of discoidal to spherical HDL (Jayaraman et al. 2008). In Catte et al. (2008), all-atom simulations of smaller spherical HDL particles were reported to contain between 10 and 12 salt bridges. The discrepancy between Catte et al. (2008) and our simulation could be due to those authors' use of all-atom MD without prior coarse-grained simulations, which may not allow for the proteins or lipids to fully relax.
In a recent experimental study, Silva et al. (2008) reported results from chemical cross-linking/mass spectroscopy experiments, in which the authors propose a ‘trefoil’ model of spherical HDL. In their model, the spherical HDL particle is partitioned by the proteins in a highly symmetrical fashion into three equal slices. The model is clearly at variance with the disordered form of the mature HDL particle seen in our simulations. Interestingly, the chemical cross-links between K59-K208, K77-K195 and K118-K140, reported by Silva et al. (2008), can also arise from our structure, despite the fact that our structure has only two protein strands.
In the spherical HDL particles, the N- and C-terminal helices of the apo A-I strands are more flexible and mobile than the central region (helices 3 through 7), the latter being stabilized by salt bridges. The increased mobility of the terminal region was previously observed in discoidal HDL particles when accommodating various particle sizes (Bhat et al. 2007; Shih et al. 2007b,c; Jones et al. 2009). A natural hinge occurs between the flexible terminal region and the rigid central region (figure 5). It is possible that the increased disorder of the terminal region along with the increase in the solvent-accessible surface area favours a spherical HDL particle and possibly also the incorporation of a third apo A-I strand. We note, though, that our simulations involved a truncated form of apo A-I with the N-terminal domain (residues 1–54) removed. The effect of the presence of the N-terminal domain on the stability of the proteins in spherical HDL is unknown, but has been shown not to be involved in lipid binding in the case of discoidal HDL particles as reported in Denisov et al. (2005) and Shih et al. (2005).
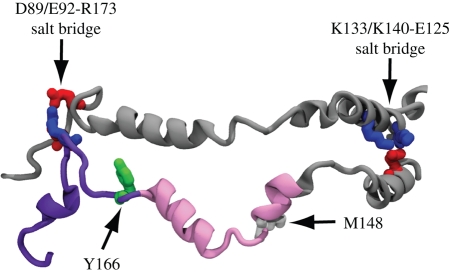
The apo A-I proteins are observed to maintain a stable central region that could be important for the binding and activation of LCAT. Indeed, LCAT is proposed to bind in the vicinity of helix 6 or 7 (Zannis et al. 2006). In recent studies, it was suggested that residue Tyr-166 in helix 7 is involved in the direct activation of LCAT (Wu et al. 2007), whereas oxidation of Met-148 in helix 6 was shown to impair the activation of LCAT by apo A-I (Shao et al. 2008). Although it is still unclear exactly which apo A-I residues are needed for LCAT activation, it is noteworthy that in our atomic level structure, the stable salt bridges effectively border the LCAT binding region (figure 6). The strategic placement of the stable salt bridges at the border of this region permits the two apo A-I proteins to maintain intermolecular contact while still allowing the residues between the protein strands to form a loop-type structure upon binding of LCAT as proposed previously (Wang et al. 1997; Shao et al. 2008).
Figure 6.
Putative LCAT binding region of apo A-I in spherical HDL. LCAT is proposed to bind to helix 6 (in pink) or 7 (in purple) of apo A-I. Stable salt bridges, D89/E92-R173 and K133/K140-E125 (shown with acidic residues coloured in red and basic residues in blue), that border this region hold the two protein strands together. Recent experimental evidence suggests Y166 (shown in green; Wu et al. 2007) and M148 (shown in white; Shao et al. 2008) to be involved in LCAT activation.
4. Conclusion
We employed coarse-grained MD to dynamically transform a discoidal HDL particle into a mature spherical particle through stepwise addition of cholesterol ester. Particle size and shape, protein helicity, as well as the distribution of cholesterol ester molecules seen in the simulated particle are consistent with prior expectations. However, we also find that the final spherical HDL is highly dynamic and rather disordered, features that serve HDL function well, but are difficult to characterize experimentally. Computational modelling in the past has proved to be a powerful tool in characterizing disordered structures such as membranes and discoidal HDL (Shelley et al. 2001a,b; Ayton & Voth 2002; Lopez et al. 2002; Marrink et al. 2004, 2007; Stevens 2004; Markvoort et al. 2005; Shih et al. 2005, 2006, 2007a,b,c, 2008b; Catte et al. 2008; Monticelli et al. 2008) and offers now the first detailed glimpse of HDL maturation.
Although models of spherical HDL had already been developed using molecular modelling (Catte et al. 2008) and mass spectrometry/chemical cross-linking (Silva et al. 2008) methods, the approach used here allowed us to not only characterize the average structure of a spherical HDL particle, but also to describe the actual maturation process. This process starts with free unesterified cholesterol interacting with LCAT and being converted at the periphery of the HDL particle into cholesterol esters. The esterified cholesterols quickly partition into the HDL particle forming a dynamic core.
The simulations presented relied on an engineered HDL system involving truncated apo A-I protein (Bayburt et al. 2002; Denisov et al. 2005). To more fully characterize HDL maturation, simulations should be extended to different lipids, as well as to different stoichiometries of full-length protein, lipid, cholesterol and cholesterol ester. Eventually, through a combination of computational and experimental approaches, the mysteries of HDL and reverse cholesterol transport should be solved.
Acknowledgements
This work is supported by grants from the National Institutes of Health (R01-GM067887 and P41-RR05969 to K.S. and R01-GM33775 to S.G.S.) and from the National Science Foundation (MCB02-34938 to K.S.). The authors gladly acknowledge supercomputer time provided by the National Center for Supercomputing Applications via Large Resources Allocation Committee grant MCA93S028. A.Y.S. acknowledges support from a Beckman Postdoctoral Fellowship.
References
- Ayton G. S., Voth G. A. 2002. Bridging microscopic and mesoscopic simulations of lipid bilayers. Biophys. J. 83, 3357–3370. ( 10.1016/S0006-3495(02)75336-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayburt T. H., Grinkova Y. V., Sligar S. G. 2002. Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2, 853–856. ( 10.1021/nl025623k) [DOI] [Google Scholar]
- Bhat S., Sorci-Thomas M. G., Tuladhar R., Samuel M. P., Thomas M. J. 2007. Conformational adaptation of apolipoprotein A-I to discretely sized phospholipid complexes. Biochemistry 46, 7811–7821. ( 10.1021/bi700384t) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catte A., et al. 2006. Novel changes in discoidal high density lipoprotein morphology: a molecular dynamics study. Biophys. J. 90, 4345–4360. ( 10.1529/biophysj.105.071456) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catte A., et al. 2008. Structure of spheroidal HDL particles revealed by combined atomistic and coarse grained simulations. Biophys. J. 94, 2306–2319. ( 10.1529/biophysj.107.115857) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabria M. T., Suhagia B. N., Brahmkshatriya P. S. 2007. HDL elevation and lipid lowering therapy: current scenario and future perspectives. Recent Pat. Cardiovasc. Drug Discov. 2, 214–227. ( 10.2174/157489007782418973) [DOI] [PubMed] [Google Scholar]
- Davidson W. S., Silva R. A. G. D. 2005. Apolipoprotein structural organization in high density lipoproteins: belts, bundles, hinges and hairpins. Curr. Opin. Lipidol. 16, 295–300. ( 10.1074/01.mol.0000169349.38321.ad) [DOI] [PubMed] [Google Scholar]
- Davidson W. S., Thompson T. B. 2007. The structure of apolipoprotein A-I in high density lipoproteins. J. Biol. Chem. 282, 22 249–22 254. ( 10.1074/jbc.R700014200) [DOI] [PubMed] [Google Scholar]
- Denisov I. G., Grinkova Y. V., Lazarides A. A., Sligar S. G. 2004. Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J. Am. Chem. Soc. 126, 3477–3487. ( 10.1021/ja0393574) [DOI] [PubMed] [Google Scholar]
- Denisov I. G., McLean M. A., Shaw A. W., Grinkova Y. V., Sligar S. G. 2005. Thermotropic phase transition in soluble nanoscale lipid bilayers. J. Phys. Chem. B 109, 15 580–15 588. ( 10.1021/jp051385g) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freddolino P. L., Arkhipov A., Shih A. Y., Yin Y., Chen Z., Schulten K. 2008. Application of residue-based and shape-based coarse graining to biomolecular simulations. In Coarse-graining of condensed phase and biomolecular systems (ed. Voth G. A.), ch. 20, pp. 299–315. Boca Raton, FL: Chapman and Hall/CRC Press, Taylor and Francis Group. [Google Scholar]
- Heller H., Schaefer M., Schulten K. 1993. Molecular dynamics simulation of a bilayer of 200 lipids in the gel and in the liquid crystal-phases. J. Phys. Chem. 97, 8343–8360. ( 10.1021/j100133a034) [DOI] [Google Scholar]
- Jayaraman S., Gantz D. L., Gursky O. 2008. Effects of protein oxidation on the structure and stability of model discoidal high-density lipoproteins. Biochemistry 47, 3875–3882. ( 10.1021/bi7023783) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas A., Wald J. H., Toohill K. L. H., Krul E. S., Kézdy K. E. 1990. Apolipoprotein A-I structure and lipid properties in homogeneous reconstituted spherical and discoidal high density lipoproteins. J. Biol. Chem. 265, 22 123–22 129. [PubMed] [Google Scholar]
- Jones M. K., Catte A., Patterson J. C., Gu F., Chen J., Li L., Segrest J. P. 2009. Thermal stability of apolipoprotein A-I in high-density lipoproteins by molecular dynamics. Biophys. J. 96, 354–371. ( 10.1016/j.bpj.2008.09.041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joy T., Hegele R. A. 2008. Is raising HDL a futile strategy for atheroprotection? Nat. Rev. Drug Discov. 7, 143–155. ( 10.1038/nrd2489) [DOI] [PubMed] [Google Scholar]
- Kalé L., et al. 1999. NAMD2: greater scalability for parallel molecular dynamics. J. Comp. Phys. 151, 283–312. ( 10.1006/jcph.1999.6201) [DOI] [Google Scholar]
- Klon A. E., Segrest J. P., Harvey S. C. 2002a. Comparative models for human apolipoprotein A-I bound to lipid in discoidal high-density lipoprotein particles. Biochemistry 41, 10 895–10 905. ( 10.1021/bi020315m) [DOI] [PubMed] [Google Scholar]
- Klon A. E., Segrest J. P., Harvey S. C. 2002b. Molecular dynamics simulations on discoidal HDL particles suggest a mechanism for rotation in the apo A-I belt model. J. Mol. Biol. 324, 703–721. ( 10.1016/S0022-2836(02)01143-9) [DOI] [PubMed] [Google Scholar]
- Laggner P., Kostner G. M., Degovics G., Worcester D. L. 1984. Structure of the cholesterol ester core of human plasma low density lipoproteins: selective deuteration and neutron small-angle scattering. Proc. Natl Acad. Sci. USA 81, 4389–4393. ( 10.1073/pnas.81.14.4389) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Kijac A. Z., Sligar S. G., Rienstra C. M. 2006. Structural analysis of nanoscale self-assembled discoidal lipid bilayers by solid-state NMR spectroscopy. Biophys. J. 91, 3819–3828. ( 10.1529/biophysj.106.087072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez C., Moore P., Shelley J., Shelley M., Klein M. 2002. Computer simulation studies of biomembranes using a coarse grain model. Comput. Phys. Commun. 147, 1–6. ( 10.1016/S0010-4655(02)00195-9) [DOI] [Google Scholar]
- MacKerell A., Jr, et al. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616. ( 10.1021/jp973084f) [DOI] [PubMed] [Google Scholar]
- MacKerrel A. D., Feig M., Brooks C. L., III 2004. Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comp. Chem. 25, 1400–1415. ( 10.1002/jcc.20065) [DOI] [PubMed] [Google Scholar]
- Markvoort A. J., Pieterse K., Steijaert M. N., Spijjker P., Hilbers P. A. J. 2005. The bilayer–vesicle transition is entropy driven. J. Phys. Chem. B 109, 22 649–22 654. ( 10.1021/jp053038c) [DOI] [PubMed] [Google Scholar]
- Marrink S. J., de Vries A. H., Mark A. E. 2004. Coarse grained model for semiquantitative lipid simulations. J. Phys. Chem. B 108, 750–760. ( 10.1021/jp0365089) [DOI] [Google Scholar]
- Marrink S. J., Risselada J. H., Yefimov S., Tieleman D. P., de Vries A. H. 2007. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B 111, 7812–7824. ( 10.1021/jp071097f) [DOI] [PubMed] [Google Scholar]
- Miao L., Schulten K. 2009. Transport-related structures and processes of the nuclear pore complex studied through molecular dynamics. Structure 17, 449–459. ( 10.1016/j.str.2008.12.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra V. K., Palgunachari M. N., Segrest J. P., Anantharamaiah G. M. 1994. Interactions of synthetic peptide analogs of the class A amphipathic helix with lipids. Evidence for the snorkel hypothesis. J. Biol. Chem. 269, 7185–7191. [PubMed] [Google Scholar]
- Monticelli L., Kandasamy S. K., Periole X., Larson R. G., Tieleman D. P., Marrink S. J. 2008. The MARTINI coarse-grained force field: extension to proteins. J. Chem. Th. Comp. 4, 819–834. ( 10.1021/ct700324x) [DOI] [PubMed] [Google Scholar]
- Phillips J. C., Wriggers W., Li Z., Jonas A., Schulten K. 1997. Predicting the structure of apolipoprotein A-I in reconstituted high density lipoprotein disks. Biophys. J. 73, 2337–2346. ( 10.1016/S0006-3495(97)78264-X) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J. C., et al. 2005. Scalable molecular dynamics with NAMD. J. Comp. Chem. 26, 1781–1802. ( 10.1002/jcc.20289) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman M. C., Suits F., Mackerell A. D., Jr, Feller S. E. 2004. Molecular-level organization of saturated and polyunsaturated fatty acids in a phosphatidylcholine bilayer containing cholesterol. Biochemistry 43, 15 318–15 328. ( 10.1021/bi048231w) [DOI] [PubMed] [Google Scholar]
- Segrest J. P., Jones M. K., Klon A. E., Sheldahl C. J., Hellinger M., De Loof H., Harvey S. C. 1999. A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J. Biol. Chem. 274, 31 755–31 758. ( 10.1074/jbc.274.45.31755) [DOI] [PubMed] [Google Scholar]
- Shao B., Cavigiolio G., Brot N., Oda M. N., Heinecke J. W. 2008. Methionine oxidation impairs reverse cholesterol transport by apolipoprotein A-I. Proc. Natl Acad. Sci. USA 105, 12 224–12 229. ( 10.1073/pnas.0802025105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelley J., Shelley M., Reeder R., Bandyopadhyay S., Moore P., Klein M. L. 2001a. Simulations of phospholipids using a coarse grain model. J. Phys. Chem. 105, 9785–9792. ( 10.1021/jp011637n) [DOI] [Google Scholar]
- Shelley J. C., Shelley M. Y., Reeder R. C., Bandyopadhyay S., Klein M. L. 2001b. A coarse grain model for phospholipid simulations. J. Phys. Chem. B 105, 4464–4470. ( 10.1021/jp011637n) [DOI] [Google Scholar]
- Shih A. Y., Denisov I. G., Phillips J. C., Sligar S. G., Schulten K. 2005. Molecular dynamics simulations of discoidal bilayers assembled from truncated human lipoproteins. Biophys. J. 88, 548–556. ( 10.1529/biophysj.104.046896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih A. Y., Arkhipov A., Freddolino P. L., Schulten K. 2006. Coarse grained protein–lipid model with application to lipoprotein particles. J. Phys. Chem. B 110, 3674–3684. ( 10.1021/jp0550816) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih A. Y., Arkhipov A., Freddolino P. L., Sligar S. G., Schulten K. 2007a. Assembly of lipids and proteins into lipoprotein particles. J. Phys. Chem. B 111, 11 095–11 104. ( 10.1021/jp0723206) [DOI] [PubMed] [Google Scholar]
- Shih A. Y., Freddolino P. L., Arkhipov A., Schulten K. 2007b. Assembly of lipoprotein particles revealed by coarse-grained molecular dynamics simulations. J. Struct. Biol. 157, 579–592. ( 10.1016/j.jsb.2006.08.006) [DOI] [PubMed] [Google Scholar]
- Shih A. Y., Freddolino P. L., Sligar S. G., Schulten K. 2007c. Disassembly of nanodiscs with cholate. Nano Lett. 7, 1692–1696. ( 10.1021/nb0706906) [DOI] [PubMed] [Google Scholar]
- Shih A. Y., Freddolino P. L., Arkhipov A., Sligar S. G., Schulten K. 2008a. Molecular modeling of the structural properties and formation of high-density lipoprotein particles. In Current topics in membranes: computational modeling of membrane bilayers (ed. Feller S.), ch. 11, pp. 313–342. London, UK: Elsevier. [Google Scholar]
- Shih A. Y., Sligar S. G., Schulten K. 2008b. Molecular models need to be tested: the case of a solar flares discoidal HDL model. Biophys. J. 94, L87–L89. ( 10.1529/biophysj.108.131581) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva R. A. G. D., et al. 2008. Structure of apolipoprotein (a-i) in spherical high density lipoproteins of different sizes. Proc. Natl Acad. Sci. USA 105, 12 176–12 181. ( 10.1073/pnas.0803626105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks D. L., Lund-Katz S., Phillips M. C. 1992. The charge and structural stability of apolipoprotein A-I in discoidal and spherical recombinant high density lipoprotein particles. J. Biol. Chem. 267, 25 839–25 847. [PubMed] [Google Scholar]
- Sparks D. L., Davidson W. S., Lund-Katz S., Phillips M. C. 1995. Effects of the neutral lipid content of high density lipoprotein on apolipoprotein A-I structure and particle stability. J. Biol. Chem. 270, 26 910–26 917. ( 10.1074/jbc.270.9.4280) [DOI] [PubMed] [Google Scholar]
- Stevens M. J. 2004. Coarse-grained simulations of lipid bilayers. J. Chem. Phys. 121, 11 942–11 948. ( 10.1063/3.1814058) [DOI] [PubMed] [Google Scholar]
- Thøgersen L., Schiøtt B., Vosegaard T., Nielsen N. C., Tajkhorshid E. 2008. Peptide aggregation and pore formation in a lipid bilayer: a combined coarse-grained and all atom molecular dynamics study. Biophys. J. 95, 4337–4347. ( 10.1529/biophysj.108.133330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Briggs M. R. 2004. HDL: the metabolism, function, and therapeutic importance. Chem. Rev. 104, 119–137. ( 10.1021/cr020466v) [DOI] [PubMed] [Google Scholar]
- Wang G., Sparrow J. T., Cushley R. J. 1997. The helix-hinge-helix structural motif in human apolipoprotein A-I determined by NMR spectroscopy. Biochemistry 36, 13 657–13 666. ( 10.1021/bi971151q) [DOI] [PubMed] [Google Scholar]
- Wu Z., Wagner M. A., Zheng L., Parks J. S., III, Shy J. M., Smith J. D., Gogonea V., Hazen S. L. 2007. The refined structure of nascent HDL reveals a key functional domain for particle maturation and dysfunction. Nat. Struct. Mol. Biol. 14, 861–868. ( 10.1038/nsmb1284) [DOI] [PubMed] [Google Scholar]
- Zannis V. I., Chroni A., Krieger M. 2006. Role of apoA-I, ABCA1, LCAT, and SR-BI in the biogenesis of HDL. J. Mol. Med. 84, 276–294. ( 10.1007/S00109-005-0030-4) [DOI] [PubMed] [Google Scholar]