Abstract
The engineering of biological systems is anticipated to provide effective solutions to challenges that include energy and food production, environmental quality, and health and medicine. Our ability to transmit information to and from living systems, and to process and act on information inside cells, is critical to advancing the scale and complexity at which we can engineer, manipulate, and probe biological systems. We developed a general approach for assembling RNA devices that can execute higher-order cellular information processing operations from standard components. The engineered devices can function as logic gates (AND, NOR, NAND, or OR gates) and signal filters, and exhibit cooperativity. RNA devices process and transmit molecular inputs to targeted protein outputs, linking computation to gene expression and thus the potential to control cellular function.
Genetically encoded technologies that perform information processing, communication, and control operations are needed to produce new cellular functions from the diverse molecular information encoded in the various properties of small molecules, proteins, and RNA present within biological systems. For example, genetic logic gates that process and translate multiple molecular inputs into prescribed amounts of signaling through new molecular outputs would enable the integration of diverse environmental and intracellular signals to a smaller number of phenotypic responses. Basic operations such as signal filtering, amplification, and restoration would also enable expanded manipulation of molecular information through cellular networks.
Molecular information processing systems have been constructed that perform computation with biological substrates. For example, protein-based systems can perform logic operations to convert molecular inputs to regulated transcriptional events (1–4). Information processing systems that perform computation on small-molecule and nucleic acid inputs can be constructed from nucleic acid components (5–11). RNA-based systems can process single inputs to regulated gene expression events (12, 13) and integrate multiple regulatory RNAs for combinatorial gene regulation (14, 15). We sought to combine the rich capability of nucleic acids for performing information processing, transduction, and control operations with the design advantages expected from the relative ease by which RNA structures can be modeled and designed (16, 17).
We proposed a framework for the construction of single input–single output RNA devices (18) based on the assembly of three functional components: a sensor component, made of an RNA aptamer (19); an actuator component, made of a hammerhead ribozyme (20); and a transmitter component, made of a sequence that couples the sensor and actuator components. The resulting devices distribute between two primary conformations: one in which the input cannot bind the sensor, and the other in which the input can bind the sensor as a result of competitive hybridization events within the transmitter component. Input binding shifts the distribution to favor the input-bound conformation as a function of increasing input concentration and is translated to a change in the activity of the actuator, where a “ribozyme-active” state results in self-cleavage of the ribozyme (21). The RNA device is coupled to the 3′ untranslated region (UTR) of the target gene, where ribozyme self-cleavage inactivates the transcript and thereby lowers gene expression independent of cell-specific machinery. We made simple RNA devices that function as single-input Buffer and Inverter gates that convert a molecular input to increased and decreased gene expression output, respectively (18).
The utility of a proposed composition framework depends partly on the extensibility of the framework itself. A framework that provides a general approach for the forward engineering of multi-input devices will allow the combinatorial assembly of many information processing, transduction, and control devices from a smaller number of components. Thus, we used defined points of integration to facilitate the assembly of putatively modular RNA components into sophisticated information processing devices (Fig. 1A) and specified three signal integration (SI) schemes (Fig. 1B). SI 1 was used to construct RNA devices that acted as logic gates (AND or NOR gates) and signal and bandpass filters through the assembly of independent single-input gates. SI 2 was used to construct devices that allowed other logic operations (NAND or OR gates) through the assembly of sensor-transmitter components linked to both stems of the ribozyme. SI 3 was used to construct devices that acted as logic gates (AND or OR gates) and exhibited cooperativity through the assembly of two sensor-transmitter components linked to a single ribozyme stem. The various operations were achieved by altering the function or input responsiveness of the single-input gates in SI 1 or sensor-transmitter components in SI 2 and 3. We assembled multiple RNA devices from various components for all operations to demonstrate the generality of the integration schemes.
Fig. 1.
Functional RNA device composition framework. The color scheme for all figures is as follows: brown, aptamer or sensor component; purple, catalytic core of the ribozyme or actuator component; blue, loop regions of the actuator component; green and red, strands within the transmitter component that participate in the competitive hybridization event. (A) A functional composition framework for assembling RNA devices from modular components. Information in the form of a molecular input is received by the sensor and transmitted by the transmitter to a regulated activity of the actuator, which in turn controls the translation of a target transcript as an output. (B) Three signal integration schemes represent different component assembly strategies to build higher-order RNA devices. The RNA device in SI 1 involves multiple actuator components controlled by single sensor-transmitter components, whereas those in SI 2 and 3 involve multiple sensor-transmitter components controlling a single actuator component.
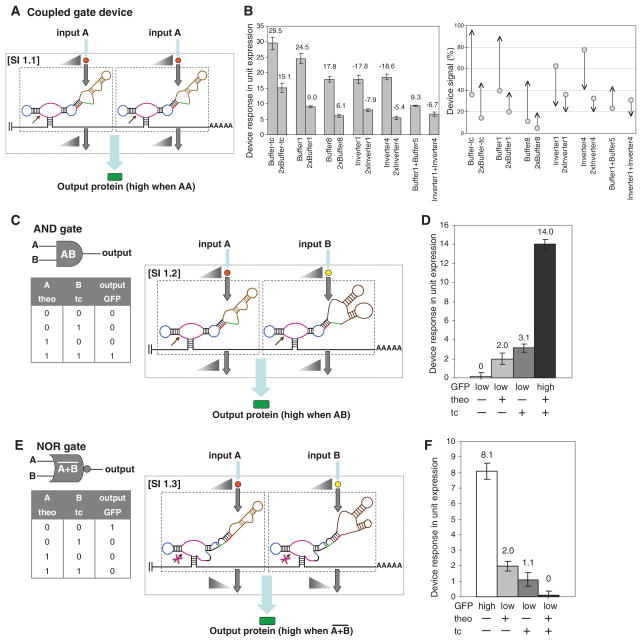
In SI 1, the single-input gates act independently such that computation is performed through integration of individual gate operations in the 3′UTR of the target transcript. Because only one of the ribozymes needs to be in an active state to inactivate the transcript, the device output (gene expression activity) is high only when both ribozymes of the single-input gates are in their inactive states. We engineered signal filters by coupling representative Buffer or Inverter gates (18) responsive to either theophylline or tetracycline (SI 1.1; Fig. 2A). Coupled-gate devices exhibited a device response that was shifted lower compared to that of the single-input gate, indicating the independent action of each single-input gate (Fig. 2B, SOM text S1 and S2, and table S1).
Fig. 2.
RNA devices based on signal integration within the 3′ UTR (SI 1). Single-input gates are indicated in dashed boxes, and triangles indicate relationships between associated gate inputs and outputs. (A) An RNA device composed of two Buffer gates responsive to the same input functions to shift the device response lower than that of the single-input gate. (B) The device output of RNA devices composed of two single-input gates and their single-input gate counterparts. (Left) Device response (bars) is reported as the difference between gene expression activities in the absence and presence of the appropriate inputs [10 mM theophylline (theo) or 1 mM tetracycline (tc)] (21). (Right) Device signal (arrows) is reported over the full transcriptional range of the promoter system used as a percentage of the expression activity relative to that of an inactive ribozyme control, where circles and arrowheads indicate device signals in the absence and presence of input, respectively. The negative sign indicates the down-regulation of target gene expression by the Inverter gates. (C) An RNA device that performs an AND operation by coupling two Buffer gates responsive to different inputs and the associated truth table. (D) The device response of an AND gate (L2bulge1 + L2bulge1tc). Device response under different input conditions [theo or tc (−), 0 mM; theo (+), 5 mM; tc (+), 0.25 mM] is reported as the difference between expression activity in the absence of both inputs and that at the indicated input conditions. (E) An RNA device that performs a NOR operation by coupling two Inverter gates responsive to different inputs and the associated truth table. (F) The device response of a NOR gate (L2bulgeOff1 + L2bulgeOff1tc). Device response under different input conditions [theo or tc (−), 0 mM; theo (+), 10 mM; tc (+), 0.5 mM] is reported as the difference between expression activity in the presence of both inputs and that at the indicated input conditions. Error bars represent the SD from at least three independent experiments.
We constructed an AND gate that exhibited high output only when both inputs were present by coupling a theophylline-responsive Buffer gate and a tetracycline-responsive Buffer gate (SI 1.2; Fig. 2C). In this composition, only in the presence of both molecular inputs (theophylline and tetracycline) did both Buffer gates favor the ribozyme-inactive state, resulting in high device output (Fig. 2D and fig. S1).
We constructed a NOR gate that exhibited high output only when both inputs were absent by coupling a theophylline-responsive Inverter gate and a tetracycline-responsive Inverter gate (SI 1.3; Fig. 2E and fig. S2). In this composition, only in the absence of both inputs did both Inverter gates favor the ribozyme-inactive state, resulting in high device output (Fig. 2F and fig. S3). We also engineered a bandpass filter that exhibited high output only over intermediate input concentrations by coupling theophylline-responsive Buffer and Inverter gates (fig. S4). The various devices demonstrated that diverse information processing operations can be assembled through SI 1, where layering strategies can extend the attainable operations (SOM text S3).
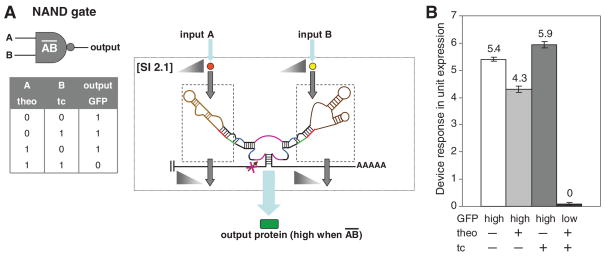
Devices constructed through SI 2 and 3 consisted of multiple sensor-transmitter components, or internal gates (Fig. 1B). An internal Inverter or Buffer gate is defined as a sensor-transmitter component that activates or inactivates, respectively, a coupled component, such as an actuator or other internal gate, in the presence of input. In SI 2, the internal gates act independently through the linked ribozyme stems and therefore computation is performed through the integration of individual internal gate operations in the ribozyme core. The single ribozyme is only in the active state, corresponding to low device output, when both sensor-transmitter components are in states that activate the coupled ribozyme. We constructed a NAND gate by coupling a theophylline-responsive internal Inverter gate through stem I and a tetracycline-responsive internal Inverter gate (fig. S2) through stem II (SI 2.1; Fig. 3A). The device exhibited low output only in the presence of both inputs, because both internal Inverter gates favored the ribozyme-active state (Fig. 3B and fig. S5). Other logic operations can be performed by SI 2 devices, such as an OR operation, through the coupling of two internal Buffer gates (SOM text S4).
Fig. 3.
RNA devices based on signal integration at the ribozyme core (SI 2). Internal gates are indicated in dashed boxes, and triangles indicate relationships between associated internal gate inputs and outputs. (A) An RNA device that performs a NAND operation by coupling two internal Inverter gates responsive to different inputs to different ribozyme stems and the associated truth table. (B) The device response of a NAND gate (L1cm10 – L2bulgeOff3tc). Device response under different input conditions [theo or tc (−), 0 mM; theo (+), 10 mM; tc (+), 1 mM] is reported as in Fig. 2F. Error bars represent the SD from at least three independent experiments.
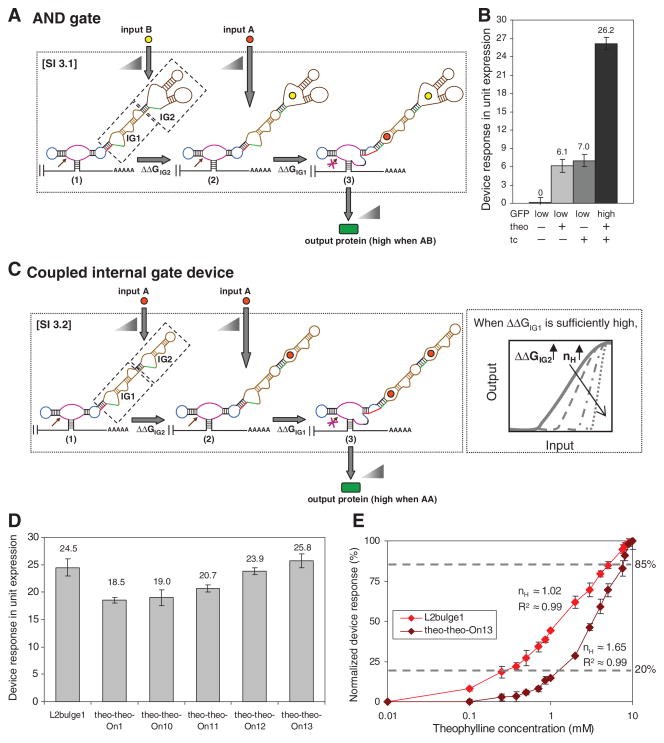
In SI 3, the sensor-transmitter components are coupled within a ribozyme stem, and computation occurs via the integrated operations of the internal gates. Internal gates were linked through the aptamer loop of the lower gate, IG(n), and the transmitter of the higher gate, IG(n + 1). The operation of the higher internal gate determines the state of the lower internal gate, where an internal gate can perform its encoded operation when it is in an active state, and the state of the internal gate linked to the ribozyme (IG1) determines the state of the device. We constructed an alternative AND gate by coupling a theophylline-responsive internal Buffer gate (IG1) and a tetracycline-responsive internal Inverter gate (IG2) at stem II (SI 3.1; Fig. 4A). In this composition, only in the presence of both inputs did IG1 change the state of the RNA device to favor the ribozyme-inactive state, resulting in high device output (Fig. 4B and fig. S6). We also constructed RNA devices that perform an OR operation through SI 3 (SOM text S4).
Fig. 4.
RNA devices based on signal integration at a single ribozyme stem (SI 3). Internal gates (IGn) are indicated in dashed boxes, and triangles indicate relationships between associated internal gate inputs and the device output. (A) An RNA device that performs an AND operation by coupling internal Buffer (IG1) and Inverter (IG2) gates responsive to different inputs to a single ribozyme stem. (B) The device response of an AND gate (tc-theo-On1). Device response under different input conditions [theo or tc (−), 0 mM; theo (+), 2.5 mM; tc (+), 0.5 mM] is reported as in Fig. 2D. (C) An RNA device composed of internal Buffer (IG1) and Inverter (IG2) gates responsive to the same input coupled to a single ribozyme stem. (D) The device response of RNA devices composed of internal Buffer and Inverter gates and their single-internal gate device counterpart (L2bulge1). Device response is reported as in Fig. 2B. Theo-theo-On10, -On11, -On12, and -On13 exhibit varying degrees of cooperativity, as quantified by Hill coefficients (nH) greater than 1 (26). (E) The device output response of theo-theo-On13 shows a high degree of programmed cooperativity. The device response is normalized to the response at 10 mM theophylline (21). Error bars represent the SD from at least three independent experiments.
We engineered RNA devices that exhibited programmed cooperativity through SI 3 by manipulating the relative energies required to switch the device between different states (SOM text S5). RNA devices were composed of theophylline-responsive internal Buffer (IG1) and Inverter (IG2) gates (SI 3.2; Fig. 4C), in which the energetic differences between the input-unbound (1) and single-input–bound (2) states were varied (programmed through IG2; ΔΔGIG2; table S2) and the differences between the single-input–bound and two-input–bound (3) states were kept constant (programmed through IG1; ΔΔGIG1 = 1 kcal/mol). The devices exhibited Buffer operations and substantial degrees of cooperativity (Fig. 4D and fig. S7), where one device exhibited a degree of cooperativity [Hill coefficient (nH) ≈ 1.65; Fig. 4E] similar to that of a naturally occurring cooperative riboswitch (22). We also placed internal Inverter gates into IG1 to construct a device that performed an Inverter operation and exhibited cooperativity (figs. S8 and S9). Control studies indicated that the value of ΔΔGIG1 was important to the observed cooperative response (figs. S10 and S11) and verified that the response was achieved through input binding to both sensors (figs. S12 to S15).
We have developed a composition framework for constructing higher-order RNA devices. Functional modularity is a critical element of any composition framework and was achieved in this study partly through the separation of device functions into distinct components. Although the functions of sensing and actuation frequently rely on tertiary interactions, which are not accounted for in this framework, the integration of these functions into a device is simplified via a transmitter that insulates component functions and controls the interactions between components through predictive hybridization interactions. The variety of information processing operations demonstrated from a small number of standard components emphasizes the utility of modular assembly. In addition, three of the devices have naturally occurring functional counterparts (22–24), supporting the biological relevance of such information processing operations. The framework may be further extended to more complex devices by combining multiple SI schemes within a device and implementing layering strategies. We anticipate that further insight into RNA structure-function relationships (25), and improved predictions of RNA secondary and tertiary structures (16), may allow the development of improved modular assembly schemes, in which an important design challenge will be to insulate device functions across distinct components and control interactions between these components.
Supplementary Material
Supporting Online Material
www.sciencemag.org/cgi/content/full/322/5900/456/DC1
Materials and Methods
SOM Text S1 to S6
Figs. S1 to S17
Tables S1 and S2
References
References and Notes
- 1.Guet CC, Elowitz MB, Hsing W, Leibler S. Science. 2002;296:1466. doi: 10.1126/science.1067407. [DOI] [PubMed] [Google Scholar]
- 2.Kramer BP, Fischer C, Fussenegger M. Biotechnol Bioeng. 2004;87:478. doi: 10.1002/bit.20142. [DOI] [PubMed] [Google Scholar]
- 3.Cox RS, III, Surette MG, Elowitz MB. Mol Syst Biol. 2007;3:145. doi: 10.1038/msb4100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson JC, Voigt CA, Arkin AP. Mol Syst Biol. 2007;3:133. doi: 10.1038/msb4100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seelig G, Soloveichik D, Zhang DY, Winfree E. Science. 2006;314:1585. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]
- 6.Benenson Y, Gil B, Ben-Dor U, Adar R, Shapiro E. Nature. 2004;429:423. doi: 10.1038/nature02551. [DOI] [PubMed] [Google Scholar]
- 7.Dirks RM, Pierce NA. Proc Natl Acad Sci USA. 2004;101:15275. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stojanovic MN, Stefanovic D. Nat Biotechnol. 2003;21:1069. doi: 10.1038/nbt862. [DOI] [PubMed] [Google Scholar]
- 9.Penchovsky R, Breaker RR. Nat Biotechnol. 2005;23:1424. doi: 10.1038/nbt1155. [DOI] [PubMed] [Google Scholar]
- 10.Breaker RR. Curr Opin Biotechnol. 2002;13:31. doi: 10.1016/s0958-1669(02)00281-1. [DOI] [PubMed] [Google Scholar]
- 11.Robertson MP, Ellington AD. Nat Biotechnol. 1999;17:62. doi: 10.1038/5236. [DOI] [PubMed] [Google Scholar]
- 12.Isaacs FJ, Dwyer DJ, Collins JJ. Nat Biotechnol. 2006;24:545. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- 13.Suess B, Weigand JE. RNA Biol. 2008;5:24. doi: 10.4161/rna.5.1.5955. [DOI] [PubMed] [Google Scholar]
- 14.Rinaudo K, et al. Nat Biotechnol. 2007;25:795. doi: 10.1038/nbt1307. [DOI] [PubMed] [Google Scholar]
- 15.Brown BD, et al. Nat Biotechnol. 2007;25:1457. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 16.Parisien M, Major F. Nature. 2008;452:51. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 17.Mathews DH, Turner DH. Curr Opin Struct Biol. 2006;16:270. doi: 10.1016/j.sbi.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 18.Win MN, Smolke CD. Proc Natl Acad Sci USA. 2007;104:14283. doi: 10.1073/pnas.0703961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hermann T, Patel DJ. Science. 2000;287:820. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 20.Khvorova A, Lescoute A, Westhof E, Jayasena SD. Nat Struct Biol. 2003;10:708. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- 21.Materials and methods are available as supporting material on Science Online.
- 22.Mandal M, et al. Science. 2004;306:275. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 23.Sudarsan N, et al. Science. 2006;314:300. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- 24.Welz R, Breaker RR. RNA. 2007;13:573. doi: 10.1261/rna.407707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodside MT, et al. Proc Natl Acad Sci USA. 2006;103:6190. doi: 10.1073/pnas.0511048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson DL, Cox MM. Lehninger Principles of Biochemistry. 4. Freeman; New York: 2005. pp. 167–174. [Google Scholar]
- 27.We thank Y. Chen, J. Liang, and D. Endy for critical reading of the manuscript and A. Babiskin for pRzS. This work was supported by the Center for Biological Circuit Design at the California Institute of Technology, the Arnold and Mabel Beckman Foundation, and the NIH. The authors declare competing financial interests in the form of a pending patent application.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Online Material
www.sciencemag.org/cgi/content/full/322/5900/456/DC1
Materials and Methods
SOM Text S1 to S6
Figs. S1 to S17
Tables S1 and S2
References