Abstract
The removal of DNA interstrand cross-links (ICLs) has proven to be notoriously complicated due to the involvement of multiple pathways of DNA repair, which include the Fanconi anemia/BRCA pathway, homologous recombination, and components of the nucleotide excision and mismatch repair pathways. Members of the SNM1 gene family have also been shown to have a role in mediating cellular resistance to ICLs, although their precise function has remained elusive. Here we show that knockdown of Snm1B/Apollo in human cells results in hypersensitivity to mitomycin C (MMC), but not to IR. We also show that Snm1B-deficient cells exhibit a defective S phase checkpoint in response to MMC, but not to IR, and this finding may account for the specific sensitivity to the cross-linking drug. Interestingly, although previous studies have largely implicated ATR as the major kinase activated in response to ICLs, we show that it is activation of the ATM-mediated checkpoint that is defective in Snm1B-deficient cells. The requirement for Snm1B in ATM checkpoint activation specifically after ICL damage is correlated with its role in promoting double-strand break formation, and thus replication fork collapse. Consistent with this result Snm1B was found to interact directly with Mus81-Eme1 an endonuclease previously implicated in fork collapse. In addition, we also show that Snm1B interacts with the Mre11-Rad50-Nbs1 (MRN) complex and with FancD2 further substantiating its role as a checkpoint/DNA repair protein.
Keywords: Snm1B/Apollo, interstrand cross-links, cell cycle checkpoint, ATM
Introduction
The budding yeast snm1/pso2 mutant was first isolated over 25 years ago and exhibits a singular sensitivity to DNA interstrand cross-linking agents, but not to other forms of DNA damage such as IR, UV, or monofunctional alkylating agents (Haase et al., 1989; Henriques and Moustacchi, 1980; Ruhland et al., 1981). The molecular function of yeast Snm1 remains poorly defined. Mutants of snm1/pso2 appear normal in the initial processing steps of interstrand cross-link (ICL) repair, but are defective in the resolution of double-strand breaks that occur presumably as a consequence of replication fork collapse in response to ICLs (Barber et al., 2005; Li and Moses, 2003; Magana-Schwencke et al., 1982; Wilborn and Brendel, 1989). Interestingly, Snm1 has been shown to have an overlapping role with the 5’-3’ mismatch repair exonuclease Exo1 during processing of collapsed replication forks via homologous recombination (Barber et al., 2005). It was also shown in this same report that Snm1 has a separate role in G1 phase repair of ICLs that requires the nucleotide excision repair (NER) pathway, but not homologous recombination. Thus, Snm1 plays a role in both homology-dependent and homology-independent pathways of ICL repair in S. cerevisiae (Grossmann et al., 2001).
In chicken and mammalian cells three orthologues of SNM1 have been identified that are involved in the cellular response to genotoxic agents (Dronkert et al., 2000; Ishiai et al., 2004). These genes include SNM1A, SNM1B/Apollo and Artemis. All of the SNM1 orthologues have in common a metallo-β-lactamase fold and an appended β-CASP (CPSF-Artemis-Snm1-Pso2) domain (Callebaut et al., 2002), which together are sometimes referred to as the SNM1 domain. The β-CASP domain is predicted to be a nucleic acid binding domain, and together with the metallo-β-lactamase fold has been shown to constitute a nuclease function in the Snm1 proteins (Chan et al., 2002; Lenain et al., 2006; Li et al., 2005; Pannicke et al., 2004). Outside of the SNM1 domain the sequence of each of the proteins is distinct. Artemis, which is the most intensively studied member of the SNM1 gene family, is known to be required in partnership with DNA-PKcs for the cleavage of hairpins that occur at coding joints during V(D)J recombination (Ma et al., 2002). The inability to complete V(D)J recombination in Artemis-deficient cells has been shown to lead to a severe combined immunodeficiency (SCID) syndrome (Moshous et al., 2001; Moshous et al., 2000). In addition, Artemis-deficient cells are radiosensitive, and this phenotype has been ascribed to both a deficiency in nonhomologous end-joining and cell cycle checkpoint responses (Geng et al., 2007; Ma et al., 2002; Riballo et al., 2004; Wang et al., 2005; Zhang et al., 2004). A knockout of Artemis in the mouse has recapitulated the SCID syndrome and has also shown that it is a tumor suppressor, but only when combined with p53 deficiency (Rooney et al., 2004; Rooney et al., 2002). Studies of SNM1A have been somewhat less revealing. Snm1A has been shown to colocalize with Mre11 foci after exposure of cells to IR or ICL-inducing agents, and to interact with the checkpoint protein 53BP1 (Richie et al., 2002). However, mammalian SNM1A-deficient cells exhibit no hypersensitivity to IR and only a modest hypersensitivity to interstrand cross-linking agents (Ahkter et al., 2005; Dronkert et al., 2000), although sensitivity to cisplatin has been observed in chicken DT40 cells (Ishiai et al., 2004; Nojima et al., 2005). Intriguingly, Snm1A-deficient mouse embryonic fibroblasts are highly sensitive to spindle poisons such as nocodazole and taxol, and Snm1A has been shown to be involved in an early mitotic checkpoint pathway in response to these drugs (Akhter et al., 2004). This mitotic checkpoint pathway appears to be congruent with that involving the Chfr tumor suppressor gene (Chaturvedi et al., 2002; Matsusaka and Pines, 2004; Scolnick and Halazonetis, 2000; Summers et al., 2005; Yu et al., 2005). A knockout of Snm1A in the mouse has shown that it too is a tumor suppressor gene (Ahkter et al., 2005).
Only a few studies have been conducted on the DNA repair function of SNM1B in vertebrate cells. In chicken DT40 cells lack of SNM1B results in a slight to moderate increased sensitivity to cisplatin and mitomycin C (MMC), but not to IR (Ishiai et al., 2004; Nojima et al., 2005). In human cells siRNA-mediated knockdown of SNM1B has been shown to result in moderate hypersensitivity to cisplatin, MMC and IR (Demuth et al., 2004). Also, several recent papers have demonstrated a novel function for Snm1B/Apollo namely that it interacts with the telomere protein TRF2 and protects telomeres from the DNA repair machinery during S phase (Freibaum and Counter, 2006; Lenain et al., 2006; van Overbeek and de Lange, 2006).
The mechanisms of ICL repair are still poorly understood in mammalian cells. This situation is particularly true for the early stages of repair involving replication fork collapse and uncoupling of the ICL. In our work reported here, we found, consistent with a previous report (Demuth et al., 2004), that Snm1B-deficient cells are hypersensitive to interstrand cross-linking agents, however, we did not observe an increased sensitivity to IR. Significantly, we find that Snm1B is required for the induction of an S phase checkpoint after exposure of cells to MMC, but not to IR. The loss of the S phase checkpoint in Snm1B-deficient cells is due to an inability to activate ATM, Chk2, and Nbs1. This failure to activate ATM and downstream targets is due to the absence of replication fork collapse and the production of double-strand breaks (DSBs). We also show that Snm1B physically associates with Mre11, FancD2, and Mus81, suggesting that it may be involved in recruiting or maintaining these ICL response proteins at the site of the lesion and /or stalled replication fork.
Results
The Role of Snm1B in DNA Repair Processing of ICLs
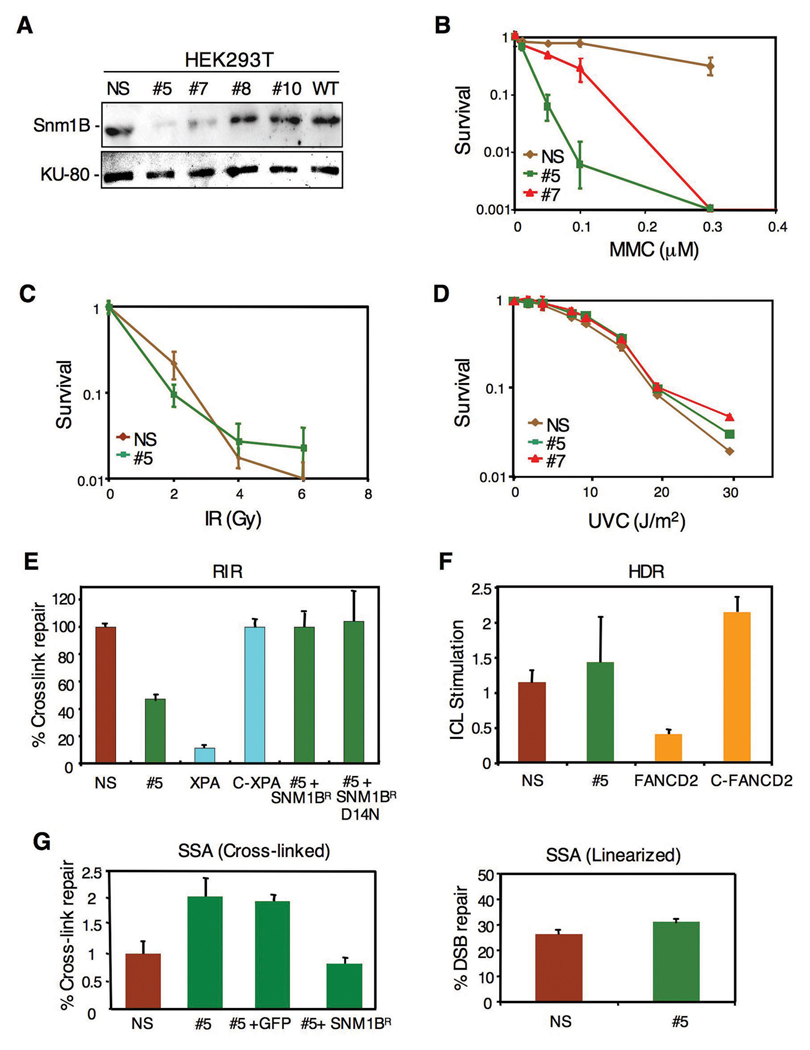
To initiate our studies of SNM1B, we isolated stable clones of HEK293T cells expressing a shRNA targeted to this gene. Two clones, designated #5 and #7, were identified with significant knockdown of Snm1B as indicated by immunoblotting with clone #5 exhibiting the greater depletion of the protein (Fig. 1A). A clone expressing a nonspecific shRNA was also derived as a control. These clones were analyzed for sensitivity to agents that introduce DNA ICLs or to IR. Consistent with a previous report (Demuth et al., 2004), the Snm1-deficient cells exhibited hypersensitivity to three cross-linking drugs tested, although a significantly higher degree of cell killing was observed with MMC as opposed to cisplatin or psoralen plus UVA (Fig. 1B and results not shown). However, no increased sensitivity was observed upon exposure to IR (Fig. 1C) or UV (Fig. 1D). Consistently, clone #5 showed the greatest hypersensitivity in agreement with the lower level of Snm1B in this clone.
Figure 1. Knockdown of Snm1B by RNAi causes sensitivity to DNA interstrand cross-linking agents, but Snm1B is not involved in the recombination steps of ICL repair.
(A) Stable knockdown of Snm1B by shRNA in HEK293T cells as shown by immunoblotting. Numbered clones with various levels of knockdown are shown. WT indicates untransfected wild-type cells. NS indicates a clone expressing a nonspecific shRNA. (B–D) Colony survival assays for clones #5, #7, and NS following exposure to the indicated DNA damaging treatments. (E) SNM1B-deficient cells are partially defective in recombination-independent DNA repair (RIR). Reactivation of a luciferase reporter gene by repair of a single site-specific psoralen ICL in the indicated cell lines is shown. The relative efficiencies were calculated as the percentage of luciferase activity of the cross-linked reporter gene normalized to that of unmodified reporter gene. C-XPA indicates a clone with stable correction of the XPA cell line. SNM1BR indicates an allele refractory to the SNM1B shRNA. D14N indicates a point mutation of SNM1B in the metallo-β-lactamase domain. All assays were carried out in triplicate and standard deviations are indicated. (F) SNM1B-deficient cells show no defect in homology dependent recombination (HDR) of a linearized cross-linked substrate. pECFPHR contains a donor ECFP gene without a start codon and an interrupted ECFP gene due to the insertion of an oligonucleotide, with or without a psoralen cross-link, within the coding region (Zhang et al., 2007). The ratio of homologous recombination in cross-linked plasmids to homologous recombination in noncross-linked plasmids is presented as ICL stimulation of HDR. (G) SNM1B-deficient cells exhibit increased levels of repair by SSA. Psoralen-crosslinked (left panel) or non-crosslinked, linearized (right panel) pSupN plasmids were transfected into the indicated cell lines. For the rescue experiment the control (GFP) and SNM1BR DNAs were co-transfected with the pSupN plasmid. Recombination frequency refers to percentage of blue colonies derived from more than 10,000 total colonies.
To directly assess the role of putative DNA repair genes in ICL repair, we have developed a number of plasmid-based in vivo assays that measure the involvement of various DNA repair pathways including NER, homology dependent recombination (HDR), and single-strand annealing (SSA). These assays utilize plasmids that contain site-specific psoralen ICLs, and have been designed to interrogate a particular DNA repair pathway (Shen et al., 2006; Wang et al., 2001; Zhang et al., 2007; Zheng et al., 2006; Zheng et al., 2003). In the recombination-indepenent repair (RIR) assay, which measures repair of ICLs mediated by NER and translesion bypass synthesis, the #5 clone showed an approximate two-fold reduction in the reactivation of cross-linked plasmids compared to control cells (Fig. 1E). As noted above, the yeast snm1 mutant also exhibits a defect in G1 phase repair of ICLs (Barber et al., 2005). This result is to be compared to XPA-deficient cells that showed an approximate ten-fold reduction in reactivation. As a control for the specificity of this effect, a construct expressing a SNM1 allele resistant to the shRNA-mediated knockdown was able to fully rescue the reactivation assay. Somewhat surprisingly, a point mutant (D14N) of SNM1B was also able to fully rescue the reactivation assay. In Artemis, mutation of this residue, located in the metallo-β-lactamase domain and conserved in all SNM1 family members, has been shown to inactivate the nuclease activity of the protein (Pannicke et al., 2004). In contrast to the results with the RIR assay, the #5 clone showed no defect in the HDR assay (Fig. 1F). This assay is performed with a linearized plasmid DNA that contains an ICL near the broken end thus mimicking a collapsed replication fork (Zhang et al., 2007). FANCD2-deficient cells, which are defective in HDR of ICLs (Nakanishi et al., 2005; Thompson et al., 2005), are also highly defective in this assay (Zhang et al., 2007). Thus, these results suggest that Snm1B is not directly involved in the recombination steps of fork restoration during ICL repair. We also examined the #5 clone in an assay that measures the single-strand annealing (SSA) pathway during repair of ICLs (Zheng et al., 2006). Interestingly, the #5 clone showed an approximate three-fold increase in ICL repair by SSA, suggesting that the lack of Snm1B results in a funneling of these lesions into the SSA pathway (Fig. 1G). Expression of the shRNA refractory construct reverted the SSA response to normal levels.
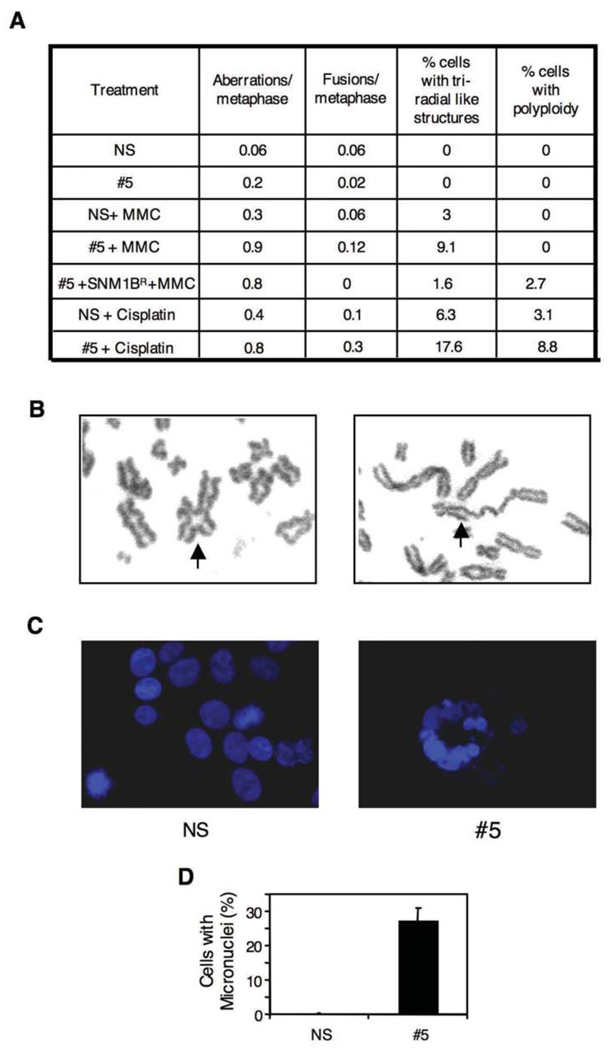
To further define the phenotype of cells deficient in Snm1B, we conducted a number of assays to assess its role in maintenance of genomic stability. Mutants defective in ICL repair typically exhibit chromosomal aberrations upon exposure to cross-linking drugs. Treatment of clone #5 with either MMC or cisplatin caused an approximate 2–3 fold increase in the total number of chromosomal aberrations (Fig. 2A, B), which is similar to the levels observed in the highly MMC-sensitive Mus81 nullizygous mouse cells (McPherson et al., 2004). Abnormal chromosome structures can result in an aberrant mitosis leading to mitotic catastrophe and the formation of micronuclei. Exposure of the #5 clone to MMC showed that approximately a quarter of the cells exhibited micronuclei, while no such aberrant nuclei were observed in the control cell line (Fig. 2C, D). Taken together, our findings suggest that Snm1B is involved in repair processing of ICLs since lowered levels of the protein result in increased chromosomal aberrations in response to ICL-inducing drugs. However, the DNA repair assays suggest that Snm1B likely acts upstream of the recombinational processing steps of ICL repair.
Figure 2. Knockdown of Snm1B causes chromosomal abnormalities.
(A) Quantification of various chromosomal aberrations after MMC (50 ng/ml) or cisplatin (2 µM) treatment. Cells were treated with the indicated agent for 8 hrs and then allowed to recover for 24 hrs in fresh medium. Colcemid was then added for 1 hr to accumulate mitotic cells. (B) Examples of aberrant chromosomes scored as indicated in (A). Arrows indicate a triradial (left panel) and a fused chromosome (right panel). (C) Knockdown of Snm1B induces micronuclei formation after treatment with MMC (50 ng/ml). Nuclei are stained with DAPI (blue). (D) Quantification of micronuclei shown in (C).
Snm1B is Required for the S Phase Cell Cycle Checkpoint in Response to ICL Damage
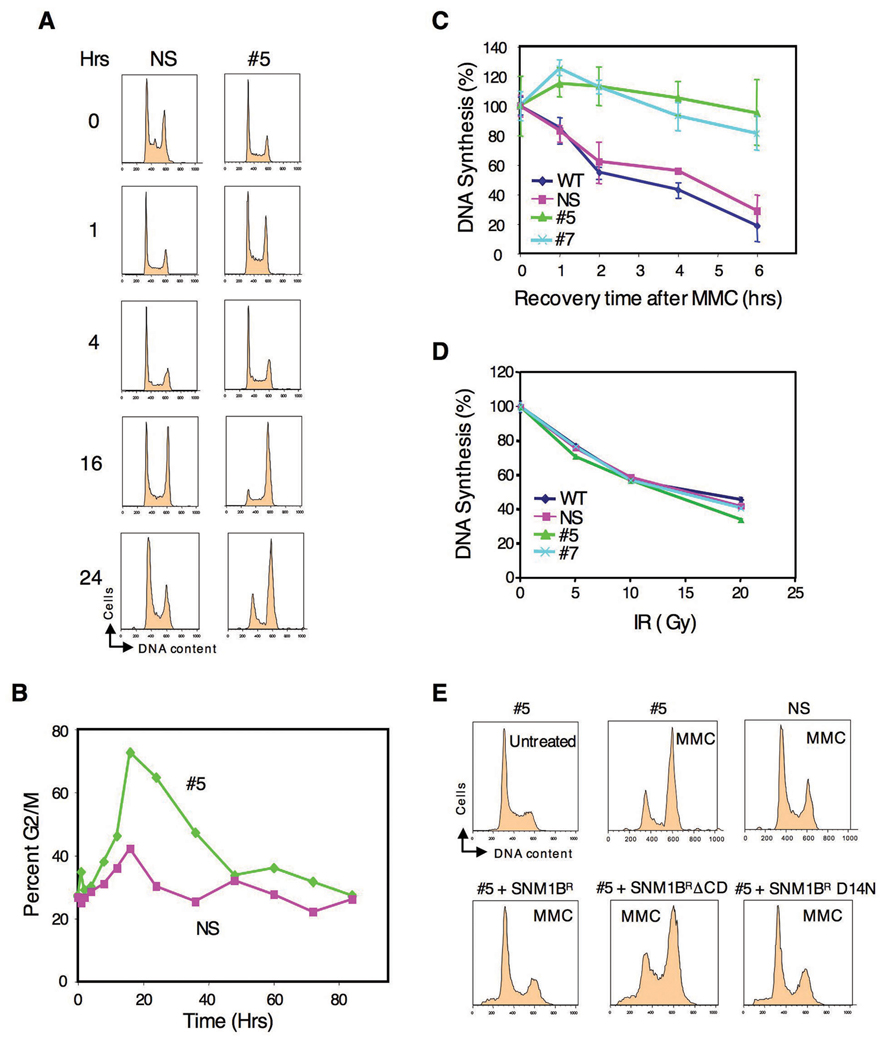
We next examined the cell cycle response after exposure of Snm1-deficient cells to ICLs. FACS analysis after treatment with MMC resulted in a strong G2/M accumulation in clone #5 cells compared to the control cells (Fig. 3A, B). To verify the generality of this effect, we transiently knocked down Snm1B in HeLa cells using an siRNA that targets a different region of SNM1B than the shRNA used to develop the stable knockdown clones in HEK293T cells. Treatment of these cells with MMC again resulted in an increased G2/M accumulation compared to control cells (Fig. S1). G2/M or late S phase accumulation after DNA damage is also observed in FA cells (Akkari et al., 2001; Seyschab et al., 1995), and may be due to a failure to repair, and/or to a defect in the S phase checkpoint. Such a phenotype has been observed in ATM- and BRCA1-deficient cells after IR-induced DNA damage and has been attributed to a S phase checkpoint defect (Xu et al., 2002). To determine if Snm1B-deficient cells are defective in an S phase checkpoint, we examined the effect on DNA synthesis of exposure of cells to MMC. As shown (Fig. 3C), treatment of either clone #5 or #7 with MMC did not result in decreased DNA synthesis as observed in the control cells indicating a failure to arrest cells in S phase, and thus a defective checkpoint. A similar defect in the intra-S phase checkpoint has been observed in FA cells after treatment with cross-linking drugs (Ho et al., 2006; Nakanishi et al., 2002; Pichierri and Rosselli, 2004b; Sala-Trepat et al., 2000; Sobeck et al., 2006; Taniguchi et al., 2002). Significantly, we did not observe a failure to arrest DNA synthesis in the Snm1B-deficient cells after exposure to IR (Fig. 3D), although this has been observed in FA cells (Ho et al., 2006; Taniguchi et al., 2002). These latter experiments were conducted as a function of IR dose in order to examine the S phase checkpoint over a wide degree of DNA damage. These findings indicate that Snm1B is required for the enforcement of an S phase checkpoint specifically in response to ICLs, but not to DSBs introduced by IR. To demonstrate that these observed cell cycle effects are specific to Snm1B knockdown, we attempted a rescue with shRNA-refractory constructs. As shown (Fig. 3E), both wild-type SNM1B and the D14N mutant were able to rescue the G2/M accumulation phenotype, whereas, a mutant (ΔCD) with the entire SNM1 conserved domain deleted was unable to achieve a rescue.
Figure 3. Cell cycle analysis of SNM1B knockdown cells after MMC treatment.
(A) Knockdown of Snm1B results in G2/M accumulation in the presence of MMC. The indicated clones were continuously exposed to MMC (100 ng/ml) and examined at the indicated time points for DNA content by FACS analysis. (B) Quantification of the G2/M accumulation shown in (A). (C,D) SNM1B-deficient cells are defective in an S phase checkpoint after treatment with MMC, but not with IR. DNA synthesis was evaluated (as described in Experimental Procedures) with the indicated clones after treatment with MMC (10 µg/ml for 1 hr), or as a function of IR dose. (E) The hydrolase activity of Snm1B is not required for rescue of the G2/M accumulation phenotype. The indicated clones were either treated with MMC (50 ng/ml) for 24 hrs or not treated, and then analyzed by FACS. For the rescue experiments (lower panels), the indicated SNM1B constructs were transfected into #5 cells, and 24 hrs later MMC was added for an additional 24 hrs. The Snm1B proteins were tagged with EGFP and only the GPF positive cell populations as determined by FACS analysis are shown. The SNM1B alleles are as described in Fig. 3A, except for SNM1ΔCD which lacks the entire conserved domain (metallo-β-lactamase plus β-CASP).
Snm1B is Required for Checkpoint Signaling in Response to ICL Damage
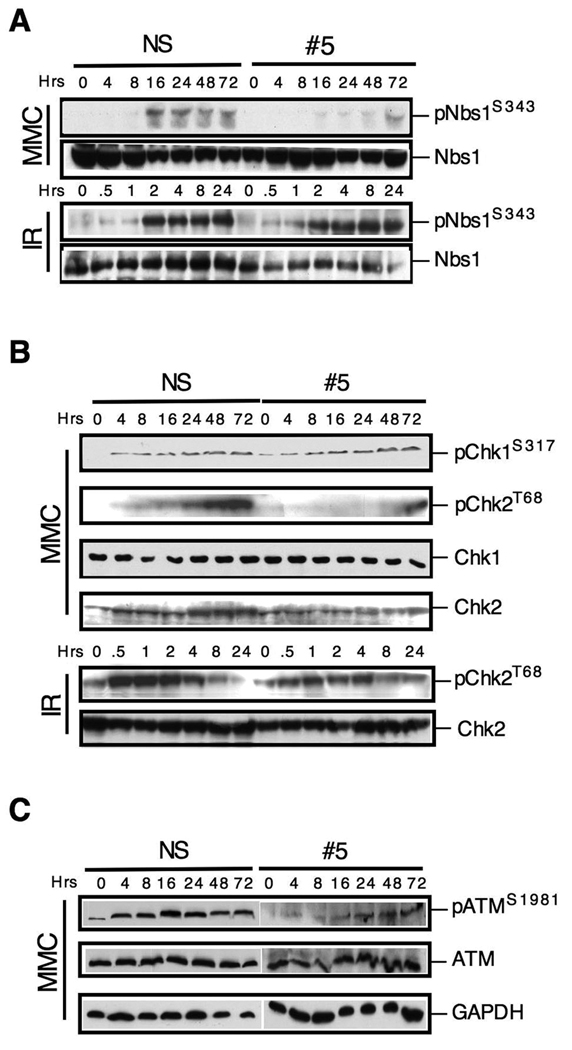
Since Snm1B is required for the S phase checkpoint in response to ICL damage, we next examined whether known markers of checkpoint activation were affected by its knockdown. Phosphorylation of the Nbs1 serine 343 residue (S343) is known to occur in response to both IR and ICL damage. Examination of clone #5 showed that in response to MMC, but not to IR, there was defective phosphorylation of this residue (Fig. 4A). This result is consistent with the findings shown above (Fig. 3) indicating that Snm1B-deficient cells are defective in an S phase checkpoint in response to ICL damage, but not IR-induced DSBs. Chk1 is also known to be phosphorylated by ATR after ICL damage (Pichierri and Rosselli, 2004b). We, therefore, examined the phosphorylation of Chk1 and as a control Chk2 after MMC treatment. Interestingly, the phosphorylation of Chk1 was unaffected in the clone #5 cells, whereas, the phosphorylation of Chk2 was strongly reduced (Fig. 4B, upper panel). However, Chk2 phosphorylation was not affected after exposure of cells to IR (Fig. 4B, lower panel). Chk2 is typically a substrate of ATM as opposed to ATR, we therefore examined the activation of ATM as indicated by phosphorylation at the S1981 residue, which is a marker for activation of this kinase (Bakkenist and Kastan, 2003). As shown (Fig. 4C), activation of ATM after MMC treatment is reduced in clone #5 compared to control cells. Finally, monoubiquitination of FancD2 by the FA core complex is a hallmark of fork stalling in response to ICLs. We, however, found no defect in this modification in Snm1B-depleted cells after treatment with MMC indicating that this branch of the signaling pathways is intact (results not shown), consistent with the finding reported by Demuth et al. (Demuth et al., 2004).
Figure 4. SNM1B is required for checkpoint signaling through the ATM pathway.
(A) Phosphorylation of Nbs1 is defective in SNM1B knockdown cells upon exposure to MMC. The NS and #5 clones were treated with MMC (50 ng/ml) continuously or to IR (2 Gy), and harvested at the indicated time points for immunoblotting. (B) Phosphorylation of Chk2, but not Chk1, is defective in SNM1B knockdown cells upon exposure to MMC. The NS and #5 clones were continuously exposed to MMC (300 ng/ml) or to IR (2 Gy) for the indicated times. (C) Activation of ATM is defective in SNM1B knockdown cells. The NS and #5 clones were continuously exposed to MMC (300 ng/ml) for the indicated times.
Snm1B Interacts with MRN and FancD2
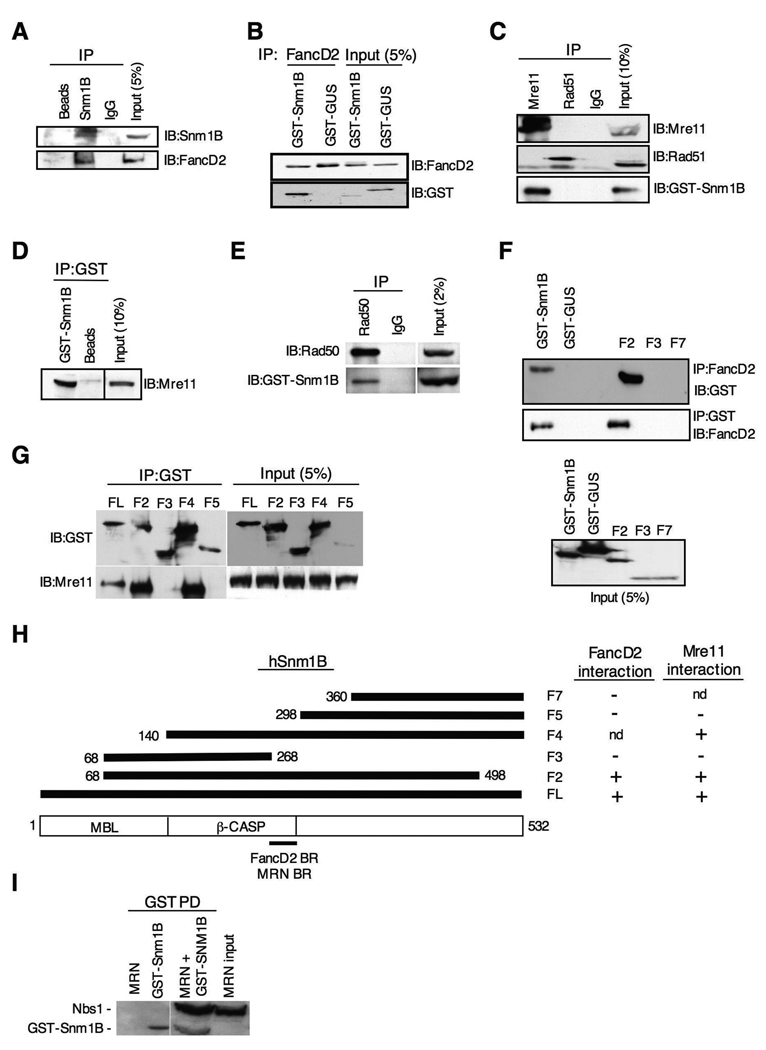
Mre11, Rad50 and Nbs1 form a complex, referred to as MRN, that has been implicated in both DNA repair processing and cell cycle checkpoint signaling in response to ICLs (Nakanishi et al., 2002; Pichierri et al., 2002; Pichierri and Rosselli, 2004b). In addition, FancD2 and Nbs1 have been shown to colocalize in MMC-induced nuclear foci, and to co-IP together (Nakanishi et al., 2002). To determine whether Snm1B interacts with proteins known to be involved in the cellular ICL response, we performed co-IP experiments. A positive interaction was found for FancD2 (Fig. 5A,B), and Mre11 and Rad50 (Fig. 5C–E), but not for other proteins such as Brca1, FancA, FancG, and Rad51 (Fig. 5C and results not shown). Reciprocal co-IPs confirmed the interactions with Mre11, Rad50 and FancD2. These positive interactions were not mediated by DNA as the inclusion of ethidium bromide did not affect the co-IP experiments (results not shown). To ascertain the regions of Snm1B that mediated these interactions, several deletion constructs of Snm1B were prepared and examined in the co-IP assay. Interestingly, both the Mre11 and FancD2 interaction domains mapped to the same region of Snm1B located near the carboxy terminal end of the β-CASP domain (Fig. 5F–H), although it should be noted that while this region is necessary, it may not be sufficient for these interactions. Co-IP assays performed with purified recombinant proteins showed that Snm1B does not directly interact with FancD2 (results not shown), but does appear to directly interact with the MRN complex (Fig. 5I).
Figure 5. Snm1B interacts with FancD2 and the MRN complex.
(A) Antibodies to Snm1B co-IP FancD2. GST-Snm1B was transiently expressed in HEK293 cells, and the indicated co-IP assays were performed from lysates. “Beads” indicates IP with sepharose A beads only. “IgG” indicates IP with nonspecific antibody. (B) Antibodies to FancD2 co-IP Snm1B. GST-Snm1B or the control GST-GUS were transiently expressed in HEK293 cells, and co-IP assays were performed from lysates. (C) Antibodies to Mre11, but not Rad51, co-IP Snm1B. GST-Snm1B was transiently expressed in HEK293 cells, and the indicated co-IP assays were performed from lysates. “IgG” indicates IP with nonspecific antibody. (D) Reciprocal co-IP of Mre11 with antibodies to GST. “Beads” indicates IP with sepharose A beads only. (E) Snm1B co-IPs with Rad50. GST-Snm1B was transiently expressed in HEK293 cells, and the indicated co-IP assays were performed from lysates. (F) The FancD2 interaction domain maps to the carboxy terminal end of the β-CASP domain of Snm1B. The indicated GST-SNM1B deletion constructs were transiently expressed in HEK293 cells, and the indicated co-IP assays were performed. “FL” indicates full-length Snm1B. (G) The Mre11 interaction domain maps to the carboxy terminal end of the β-CASP domain of Snm1B. The indicated GST-SNM1B deletion constructs were transiently expressed in HEK293 cells and the indicated co-IP assays were performed. (H) Schematic depicting the interaction mapping results from (F) and (G). “nd” indicates not determined. “BR” indicates binding region. (I) Snm1B interacts directly with the MRN complex. Purified recombinant GST-Snm1B and MRN complex were incubated together, and then subjected to the indicated pull-down (PD) assay. Immunoblotting was performed with anti-GST and anti-Nbs1.
Snm1B is Required for Replication Fork Collapse at ICLs
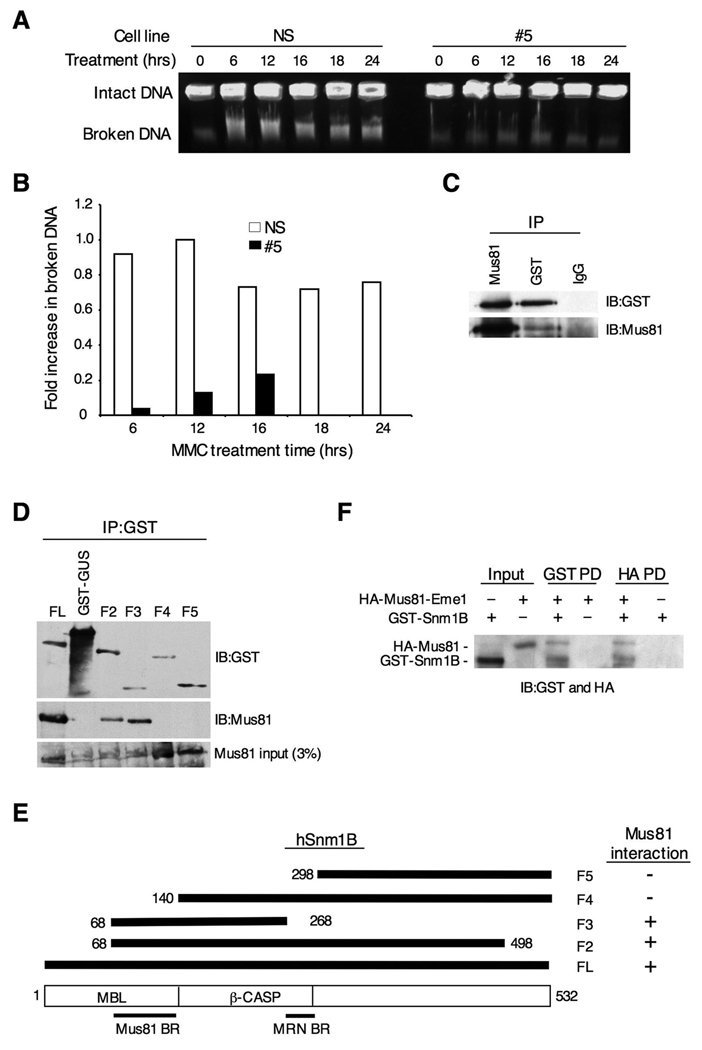
Our findings thus far indicate that Snm1B is required for the activation of ATM and Chk2 upon treatment with MMC, but not after exposure to IR. ATM is principally activated after induction of DSBs, thus, one possible explanation for our results is that Snm1B is required for the introduction of DSBs that occur during replication fork collapse in response to ICLs. To test this hypothesis, we used pulsed-field gel electrophoresis to separate intact chromosomes from broken DNA after exposure of cells to MMC (Hanada et al., 2006; Mladenov et al., 2007). As shown, clone #5 cells showed both a delay and an overall reduction in the amount of broken DNA compared to control cells indicating that Snm1B is required for replication fork collapse (Fig. 6A,B), thus validating our hypothesis.
Figure 6. Snm1B is required for fork collapse after MMC treatment.
(A) Double-strand break formation was analyzed by pulsed field gel electrophoresis. Intact DNA stays in the well while broken DNA migrates into the agarose gel. The NS and #5 clones were incubated with MMC (1 ug/ml) for the indicated times, and cells were collected in agarose plugs for gel electrophoresis. (B) Quantitation of results shown in (A). Images were quantitated using ImageJ 1.37v software (developed by Wayne Rasband, NIH). The background present at the 0 hr time point was subtracted from the other time points, and the band with the highest intensity was normalized to a value of 1.0. (C) Snm1B co-IPs with Mus81. GST-Snm1B was transiently expressed in HEK293 cells, and the indicated co-IP assays were performed from lysates. (D) The Mus81 interaction domain maps to the metallo-β-lactamase domain of Snm1B. The indicated GST-SNM1B deletion constructs were expressed in HEK293 cells and the indicated co-IP assays were performed and analyzed by immunoblotting. (E) Schematic depicting the results from (D). (F) Snm1B directly interacts with Mus81-Eme1. Immunoblots showing results of reciprocal pull-down (PD) assays between GST-Snm1B and HA-Mus81-Eme1. HA indicates the hemagglutinin tag.
Recently, it has been shown that the structure-specific endonuclease Mus81-Eme1 is required for replication fork collapse after treatment with MMC (Hanada et al., 2006). We therefore tested whether Snm1B and Mus81-Eme1 interact with each other. Antibodies to Snm1B were found to robustly co-IP Mus81, and this interaction was confirmed by a reciprocal co-IP with Mus81 antibodies (Fig. 6C). To further define this interaction, we mapped the Mus81-Eme1 interaction site on Snm1B, and were able to limit it to within the metallo-β-lactamase domain (Fig. 6D,E), although it is possible that other regions of Snm1B are involved in mediating this interaction. Nevertheless, the interaction between Mus81-Eme1 and Snm1B is distinct from the interaction described above for Snm1B with FancD2 and the MRN complex. Finally, we used affinity-purified recombinant preparations of GST-Snm1B and HA-Mus81-Eme1 to demonstrate that the interaction between these proteins is direct (Fig. 6F). Taken together, these findings suggest that Snm1B and Mus81-Eme1 cooperate together in mediating replication fork collapse in response to ICLs.
Discussion
In this study, we have shown that Snm1B is uniquely involved in the cellular response to ICLs in DNA, but is not involved in the response to frank DSBs created by IR. This unique role is explained by our finding that Snm1B is required for induction of DSBs that occur at replication forks as they encounter an ICL. Until recently it was not clear whether replication fork collapse was a regulated biochemical process or an unintended consequence of long-term fork stalling mediated by mechanical processes. The discovery that the structure specific endonuclease Mus81-Eme1 is specifically required for DSB formation suggested that the former view is likely correct (Hanada et al., 2006). Moreover, it seems highly probable that fork collapse is a necessary, if drastic, step for the eventual removal of the ICL. The finding that Mus81-Eme1 and Snm1B directly interact with each other strengthens the conclusion that both of these proteins participate in actively promoting DSBs in response to ICLs. Interestingly, although the SNM1 gene family has been shown to possess nucleic acid processing activity mediated through its conserved metallo-β-lactamase and β-CASP domains (Dominski, 2007; Lenain et al., 2006; Li et al., 2005; Ma et al., 2002; Pannicke et al., 2004), inactivation of the hydrolase domain of Snm1B by a point mutation did not affect the normal cell cycle response to fork collapse. This result suggests that Mus81-Eme1 is likely the actual endonuclease forming the DSB, while Snm1B may be required to either recruit or maintain Mus81-Eme1 at the site of the stalled replication fork. Interestingly, the metallo-β-lactamase domain of Snm1B is required for the interaction with Mus81-Eme1 indicating that in addition to its hydrolase function this domain also mediates protein-protein interactions. Our results also suggest that Snm1B may not be involved in the subsequent recombinational steps of ICL repair since it was not required in the HDR assay (Zhang et al., 2007), which measures ICL-stimulated gene conversion in conjunction with a DSB. Furthermore, Snm1B did not interact with Rad51. These results are in contrast to Mus81, which interacts directly with the recombination protein Rad54, and appears to act in the same pathway of ICL repair (Hanada et al., 2006; Interthal and Heyer, 2000). Nevertheless, since fork collapse is likely required for the ultimate excision and uncoupling of the ICL, Snm1B would be a necessary element for the repair of these lesions via an S phase repair pathway.
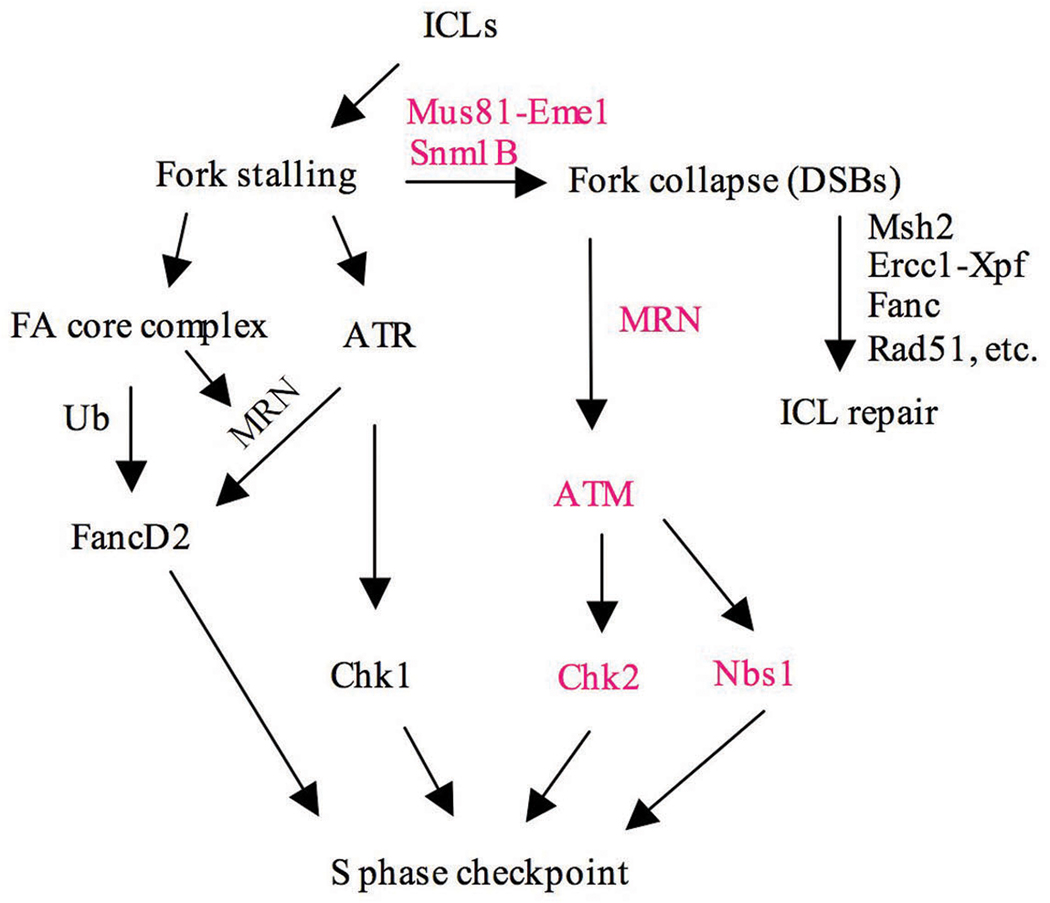
Previous studies have shown that the ATR-mediated checkpoint signaling pathway is activated in response to ICLs detected in the S phase of the cell cycle. As shown (Fig. 7), ATR signaling is composed of two branches represented by the ATR-Chk1 subpathway and a second branch involving ATR-Nbs1-FancD2 (Lukas et al., 2004; Nakanishi et al., 2005; Pichierri and Rosselli, 2004b). The former pathway, whose target is Cdc25A, is well understood, while less is known about the second branch, which involves ATR-mediated Nbs1-dependent phosphorylation of FancD2 and subsequent ubiquitylation by the FA core complex (Andreassen et al., 2004; Stiff et al., 2005). How this latter pathway interacts with the cell cycle machinery is not clear at present. Our findings indicate that, in addition to the ATR-mediated pathways, subsequent to DSB induction and fork collapse, ATM-mediated checkpoint pathways are also activated. Similar to the ATR pathways, ATM signaling is also mediated by two parallel pathways involving ATM-Chk2 and ATM-Nbs1-Smc1 (Bartek et al., 2004; Lambert and Carr, 2005). The involvement of ATM in checkpoint signaling in response to ICLs has not been previously described, and in fact experiments to detect such an involvement have largely proven negative (Mladenov et al., 2007; Pichierri and Rosselli, 2004b). This situation is likely due to the fact that activation of ATM only occurs after fork collapse, which usually takes place many hours after introduction of the ICL-inducing drug, while most checkpoint assays are performed within a few hours of drug treatment. High drug concentrations can likely accelerate this process, which is presumably why we were able to detect a defective checkpoint response at early times in the absence of Snm1B. Thus, in response to ICLs there appears to be an early and possibly transient checkpoint activation mediated by ATR in response to fork stalling (Pichierri and Rosselli, 2004a; Pichierri and Rosselli, 2004b), and a later checkpoint activation mediated by ATM in response to formation of DSBs at stalled forks (Fig. 7). Snm1B is involved in this later checkpoint activation through its role in mediating fork collapse in collaboration with Mus81-Eme1. We also did not observe an effect on the monoubiquitylation of FancD2 in the Snm1B knockdown cells which is consistent with this modification being ATR- but not ATM-dependent in response to ICL damage (Taniguchi et al., 2002).We cannot rule out that Snm1B might have additional direct role(s) in checkpoint signaling such as the activation of Nbs1 by either ATM or ATR, since Nbs1 phosphorylation appears to be highly compromised after ICL damage in the Snm1B knockdown cells. This contention is supported by our finding that Snm1B directly interacts with the MRN complex. The functional significance of this interaction requires further investigation, however, there are at least two possibilities. One is that the MRN complex is involved in recruiting Snm1B specifically to stalled replication forks at ICLs to promote fork collapse, and a second is that Snm1B arrives early at the stalled fork and subsequently is involved in recruiting MRN, FancD2, and Mus81-Eme1.
Figure 7. Model depicting the role of Snm1B in the cellular response to ICLs.
Items in bold indicate pathways likely affected by Snm1B.
Together our findings demonstrate that Snm1B is specifically involved in mediating resistance to ICL-inducing compounds by its role in promoting replication fork collapse, which leads to activation of ATM-mediated cell cycle arrest and likely initiation of removal of the lesion. Since Snm1B/Apollo has also been implicated in telomere protection during S phase (Freibaum and Counter, 2006; Lenain et al., 2006; van Overbeek and de Lange, 2006), it is clear that it has a multifunctional role in maintenance of genomic stability, and experiments designed to assess its role as a tumor suppressor merit further investigation.
Materials and Methods
shRNA-Mediated Stable Knockdown of Snm1B Expression
HEK293T cells were transfected with an shRNA vector (Open Biosystems) targeted for SNM1B (target sequence: 5’gaaacagatccatacttta) and a nonspecific shRNA control vector. Positive clones were selected in Puromycin (2.5 mg/ml), and knockdown of Snm1B was confirmed by immunoblotting. For immunoblotting rabbit polyclonal antibodies against Snm1B were generated using a polypeptide encompassing the region (residues 304 to 532) of the protein not conserved in other family members.
Clonogenic Cell Survival Assay
Cells were plated to achieve a final colony density of approximately 100–300 colonies per 10 cm dish. One day after plating irradiation was performed or drugs were added, and the medium was left unchanged until colonies appeared. Cells were fixed and stained with 10% Crystal Violet in 100% ethanol.
Chromosome Analysis
Chromosome analyses were performed as previously described (Multani et al., 2000). At least 35 metaphases were analyzed from each sample for chromosome aberrations.
In Vivo Plasmid-Based DNA Repair Assays
Preparation of cross-linked plasmids, and assays for recombination-independent repair (Wang et al., 2001; Zheng et al., 2003), single-strand annealing (Zheng et al., 2006), and homology-dependent repair (Zhang et al., 2007) were carried out as described.
Site directed mutagenesis
The QuikChange™ Site Directed Mutagenesis Kit (Invitrogen) was used to prepare mutated SNM1B constructs. Primers used for the shRNA resistant clone (SNM1BR) were: 5’gggaagcaaatccacaccttatacc and 5’ggtataaggtgaggatttgcttccc. Primers used for the D14N point mutation were: 5’gcc catcgcagtgaacttctggagcc and 5’ggctccagaagttcactgcgatgggc.
Co-Immunoprecipitation and Pull-Down Assays
Cells were grown in 60 mm plates, washed with cold PBS twice, and lysed by adding 500 µl of EBC buffer (50 mM Tris-HCl [pH 8.0], 120 mM NaCl, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride) on ice for 20 min. In some cases, cells were transfected with a construct (pDEST27-SNM1B) expressing a glutathione S-transferase (GST)-Snm1B fusion protein and incubated for 2 days prior to extract preparation. Lysate preparation and co-IP assays were performed as previously described (Zhang et al., 2004).
Pull-down assays with purified recombinant proteins were conducted essentially as previously described (Zhang et al., 2005). GST-Snm1B and HA-Mus81-Eme1 were expressed in E. coli and Sf9 insect cells, respectively. Both proteins were purified by affinity chromatography.
Evaluation of DNA Synthesis
To analyze inhibition of DNA synthesis by MMC and IR (Painter and Young, 1980), cells were prelabeled with 14C-thymidine (50 nCi/ml) for 1 day. Cells were treated either with MMC (10 µg/ml) for 1 hr or different doses of IR, and pulse-labeled with 3H-thymidine (10 µCi/ml) for 15 min before harvesting. Cells were fixed with 70% methanol and radioactivity was measured by scintillography. The degree of DNA synthesis was derived from the resulting ratios of 3H to 14C counts per minute and expressed as a percentage of untreated cells.
Detection of DSBs by Pulsed-Field Gel Electorphoresis
Subconfluent cell cultures were treated with 1 µg/ml of MMC for different times. Cells were harvested after trypsinization, and agarose plugs containing 106 cells were prepared and DNAs separated by pulse-field gel electrophoresis as previously described (Hanada et al., 2006; Mladenov et al., 2007).
Acknowledgements
We thank Tanya Paull and Steve Patrick for the gift of MRN protein, and Jeffrey Parvin for the gift of FancD2 protein. This work was supported by NCI grants CA052461 and CA097175. DNA sequencing resources were supported by the Cancer Center Support (Core) Grant CA16672.
References
- Ahkter S, Richie CT, Zhang N, Behringer RR, Zhu C, Legerski RJ. Snm1-deficient mice exhibit accelerated tumorigenesis and susceptibility to infection. Mol Cell Biol. 2005;25:10071–10078. doi: 10.1128/MCB.25.22.10071-10078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhter S, Richie CT, Deng JM, Brey E, Zhang X, Patrick, et al. Deficiency in SNM1 abolishes an early mitotic checkpoint induced by spindle stress. Mol Cell Biol. 2004;24:10448–10455. doi: 10.1128/MCB.24.23.10448-10455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkari YM, Bateman RL, Reifsteck CA, D'Andrea AD, Olson SB, Grompe M. The 4N cell cycle delay in Fanconi anemia reflects growth arrest in late S phase. Mol Genet Metab. 2001;74:403–412. doi: 10.1006/mgme.2001.3259. [DOI] [PubMed] [Google Scholar]
- Andreassen PR, D'Andrea AD, Taniguchi T. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 2004;18:1958–1963. doi: 10.1101/gad.1196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Barber LJ, Ward TA, Hartley JA, McHugh PJ. DNA interstrand cross-link repair in the Saccharomyces cerevisiae cell cycle: overlapping roles for PSO2 (SNM1) with MutS factors and EXO1 during S phase. Mol Cell Biol. 2005;25:2297–2309. doi: 10.1128/MCB.25.6.2297-2309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat Rev Mol Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- Callebaut I, Moshous D, Mornon JP, de Villartay JP. Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res. 2002;30:3592–3601. doi: 10.1093/nar/gkf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DW, Chen BP, Prithivirajsingh S, Kurimasa A, Story MD, Qin J, et al. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002;16:2333–2338. doi: 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi P, Sudakin V, Bobiak ML, Fisher PW, Mattern MR, Jablonski S, et al. Chfr regulates a mitotic stress pathway through its RING-finger domain with ubiquitin ligase activity. Cancer Res. 2002;62:1797–1801. [PubMed] [Google Scholar]
- Demuth I, Digweed M, Concannon P. Human SNM1B is required for normal cellular response to both DNA interstrand crosslink-inducing agents and ionizing radiation. Oncogene. 2004;23:8611–8618. doi: 10.1038/sj.onc.1207895. [DOI] [PubMed] [Google Scholar]
- Dominski Z. Nucleases of the metallo-beta-lactamase family and their role in DNA and RNA metabolism. Crit Rev Biochem Mol Biol. 2007;42:67–93. doi: 10.1080/10409230701279118. [DOI] [PubMed] [Google Scholar]
- Dronkert ML, de Wit J, Boeve M, Vasconcelos ML, van Steeg H, Tan TL, et al. Disruption of mouse SNM1 causes increased sensitivity to the DNA interstrand cross-linking agent mitomycin C. Mol Cell Biol. 2000;20:4553–4561. doi: 10.1128/mcb.20.13.4553-4561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Counter CM. hSnm1B is a novel telomere-associated protein. J Biol Chem. 2006;281:15033–15036. doi: 10.1074/jbc.C600038200. [DOI] [PubMed] [Google Scholar]
- Geng L, Zhang X, Zheng S, Legerski RJ. Artemis Links ATM to G2/M Checkpoint Recovery Via Regulation of Cdk1-Cyclin B. Mol Cell Bio. 2007;27:2625–2635. doi: 10.1128/MCB.02072-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann KF, Ward AM, Matkovic ME, Folias AE, Moses RE. S. cerevisiae has three pathways for DNA interstrand crosslink repair. Mutat Res. 2001;487:73–83. doi: 10.1016/s0921-8777(01)00106-9. [DOI] [PubMed] [Google Scholar]
- Haase E, Riehl D, Mack M, Brendel M. Molecular cloning of SNM1, a yeast gene responsible for a specific step in the repair of cross-linked DNA. Mol Gen Genet. 1989;218:64–71. doi: 10.1007/BF00330566. [DOI] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, Essers J, et al. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. Embo J. 2006;25:4921–4932. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques JA, Moustacchi E. Isolation and characterization of pso mutants sensitive to photo-addition of psoralen derivatives in Saccharomyces cerevisiae. Genetics. 1980;95:273–288. doi: 10.1093/genetics/95.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho GP, Margossian S, Taniguchi T, D'Andrea AD. Phosphorylation of FANCD2 on two novel sites is required for mitomycin C resistance. Mol Cell Biol. 2006;26:7005–7015. doi: 10.1128/MCB.02018-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Interthal H, Heyer WD. MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol Gen Genet. 2000;263:812–827. doi: 10.1007/s004380000241. [DOI] [PubMed] [Google Scholar]
- Ishiai M, Kimura M, Namikoshi K, Yamazoe M, Yamamoto K, Arakawa H, et al. DNA cross-link repair protein SNM1A interacts with PIAS1 in nuclear focus formation. Mol Cell Biol. 2004;24:10733–10741. doi: 10.1128/MCB.24.24.10733-10741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S, Carr AM. Checkpoint responses to replication fork barriers. Biochimie. 2005;87:591–602. doi: 10.1016/j.biochi.2004.10.020. [DOI] [PubMed] [Google Scholar]
- Lenain C, Bauwens S, Amiard S, Brunori M, Giraud-Panis MJ, Gilson E. The Apollo 5' exonuclease functions together with TRF2 to protect telomeres from DNA repair. Curr Biol. 2006;16:1303–1310. doi: 10.1016/j.cub.2006.05.021. [DOI] [PubMed] [Google Scholar]
- Li X, Hejna J, Moses RE. The yeast Snm1 protein is a DNA 5'-exonuclease. DNA Repair (Amst) 2005;4:163–170. doi: 10.1016/j.dnarep.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Li X, Moses RE. The beta-lactamase motif in Snm1 is required for repair of DNA double-strand breaks caused by interstrand crosslinks in S. cerevisiae. DNA Repair (Amst) 2003;2:121–129. doi: 10.1016/s1568-7864(02)00192-1. [DOI] [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J. Mammalian cell cycle checkpoints: signalling pathways and their organization in space and time. DNA Repair (Amst) 2004;3:997–1007. doi: 10.1016/j.dnarep.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- Magana-Schwencke N, Henriques JA, Chanet R, Moustacchi E. The fate of 8-methoxypsoralen photoinduced crosslinks in nuclear and mitochondrial yeast DNA: comparison of wild-type and repair-deficient strains. Proc Natl Acad Sci U S A. 1982;79:1722–1726. doi: 10.1073/pnas.79.6.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsusaka T, Pines J. Chfr acts with the p38 stress kinases to block entry to mitosis in mammalian cells. J Cell Biol. 2004;166:507–516. doi: 10.1083/jcb.200401139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson JP, Lemmers B, Chahwan R, Pamidi A, Migon E, Matysiak-Zablocki E, et al. Involvement of mammalian Mus81 in genome integrity and tumor suppression. Science. 2004;304:1822–1826. doi: 10.1126/science.1094557. [DOI] [PubMed] [Google Scholar]
- Mladenov E, Tsaneva I, Anachkova B. Activation of the S phase DNA damage checkpoint by mitomycin C. J Cell Physiol. 2007;211:468–476. doi: 10.1002/jcp.20957. [DOI] [PubMed] [Google Scholar]
- Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- Moshous D, Li L, Chasseval R, Philippe N, Jabado N, Cowan MJ, et al. A new gene involved in DNA double-strand break repair and V(D)J recombination is located on human chromosome 10p. Hum Mol Genet. 2000;9:583–588. doi: 10.1093/hmg/9.4.583. [DOI] [PubMed] [Google Scholar]
- Multani AS, Ozen M, Narayan S, Kumar V, Chandra J, McConkey DJ, et al. Caspase-dependent apoptosis induced by telomere cleavage and TRF2 loss. Neoplasia. 2000;2:339–345. doi: 10.1038/sj.neo.7900105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Taniguchi T, Ranganathan V, New HV, Moreau LA, Stotsky M, et al. Interaction of FANCD2 and NBS1 in the DNA damage response. Nat Cell Biol. 2002;4:913–920. doi: 10.1038/ncb879. [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D'Andrea AD, et al. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci U S A. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima K, Hochegger H, Saberi A, Fukushima T, Kikuchi K, Yoshimura M, et al. Multiple repair pathways mediate tolerance to chemotherapeutic cross-linking agents in vertebrate cells. Cancer Res. 2005;65:11704–11711. doi: 10.1158/0008-5472.CAN-05-1214. [DOI] [PubMed] [Google Scholar]
- Painter RB, Young BR. Radiosensitivity in ataxia-telangiectasia: a new explanation. Proc Natl Acad Sci U S A. 1980;77:7315–7317. doi: 10.1073/pnas.77.12.7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannicke U, Ma Y, Hopfner KP, Niewolik D, Lieber MR, Schwarz K. Functional and biochemical dissection of the structure-specific nuclease ARTEMIS. Embo J. 2004;23:1987–1997. doi: 10.1038/sj.emboj.7600206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichierri P, Averbeck D, Rosselli F. DNA cross-link-dependent RAD50/MRE11/NBS1 subnuclear assembly requires the Fanconi anemia C protein. Hum Mol Genet. 2002;11:2531–2546. doi: 10.1093/hmg/11.21.2531. [DOI] [PubMed] [Google Scholar]
- Pichierri P, Rosselli F. Fanconi anemia proteins and the s phase checkpoint. Cell Cycle. 2004a;3:698–700. [PubMed] [Google Scholar]
- Pichierri P, Rosselli F. The DNA crosslink-induced S-phase checkpoint depends on ATR-CHK1 and ATR-NBS1-FANCD2 pathways. Embo J. 2004b;23:1178–1187. doi: 10.1038/sj.emboj.7600113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Richie CT, Peterson C, Lu T, Hittelman WN, Carpenter PB, Legerski RJ. hSnm1 colocalizes and physically associates with 53BP1 before and after DNA damage. Mol Cell Biol. 2002;22:8635–8647. doi: 10.1128/MCB.22.24.8635-8647.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney S, Sekiguchi J, Whitlow S, Eckersdorff M, Manis JP, Lee C, et al. Artemis and p53 cooperate to suppress oncogenic N-myc amplification in progenitor B cells. Proc Natl Acad Sci U S A. 2004;101:2410–2415. doi: 10.1073/pnas.0308757101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney S, Sekiguchi J, Zhu C, Cheng HL, Manis J, Whitlow S, et al. Leaky Scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol Cell. 2002;10:1379–1390. doi: 10.1016/s1097-2765(02)00755-4. [DOI] [PubMed] [Google Scholar]
- Ruhland A, Kircher M, Wilborn F, Brendel M. A yeast mutant specifically sensitive to bifunctional alkylation. Mutat Res. 1981;91:457–462. doi: 10.1016/0165-7992(81)90052-x. [DOI] [PubMed] [Google Scholar]
- Sala-Trepat M, Rouillard D, Escarceller M, Laquerbe A, Moustacchi E, Papadopoulo D. Arrest of S-phase progression is impaired in Fanconi anemia cells. Exp Cell Res. 2000;260:208–215. doi: 10.1006/excr.2000.4994. [DOI] [PubMed] [Google Scholar]
- Scolnick DM, Halazonetis TD. Chfr defines a mitotic stress checkpoint that delays entry into metaphase. Nature. 2000;406:430–435. doi: 10.1038/35019108. [DOI] [PubMed] [Google Scholar]
- Seyschab H, Friedl R, Sun Y, Schindler D, Hoehn H, Hentze S, et al. Comparative evaluation of diepoxybutane sensitivity and cell cycle blockage in the diagnosis of Fanconi anemia. Blood. 1995;85:2233–2237. [PubMed] [Google Scholar]
- Shen X, Jun S, O'Neal LE, Sonoda E, Bemark M, Sale JE, et al. REV3 and REV1 play major roles in recombination-independent repair of DNA interstrand cross-links mediated by monoubiquitinated proliferating cell nuclear antigen (PCNA) J Biol Chem. 2006;281:13869–13872. doi: 10.1074/jbc.C600071200. [DOI] [PubMed] [Google Scholar]
- Sobeck A, Stone S, Costanzo V, de Graaf B, Reuter T, de Winter J, et al. Fanconi anemia proteins are required to prevent accumulation of replication-associated DNA double-strand breaks. Mol Cell Biol. 2006;26:425–437. doi: 10.1128/MCB.26.2.425-437.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiff T, Reis C, Alderton GK, Woodbine L, O'Driscoll M, Jeggo PA. Nbs1 is required for ATR-dependent phosphorylation events. Embo J. 2005;24:199–208. doi: 10.1038/sj.emboj.7600504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers MK, Bothos J, Halazonetis TD. The CHFR mitotic checkpoint protein delays cell cycle progression by excluding Cyclin B1 from the nucleus. Oncogene. 2005;24:2589–2598. doi: 10.1038/sj.onc.1208428. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Garcia-Higuera I, Xu B, Andreassen PR, Gregory RC, Kim ST, et al. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002;109:459–472. doi: 10.1016/s0092-8674(02)00747-x. [DOI] [PubMed] [Google Scholar]
- Thompson LH, Hinz JM, Yamada NA, Jones NJ. How Fanconi anemia proteins promote the four Rs: replication, recombination, repair, and recovery. Environ Mol Mutagen. 2005;45:128–142. doi: 10.1002/em.20109. [DOI] [PubMed] [Google Scholar]
- van Overbeek M, de Lange T. Apollo, an Artemis-related nuclease, interacts with TRF2 and protects human telomeres in S phase. Curr Biol. 2006;16:1295–1302. doi: 10.1016/j.cub.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Wang J, Pluth JM, Cooper PK, Cowan MJ, Chen DJ, Yannone SM. Artemis deficiency confers a DNA double-strand break repair defect and Artemis phosphorylation status is altered by DNA damage and cell cycle progression. DNA Repair (Amst) 2005;4:556–570. doi: 10.1016/j.dnarep.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Wang X, Peterson CA, Zheng H, Nairn RS, Legerski RJ, Li L. Involvement of nucleotide excision repair in a recombination-independent and error-prone pathway of DNA interstrand cross-link repair. Mol Cell Biol. 2001;21:713–720. doi: 10.1128/MCB.21.3.713-720.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilborn F, Brendel M. Formation and stability of interstrand cross-links induced by cis- and trans-diamminedichloroplatinum (II) in the DNA of Saccharomyces cerevisiae strains differing in repair capacity. Curr Genet. 1989;16:331–338. doi: 10.1007/BF00340711. [DOI] [PubMed] [Google Scholar]
- Xu B, Kim ST, Lim DS, Kastan MB. Two molecularly distinct G(2)/M checkpoints are induced by ionizing irradiation. Mol Cell Biol. 2002;22:1049–1059. doi: 10.1128/MCB.22.4.1049-1059.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Minter-Dykhouse K, Malureanu L, Zhao WM, Zhang D, Merkle CJ, et al. Chfr is required for tumor suppression and Aurora A regulation. Nat Genet. 2005;37:401–406. doi: 10.1038/ng1538. [DOI] [PubMed] [Google Scholar]
- Zhang N, Kaur R, Lu X, Shen X, Li L, Legerski RJ. The Pso4 mRNA splicing and DNA repair complex interacts with WRN for processing of DNA interstrand cross-links. J Biol Chem. 2005;280:40559–40567. doi: 10.1074/jbc.M508453200. [DOI] [PubMed] [Google Scholar]
- Zhang N, Liu X, Li L, Legerski R. Double-strand breaks induce homologous recombinational repair of interstrand cross-links via cooperation of MSH2, ERCC1-XPF, REV3, and the Fanconi anemia pathway. DNA Repair (Amst) 2007;6:1670–1678. doi: 10.1016/j.dnarep.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Succi J, Feng Z, Prithivirajsingh S, Story MD, Legerski RJ. Artemis is a phosphorylation target of ATM and ATR and is involved in the G2/M DNA damage checkpoint response. Mol Cell Biol. 2004;24:9207–9220. doi: 10.1128/MCB.24.20.9207-9220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Wang X, Legerski RJ, Glazer PM, Li L. Repair of DNA interstrand cross-links: interactions between homology-dependent and homology-independent pathways. DNA Repair (Amst) 2006;5:566–574. doi: 10.1016/j.dnarep.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Zheng H, Wang X, Warren AJ, Legerski RJ, Nairn RS, Hamilton JW, et al. Nucleotide excision repair- and polymerase eta-mediated error-prone removal of mitomycin C interstrand cross-links. Mol Cell Biol. 2003;23:754–761. doi: 10.1128/MCB.23.2.754-761.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]