Abstract
Defining human B cell repertoires to viral pathogens is critical for design of vaccines that induce broadly protective antibodies to infections such as HIV-1 and influenza. Single B cell sorting and cloning of immunoglobulin (Ig) heavy- and light-chain variable regions (VH and VL) is a powerful technology for defining anti-viral B cell repertoires. However, the Ig-cloning step is time-consuming and prevents high-throughput analysis of the B cell repertoire. Novel linear Ig heavy- and light-chain gene expression cassettes were designed to express Ig VH and VL genes isolated from sorted single B cells as IgG1 antibody without a cloning step. The cassettes contain all essential elements for transcriptional and translational regulation, including CMV promoter, Ig leader sequences, constant region of IgG1 heavy- or Ig light-chain, poly(A) tail and substitutable VH or VL genes. The utility of these Ig gene expression cassettes was established using synthetic VH or VL genes from an anti-HIV-1 gp41 mAb 2F5 as a model system, and validated further using VH and VL genes isolated from cloned EBV-transformed antibody-producing cell lines. Finally, this strategy was successfully used for rapid production of recombinant influenza mAbs from sorted single human plasmablasts after influenza vaccination. These Ig gene expression cassettes constitute a highly efficient strategy for rapid expression of Ig genes for high-throughput screening and analysis without cloning.
Keywords: Monoclonal antibody, single B cells, immunoglobulin gene, RT-PCR, linear gene expression cassette
1. Introduction
Immunoglobulin (Ig) is comprised of 2 identical heavy- and 2 identical light-chains. Ig heavy- and light-chain genes are produced by rearrangement of germline variable (V) and joining (J) gene segments at the light-chain locus, and by rearrangement of V, diversity (D) and J gene segments at the heavy-chain locus, respectively (Tonegawa, 1983; Diaz and Casali, 2002; Di Noia and Neuberger, 2007). Ig diversity is enhanced by somatic hypermutation of the rearranged genes (Kim et al., 1981; Di Noia and Neuberger, 2007). Antibody diversity allows the immune system to recognize a wide array of antigens (Honjo and Habu, 1985; Market and Papavasiliou, 2003). Antibodies represent the correlates of protective immunity to most infectious agents (Barreto et al., 2006). Monoclonal antibodies (mAbs) are important tools for studying pathogenesis, the protein structure of infectious agents and the correlates of protective immunity, and are essential to the development of passive immunotherapy and diagnostics against infectious agents. Defining the molecular aspects of human B cell repertoires to viral pathogens is critical for designing vaccines to induce broadly protective antibody responses to infections such as HIV-1 and influenza. The traditional methods used for generating human mAbs include screening Epstein-Barr virus (EBV)-transformed human B cell clones or antibody phage display libraries. These methods are often time-consuming and can have low yields of pathogen-specific mAbs. Although electroporation (Yu et al., 2008) and use of B cell activation by oCPGs (Traggiai et al., 2004) have improved the efficiency for development of EBV-transformed antibody-secretion B cell lines, techniques for the isolation, sequencing and cloning of rearranged heavy- and light-chain genes directly from human B cells are of interest because they provide a means to produce higher numbers of specific human mAbs. It has been shown that rearranged Ig heavy- and light-chain variable regions (VH and VL) can be amplified from single B cells using RT-PCR (Tiller et al., 2008; Volkheimer et al., 2007; Wrammert et al., 2008), thus making it possible to produce mAbs recombinantly (Wardemann et al., 2003; Koelsch et al., 2007; Tiller et al., 2008; Wrammert et al., 2008). Generally, the expression of rearranged Ig genes as antibodies requires laborious cloning of the amplified Ig VH and VL into eukaryotic cell expression plasmids containing a transcription regulation control element such as the CMV promoter (Boshart et al., 1985), sequences encoding the Ig leader, heavy- and light-chain Ig constant regions and a poly(A) signal sequence (McLean et al., 2000; Connelly and Manley, 1988; Norderhaug et al., 1997). Thus, what is needed to profile the Ig repertoire following immunization or an infection is the ability to amplify large numbers of Ig genes using a strategy that circumvents the Ig cloning step and yields sufficient quantities of transiently expressed Ig to allow functional characterization of expressed Igs. Linear expression constructs generated by one-step PCR have been used for expression of vaccinia DNA topoisomerase I (Xiao, 2007) and HIV-1 envelope proteins (Kirchherr JL, 2007). To facilitate high throughput testing of amplified Ig VH and VL genes for antibody expression and specificity analysis, a strategy was designed that uses PCR and novel linear Ig heavy- and light-chain gene expression cassettes for rapid expression of Ig VH and VL genes as recombinant antibodies without cloning procedures.
2. Materials and methods
2.1. Antibodies, cell lines and Ig heavy- and light-chain genes
Anti-HIV-1 membrane proximal gp41 mAb 2F5 was purchased from Polymun Scientific (Vienna, Austria). DNA sequences encoding the variable region of 2F5 heavy- and light-chain (Ofek et al., 2004) were reconstructed using the amino acid sequences from PDB (PDBID:1TJG:H and 1TJG:L) and the published DNA sequence (Kunert et al., 1998). Sequences of a full-length IgG1 heavy gene (Strausberg et al., 2002) and a full-length kappa chain gene (Strausberg et al., 2002) that were modified to contain sequences encoding for the VH and VL of mAb 2F5 were de novo synthesized (Blue Heron, Bothell, WA). The full-length 2F5 heavy- and light-chain genes were separately cloned into pCDNA3.1+ plasmid/hygro (Invitrogen, Carlbad, CA) that contains hygromycin resistant gene to facilitate screening of stably transfected-mammalian cell clones and resulted in plasmids HV13221 and HV13501, respectively. The HV13221 and HV13501 plasmids were used as sources of VH and VL chain sequences for method development and also used to generate stably transfected-293T cell line for producing purified recombinant 2F5 antibody as positive controls. A human embryonic kidney cell line, 293T, was obtained from the ATCC (Manassas, VA), cultured in DMEM supplemented with 10% FCS and used for DNA transfections. A stably transfected-293T cell line was generated by co-transfection with plasmids HV13221 and HV13501 using PolyFect (Qiagen, Valencia, CA), grown in DMEM supplemented with 10% FCS and maintained in DMEM supplemented with 2% FCS for production of recombinant 2F5 antibody. Recombinant 2F5 antibody was purified from culture supernatants of the stably transfected-293T cells by anti-human Ig heavy chain specific antibody-agarose beads (Sigma, St. Louis, MO). A human B cell line, G8, that secretes antibody recognizing the HIV-1 gp41 immunodominant epitope, was generated by EBV-transformation of B cells in terminal ileum biopsy obtained from an acute/early HIV-1 positive subject (Hwang, unpublished). An EBV-transformed human B cell line, 7B2, that produces an anti-HIV-1 gp41 antibody (Binley et al., 2000) was kindly provided by James Robinson. G8 and 7B2 cell lines were grown in Hybridoma-SFM (Invitrogen, Carlsbad, CA). mAbs were purified from culture supernatants using a ProPur Protein G column (NuNC, Rochester, NY).
2.2. Flow cytometry and cell sorting
Blood samples were collected as part of an IRB-approved protocol from a volunteer who received Fluzone® 2007-2008 vaccination. Peripheral blood mononuclear cells (PBMC) were isolated from blood that was collected on day 0, 7 and 21. PBMC were suspended in RPMI culture medium containing 20% FCS and 7.5% DMSO and stored in vapor phase liquid nitrogen until use. Antibodies used for flow cytometry were anti-human IgG- PE, CD3 PE-Cy5, CD16 PE-Cy5, CD19 APC-Cy7, CD20 PE-Cy7, CD27 Pacific Blue, CD235a PE-Cy5, IgD PE, IgM FITC (BD Biosciences, San Jose, CA), CD14 PE-Cy5 and CD38 APC-Cy5.5 (Invitrogen, Carlsbad, CA). All antibodies were titered in advance and used at optimal concentrations for flow cytometry. Plasma cells gated as CD3-, CD14-, CD16-, CD235a-, CD19+, CD20low-neg, CD27hi, and CD38hi were sorted as single cells into 96-well PCR plates containing 20 μl/well of RT reaction buffer that included 5 μl of 5× First strand cDNA buffer, 0.5 μl of RNAseOut (Invitrogen, Carlsbad, CA), 1.25 μl of DTT, 0.0625 μl of Igepal and 13.25 μl of dH2O (Invitrogen, Carlsbad, CA). The plates were stored at -80°C until use. Flow cytometric analysis and cell sorting were performed on a BD FACSAria (BD Biosciences, San Jose, CA) and the data were analyzed using FlowJo (Tree Star, Ashland, OR).
2.3. Isolation of Ig variable region transcripts from EBV-transformed B cells and sorted single plasmablasts by RT-PCR
The genes encoding Ig VH and VL chains were amplified by RT and nested PCR using a modification of a previously reported method (Tiller et al., 2008). Briefly, synthesis of Ig VH and VL was performed in 96-well PCR plates containing cloned EBV-transformed B cells or sorted single human plasmablasts. The RT reaction was carried out at 37°C for 1 hour after addition of 50 units/reaction Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA) and 0.5 μM human IgG, IgM, IgD and IgA1, IgA2, Igκ and Igλ constant region primers (Supplementary Table 1). After cDNA synthesis, VH, Vκ and Vλ genes were amplified separately by two rounds of PCR in 96-well PCR plates in 50 μL reaction mixtures. The first-round of PCR contained 5 μL of RT reaction products, 5 units of HotStar Taq Plus (Invitrogen; Carlsbad, CA), 0.2 mM dNTPs, and 0.5 μM of either IgM, IgG, IgD, IgA1 and IgA2, or Igκ or Igλ constant region primers and sets of IgH, Igκ or Igλ variable region primers (Supplementary Tables 2 and 3). The first round of PCR was performed at 95°C × 5 min followed by 35 cycles of 95°C × 30 sec, 55°C (VH and Vκ) or 50°C (Vλ) × 60 sec, 72°C × 90 sec, and one cycle at 72°C × 7 min. Nested second round PCR was performed with 2.5 μL of first-round PCR product, 5 units of HotStar Taq Plus, 0.2 mM dNTPs, 0.5 μM of either IgM, IgG, IgD, IgA1 and IgA2, or Igκ and Igλ nested constant region primers and sets of IgH, Igκ or Igλ nested variable region primers (Supplementary Tables 4-6). During the second round of nested PCR, the IgH, Igκ and Igλ variable region primers were amplified in separate reaction mixes for each variable region primer. The second-round of PCR was performed at 95°C × 5 min followed by 35 cycles of 95°C × 30 sec, 58°C (VH), 60°C (Vκ) or 64°C (Vλ) × 60 sec, 72°C × 90 sec, and one cycle at 72°C × 7 min. Samples of VH, Vκ and Vλ chain PCR products were analyzed on 1.2% agarose gels. Bone marrow RNA (Clontech, Mountain View, CA) samples were included during all RT-PCR runs as positive controls. All primers used for the 2nd round of PCR included tag sequences at the 5′ end of each primer (Supplemental Tables 4-6). This permits assembly of the VH and VL genes into functional linear Ig gene expression cassettes as described below. All PCR products were purified using a Qiagen (Valencia, CA) PCR purification kit and sequenced in forward and reverse directions using an ABI 3700 instrument and BigDye® sequencing kit (Applied Biosystems, Foster City, CA). Sequences were analyzed using the IMGT information system (http://imgt.cines.fr/) to identify variable region gene segments and somatic mutations.
2.4. Design and construction of the linear Ig expression cassettes
The linear Ig expression cassettes were assembled by overlapping PCR for facilitating high throughput testing of the Ig VH and VL genes for antibody expression and specificity analysis without cloning steps (Fig. 1). Each cassette was PCR-amplified from 3 overlapping DNA fragments including 1.) the C fragment made of the CMV promoter (705 bp) (Boshart et al., 1985) and sequence encoding for an Ig leader (METDTLLLWVLLLWVPGSTGD) (Burstein, 1978), 2.) either the H fragment (1,188 bp) made of the IgG1 constant region (315 aa) (Genbak Accession no. BC041037) (Strausberg et al., 2002) and bovine growth hormone (BGH) poly(A) signal sequences (Gimmi et al., 1989), K fragment (569 bp) made of the Ig kappa constant region (107 aa) (Strausberg et al., 2002) (GenBank Accession no. BC073791) and BHG poly(A) signal sequences, or L fragment (552 bp) made of the Ig lambda constant region (102 aa) (GenBank Accession no. BC073769) (Strausberg et al., 2002) and BGH poly(A) signal sequences (Gimmi et al., 1989) and 3.) either the VH, Vκ or Vλ genes amplified from single B cells as described above (Fig.1). The linear Ig gene cassettes contained the 5′ end restriction enzyme (Nhe I) site between the CMV promoter and Ig leader and the 3′ end restriction enzyme (Xba I) site between the Ig constant region stop codon and the poly(A) signal sequence (Fig.1). The purpose of these restriction enzyme sites was for potential cloning of Ig genes into expression plasmids for development of stable cell lines to produce recombinant antibodies of interest.
Fig. 1.

Schematic diagram for generation of linear full-length Ig heavy- and light-chain genes. Shown is a schematic diagram for the assembly by overlapping PCR of linear full-length Ig heavy-chain gene (A), Ig kappa light-chain gene (B) and lambda light-chain gene (C) expression cassettes. Sequences in the Ig leader region at the 3′ end of the C fragment overlapping with the sequences at the 5′ end of the VH, Vκ and Vλ fragments are indicated. Sequences at the 5′ end of the H fragment, K fragment and L fragments overlapping with the sequences at the 3′ end of the corresponding VH, Vκ and Vλ fragments are also indicated. The same forward and reverse primers (CMV-F262 and BGH-R1235, Supplementary Table 7) used in the overlapping PCR for all Ig heavy-chain genes, Ig kappa light-chain genes and lambda light-chain genes are indicated with arrows.
The C, H, K and L fragments were de novo synthesized (Blue Heron, Bothell, WA) and cloned into pCR2.1 plasmids (Invitrogen, Carlsbad, CA) resulting in plasmids HV0024, HV0023, HV0025 and HV0026, respectively. For use in assembling linear Ig gene cassettes, these DNA fragments were generated from these plasmids by PCR using the primers as shown in Supplementary Table 7. The PCR was carried out in a total volume of 50 μl with 1 unit of AccuPrime pfx polymerase (Invitrogen, Carlsbad, CA), 5 μl of 10× AccuPrime PCR buffer, 1ng plasmid, and 10 pmol of each primer. The PCR cycle conditions were one cycle at 94°C for 2 min, 25 cycles of a denaturing step at 94°C for 30 seconds, an annealing step at 60°C for 30 seconds, an extension step at 68°C for 40 seconds for the C, K and L fragments or 80 seconds for the H fragment, and one cycle of an additional extension at 68°C for 5 min.
The linear full-length Ig heavy- and light-chain gene expression cassettes were assembled by PCR from the C, VH and H fragments for heavy-chain, the C, Vκ and K fragments for kappa chain, and the C, Vλ and L fragments for lambda chain (1 ng of each). The PCR reaction was carried out in a total volume of 50 μl with 1 unit of KOD DNA polymerase (Novagen, Gibbstown, NJ), 5 μl of polymerase 10× PCR buffer, 200 μM of dNTP, 10 pmol of 5′ primer CMV-F262 and 3′ primer BGH-R1235 (Supplementary Table 7). The PCR cycle program consisted of one cycle at 98°C for 1 min, 25 cycles of a denaturing step at 98°C for 15 seconds, an annealing step at 60°C for 5 seconds, an extension step at 72°C for 35 seconds and one extension cycle for 10 min at 68°C.
2.5. Expression of recombinant antibodies
PCR products of the linear Ig expression cassettes were purified using a Qiagen PCR Purification kit (Qiagen, Valencia, CA). The purified PCR products of the paired Ig heavy- and light-chain gene expression cassettes were co-transfected into 80-90% confluent 293T cells grown in 12-well (1μg of each per well) tissue culture plates (Becton Dickson, Franklin Lakes, NJ) using PolyFect (Qiagen, Valencia, CA) and the protocol recommended by the manufacturer. Plasmids HV13221 and HV13501 (1μg of each per well) expressing Ig heavy or light-chain genes derived from the 2F5 mAb were used under the same conditions as positive controls. Six to eight hours after transfection, the 293T cells were fed with fresh culture medium supplemented with 2% FCS and were incubated for 72 hours at 37°C in a 5% CO2 incubator.
2.6. ELISA to determine the specificity and quantity of antibodies
To measure the concentration of recombinant mAbs in transfected culture supernatants, mouse anti-human Ig (Invitrogen, Carlsbad, CA) at 200 ng/well was used to coat 96-well high-binding ELISA plates (Costar/Corning; Lowell, MA) using carbonate bicarbonate buffer at pH 9.6 incubated overnight at 4°C. Plates were blocked at room temperature (RT) for 2 hours with PBS containing 4% wt/vol whey protein, 15% goat serum, 0.5% Tween-20, and 0.05% NaN3. 100 μL of supernatant from transfected cell cultures or control human IgG1 antibodies were incubated at RT for 2 hours. Goat-anti-human IgG specific (heavy- and light-chain)-alkaline phosphatase (AP) (1:3000 dilution) (Sigma, St. Louis, MO) diluted in blocking buffer was used as the secondary antibody and incubated at RT for 1 hour. For color development, the AP substrate was 2 mM MgCl2 and 1 mg/ml 4-nitrophenyl phosphate di(2-amino-2-ethyl-1,3-propanediol) salt in 50 mM Na2CO3 buffer (pH 9.6) incubated for 45 minutes. Plates were read in an ELISA reader at 405 nm. Amounts of IgG secreted in the transfected 293T cells were determined by comparison to a standard curve generated using known concentration of the control human IgG1.
Similar ELISA procedures as described above were used for detecting the binding of recombinant mAbs to specific antigens. Antigens for detection of anti-HIV-1 antibodies included HIV-1 Env MPER peptide, SP62 (QQEKNEQELLELDKWASLWN) (Alam et al., 2008), HIV-1 Env immunodominant epitope peptide (PrimmBiotech, Cambridge, MA), SP400 (RVLAVERYLRDQQLLGIWGCSGKLICTTAVPWNASWSNKSLNKI) (CPC Scientific, San Jose, CA), SP62-scrambled peptide (NKEQDQAEESLQLWEKLNWL) as a negative control (Alam et al., 2008), HIV-1 gp41 and HIV-1 JRFL gp140 protein (Liao, 2006). Fluzone® 2007-2008 (Sanofi Pasteur, Lyon, France), a trivalent inactivated influenza vaccine containing an A/Solomon Islands/3/2006 (H1N1)-like virus, an A/Wisconsin/67/2005 (H3N2)-like virus and a B/Malaysia/2506/2004-like virus, HA of H1 A/Solomon Islands, H3 A /Wisconsin, H3 A/Johannesburg and H5 A/Vietnam (Protein Sciences; Meriden, CT) were used as coating antigens in ELISA for detection of anti-influenza antibodies. Individual antigens at 200 ng/well were used to coat 96-well high-binding ELISA plates.
2.7. SDS-polyacrylamide gel electrophoresis and Western blot blot analysis of expressed recombinant mAb
Transfected culture supernatant samples (16 μl per lane) and controls were fractionated on precasted 4-12% Bis-Tris SDS-PAGE gels (Invitrogen, Carlsbad, CA) under non-reducing conditions, transferred onto nitrocellulose filters and probed with goat-anti-human IgG specific (heavy- and light-chain)-AP (1:3000 dilution) (Sigma, St. Louis, MO). The immunoblots were developed with Western-blue substrate (Promega; Madison, WI).
3. Results
3.1. Expression of VH and VL genes without cloning
Synthetic recombinant mAb 2F5 VH and VL genes (Ofek et al., 2004) were used as a model system for method development. Synthetic IgG1 heavy-chain and kappa chain genes were first cloned into pcDNA3.1/hygro plasmids and used to produce functional recombinant 2F5 antibody by stable transfection. Purified recombinant 2F5 antibody was compared with the commercial mAb 2F5 for their neutralizing activity in pseudotype HIV-1 neutralization assays (Montefiori, 2005). It was found that the recombinant 2F5 neutralized HIV-1 isolates with a similar potency as the commercial mAb 2F5 (Table 1). Next, the 2F5 VH and VL genes were amplified from 2F5 Ig heavy- and light-chain plasmids using the primer pair of CL-F681 and H-R474 for VH and the pair of CL-F681 and K-R405 for VL as shown in Supplementary Table 7. Assembly of 2F5 VH and VL genes into linear Ig gene cassettes was performed by overlapping PCR of the 2F5 VH and VL genes and the C, H and K, DNA fragments, and analyzed using agarose gel electrophoresis (Fig. 2A). A pair of VH and VL genes (rH42) isolated from a sorted single B cell were used as negative controls and assembled into the linear full-length Ig gene cassettes using the same procedure as for 2F5 Ig genes. The linear full-length 2F5 heavy- and light-chain gene cassettes were co-transfected into 293T cells. Plasmids HV13221 expressing the 2F5 heavy-chain gene and HV13501 expressing the 2F5 light-chain gene were used in parallel cultures as positive controls during transfection. The culture supernatants of the transfected-293T cells were harvested 3 days after transfection, analyzed by Western blot for the presence of Ig (Fig. 2B), and assayed by ELISA to measure the concentration of IgG (Fig. 2C) and determine the specificity of Igs against the HIV-1 Env MPER peptide SP62 (Alam et al., 2008), HIV-1 gp41 and HIV-1 JRFL gp140 (Fig. 2D). Co-transfection of the 2F5 heavy- and light-chain genes in the form of either plasmids or linear Ig gene cassettes not only produced whole IgG molecules with molecular weights of 150kDa as well as IgG molecules containing extra heavy- or light-chains with molecular weights of more than 150kDa as detected by both anti human Ig heavy- and light-chain antibodies (Fig. 2B). As shown in Fig. 2B, co-transfection of the 2F5 heavy- and light-chain genes also produced 1) mixtures of monomers of Ig heavy-chains (50kDa) detected only by an anti-human heavy-chain antibody; 2) monomers (23kDa), dimers (46kDa) and trimers (69kDa) of light-chains detected by an anti-light-chain antibody and 3) Ig molecules with different combinations (1:1 or 2:1 ratio) of heavy- and light-chains. From six independent transfection experiments, the average amounts of IgG produced by 293T cells transfected with the linear 2F5 heavy- and light-chain Ig gene cassettes were comparable to that produced in 293T cells transfected with plasmids of the 2F5 heavy- and light-chain genes (1.9 μg/ml IgG + 0.7 μg/ml (mean ± SEM), n=6 versus 1.7 μg/ml + 0.4 μg/ml, n=6, respectively) (Fig. 2C). As expected, similar to commercial mAb 2F5, the recombinant 2F5 IgG antibodies produced in 293T cells by transfection with either the linear 2F5 Ig gene cassettes or plasmids expressing 2F5 Ig genes reacted with HIV-1 MPER peptide SP62, HIV-1 gp41 and HIV-1 gp140 proteins but did not react with the negative control scrambled SP62 peptide (Fig. 2D). Supernatants generated by transfection of 293T cells with linear Ig heavy- and light-chain cassettes from the control antibody (rH42) and supernatants from mock-transfected 293T cells did not react with any of the HIV-1 proteins or peptides (Fig. 2D).
Table 1.
HIV-1 neutralization activity of mAb 2F5 and recombinant (r) 2F5 antibody.
| Antibody | 50% neutralization level (ug/ml) against HIV-1 Isolates | ||
|---|---|---|---|
| B.SF162 | B.BG1168 | C.TV-1 | |
| mAb 2F5 | 0.1 | 0.32 | 10.87 |
| r2F5 | 0.3 | 1.17 | 34.48 |
Fig. 2.


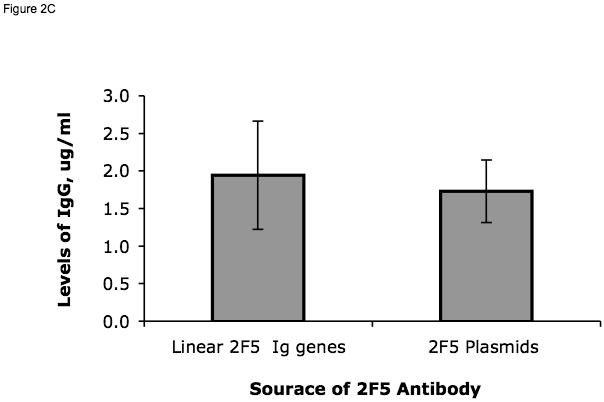

Expression of 2F5 VH and VL. In panel A, lane M: DNA ladders marked in kilobases (kb) next to the lane, lanes 1-8, respectively: DNA fragments C (705 bp), H (1,188 bp), K (569 bp), 2F5 VH (489 bp) and VL (370 bp) as well as the full-length Ig heavy- (2,339 bp) and light-chain (1,595 bp) gene expression cassettes generated by PCR and analyzed on a 1% agarose gel. Arrows indicate the expected DNA fragments. Panel B shows the results of Western blots of commercial mAb 2F5 (lane 1), supernatant harvested from 293T cells transfected with plasmids expressing 2F5 Ig genes (lane 2), with the linear full-length 2F5 Ig heavy- and light-chain gene cassettes (lanes 3) and mock-transfected 293T cells (lane 4). Igs on the blots were detected by either an anti-human heavy-chain specific antibody or an anti-human kappa light-chain specific antibody as indicated at the bottom of the blots. The arrows with short notations indicate the possible composition of antibody heavy-chains (HC) and light-chains (LC). Panel C is a comparison of the amounts of Ig secreted from 293T cells transiently transfected with either linear full-length 2F5 heavy- and light-chain Ig gene constructs (1ug of each) or plasmids (1ug of each) expressing the 2F5 heavy- or light-chain Ig genes as indicated. Average amounts (n=6) of IgG secreted in the transfected 293T cells are shown on the y axis and were determined by comparison to a standard curve generated using known concentrations of IgG1. Panel D shows the measurement of antibody binding by ELISA to the following antigens: HIV-1 Env MPER epitope peptide, HIV-1 gp41 and HIV-1 gp140 or scrambled SP62 peptides as negative controls. Supernatants from the 293T cells transfected with each DNA construct (indicated on the x axis) were assayed by ELISA for binding to HIV-1 antigens and the results were compared to the purified mAb 2F5 at 1.25 μg/ml.
3.2. Expression of Ig VH and VL genes derived from cloned EBV-transformed B cell lines
A major problem with available techniques for EBV transformation of B cells for generation of human mAbs is the low rate of B cell clone rescue. To determine whether the utility of the Ig linear cassette method for isolation and functional characterization of Ig genes could be used for rapid Ig gene profiling of EBV transformed B cells, this approach was tested on two cloned EBV-transformed human B cell lines, 7B2 (Binley et al., 2000) and G8 (Hwang, unpublished), that produce mAbs against HIV-1 gp41 and HIV-1 Env immunodominant epitope, respectively. Ig sequence information was not available from the 7B2 and G8 cell lines, therefore, the VH and VL genes of 7B2 and G8 were amplified using the RT-PCR method as described above. It was found that the Ig genes for 7B2 consisted of an IgG1 heavy-chain and a kappa light-chain and the Ig genes for G8 consisted of an IgG1 heavy-chain and a lambda light-chain. Assembly of the 7B2 and G8 VH and VL genes into linear full-length Ig gene cassettes was performed by overlapping PCR using the same method as for 2F5 Ig genes. The resulting linear Ig gene cassettes were transfected into 293T cells for expression of recombinant mAbs. By ELISA, the recombinant 7B2 IgG antibodies produced by transfection using linear Ig gene cassettes performed just like the mAb produced by the 7B2 EBV-transformed B cell line. Both preparations of 7B2 mAb reacted with HIV-1 gp41 and gp140 proteins, while the control antibody (rH70) or supernatant of mock-transfected 293T cells was non-reactive with these same proteins (Fig. 3A). Similar results were obtained using linear Ig gene cassettes generated from the G8 human B cell line (Fig. 3B). These results demonstrated that the linear Ig gene cassette method could be used to produce mAbs from B cells.
Fig. 3.


Measurement of the reactivity of recombinant antibodies. Panel A compares the reactivity of recombinant antibody produced in 293T cells by transfection of 7B2 Ig genes and mAb 7B2 produced by the EBV-transformed 7B2 B cell line. Supernatants of 293T cells transfected with linear rH70 Ig genes and supernatants from mock transfections were used as negative controls. Panel B compares the reactivity of the recombinant antibody produced by transfection with linear G8 Ig genes with antibody produced by the EBV-transformed G8 B cell line and with purified mAb G8. Supernatant from 293T cells transfected with linear rH0045 Ig genes with unknown specificity and from mock-transfection were used as negative controls.
3.3. Isolation and expression of Ig VH and VL genes derived from sorted single plasma cells
To demonstrate the utility of linear Ig gene cassettes for producing and screening mAbs from the VH and VL genes from sorted single primary human B cells, this strategy was tested using plasmablasts from a subject immunized with killed influenza vaccine Fluzone® 2007-2008. PBMC were isolated from a subject at day 0, 7 and 21 post-vaccination with Fluzone® 2007-2008 and were analyzed by flow cytometry. It was found that at day 7 after the Fluzone® vaccination, peripheral blood cells with a plasmablast phenotype (CD19+, CD20low-neg, CD27++ and CD38++) were increased compared to baseline (day 0); plasmablasts returned to baseline by day 21 after vaccination (Fig. 4). These results were consistent with studies reported by Wrammert and colleagues (Wrammert et al., 2008). Using BSL-3 BD FACSAria-based preparative cell sorting, single plasma cells from day 7 PBMC were sorted into 96-well plates (Wrammert et al., 2008). Nine Ig VH and VL pairs were isolated from day 7 plasmablasts by RT-PCR amplification of 24 wells of sorted single cells. The Ig VH and VL pairs were assembled into linear Ig gene cassettes and used to produce mAbs in 293T cells by transient transfection. It was found that 5 of the 9 recombinant mAbs were strongly reactive high-affinity anti-HA mAbs that reacted with inactivated influenza viruses in Fluzone® 2007-2008 (Fig. 5A) and with H1 A/Solomon Islands hemagglutinin (HA) (Fig. 5B) but not with H3 A/Wisconsin HA (Fig. 5C) that was also in the vaccine. The Ig concentration of these 5 antibodies ranged from 0.2 μg/ml to 1.3 ug/ml in the transfected culture supernatants. Sequence analysis of the VH and VL genes indicated that these 5 HA binding antibodies were distinct from each other (Table 2). The antibody B6 was an IgA antibody and the other 4 HA binding antibodies were IgG (Table 2). Importantly, the spectrum of the reactivity of these antibodies was reflective of serum antibody responses in the vaccinee. ELISA assays on serum samples collected at day 0 and 21 days after vaccination showed that there were preexisting, high levels of antibody to Fluzone® 2007-2008 and only low levels of antibody to H1 A/Solomon Islands HA or to H3 A/Wisconsin HA (Fig. 6). As such, Fluzone® 2007-2008 vaccination boosted antibody responses to the Fluzone®-2007-2008 and to A/Solomon Islands HA, and only weakly boosted antibody responses to H3 A/Wisconsin HA (Fig. 6). Not only did the results of the Fluzone® 2007-2008 plasmablast analysis support the utility of the linear Ig gene cassette method for producing human antibodies from human B cells but these results also demonstrated that this method of producing human mAbs reflects the range of human antibody responses.
Fig. 4.

Changes in plasmablast populations induced by vaccination with Fluzone. Shown is the frequency of plasmablasts (located in the upper right and defined as CD19+, CD20low-neg, CD3-, CD14-, CD16-, CD235-, CD38hi and CD27hi) in PB collected at day 0, 7 and 21 after vaccination from a subjecte vaccinated with FluzoneR 2007-2008 and analyzed by flow cytometry.
Fig. 5.
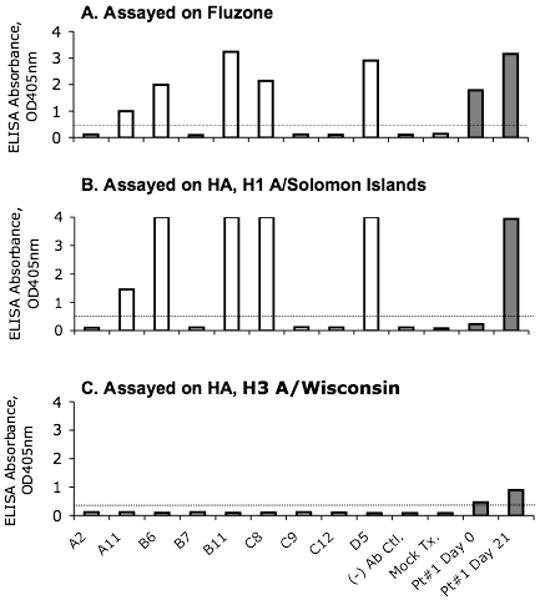
Reactivity of recombinant human mAbs from single plasma cells after Fluzone® 2007-2008 vaccination. Shown are the results of ELISA assays for detection of the reactivity of the antibodies derived from the individual Ig heavy- and light-chain gene pairs isolated from sorted single plasma cells (first 9 bars in the x-axis) or a negative control Ig pair (- Ab Ctl) and mock transfection control (Mock Tx.). Antibody reactivity to the inactivated influenza viruses (Panel A), with H1 A/Solomon islands HA (Panel B) and H3 A/Wisconsin HA (Panel C) are shown. Serum samples collected from the vaccinee at day 0 and 21 (grey columns) were used as positive controls in these assays. Data are representative of 2 independent experiments.
Table 2.
Variable regions of Ig heavy and light chain genes isolated from sorted single plasmablasts that reacted with Fluzone and H1 HA.
| Antibody ID | VH | VL | |||||
|---|---|---|---|---|---|---|---|
| VH ID | Family | CDR3 Length | Ig Isotype | VL ID* | Family | CDR3 Length | |
| A11 | H0076 | 4-402 | 23 | G | K0069 | 1-39 | 10 |
| B6 | H0077 | 3-30 | 17 | A | K0070 | 3-20 | 10 |
| B11 | H0079 | 3-43 | 17 | G | L0020 | 1-44 | 11 |
| C8 | H0080 | 4-39 | 19 | G | L0021 | 6-57 | 11 |
| D5 | H0082 | 2-04 | 19 | G | L0024 | 3-21 | 11 |
Fig. 6.

Serum antibody responses to Fluzone vaccination. Serum samples were collected from the vaccinee at day 0 and 21 days after vaccination with Fluzone and assayed against a panel of influenza antigens as indicated on the x-axis. Shown is the reactivity of serum samples at 1:800 dilution to the indicated antigens. Data are representative 2 independent experiments.
4. Discussion
In this study, a novel system was tested for Ig gene expression without prior cloning of VH and VL genes into expression vectors. In vitro expression of rearranged Ig genes as antibodies, requires cloning of amplified Ig VH and VL into eukaryotic cell expression plasmids containing a transcription regulation control element such as the CMV promoter, an Ig leader sequence, a poly(A) signal sequence and the constant region of the Ig heavy- or light-chain (Persic et al., 1997; Tiller et al., 2008; Wrammert et al., 2008). Several Ig expression vectors have been developed that produce functional Ig (Norderhaug et al., 1997; Persic et al., 1997; McLean et al., 2000; Tiller et al., 2008). However, cloning procedures are often the bottleneck for expression of recombinant antibodies for antibody selection. In this report, functional linear Ig gene cassettes assembled from three DNA fragments with overlapping sequences by PCR were described. The feasibility of the Ig production approach was demonstrated in 3 ways. First, the VH and VL genes derived from the anti-HIV-1 gp41 mAb 2F5 were used to produce functional 2F5 antibody. Second, it was demonstrated that the linear Ig gene cassette method could be used to produce functional HIV-1 antibodies from 2 EBV transformed cell lines, thus providing a powerful method of rescue of human mAbs from EBV-transformed B cell cultures. Finally, it was demonstrated that the linear Ig gene cassette method could be used to produce functional antibodies that bind influenza HA from peripheral blood plasmablasts from subjects vaccinated for influenza.
The linear Ig gene cassettes described in this paper contain all the essential elements necessary to produce functional antibodies. The cassettes contain a promoter (Boshart et al., 1985), Ig leader (Burstein, 1978), the constant region of IgG1 heavy-chain (Strausberg et al., 2002) or Ig light-chains (kappa and lambda) (Strausberg et al., 2002), poly(A) tail (Gimmi et al., 1989) and VH or VL genes. The VH and VL genes can be easily substituted with any VH and VL genes of humans, mouse or other origin (data not shown). Given the different forms of VH and VL that might be derived from different sources such as human or mouse, guidelines for designing the primers have been given in Supplementary Table 7 for creating the overlapping sequences. The constant region of the linear Ig heavy-chain gene cassette was derived from IgG1 because IgG1 is the most common Ig isotype among all Ig types. It was demonstrated that the chimeric IgG1 antibodies derived from B cells that expressed IgG (G8 and 7B2) had the same specificity and similar binding affinity as the original antibodies. Importantly, functional linear Ig gene cassettes produced Ig by transient transfection in 293T cells at levels that were comparable to that produced by transfection with plasmid DNA (Fig. 2) or by EBV-transformed B cell lines (Fig. 3b). The amounts (1 ml per well) of antibody samples generated in 12-well plates by transfection with linear Ig gene cassettes would be sufficient for most binding or neutralization assays, especially in multiplexed luminex systems (Croft et al., 2008) or antigen microarrays (Robinson, 2006).
The isolation of VH and VL genes from sorted single cells makes it possible to analyze Ig genes from single B cells and to produce recombinant mAbs (Babcook et al., 1996; Wardemann et al., 2003; Volkheimer et al., 2007; Tiller et al., 2008; Wrammert et al., 2008). The analysis of single B cells and linkage of the Ig reactivity profile with Ig gene sequences can provide valuable insight into the molecular basis of Ig gene rearrangement, allelic exclusion and Ig selection in the antibody repertoire (Kuppers et al., 1993; Brezinschek et al., 1995; Babcook et al., 1996; Wang and Stollar, 2000; Owens et al., 2003). Sorting of single cells into 96-well PCR plates followed by RT-PCR has been demonstrated as a very efficient process for isolation of small numbers of single cells with paired VH and VL genes (Tiller et al., 2008; Wrammert et al., 2008). By using the linear Ig expression cassettes method, it took only 6 working days from the time of flow cytometry analysis and single cell sorting of the PBMC from an influenza vaccinee to obtain 5 recombinant mAbs that were specific for influenza viruses. For production of mAb-expressing cell lines by stable transfection, once mAbs are obtained with the desired specificity, the Ig gene expression cassettes can be readily cloned into an expression plasmid like pcDNA3.3-TOPO usingTA cloning (Invitrogen, Carlsbad, CA) or pcDNA3.1 (Invitrogen, Carlsbad, CA) using restriction enzyme digestion-ligation, because the Ig gene expression cassettes were designed to contain unique Nhe I–Xbo I sites (5′-3′) that are extremely rare cutters for Ig genes (Persic et al., 1997) of the full-length of Ig heavy- and light-chain constructs (Fig. 1).
Thus, by combining the isolation of Ig VH and VL genes from single cells by RT-PCR (Tiller et al., 2008; Wrammert et al., 2008) and the use of novel linear Ig gene expression cassettes described here, a rapid strategy for expressing Ig genes was developed for screening and analysis within days of B cell isolation. Importantly, this system has the advantage that it can be scaled up for high-throughput human mAb production as we have recently generated more than 600 recombinant antibodies derived from sorted human plasmablasts by using this approach for screening against HIV-1 and other antigens to profile B cells responses to acute HIV-1 infection (manuscript in preparation, H-X Liao and B. F. Haynes). This strategy could also be adapted to generate recombinant high affinity human or non-human antibodies, for use as therapeutic agents, for development of mutant antibodies, for use in mechanistic studies of antibody-antigen interactions, and for rescuing antibodies from EBV-transformed cell lines or mycoplasma-contaminated antibody-producing B cell or hybridoma cell lines.
Supplementary Material
Acknowledgments
This work was supported by the Center for HIV/AIDS Vaccine Immunology NIAID grant U19 AI067854, NIAID grant P01 AI061734 and a Collaboration for HIV Vaccine Discovery grant from the Bill and Melinda Gates Foundation. Flow cytometry was performed in the Duke Center for AIDS Research BSL3 Flow Cytometry Core Facility supported by the NIH grants S10RR019145, UC6 AI058607, AI64518 and P30 AI051445.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alam SM, Scearce RM, Parks RJ, Plonk K, Plonk SG, Sutherland LL, Gorny MK, Zolla-Pazner S, Vanleeuwen S, Moody MA, Xia SM, Montefiori DC, Tomaras GD, Weinhold KJ, Karim SA, Hicks CB, Liao HX, Robinson J, Shaw GM, Haynes BF. Human immunodeficiency virus type 1 gp41 antibodies that mask membrane proximal region epitopes: antibody binding kinetics, induction, and potential for regulation in acute infection. J Virol. 2008;82:115–25. doi: 10.1128/JVI.00927-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcook JS, Leslie KB, Olsen OA, Salmon RA, Schrader JW. A novel strategy for generating monoclonal antibodies from single, isolated lymphocytes producing antibodies of defined specificities. Proc Natl Acad Sci USA. 1996;93:7843–8. doi: 10.1073/pnas.93.15.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto ML, Teixeira MG, Carmo EH. Infectious diseases epidemiology. J Epidemiol Community Health. 2006;60:192–5. doi: 10.1136/jech.2003.011593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, Moore JP. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74:627–43. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshart M, Weber F, Jahn G, Dorsch-Hasler K, Fleckenstein B, Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985;41:521–30. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- Brezinschek HP, Brezinschek RI, Lipsky PE. Analysis of the heavy chain repertoire of human peripheral B cells using single-cell polymerase chain reaction. J Immunol. 1995;155:190–202. [PubMed] [Google Scholar]
- Burstein Y, Schechter I. Primary structures of N-terminal extra peptide segments linked to the variable and constant regions of immunoglobulin light chain precursors: implications on the organization and controlled expression of immunoglobulin gene. Biochemistry. 1978;17:2392–2400. doi: 10.1021/bi00605a022. [DOI] [PubMed] [Google Scholar]
- Connelly S, Manley JL. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev. 1988;2:440–52. doi: 10.1101/gad.2.4.440. [DOI] [PubMed] [Google Scholar]
- Croft H, Malinowski T, Krizbai L, Mikec I, Kajic V, Reed C, Varga A, James D. Use of Luminex xMAP-derived Bio-Plex bead-based suspension array for specific detection of PPV W and characterization of epitopes on the coat protein of the virus. J Virol Methods. 2008;153:203–13. doi: 10.1016/j.jviromet.2008.07.016. [DOI] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- Diaz M, Casali P. Somatic immunoglobulin hypermutation. Curr Opin Immunol. 2002;14:235–40. doi: 10.1016/s0952-7915(02)00327-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimmi ER, Reff ME, Deckman IC. Alterations in the pre-mRNA topology of the bovine growth hormone polyadenylation region decrease poly(A) site efficiency. Nucleic Acids Res. 1989;17:6983–98. doi: 10.1093/nar/17.17.6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo T, Habu S. Origin of immune diversity: genetic variation and selection. Annu Rev Biochem. 1985;54:803–30. doi: 10.1146/annurev.bi.54.070185.004103. [DOI] [PubMed] [Google Scholar]
- Kim S, Davis M, Sinn E, Patten P, Hood L. Antibody diversity: somatic hypermutation of rearranged VH genes. Cell. 1981;27:573–81. doi: 10.1016/0092-8674(81)90399-8. [DOI] [PubMed] [Google Scholar]
- Kirchherr JL, L X, Kasongo W, Chalwe V, Mwananyanda L, Musonda RM, Xia SM, Scearce RM, Liao HX, Montefiori DC, Haynes BF, Gao F. High throughput functional analysis of HIV-1 env genes without cloning. J Virol Methods. 2007;143:104–11. doi: 10.1016/j.jviromet.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelsch K, Zheng NY, Zhang Q, Duty A, Helms C, Mathias MD, Jared M, Smith K, Capra JD, Wilson PC. Mature B cells class switched to IgD are autoreactive in healthy individuals. J Clin Invest. 2007;117:1558–65. doi: 10.1172/JCI27628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunert R, Ruker F, Katinger H. Molecular characterization of five neutralizing anti-HIV type 1 antibodies: identification of nonconventional D segments in the human monoclonal antibodies 2G12 and 2F5. AIDS Res Hum Retroviruses. 1998;14:1115–28. doi: 10.1089/aid.1998.14.1115. [DOI] [PubMed] [Google Scholar]
- Kuppers R, Zhao M, Hansmann ML, Rajewsky K. Tracing B cell development in human germinal centres by molecular analysis of single cells picked from histological sections. Embo J. 1993;12:4955–67. doi: 10.1002/j.1460-2075.1993.tb06189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Xia SM, Scearce RM, Sutherland LL, Brock ME, Vanleeuwen S, Alam SM, McAdams M, Ma BJ, Weaver EA, Plonk K, Montefiori DC, Hahn BH, Korber BT, Gao F, Haynes BF. Group M consensus Env oligomers induce antibodies that neutralize subtype C HIV- primary isolates. Virology. 2006;353:268–282. doi: 10.1016/j.virol.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Market E, Papavasiliou FN. V(D)J recombination and the evolution of the adaptive immune system. PLoS Biol. 2003;1:E16. doi: 10.1371/journal.pbio.0000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean GR, Nakouzi A, Casadevall A, Green NS. Human and murine immunoglobulin expression vector cassettes. Mol Immunol. 2000;37:837–45. doi: 10.1016/s0161-5890(00)00101-2. [DOI] [PubMed] [Google Scholar]
- Montefiori DC. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protoc Immunol. 2005;Chapter 12(Unit 1211) doi: 10.1002/0471142735.im1211s64. [DOI] [PubMed] [Google Scholar]
- Norderhaug L, Olafsen T, Michaelsen TE, Sandlie I. Versatile vectors for transient and stable expression of recombinant antibody molecules in mammalian cells. J Immunol Methods. 1997;204:77–87. doi: 10.1016/s0022-1759(97)00034-3. [DOI] [PubMed] [Google Scholar]
- Ofek G, Tang M, Sambor A, Katinger H, Mascola JR, Wyatt R, Kwong PD. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol. 2004;78:10724–37. doi: 10.1128/JVI.78.19.10724-10737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens GP, Ritchie AM, Burgoon MP, Williamson RA, Corboy JR, Gilden DH. Single-cell repertoire analysis demonstrates that clonal expansion is a prominent feature of the B cell response in multiple sclerosis cerebrospinal fluid. J Immunol. 2003;171:2725–33. doi: 10.4049/jimmunol.171.5.2725. [DOI] [PubMed] [Google Scholar]
- Persic L, Roberts A, Wilton J, Cattaneo A, Bradbury A, Hoogenboom HR. An integrated vector system for the eukaryotic expression of antibodies or their fragments after selection from phage display libraries. Gene. 1997;187:9–18. doi: 10.1016/s0378-1119(96)00628-2. [DOI] [PubMed] [Google Scholar]
- Robinson WH. Antigen arrays for antibody profiling. Curr Opin Chem Biol. 2006;10:67–72. doi: 10.1016/j.cbpa.2005.12.028. [DOI] [PubMed] [Google Scholar]
- Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer AA, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Brownstein MJ, Usdin TB, Toshiyuki S, Carninci P, Prange C, Raha SS, Loquellano NA, Peters GJ, Abramson RD, Mullahy SJ, Bosak SA, McEwan PJ, McKernan KJ, Malek JA, Gunaratne PH, Richards S, Worley KC, Hale S, Garcia AM, Gay LJ, Hulyk SW, Villalon DK, Muzny DM, Sodergren EJ, Lu X, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Madan A, Young AC, Shevchenko Y, Bouffard GG, Blakesley RW, Touchman JW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Krzywinski MI, Skalska U, Smailus DE, Schnerch A, Schein JE, Jones SJ, Marra MA. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002;99:16899–903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods. 2008;329:112–24. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–81. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Traggiai E, Becker S, Subbarao K, Kolesnikova L, Uematsu Y, Gismondo MR, Murphy BR, Rappuoli R, Lanzavecchia A. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat Med. 2004;10:871–5. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkheimer AD, Weinberg JB, Beasley BE, Whitesides JF, Gockerman JP, Moore JO, Kelsoe G, Goodman BK, Levesque MC. Progressive immunoglobulin gene mutations in chronic lymphocytic leukemia: evidence for antigen-driven intraclonal diversification. Blood. 2007;109:1559–67. doi: 10.1182/blood-2006-05-020644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Stollar BD. Human immunoglobulin variable region gene analysis by single cell RT-PCR. J Immunol Methods. 2000;244:217–25. doi: 10.1016/s0022-1759(00)00260-x. [DOI] [PubMed] [Google Scholar]
- Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–7. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng NY, Mays I, Garman L, Helms C, James J, Air GM, Capra JD, Ahmed R, Wilson PC. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–71. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao JH, Wen X, Liu Yong-Jie, Murphy Robert, Huang Da-Wei. eneration of linear expression constructs by one-step PCR with vaccinia DNA topoisomerase I. Molecular Biotechnology. 2007;35 doi: 10.1385/mb:35:1:15. [DOI] [PubMed] [Google Scholar]
- Yu X, McGraw PA, House FS, Crowe JE., Jr An optimized electrofusion-based protocol for generating virus-specific human monoclonal antibodies. J Immunol Methods. 2008;336:142–51. doi: 10.1016/j.jim.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


