Abstract
DNA glycosylases/AP lyases initiate repair of oxidized bases in the genomes of all organisms by excising these lesions and then cleaving the DNA strand at the resulting abasic (AP) sites and generate 3′ phospho α,β-unsaturated aldehyde (3′ PUA) or 3′ phosphate (3′ P) terminus. In Escherichia coli, the AP-endonucleases (APEs) hydrolyze both 3′ blocking groups (3′ PUA and 3′ P) to generate the 3′-OH termini needed for repair synthesis. In mammalian cells, the previously characterized DNA glycosylases, NTH1 and OGG1, produce 3′ PUA, which is removed by the only AP-endonuclease, APE1. However, APE1 is barely active in removing 3′ phosphate generated by the recently discovered mammalian DNA glycosylases NEIL1 and NEIL2. We showed earlier that the 3′ phosphate generated by NEIL1 is efficiently removed by polynucleotide kinase (PNK) and not APE1. Here we show that the NEIL2-initiated repair of 5-hydroxyuracil (5-OHU) similarly requires PNK. We have also observed stable interaction between NEIL2 and other BER proteins DNA polymerase β (Pol β), DNA ligase IIIα (Lig IIIα) and XRCC1. In spite of their limited sequence homology, NEIL1 and NEIL2 interact with the same domains of Pol β and Lig IIIα. Surprisingly, while the catalytically dispensable C-terminal region of NEIL1 is the common interacting domain, the essential N-terminal segment of NEIL2 is involved in analogous interaction. The BER proteins including NEIL2, PNK, Pol β, Lig IIIα and XRCC1 (but not APE1) could be isolated as a complex from human cells, competent for repair of 5-OHU in plasmid DNA.
Keywords: Base excision repair, DNA glycosylase, Oxidative DNA damage, NEIL2, PNK-dependent repair
1. Introduction
Sporadic mutations due to oxidatively induced lesions in mammalian genomes have been implicated in the etiology of many diseases including cancer, and in aging [1–3]. Among many mutagenic base lesions induced by reactive oxygen species (ROS), 5-hydroxyuracil (5-OHU), and 8-oxoguanine (8-oxoG), with propensity for pairing with adenine could cause GC → AT and GC → TA mutations respectively [4,5]. Repair of oxidatively damaged bases in all organisms occurs primarily via the DNA base excision repair (BER) pathway, initiated with the lesion excision by DNA glycosylases/AP lyases. These dual function glycosylases excise the damaged base and then cleave the DNA strand via AP lyase reaction [6].
Only two mammalian DNA glycosylases, OGG1 and NTH1, were previously characterized which excise the majority of oxidatively damaged lesions. NTH1, the human ortholog of Escherichia coli Nth, primarily removes pyrimidine damage; OGG1, the functional counterpart of E. coli Fpg (MutM), removes mostly purine lesions [7]. Based on the structure and reaction mechanism, these enzymes belong to the E. coli Nth family which utilizes an internal Lys residue as the active site nucleophile, and carries out AP lyase reaction via β-elimination [8]. E. coli Fpg and its paralog Nei comprise the second family of oxidized base-specific DNA glycosylases that use the N-terminal Pro as the active site to carry out βδ-elimination at the AP site, and produce 3′ P and 5′ P termini [9,10]. The 3′ blocked products of β- and βδ-elimination are both processed by the APEs, Xth and Nfo in E. coli [11], to provide the 3′-OH terminus required for subsequent repair synthesis by a DNA polymerase. In contrast to E. coli or yeast, mammalian cells express only one APE, named APE1. While APE1 can efficiently process the β-elimination product, its DNA 3′-phosphatase activity needed for βδ-elimination product is extremely weak [12]. Earlier, the weak 3′-phosphatase activity of APE1 was considered inconsequential in oxidized base repair because no ortholog of E. coli Fpg/Nei was known in mammalian cells that could generate the DNA 3′ phosphate termini. However, we had detected βδ-elimination activity in HeLa extracts [13], and more recently discovered and characterized two human orthologs of E. coli Nei, which we named NEIL (Nei-like)-1 and -2. The NEILs with N-terminal Pro catalyze βδ-elimination like Fpg/Nei to produce 3′ phosphate termini [14–17]. Thus NEILs are distinct from NTH1 and OGG1 in structural features and reaction mechanism.
Polynucleotide kinase (PNK), which is present in mammalian cells but not in E. coli, has two distinct activities; a 5′ DNA kinase, and a 3′ DNA phosphatase [18,19]. PNK was shown to function in the repair of DNA single-strand breaks, generated by ionizing radiation [20], some of which are likely to contain 3′ phosphate termini. We have recently shown that NEIL1-initiated repair is PNK-dependent [12]; here we show that NEIL2-mediated repair of 5-OHU similarly utilizes PNK, and that APE1 is dispensable for repair. Additionally, NEIL2 stably interacts with DNA polymerase β (Pol β), DNA ligase IIIα (Lig IIIα) and XRCC1 using its essential N-terminal region, unlike NEIL1. An active repair complex containing NEIL2 with XRCC1, Pol β, PNK and Lig IIIα could be isolated from cell extracts, suggesting NEIL2-directed repair coordination.
2. Materials and methods
2.1. Purification and characterization of enzymes
Recombinant NEIL2, PNK, APE1, Polβ and Lig IIIα were separately purified to homogeneity from E. coli transformed with the corresponding expression plasmids [15,20–24]. NTH1 was a gift from R. Roy (Georgetown University, Washington, DC). For some biochemical studies, full-length NEIL2 (residues 1–331) with C-terminal 6XHis tag was expressed and purified from Sf9 insect cells as follows.
NEIL2 cDNA was amplified by PCR (the forward primer, 5′-CCGGGATCCACCATGCCAGAAGGGCCGTTGGTGAGG-3′ and reverse primer, 5′-CCCGGGAAGCTTTTAATGATG- ATGATGAT-GATGGGAGAAC TGGCACTGCTCTGG-3′) and subcloned into pFastBacDual (Invitrogen) at the BamHI and HindIII sites. The recombinant bacmid was then transfected in Sf9 cells and expression of soluble NEIL2 was optimized at 72 h post infection. Expression of NEIL2 was examined by Coomassie blue staining after SDS-PAGE and verified by immunoblotting with an anti-His monoclonal antibody (1:1000 dilution, Qiagen). His-tag NEIL2 (soluble extract from 1l Sf9 cells) was bound to the Ni-NTA resin (Qiagen) at 4°C for 1h. After washing extensively with 25 mM Tris–HCl (pH 7.5), 500mM NaCl containing 20 mM imidazole, the bound proteins were eluted with the same buffer but containing 250 mM imidazole. NEIL2 was further purified by chromatography on SP-sepharose, then dialyzed in phosphate-buffered saline (PBS) containing 50% glycerol and 1 mM DTT, and stored at −20 °C.
To examine the domain organization of NEIL2, we carried out limited proteolytic digestion of NEIL2 (E. coli purified) with endoproteinase Asp-N (Roche Applied Science). Two major non-overlapping domains consisting of N-terminal (residues 1–198) and C-terminal (residues 199–331) fragments were identified by N-terminal sequencing and mass spectroscopic analysis in the UTMB core facility. The coding sequences corresponding to these domains were subcloned in frame with a C-terminal His-tag in pET22b, expressed in E. coli and purified on Ni-NTA agarose, followed by SP-sepharose chromatography as described above.
2.2. Far-Western analysis
Purified proteins were separated by SDS-PAGE (12% polyacrylamide) and transferred to nitrocellulose membranes. After incubation in a denaturation buffer (6 M guanidine-HCl in PBS) twice for 5 min at 4°C, the proteins were renatured on the membranes with successive dilutions (1:1) of guanidine-HCl in PBS containing 1mM DTT [25]. The membranes were blocked with PBS containing 0.5% Tween 20 and 5% non-fat dry milk (NFDM) for 45 min at 20 °C. After two washes with the blocking buffer, the blots were incubated with NEIL2 (10 pmol/ml) for 3h at 4 °C in PBS/Tween 20 (0.5%), NFDM containing 1 mM DTT, 0.5 mM PMSF. The filters were washed (4 × 10 min) in PBS/Tween 20 containing 0.25% NFDM and then incubated with affinity-purified anti-NEIL2 antibody prior to secondary goat anti-rabbit antibody incubation, followed by ECL detection (Amersham Pharmacia).
2.3. His-pulldown assay
Ni-NTA magnetic bead (Qiagen) suspension (50 µl) was added separately to purified His-tagged XRCC1 and His-tagged PNK proteins (250 ng), and the suspensions (500 µl) were incubated on an end-over-end shaker for 30 min at 4°C. The Ni-NTA beads bound to XRCC1 or PNK were washed using a magnetic separator three times with buffer A (50 mM Tris–HCl, pH 7.5, 10 mM imidazole, 0.005% Tween-20) containing 150 mM NaCl, resuspended in 500 µl of fresh buffer A containing non-tagged NEIL2 protein (500 ng), and incubated on the shaker for 1 h at 4 °C. Finally the supernatant was removed and the beads were washed twice with buffer A containing 300 mM NaCl and 20 mM imidazole. The complexes were then eluted from the beads with 50 µl of 1× SDS–PAGE loading buffer. The presence of NEIL2 was examined by Western analysis using anti-NEIL2 antibody and presence of the bait proteins (XRCC1 and PNK) were probed with the anti-His antibody.
2.4. Immunoprecipitation of FLAG-NEIL2
To identify NEIL2-associated proteins, HCT116 (human colon carcinoma) cells were transfected with either a FLAG-NEIL2-expression vector or control empty vector. Forty-five hours after transfection, the cells were washed thoroughly with PBS, and then with Tris-buffered saline (TBS; 50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA) as described previously [26]. After lysing cells with cold TBS containing 1% TritonX-100 and protease inhibitor cocktail (Roche) followed by centrifugation, the supernatants were immunoprecipitated with anti-FLAG M2 antibody (Sigma). Immunoprecipitates (IP) of both control and FLAG-NEIL2-transfected cells were washed with TBS and then probed for the presence of XRCC1, PNK, Lig IIIα and Polβ by immunoblotting.
2.5. Co-transfection studies
HCT116 cells were lysed in cold lysis buffer 40 h after cotransfection with plasmids for PNK and FLAG-tagged NEIL2 or OGG1. Proteins in the cleared lysates were immunoprecipitated with anti-FLAG M2 antibody (Sigma), as described previously [26]. Reciprocal IP was performed after co-transfection of untagged NEIL2 with either FLAG-PNK, FLAG-OGG1 or vector alone. The presence of PNK and NEIL2 in the immunoprecipitates was detected by immunoblotting with anti-PNK, anti-NEIL2 or anti-FLAG antibodies.
2.6. Yeast two-hybrid analysis
NEIL2 and Lig IIIα cDNAs were cloned into the two-hybrid vectors pGADT7 and pGBKT7, respectively (Clontech) and the plasmids were individually introduced into the isogenic haploid yeast strains PJ69–4α and PJ69–4α by transfection with LiCl [27]. After selecting the diploids in synthetic medium lacking Trp and Leu, the cells were plated in synthetic medium supplemented with 30 mM 3-amino-1,2,4-triazole, but lacking Trp, Leu and His. After growth for 3 days at 30 °C, the colonies were transferred onto synthetic medium lacking Trp, and Leu to test for protein–protein interaction.
2.7. Plasmid DNA substrates
Covalently closed Form I plasmid DNA containing a single 5-OHU opposite A, used as the substrate in NEIL2-initiated BER assays, was generated with some modification from pUC19CPD, as previously described [28]. Briefly, pUC19CPD was completely digested with single-strand specific endonuclease, NBstNBI. The 30 nt oligomer product (oligo A: 5′-GCG GAT ATT AAT GTG ACG GTA GCG AGT CGC TC-3′) was removed from the plasmid by heating at 65 °C and passage through a streptavidin–agarose column containing a biotinylated complementary oligo (5′-GAG CGA CTC GCT ACC GTC ACA TTA ATA TCC GC-biotin-3′). The resulting gapped plasmid was extracted with phenol/chloroform and ethanol-precipitated. Form I DNA was generated by annealing the plasmid with 5-OHU-containing 5′-phosphorylated oligo (5′-PGCG GAT ATT AAT GTG ACG G 5-OHUA GCG AGT CGC TC-3′) followed by ligation and then purified by electrophoresis in 1% agarose containing 0.4 µg/ml ethidium bromide. The presence of 5-OHU in the plasmid DNA was confirmed by treating the plasmid DNA with E. coli Nth, which converted the plasmid to the nicked circle (Form II) due to lyase reaction.
2.8. Repair synthesis assay
Repair of 5-OHU-containing plasmid was carried out in a reconstituted system containing purified proteins (NEIL2, Lig IIIα, Polβ, APE1 and PNK). Briefly, the reaction mixture (50 µl) contained 1nM plasmid DNA, 50 mM HEPES-KOH, pH 7.5, 70 mM KCl, 5 mM MgCl2, 0.5 mM DTT, 50 µM each of dATP, dCTP, dGTP and 5 µM dTTP (with 2 µCi32P), 2 mM ATP, 1 µg BSA and various combinations of repair proteins. The protein concentrations were optimized for maximum repair synthesis.
To test whether the FLAG-NEIL2 immunocomplex was capable of complete repair, the immunoprecipitates (IP) isolated from control and the FLAG-NEIL2 transfected cells were washed with a buffer (50 mM Tris–HCl, pH 7.5,150 mM NaCl, protease inhibitor cocktail) containing 10% glycerol, and the proteins were eluted with FLAG peptide. The eluate was subsequently used in repair reactions as described above with the 5-OHU-containing plasmid DNA. After 60 min at 30°C, the plasmid DNA was recovered and digested with AhdI to linearize the plasmid which was then resolved by agarose gel (1%) electrophoresis. A 1 kb DNA ladder was 5′ 32P-labeled and used as a marker. The BER products were visualized and analyzed by a PhosphorImager.
3. Results
3.1. Identification of NEIL2-associated proteins
To identify NEIL2-associated proteins in vivo, HCT 116 cells were transfected with either FLAG-NEIL2 or vector alone, and the cell extracts were immunoprecipitated with anti-FLAG antibody. As shown in Fig. 1, XRCC1, Pol β, PNK and Lig IIIα, but not APE1, are present in NEIL2-immunocomplex. This is the first evidence for the presence of these proteins in the same cellular complex with NEIL2. Because these proteins were shown earlier to be involved in NEIL1-mediated repair [12], it can be envisaged that these proteins also act as critical partners of NEIL2 to form complex essential to the functional BER machinery in vivo.
Fig. 1.
Co-immunoprecipitation of NEIL2 and other BER proteins. Extracts of HCT 116 cells transfected with either a FLAG-tagged NEIL2 or control plasmid were immunoprecipitated with anti-FLAG antibody. The blots were then immunoblotted for PNK (A), Lig IIIα (B), XRCC1 (C), Pol β (D) or APE1 (E).
3.2. Interaction of NEIL2 with other BER proteins
Several previous studies suggested that the sequential steps in BER are coordinated via specific physical interactions among the BER proteins [29,30]. We already showed binary interaction of NEIL1 with Pol β, Lig IIIα and XRCC1 [12]. Because NEIL2 and NEIL1 share limited sequence homology, we asked whether NEIL2 associates with these proteins as NEIL1. To detect interactions between NEIL2 and other components of the BER pathway, we performed Far-Western analyses using purified proteins and observed that NEIL2 interacted directly with Pol β and Lig IIIα (Fig. 2A, lanes 3 and 4), but not with PNK or XRCC1 (lanes 2 and 5). We used BSA and OGG1 as negative control (lanes 1 and 6). Furthermore, DNA ligase l, though active in repair in vitro (data not shown), did not interact with NEIL2 (Fig. 2A, lane 7). Next, we were able to map the NEIL2-interacting region of Lig IIIα. Coomassie staining (Fig. 2A) showed two bands for Lig IIIα (lane 4); the lower band was a truncation product lacking 175 residues, as characterized by mass spectrometry, presumably generated during expression and purification of the recombinant protein [12]. Because the protein was purified based on affinity chromatography of an N-terminal His-tag, the deletion must have occurred at the C-terminus. These results indicate that NElL2 interacts with the C-terminal 175 amino acid residues of Lig IIIα. Interaction of NEIL2 with Lig IIIα was further confirmed by yeast two-hybrid analysis (Fig. 2B). Though we did not observe a direct interaction between NEIL2 and XRCC1 in the Far-Western analysis, XRCC1 was found to interact with NEIL2 when the native proteins were used in an in vitro His-pull down experiment (Fig. 2C, lane 1); this discrepancy in the Far-Western results (Fig. 2A) might be due to improper refolding of XRCC1 during the renaturation step. In a parallel experiment His-tagged PNK failed to pull down NEIL2 (Fig. 2C, lane 2) indicating that PNK does not physically interact with NEIL2.
Fig. 2.
Interaction of NEIL2 with BER proteins. (A) Far-Western analysis. Proteins were separated by SDS-PAGE in duplicate gels, one of which was Coomassie stained. The proteins from the other gel were transferred to a nitrocellulose membrane and probed with NEIL2 as described in Section 2. (B) Yeast two-hybrid analysis of NEIL2 and Lig IIIα interaction. Yeast expressing NEIL2 and Lig IIIα fusion proteins (lanes 2 and 4) vs. control expressing only Lig IIIα fusion protein (lanes 1 and 3) were tested on medium with (lanes 1 and 2) and without (lanes 3 and 4) His. Other details are given in Section 2. (C) Right panel, in vitro pulldown of NEIL2 with His-tagged XRCC1 (lane 4) and His-tagged PNK (lane 5), respectively. Left panel, Western analysis using anti-His Ab showing the comparable amounts of His-tagged XRCC1 (lane 1) and PNK (lane 2). (D) Interaction of NEIL2 with full length and fragments of Pol β by Far-Western analysis. Pol β and its truncated fragments (10 µg) were separated by SDS-PAGE and Coomassie-stained (lane 1–6) and tested for interaction with NEIL2 (lanes 7–12). At right, a schematic showing the interacting Pol β domains.
To identify the region of Pol β involved in interaction with NEIL2, we examined the binding of NEIL2 to recombinant fragments of Pol β with molecular masses of 8, 14, and 16 kDa, and to C-terminal fragments of 22 and 31 kDa. NEIL2 interacted with the N-terminal 16 and 14kDa fragments of Pol β, as well as with full-length Pol β (Fig. 2D, lanes 10, 11 and 7), but not with the 31, 22 and 8 kDa fragments (lanes 8, 9 and 12, respectively). These results strongly suggest that NEIL2 interacts within the N-terminal 140 amino acids of Pol β. The same regions of Lig IIIα and Pol β were shown to be involved in NEIL1 interaction [12].
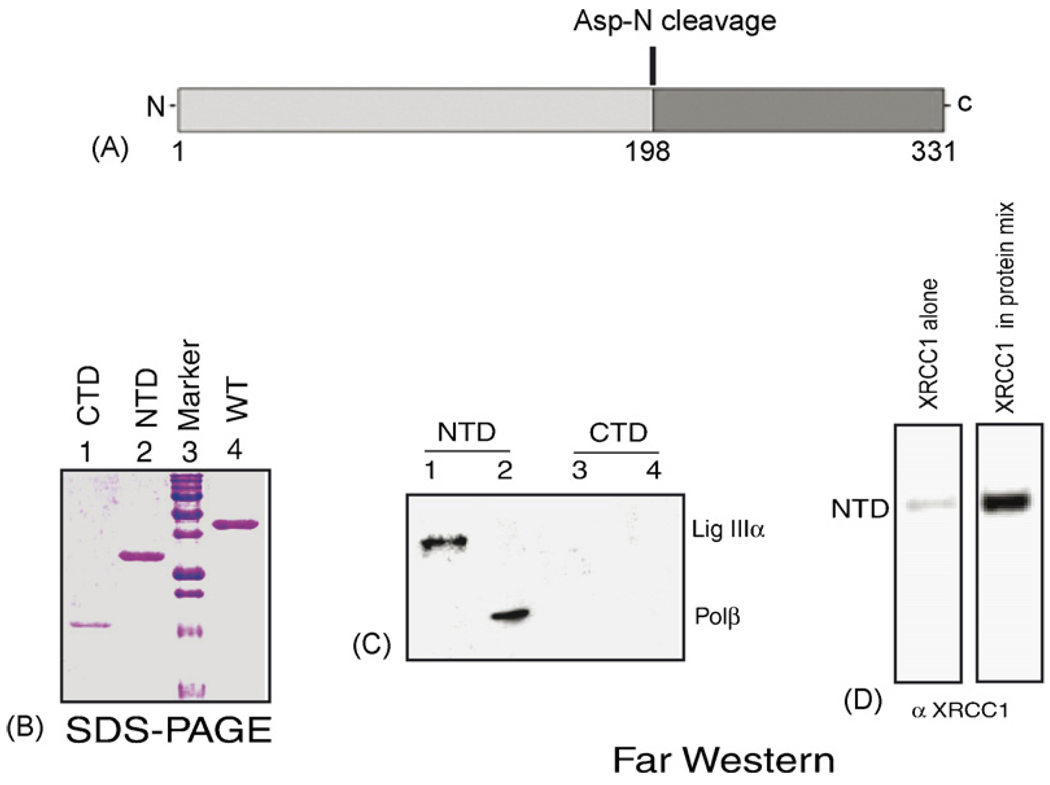
3.3. Mapping of the interacting domains of NEIL2
Limited proteolytic digestion of NEIL2 revealed the presence of two major domains (Fig. 3A); namely the N-terminal domain (NTD; 1–198 aa) and the C-terminal domain (CTD; 199–331 aa). These two domains were purified as individual His-tagged polypeptides (Fig. 3B) and tested for their interaction with immobilized Pol β and Lig IIIα by Far-Western analysis. Fig. 3C shows that the NTD but not the CTD of NEIL2 interacts with Pol β and Lig IIIα. We have found that full-length NEIL2 does not fold properly on the membrane presumably due to the disruption of the C-terminal zinc-finger motif (data not shown). However, when immobilized on the membrane, the NTD fragment of NEIL2 lacking the zinc finger, could stably fold back after the denaturation step during Far-Western analysis. The NTD showed binding with XRCC1, although weakly, and the interaction was enhanced when XRCC1 was added in a mixture containing Pol β, Lig IIIα and PNK (Fig. 3D).
Fig. 3.
(A) Schematic of proteolytic digestion of NEIL2. (B) Coomassie-stained gel of purified C-terminal (CTD, aa residues 199–331), N-terminal domain (NTD, residues 1–198) and WT NEIL2. (C) Far-Western analysis of Lig IIIα and Pol β with NTD (lanes 1 and 2) and CTD (lanes 3 and 4) of NEIL2. (D) Far-Western analysis of NTD of NEIL2 with XRCC1 alone and XRCC1 in protein mix (Pol β, Lig IIIα and PNK).
3.4. Association of NEIL2 with PNK
We have shown earlier that PNK does not interact directly with NEIL2, but is present in the FLAG-NEIL2 immunocomplex suggesting their simultaneous presence in a complex with other proteins. To further confirm the specificity of this association, we co-transfected cells with PNK expression plasmids and FLAG-tagged versions of either NEIL2 or OGG1 or empty vector. As expected, the FLAG antibody co-immunoprecipitated PNK in the presence of NEIL2, but not FLAG-OGG1 or vector control (Fig. 4A). In a reciprocal IP experiment, the FLAG antibody co-immunoprecipitated NEIL2 but not OGG1 in the presence FLAG-PNK (Fig. 4B). The absence of PNK in FLAG-OGG1 IP provided an appropriate negative control, which also showed the specificity of association of PNK with NEIL2 in the cellular lysate. Taken together, these results further support our conclusion that PNK is associated with NEIL2 in a multi-component repair complex.
Fig. 4.
Interaction of NEIL2 with PNK. (A) Extracts of HCT116 cells after co-transfection with expression plasmids of PNK and either vector, FLAG-NEIL2 or FLAG-OGG1 were immunoprecipitated with α-FLAG, and then probed with α-PNK. (A') Reprobing of the blot with α-FLAG. (B) Western analysis of FLAG-immunoprecipitate with α-NEIL2 after cotransfection with non-tagged NEIL2 and FLAG-OGG1, FLAG-PNK or vector alone. (B') Reprobing of the blot with α-FLAG.
3.5. APE-independent repair initiated by NEIL2
We showed earlier that PNK provides the primary activity in mammalian cells for processing the 3′ P generated by NEIL1 [12]. Our previous study and the results described here show that both NEIL1 and NEIL2 associate with XRCC1, PNK, Polβ and Lig IIIα. Because NEIL1 and these other proteins could efficiently repair damage contained in an oligonucleotide substrate by an APE-independent mechanism, we tested whether NEIL2 could similarly participate in APE-independent repair.
We first examined the requirement of PNK in NEIL2-initiated repair for removal of an oxidative lesion from a circular plasmid DNA. Because the damage-containing radiola-beled substrate could not be distinguished from the repaired product, we examined incorporation of labeled [α32P] dTMP in the unlabeled plasmid DNA containing a single 5-OHU opposite A (Fig. 5A). The plasmid DNA was incubated at 37°C for 1 h with NEIL2, APE1 or PNK, Pol β and Lig IIIα, as indicated (Fig. 5A), in buffer A containing 50 mM HEPES-KOH, pH 7.5, 70 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.4 mM EDTA, 1 mM ATP, 2.5% glycerol, 200 µg/ml BSA. Incorporation of radiolabeled dTMP in the 2 kb plasmid DNA in the presence of NEIL2, PNK, Lig IIIα and Polβ (lane 4) indicated complete repair of 5-OHU. Insignificant repair was observed when NEIL2 (lane 2) or PNK (lane 3) was omitted, or in the presence of APE1 substituting for PNK (lane 5). No radioactivity was incorporated in the control, undamaged plasmid (lane 1) indicating that the repair is damage-specific. NEIL2 has overlapping substrate specificity with NTH1; 5-OHU is excised from DNA by both NEIL2 and NTH1. Therefore, we compared repair initiated by NTH1 versus NEIL2. As expected, repair initiated by NTH1 required APE1 (lane 7), and PNK could not substitute for APE1 (lane 6). These experiments thus show the dependence of NEIL2 on PNK versus that of NTH1 on APE1 in repairing the same lesion.
Fig. 5.
Repair of 5-OHU in reconstituted system. (A) For α-32PdTMP incorporation into 5-OHU-containing plasmid DNA, 10 pmol substrates were incubated with combinations of 0.5 pmol of NEIL2, 25 fmol of PNK, 25 fmol APE1 and 0.25 pmol of other proteins as described in Section 2. Repaired plasmid was linearized with AhDI and analyzed on 0.8% agarose gel. Lane 1, undamaged DNA and lanes 2–8, 5-OHU-containing plasmid DNA. Complete repair is indicated by the radioactive dTMP incorporation. The agarose gel was dried and the reactions products were analyzed by PhosphorImager. Lane 9, end-labeled 1 kb DNA ladder (NEB). (B) Repair of 5-OHU-containing plasmid DNA with vector (lanes 1 and 2 µl, respectively; 5 and 10 µl, respectively) and FLAG-NEIL2 immunoprecipitate (lanes 3 and 4 µl, respectively; 5 and 10 µl, respectively). Lane 5, end-labeled 1 kb DNA ladder (New England Biolabs).
To mimic the in vivo situation, we then tested NEIL2 IP for its ability to repair the same 5-OHU-containing plasmid DNA (Fig. 5B). While there was no radioactivity incorporated in the lesion-containing plasmid DNA using control IP from empty vector expressing cells (lanes 1 and 2), the FLAG-NEIL2 IP catalyzed a modest incorporation of [α32P] dTMP in the 5-OHU-containing plasmid DNA, in a concentration-dependant manner (lanes 3 and 4). This apparent weak repair activity could be due to partial inactivation of the protein(s) during the long process of immunoprecipitation and elution of the complex from the FLAG beads. Nevertheless, these results indicate that the repair proficient complex contained at least NEIL2 together with PNK, Pol β and Lig IIIα.
4. Discussion
NTH1 and OGG1, which repair oxidized bases in mammalian cells, belong to the Nth family of DNA glycosylases, and carry out β elimination-mediated DNA strand cleavage to produce 3′-phospho α,β-unsaturated aldehyde (3′ PUA) terminus at the site of the damaged base. In contrast, the more recently characterized mammalian DNA glycosylases NEIL1 and NEIL2, which belong to the Nei family, produce 3′ phosphate containing terminus after βδ elimination reaction at the AP site. Both types of termini (3′ PUA and 3′ P) are efficiently removed by E. coli APEs to produce the 3′ OH necessary for DNA repair synthesis. APE1, the only APE in mammalian cells, was believed to play a central role in all types of BER processes, like E. coli APE [11,31,32]. However, we showed recently that APE1 is dispensable when BER is initiated by NEIL1 [12]. Here we show that APE1 is similarly dispensable in NEIL2-initiated repair.
In this study we demonstrate that NEIL2 behaves much like NEIL1 in directly interacting with the other BER components, specifically Pol β, Lig IIIα and XRCC1. Because NEIL1 and NEIL2 share only limited sequence homology, it is remarkable that the same regions of Pol β and Lig IIIα are involved in interaction with both NEILs. Far-Western analyses revealed that the interaction region in Lig IIIα lies within the C-terminal 175 amino acids, and the interaction region in Pol β is located within the N-terminal 140 amino acid residues. The physiological significance of both NEILs' binding to the same region of Pol β and Lig IIIα warrants further investigation. This is the first report showing direct physical association of human NEIL2 with Lig IIIα and Pol β. A recent report of mouse NEIL2 and Pol β's binding to microtubules further supports their close association [33]. However NEIL2 failed to show direct interaction with PNK (Fig. 3D), although PNK was stably associated with NEIL2 in a larger complex including the other BER components. Furthermore, XRCC1 alone interacts weakly with the NTD of NEIL2, but the interaction is significantly enhanced when the other BER proteins are present. Association of NEIL2 with XRCC1 has also been recently reported [34].
An X-ray crystallographic structure of an enzymatically active deletion mutant of NEIL1, lacking 56 C-terminal residues suggests that its C-terminal region is unstructured [35]. We have shown earlier that the interacting region of NEIL1 lies within its C-terminal domain (between residues 289 and 350) [12]. Thus NEIL1's interacting domain for Polβ and Lig IIIα may have a flexible structure. The tertiary structure of NEIL2 has not yet been elucidated. However, limited proteolytic digestion suggests that there are two major domains in NEIL2, the NTD (spanning residues 1–198) and the CTD (residues 199–331) (Fig. 3A). Our data further reveal that in case of NEIL2, the 133 C-terminal residues, including the zinc finger motif that is necessary for DNA binding and catalysis [36], is dispensable for interaction with Polβ, Lig IIIα (Fig. 3C) and XRCC1 polypeptides (Fig. 3D). It is intriguing that the interacting domain of NEIL1 for the same proteins is localized in its C-terminal domain, which is dispensable for its in vitro glycosylase/lyase activity [12,35]. In contrast, not only is the C-terminal domain of NEIL2 essential, but the interacting domain, which is at the N-terminus, is also indispensable for activity.
Our in vivo data (Fig. 1) further showed that NEIL2 associates with the other BER proteins, Polβ, Lig IIIα and PNK as well as XRCC1, but not with APE1. We reported previously that NEIL1-mediated repair is dependent on PNK, but not on APE1, while NTH1-initiated repair is APE1-dependent [12]. In the present study we were able to reconstitute NEIL2-initiated repair, and established this repair to be PNK-dependent. XRCC1 was not required for these in vitro BER reactions and thus was not used in the reconstitution studies; however it is most probably required for assembly of the NEIL2 repair complexes in cells [37].
Finally we made the significant observation that the FLAG-NEIL2 immuno-complex was capable of completing repair of 5-OHU-containing plasmid DNA (Fig. 5B). This multi-protein complex has to be dynamic, because the same NEIL2 molecule cannot simultaneously interact with multiple proteins using its common interacting domain. A possible explanation for such a complex is that it allows coordinated hand-off among the repair proteins. However, the stoichiometry of the component proteins in this complex needs to be determined. Taken together, the association of NEIL2 with downstream BER proteins, Pol β, Lig IIIα and PNK, like NEIL1, supports a model of repair coordination in which NEIL2 recruits these proteins, and forms a larger repair complex at the site of DNA damage.
The NEILs are thus functionally distinct from the previously characterized oxidized base specific DNA glycosylases, NTH1 and OGG1. Distinctive feature of NEILs versus NTH1/OGG1 are that NEILs prefer excising lesions from DNA bubble structures, whereas both NTH1 and OGG1 are active only on duplex DNA [38]. The dichotomy in the preference of NEILs and NTH1/OGG1 for bubble versus duplex DNA raises the possibility that NEILs have preferred function in repairing base lesions present in transient transcription or replication bubbles. The NEIL1-deficient mouse embryonic fibroblasts were found to be sensitive to radiation [39], providing support for NEIL initiated BER in preventing the genotoxicity of oxidative stress. Recently Stephen Lloyd's group generated NEIL1 knockout mice, which exhibited high levels of mitochondrial DNA damage. These knockout animals also developed severe metabolic syndrome [40]. It would be interesting to investigate the organismal consequences associated with deficiency in NEIL2. The biological relevance of the NEIL/PNK-dependant repair pathway was further supported by results showing that PNK-deficient A549 cells display an elevated mutation frequency and are sensitive to ionizing radiation [41]. Thus, PNK-dependent repair of NEILs may play a unique role in maintaining the functional integrity of mammalian genomes.
Acknowledgements
This research was supported by USPHS grants CA102271 (TKH), PHS grants CA92584 (SM and AET) and CA81063 (SM), ES012512 (AET), Canadian Institute of Health Research Grant MOP15385 (MW) P01 AG021830 (IB and SM) and P01 ES06676 (TKH, IB and SM). We are grateful to Drs. Miaw-Sheue and P.K. Cooper (Lawrence Berkeley Laboratory) for the production of recombinant NEIL2 baculoviruses. We acknowledge the generous help of Drs T. Wood and A. Kurosky of the NIEHS Center for generating the expression plasmids and MS analysis of the recombinant proteins, respectively. We also thank Dr. David Konkel for editing and Ms. Wanda Smith for secretarial help of this manuscript.
Abbreviations
- APE, AP
endonuclease
- Ab
antibody
- BER
base excision repair
- CTD
C-terminal domain
- IP
immunoprecipitate
- Lig IIIα
DNA ligase IIIα
- NEIL
Nei-like
- 5-OHU
5-hydroxyuracil
- NTD
N-terminal domain
- NTH1
endonuclease III homolog 1
- OGG1
8-oxoguanine-DNA glycosylasel
- PUA
phospho α,β-unsaturated aldehyde
- PNK
polynucleotide kinase
- Pol β
DNA polymerase β
- TBS
Tris-buffered saline
- WT
wild type
REFERENCES
- 1.Ames BN. Shigenaga MK. Hagen TM. Oxidants, antioxidants, and the degenerative diseases of aging . Proc. Natl. Acad. Sci. U.S.A. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breimer LH. Molecular mechanisms of oxygen radical carcinogenesis and mutagenesis: the role of DNA base damage. Mol. Carcinog. 1990;3:188–197. doi: 10.1002/mc.2940030405. [DOI] [PubMed] [Google Scholar]
- 3.Fraga CG. Shigenaga MK. Park JW. Degan P. Ames BN. Oxidative damage to DNA during aging: 8-hydroxy-2′-deoxyguanosine in rat organ DNA and urine. Proc. Natl. Acad. Sci. U.S.A. 1990;87:4533–4537. doi: 10.1073/pnas.87.12.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kreutzer DA. Essigmann JM. Oxidized, deaminated cytosines are a source of C → T transitions in vivo. Proc. Natl. Acad. Sci. U.S.A. 1998;95:3578–3582. doi: 10.1073/pnas.95.7.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shibutani S. Takeshita M. Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 6.Mitra S. Hazra TK. Roy R. Ikeda S. Biswas T. Lock J. Boldogh I. Izumi T. Complexities of DNA base excision repair in mammalian cells. Mol. Cells. 1997;7:305–312. [PubMed] [Google Scholar]
- 7.Krokan HE. Nilsen H. Skorpen F. Otterlei M. Slupphaug G. Base excision repair of DNA in mammalian cells. FEBS Lett. 2000;476:73–77. doi: 10.1016/s0014-5793(00)01674-4. [DOI] [PubMed] [Google Scholar]
- 8.Nash HM. Bruner SD. Scharer OD. Kawate T. Addona TA. Spooner E. Lane WS. Verdine GL. Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr. Biol. 1996;6:968–980. doi: 10.1016/s0960-9822(02)00641-3. [DOI] [PubMed] [Google Scholar]
- 9.Zharkov DO. Rieger RA. Iden CR. Grollman AP. NH2-terminal proline acts as a nucleophile in the glycosylase/AP-lyase reaction catalyzed by Escherichia coli formamidopyrimidine-DNA glycosylase (Fpg) protein. J. Biol. Chem. 1997;272:5335–5341. doi: 10.1074/jbc.272.8.5335. [DOI] [PubMed] [Google Scholar]
- 10.Jiang D. Hatahet Z. Blaisdell JO. Melamede RJ. Wallace SS. Escherichia coli endonuclease. VIII. Cloning, sequencing, and overexpression of the nei structural gene and characterization of nei and nei nth mutants. J. Bacteriol. 1997;179:3773–3782. doi: 10.1128/jb.179.11.3773-3782.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demple B. Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 12.Wiederhold L. Leppard JB. Kedar P. Karimi-Busheri F. Rasouli-Nia A. Weinfeld M. Tomkinson AE. Izumi T. Prasad R. Wilson SH. Mitra S. Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Hazra TK. Izumi T. Maidt L. Floyd RA. Mitra S. The presence of two distinct 8-oxoguanine repair enzymes in human cells: their potential complementary roles in preventing mutation. Nucleic Acids Res. 1998;26:5116–5122. doi: 10.1093/nar/26.22.5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hazra TK. Izumi T. Boldogh I. Imhoff B. Kow YW. Jaruga P. Dizdaroglu M. Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hazra TK. Kow YW. Hatahet Z. Imhoff B. Boldogh I. Mokkapati SK. Mitra S. Izumi T. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J. Biol. Chem. 2002;277:30417–30420. doi: 10.1074/jbc.C200355200. [DOI] [PubMed] [Google Scholar]
- 16.Bandaru V. Sunkara S. Wallace SS. Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amsterdam) 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 17.Hailer MK. Slade PG. Martin BD. Rosenquist TA. Sugden KD. Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair (Amsterdam) 2005;4:41–50. doi: 10.1016/j.dnarep.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 18.Caldecott KW. Polynucleotide kinase: a versatile molecule makes a clean break. Structure. 2002;10:1151–1152. doi: 10.1016/s0969-2126(02)00833-x. [DOI] [PubMed] [Google Scholar]
- 19.Habraken Y. Verly WG. Further purification and characterization of the DNA 3′-phosphatase from rat-liver chromatin which is also a polynucleotide 5′-hydroxyl kinase. Eur. J. Biochem. 1988;171:59–66. doi: 10.1111/j.1432-1033.1988.tb13758.x. [DOI] [PubMed] [Google Scholar]
- 20.Whitehouse CJ. Taylor RM. Thistlethwaite A. Zhang H. Karimi-Busheri F. Lasko DD. Weinfeld M. Caldecott KW. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001;104:107–117. doi: 10.1016/s0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 21.Izumi T. Malecki J. Chaudhry MA. Weinfeld M. Hill JH. Lee JC. Mitra S. Intragenic suppression of an active site mutation in the human apurinic/apyrimidinic endonuclease. J. Mol. Biol. 1999;287:47–57. doi: 10.1006/jmbi.1999.2573. [DOI] [PubMed] [Google Scholar]
- 22.Prasad R. Kumar A. Widen SG. Casas-Finet JR. Wilson SH. Identification of residues in the single-stranded DNA-binding site of the 8-kDa domain of rat DNA polymerase beta by UV cross-linking. J. Biol. Chem. 1993;268:22746–22755. [PubMed] [Google Scholar]
- 23.Caldecott KW. Tucker JD. Stanker LH. Thompson LH. Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res. 1995;23:4836–4843. doi: 10.1093/nar/23.23.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karimi-Busheri F. Daly G. Robins P. Canas B. Pappin DJ. Sgouros J. Miller GG. Fakhrai H. Davis EM. Le Beau MM. Weinfeld M. Molecular characterization of a human DNA kinase. J. Biol. Chem. 1999;274:24187–24194. doi: 10.1074/jbc.274.34.24187. [DOI] [PubMed] [Google Scholar]
- 25.Jayaraman L. Moorthy NC. Murthy KG. Manley JL. Bustin M. Prives C. High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes Dev. 1998;12:462–472. doi: 10.1101/gad.12.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhakat KK. Izumi T. Yang SH. Hazra TK. Mitra S. Role of acetylated human AP-endonuclease (APE1/Ref-1) in regulation of the parathyroid hormone gene. EMBO J. 2003;22:6299–6309. doi: 10.1093/emboj/cdg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leppard JB. Dong Z. Mackey ZB. Tomkinson AE. Physical and functional interaction between DNA ligase IIIalpha and poly(ADP-Ribose) polymerase 1 in DNA single-strand break repair. Mol. Cell Biol. 2003;23:5919–5927. doi: 10.1128/MCB.23.16.5919-5927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H. Hays JB. Mismatch repair in human nuclear extracts. Quantitative analyses of excision of nicked circular mismatched DNA substrates, constructed by a new technique employing synthetic oligonucleotides. J. Biol. Chem. 2002;277:26136–26142. doi: 10.1074/jbc.M200357200. [DOI] [PubMed] [Google Scholar]
- 29.Prasad R. Singhal RK. Srivastava DK. Molina JT. Tomkinson AE. Wilson SH. Specific interaction of DNA polymerase beta and DNA ligase I in a multiprotein base excision repair complex from bovine testis. J. Biol. Chem. 1996;271:16000–16007. doi: 10.1074/jbc.271.27.16000. [DOI] [PubMed] [Google Scholar]
- 30.Wilson SH. Kunkel TA. Passing the baton in base excision repair. Nat. Struct. Biol. 2000;7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- 31.Izumi T. Brown DB. Naidu CV. Bhakat KK. Macinnes MA. Saito H. Chen DJ. Mitra S. Two essential but distinct functions of the mammalian abasic endonuclease. Proc. Natl. Acad. Sci. U.S.A. 2005;102:5739–5743. doi: 10.1073/pnas.0500986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evans AR. Limp-Foster M. Kelley MR. Going APE over ref-1. Mutat. Res. 2000;461:83–108. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]
- 33.Conlon KA. Miller H. Rosenquist TA. Zharkov DO. Berrios M. The murine DNA glycosylase NEIL2 (mNEIL2) and human DNA polymerase beta bind microtubules in situ and in vitro. DNA Repair (Amsterdam) 2005;4:419–431. doi: 10.1016/j.dnarep.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 34.Campalans A. Marsin S. Nakabeppu Y. O'Connor R. Boiteux TS. Radicella JP. XRCC1 interactions with multiple DNA glycosylases: a model for its recruitment to base excision repair. DNA Repair (Amsterdam) 2005;4:826–835. doi: 10.1016/j.dnarep.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 35.Doublie S. Bandaru V. Bond JP. Wallace SS. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10284–10289. doi: 10.1073/pnas.0402051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Das A. Rajagopalan L. Mathura VS. Rigby SJ. Mitra S. Hazra TK. Identification of a zinc finger domain in the human NEIL2 (Nei-like-2) protein. J. Biol. Chem. 2004;279:47132–47138. doi: 10.1074/jbc.M406224200. [DOI] [PubMed] [Google Scholar]
- 37.Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair (Amsterdam) 2003;2:955–969. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 38.Dou H. Mitra S. Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J. Biol. Chem. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- 39.Rosenquist TA. Zaika E. Fernandes AS. Zharkov DO. Miller H. Grollman AP. The novel DNA glycosylase, NEIL1, protects mammalian cells from radiation-mediated cell death. DNA Repair (Amsterdam) 2003;2:581–591. doi: 10.1016/s1568-7864(03)00025-9. [DOI] [PubMed] [Google Scholar]
- 40.Vartanian V. Lowell B. Minko IG. Wood TG. Ceci JD. George S. Ballinger SW. Corless CL. McCullough AK. Lloyd RS. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1864–1869. doi: 10.1073/pnas.0507444103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rasouli-Nia A. Karimi-Busheri F. Weinfeld M. Stable down-regulation of human polynucleotide kinase enhances spontaneous mutation frequency and sensitizes cells to genotoxic agents. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6905–6910. doi: 10.1073/pnas.0400099101. [DOI] [PMC free article] [PubMed] [Google Scholar]