Abstract
We previously isolated 25 temperature-sensitive gsp1 alleles of Saccharomyces cerevisiae Ran homologue, each of which possesses amino acid changes that differ from each other. We report here isolation of three multicopy suppressors—PDE2, NTF2, and a gene designated MOG1—all of which rescued a growth defect of these gsp1 strains. The gsp1 suppression occurred even in the absence of GSP2, another S. cerevisiae GSP1-like gene. Previously, NTF2 was reported to suppress gsp1 but not PDE2. Mog1p, with a calculated molecular mass of 24 kDa, was found to be encoded by the yeast ORF YJR074W. Both MOG1 and NTF2 suppressed a series of gsp1 alleles with similar efficiency, and both suppressed gsp1 even with a single gene dose. Consistent with the high efficiency of gsp1 suppression, Mog1p directly bound to GTP, but not to GDP-Gsp1p. The disruption of MOG1 made yeast temperature-sensitive for growth. Δmog1, which was suppressed by overexpression of NTF2, was found to have a defect in both classic and nonclassic nuclear localization signal-dependent nuclear-protein imports, but not in mRNA export. Thus, Mog1p, which was localized in the nucleus, is a Gsp1p-binding protein involved in nuclear-protein import and that functionally interacts with Ntf2p. Furthermore, the finding that PDE2 suppressed both gsp1 and rna1–1 indicates that the Ran GTPase cycle is regulated by the Ras-cAMP pathway.
Gsp1p is the Saccharomyces cerevisiae homologue of the Ran GTPase, which is essential for nucleocytoplasmic exchange of macromolecules (1–9). Indeed, all temperature-sensitive (ts) gsp1 alleles have a defect in nuclear-protein import (10, 11). However, mutants of Rcc1p, which is the guanine nucleotide-exchanging factor (GEF) of Ran (12), show a pleiotropic phenotype, such as cell cycle-specific arrest and defects in mRNA export and mRNA splicing (13–18). Furthermore, rna1–1, a ts mutant of yeast Ran GTPase activator (RanGAP; ref. 19) was initially identified as a mutant defective in RNA processing (20) and was subsequently found to have defects in mRNA metabolism similar to prp20, a ts mutant of the S. cerevisiae RCC1 homologue (21–23) and in nuclear-protein import (24). It is a matter of debate how Ran controls all of the biological defects observed.
To clarify the pathways downstream of Gsp1p, we previously isolated a series of ts mutants of GSP1 (11). In this report, we isolated multicopy suppressor genes NTF2, PDE2, and MOG1 from S. cerevisiae that conferred the temperature resistance (ts+) phenotype to these gsp1 strains. The suppression of gsp1 with these genes occurred even in the absence of another S. cerevisiae GSP1-like gene, GSP2, that encodes a protein 97% identical to Gsp1p and is expressed in a nonglucose medium (25). Ntf2p has been reported to rescue gsp1–1 and gsp1–2 (10). It is a highly conserved protein essential for nuclear-protein import (26, 27) and specifically binds to GDP-Gsp1p but not to GTP-Gsp1 (28, 29). The role of Ntf2p in nuclear-protein import, however, is still unknown. Pde2p, which is a high-affinity phosphodiesterase (30), has been reported to suppress rna1–1 (31). In contrast, Mog1p is encoded by the S. cerevisiae ORF YJR074W. Both NTF2 and MOG1 genes suppressed gsp1 with a similar efficiency. Consistent with this finding, Mog1p was found to have a functional interaction with Ntf2p. Thus, Mog1p, which was found to be required for nuclear-protein import and to bind to GTP-Gsp1p, may help us analyze the function of Ntf2p in the nuclear protein-import process.
MATERIALS AND METHODS
Strains and Media.
All of the S. cerevisiae strains used in this study are shown in Table 1. The media used for S. cerevisiae and bacterial strains were described previously (11). The strain PSY962–479 was derived from the PSY962 strain (a gift from P. Silver, Dana–Farber Cancer Institute; ref. 10) by plasmid shuffling using 5-fluoroorotic acid (5FOA) as described (11).
Table 1.
Yeast strains and plasmids used in this study
| Relevant markers | Genotype/comment | Source | |
|---|---|---|---|
| Yeast | |||
| N43-6C-gsp1-479 | MATa Δgsp1∷HIS3∷gsp1ts∷LEU2 ade2 leu2 trp1 ura3 | (11) | |
| PSY962 | MATα Δgsp1∷HIS3 Δgsp2∷HIS3 ura3-52 leu2-Δ1 his3- Δ200 trp1-Δ63 covered with GSP1 on a ura3-based vector | (10) | |
| PSY962-479 | MATα Δgsp1∷HIS3 Δgsp2∷HIS3 ura3-52 leu2-Δ1 his3-Δ 200 trp1-Δ63 [pF314gsp1-479] | This study | |
| PSY852 | MATa Δntf2∷HIS3 ura3-52 leu2-Δ1 covered with NTF2 on a ura3-based vector | (27) | |
| NN19-5B | MATa rna1-1 ade2 leu2 his3 trp1 ura3 | (34) | |
| YPH499 | MATa ade2-101 leu2-Δ 1 trp1-Δ63 ura3-52 lys2-801 his3-Δ200 | (45) | |
| YPH500 | MATα ade2-101 leu2-Δ1 trp1-Δ63 ura3-52 lys2-801 his3-Δ200 | (45) | |
| MOY1 | MATa Δmog1∷HIS3 ade2-101 leu2-Δ 1 trp1-Δ63 ura3-52 lys2-801 his3-Δ200 | This study | |
| MOY2 | MATα Δmog1∷HIS3 ade2-101 leu2-Δ1 trp1-Δ63 ura3-52 lys2-801 his3-Δ200 | This study | |
| Yeast plasmids | |||
| p195GSP1 | 2 μm URA3 GSP1 | YEplac195 with 1.95-kb GSP1 fragment at KpnI/PstI site | This study |
| p195NTF2 | 2 μm URA3 NTF2 | YEplac195 with 1 kb NTF2 fragment at XbaI/EcoRI site | This study |
| p195MOG1 | 2 μm URA3 MOG1 | YEplac 195 with 1.3-kb MOG1 fragment at SmaI/PstI site | This study |
| p195PDE2 | 2 μm URA3 PDE2 | YEplac195 with 4.2-kb PDE2 fragment at KpnI/BamHI site | This study |
| pRSCNR1 | CEN URA3 CNR1/GSP1 | pRS316 with CNRI/GSP1 fragment at BamHI/SalI site | (17) |
| p316NTF2 | CEN URA3 NTF2 | pRS316 with 1-kb NTF2 fragment at XbaI/EcoRI site | This study |
| p316MOG1 | CEN URA3 MOG1 | pRS316 with 1.1-kb MOG1 fragment at SacI/SmaI site | This study |
| p316PDE2 | CEN URA3 PDE2 | pRS316 with 3-kb PDE2 fragment at BamHI/HindIII site | This study |
| pFB1-33C | 2 μm URA3 | H2B NLS fused in-frame to β-galactosidase | (38) |
| GAL10∷H2B∷lacZ | |||
| pPS919 | CEN LEU2 ntf2-1 | pRS315 with ntf2-1 | (27) |
| pPS920 | CEN LEU2 ntf2-2 | pRS315 with ntf2-2 | (27) |
| pPS751 | 2 μm URA3 Npl3-Myc | YEp352 with Myc-tagged Npl3p | (39) |
| E. coli plasmids | |||
| pGEX-GSP1 | GST-fused GSP1 for E. coli expression | (34) | |
| pGEX-YRB1 Δ 1-9 | GST-fused YRB1 lacking N-terminal 9 amino acid residues for E. coli expression | (34) | |
| pGEX-MOG1 | GST-fused MOG1 for E. coli expression | This study | |
| pGEX-PRP20 | GST-fused PRP20 for E. coli expression | (34) | |
| pHIS3MOG1 | Disruption of MOG1 by replacement of 0.65-kb full-size MOG1 ORF with 1.75-kb HIS3 fragment | This study |
Determination of the Critical Temperature.
The multicopy vector, YEplac195, was introduced into all of the gsp1 strains. Ura+ transformants isolated at 26°C were incubated at 26°C, 30°C, 33°C, 34°C, 35°C, 36°C, or 37°C on a synthetic medium (ura−) plate. Thus, the lowest temperature above which the gsp1 strain did not papillate with expression of the multicopy plasmids was determined for each gsp1 allele.
Isolation of Multicopy Suppressors.
The genomic library (RB236) constructed by using the vector YEp24 (32) was introduced into cultures of gsp1 strains. Transfected cells (4.5 × 105) were incubated in synthetic medium (ura−) for 1 day at 26°C and then for 3–6 days at the nonpermissive temperature for each gsp1 allele. As described (33), plasmids were isolated from transformants (ts+Ura+) of gsp1 cells which grew at the nonpermissive temperature in synthetic medium (ura−). Subsequently, DNA inserts of resulting plasmids were sequenced by using as the primers 5′-GTCCTGCTCGCTTCGCTACT-3′ and 5′-GCGATATAGGCGCCAGCAAC-3′. The nucleotide sequences of both the 5′ and 3′ ends of the DNA insert were used to search for ORFs of S. cerevisiae genomic sequence by using the Munich Information Center for Protein Sequences (MIPS) (http://www.mips.biochem.mpg.del).
Construction of Plasmids.
All of the plasmids used in this study are listed in Table 1. For pGEX-MOG1, 1 kb of DNA-fragment containing MOG1 was amplified from p195MOG1 by PCR. Resultant DNA fragments were partially digested and inserted into the NcoI/HindIII site of pGEX-KG, resulting in pGEX-MOG1. For pHIS3MOG1, 0.97 kb of the 5′ noncoding region of MOG1 was amplified by using PCR and was inserted into the XbaI/BamHI site of pBluescript II SK(+), resulting in pSKMOG1XB. On the other hand, 1.03 kb of the 3′ noncoding region of MOG1 was amplified by using PCR and was inserted into the BamHI/EcoRI site of pSKMOG1XB. The resultant pSKMOG1XBBE was digested with the BamHI enzyme and ligated with 1.75 kb of BamHI fragment containing HIS3, resulting in pHIS3MOG1.
Gsp1p Binding Assay.
Gsp1p, Yrb1p, Prp20p, and Mog1p were prepared as glutathione S-transferase (GST)-fused proteins in Escherichia coli and purified on a glutathione column and examined for binding to Gsp1p as described (34).
Immunofluorescence Microscopy.
S. cerevisiae strains possessing pFB1–33C (GAL10∷H2B∷lacZ), pPS751(NPL3-myc), or p195MOG1 were processed and fixed as described (34). β-Galactosidase was stained with a primary anti-β-galactosidase antibody (34), and a secondary Texas red-conjugated goat anti-rabbit antibody (Cappel). Myc-tagged Npl3p was stained with the mAb to Myc-tag and fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse antibody (Cappel). The anti-Mog1p antibody was raised against E. coli-produced Mog1p and purified on a Mog1p-coupled HiTrap affinity column (Pharmacia). Fixed cells were treated with the affinity-purified anti-Mog1p antibodies and stained with Texas red-conjugated goat anti-rabbit IgG antibodies (Cappel). DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI). Stained cells were examined with a Zeiss Axiophot microscope.
In Situ Hybridization for mRNA Localization.
mRNA was hybridized with biotin-labeled oligo(dT)50 and stained with FITC–avidin as described (34).
Biosensor Analysis.
Real-time interaction analysis of the binding between Mog1p and Gsp1p was performed with a Biacore 2000 biosensor instrument (Biacore, Uppsala, Sweden) as described (35). E. coli-produced GST–Gsp1p was purified on a glutathione column and mixed with guanylylimidodiphosphate or GDP. The nucleotide-bound Gsp1p was purified by using a SO3− column (Merck). Purified nucleotide-bound Gsp1p contained less than 10% of nucleotide-free Gsp1p.
RESULTS
Isolation of MOG1 Gene.
The YEp24-based yeast genomic library was introduced into three gsp1 strains, N43–6C-gsp1–479, N43–6C-gsp1–1757, and N43–6C-gsp1–322, which have either single (gsp1-479 and gsp1-1757) or double (gsp1-322) mutations in the GSP1 gene (Table 2) as described (11). To identify ts+ colonies, Ura+ transformants were incubated at the nonpermissive temperature for each gsp1 allele, above which temperature the vector containing gsp1 strain could not papillate on a synthetic medium plate (ura−; see Table 2).
Table 2.
Suppression of gsp1 alleles by p195NTF2, p195MOG1, and p195PDE2
| Mutation name | Amino acid change | Multicopy Suppression
|
Critical nonpermissive temperature | ||
|---|---|---|---|---|---|
| NTF2 | MOG1 | PDE2 | |||
| gsp1-1757 | F28S | + | + | ± | 33 |
| gsp1-1268 | A85D | + | + | ± | 36 |
| gsp1-479 | I89Y | + | + | + | 34 |
| gsp1-16 | D93E, W165R | + | + | + | 36 |
| gsp1-322 | H50Q, Y149H | ± | ± | − | 33 |
| gsp1-882 | L52S, N156D | ± | + | ± | 30 |
| gsp1-1547 | V103A, K125R | + | + | ± | 35 |
| gsp1-1907 | M91V, I117A | + | + | ± | 33 |
| gsp1-245 | K14N, K62I, I89F | + | + | + | 33 |
| gsp1-1178 | Y55H, Q86P, E160V | + | + | + | 34 |
| gsp1-1568 | H50L, Y55N, Y100F | + | − | − | 36 |
| gsp1-1598 | R112S, I151F, F178L | + | + | ± | 33 |
| gsp1-1651 | W66R, E160G, Q190P | ± | + | ± | 36 |
| gsp1-1819 | K14I, K129E, D192E | + | ± | − | 36 |
| gsp1-1894 | K14R, T34A, K62E | − | − | ± | 37 |
| gsp1-1060 | V10D, E36G, E60V, N158D | + | ± | − | 34 |
| gsp1-1486 | F58S, N84I, K144N, D150G | + | + | + | 34 |
| gsp1-640 | E9D, F63L, F92L, L121N, K129N | + | + | + | 36 |
| gsp1-1582 | K73N, I98V, K125R, K129I, L176I | − | ± | ± | 37 |
| gsp1-1778 | K39M, F58L, N102I, R108K, E193D | + | + | ± | 33 |
| gsp1-1968 | F37Y, H50R, T56S, Y82C, N102D | ± | ± | − | 35 |
| gsp1-1518 | E36V, F92Y, N116S, Y199S, D213E, T207A | + | − | ± | 36 |
| gsp1-1260 | T56S, E60A, K125R, T137A, A183G, A208G | + | + | ± | 35 |
| gsp1-1763 | L33S, V94A, N116T, T137A, K154M, N173D | ± | ± | ± | 36 |
| gsp1-1817 | F28Y, T34A, I119T, K143R, Q147R, L195S, Q200H | − | − | − | 37 |
Ura+ transformants were incubated at the nonpermissive temperature for each gsp1 allele for 3 days on a synthetic-medium (ura−) plate. +, Full suppression. ±, Partial suppression. −, No suppression.
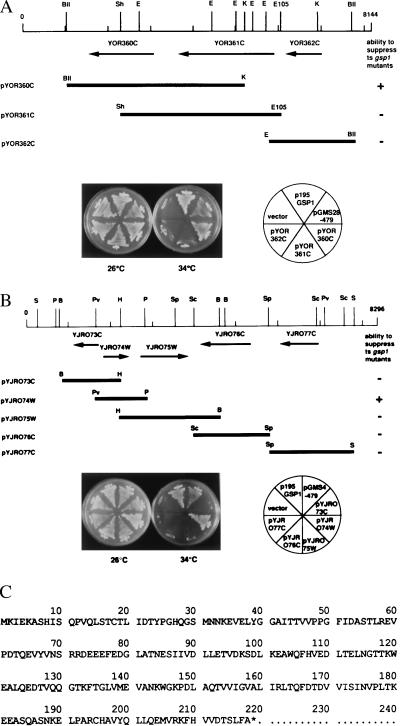
Five plasmids were recovered from ts+ transformants of the N43–6C-gsp1–479 strain. Subsequently, the yeast genomic DNA insert was sequenced and mapped to the S. cerevisiae genome by using the MIPS database. One of them contained GSP1, and the other contained NTF2, which has been reported to suppress gsp1–1 and gsp1–2 and were isolated by Wong et al. (10). Three other plasmids, pGMS1–479, pGMS4–479, and pGMS28–479, possessed new genes not reported to suppress gsp1. pGMS1–479 and pGMS4–479 possessed the same DNA insert. The ORF and the restriction-enzyme sites of the yeast genomic inserts of pGMS28–479 and pGMS4–479 are shown in Fig. 1 A and B.
Figure 1.
Suppression of gsp1 by MOG1 and PDE2. (A and B) Restriction enzyme sites and ORFs of the yeast genomic DNA insert carried on pGMS28–479 (A) and pGMS4–479 (B) are shown. The ORFs carried on these plasmids were subcloned into the indicated plasmids by using the appropriate enzyme sites of YEplac195 and introduced into the N43–6C-gsp1–479 strain. Ura+ transformants were streaked on a synthetic-medium (ura−) plate and incubated at either 26°C or 34°C for 3 days as indicated. +, full suppression. −, no suppression. The restriction enzyme sites shown are as follows. B, BamHI; H, HindIII; Pv, PvuII; P, PstI; Sc, ScaI; Sp, SpeI; S, SacI; BII, BglII; K, KpnI; Sh, SphI; E105, Eco105I; E, EcoRI. (c) Amino acid sequence of Mog1p predicted from the sequence of pYJR074W.
To determine which ORF of these plasmids suppressed gsp1–479, each ORF was subcloned into the YEplac195 vector and introduced into the N43–6C-gsp1–479 strain. pYOR360C derived from pGMS28–479 (Fig. 1A) and pYJR074W derived from pGMS4–479 (Fig. 1B) each conferred the ts+ phenotype to the N43–6C-gsp1–479 strain, but the other ORFs did not. According to the S. cerevisiae genomic sequence, YOR360C encodes Pde2p (phosphodiesterase type 2; ref. 30), and thus pYOR360C was designated as p195PDE2. YJR074W encodes a protein consisting of 218 aa with a calculated molecular mass of 24,306 Da (Fig. 1C) that was designated as Mog1p (multicopy suppressor of ts gsp1). Thereby, pYJR074W was designated as p195MOG1.
From ts+ transformants of the N43–6C-gsp1–1757 and the N43–6C-gsp1–322 strains, a total of seven plasmids were recovered. Four of these clones possessed NTF2, two clones possessed GSP2, whose product is 97% identical to Gsp1p and suppresses gsp1Δ (25), and one clone possessed MOG1. Subsequently, we determined whether the isolated suppressor genes confer the ts+ phenotype to the other gsp1 alleles that are recessive mutations. Three plasmids, p195NTF2, p195MOG1, and p195PDE2, were separately introduced into cells with all of the gsp1 alleles listed in Table 2. The abilities of these plasmids to suppress the gsp1 mutation were determined at the nonpermissive temperature for each gsp1 allele after 2–3 days incubation on synthetic-medium (ura−) plates. Of 25 gsp1 mutants, 15 gsp1 alleles were suppressed by MOG1, and 17 gsp1 alleles were suppressed by NTF2 (Table 2). On the other hand, PDE2 fully suppressed only 6 gsp1-alleles but partially suppressed 13 gsp1-alleles.
Furthermore, we investigated whether MOG1 and NTF2 suppress rna1–1, because rna1–1 was reported to be suppressed by an overexpression of Pde2p (31). p195MOG1 and p195NTF2 and (as controls) p195PDE2 and p195GSP1 were introduced into the NN19–5B (rna1–1) strain. Although PDE2 suppressed rna1–1 as reported (31), MOG1, NTF2, and GSP1 did not (data not shown).
Mog1p Directly Binds to Gsp1p.
The ability of both MOG1 and NTF2 to suppress many of the gsp1 alleles suggested that these genes were profoundly involved in the Ran GTPase cycle. However, our gsp1 strains (11) possessed GSP2, the expression of which is enhanced in glycerol medium (25) and which can rescue gsp1 strains when expressed from an exogenous promoter. To eliminate the possibility that the obtained suppressor genes suppressed gsp1 through increased expression of GSP2, the PSY962–479 (Δgsp1, Δgsp2 [pF314 gsp1–479]) strain was constructed. NTF2, MOG1, and PDE2 carried on either a multicopy vector (YEplac195) or a single-copy vector (pRS316) were separately introduced into the PSY962–479 strain. Ura+/Trp+ transformants were incubated at 34°C, the nonpermissive temperature for gsp1–479. Both MOG1 and NTF2 conferred the ts+ phenotype to the PSY962–479 strain, even with a single gene dose (Fig. 2). On the other hand, PDE2 rescued the temperature sensitivity of the PSY962–479 strain only when carried on a multicopy vector (data not shown).
Figure 2.
gsp1–479 can be suppressed by NTF2 and MOG1, but not by PDE2 carried on a single-copy plasmid. Plasmids p316NTF2, p316MOG1, and p316PDE2 and as a control, the vector alone, were introduced into the PSY962–479 (MATα Δgsp1∷HIS3 Δgsp2∷HIS3 [pF314 gsp1–479]) strain as indicated. Ura+Trp+ transformants were incubated on a synthetic-medium (ura−trp−) plate at the indicated temperature for 3 days.
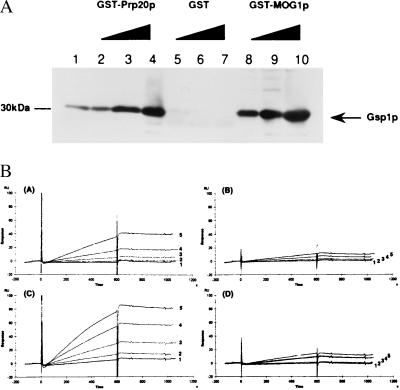
These results suggest that both Mog1p and Ntf2p have a tight functional relationship with Gsp1p. It is reported that Ntf2p directly binds to GDP–Gsp1p (28, 29). To examine whether Mog1p binds to Gsp1p as well, E. coli-produced GST-fused Mog1p and, as a control, GST-fused Prp20p (14) were mixed with increasing doses of E. coli-produced Gsp1p. After incubation in the presence of EDTA, GST-fused proteins bound to glutathione–Sepharose 4B beads were precipitated by using centrifugation and assayed for the coprecipitation of Gsp1p by immunoblotting analysis using anti-Gsp1p antibodies (Fig. 3A; ref. 34). Similar to the case of Prp20p, Gsp1p was coprecipitated with Mog1p in a dose-dependent manner. Based on this result, we addressed the question of whether Mog1p bound to GTP– or GDP–Gsp1p by using a real-time protein–protein interaction analysis using Biacore (35, 36). The mAb to GST was immobilized on the sensor chip of the Biacore to trap GST-fused Mog1p, and, as controls, GST-fused Yrb1p and GST alone. The interaction of Mog1p or Yrb1p with nucleotide-bound Gsp1p was then determined by injecting increasing amounts (μM) of GMPPNP–Gsp1p or GDP–Gsp1p. The relative response units obtained with GST alone were subtracted as background from those of GST-fused Mog1p and Yrb1p. As shown in Fig. 3B, GMPPNP–Gsp1p, but not GDP–Gsp1p significantly bound to both Mog1p and Yrb1p. The calculated association (ka) and dissociation (kd) rate constants of Yrb1p and Mog1p were as follows. ka: Yrb1p, 1.23 ± 0.17 × 104 per M⋅s−1; Mog1p, 4.43 ± 0.74 × 103 per M⋅s−1. kd: Yrb1p, 1.44 ± .07 × 10−4 s−1, Mog1p, 1.69 ± 0.74 × 10−4 s−1. Thus, the affinity constants of Gsp1-GMPPNP to Yrb1p and Mog1p were 8.54 × 107 M−1 and 2.62 × 107 M−1, respectively.
Figure 3.
Interaction of Mog1p with Gsp1p. (A). A gradient of E. coli-produced Gsp1p—4 pmol (lanes 2, 5, and 8), 40 pmol (lanes 3, 6, and 9), 200 pmol (lanes 4, 7, and 10)—was mixed with 200 pmol of GST-Prp20p (lanes 2–4), GST alone (lanes 5–7), or GST-Mog1p (lanes 8–10) bound to glutathione Sepharose 4B beads, in buffer B containing 5 mM EDTA, as reported (34). After incubation for 1 hr at 4°C, glutathione-Sepharose 4B beads were centrifuged, washed eight times with binding buffer B, and then electrophoresed in 11.25% SDS/PAGE gels, transferred onto a poly(vinylidene difluoride) membrane, and immunoblotted with anti-Gsp1p antibodies. As a control, 4 pmol of Gsp1p was electrophoresed in lane 1. (B) Real-time interaction analysis of GDP– and GTP–Gsp1p binding to Mog1p. The mAb to GST was immobilized on the sensor chip, and 0.1 μM either GST-Mog1p (A and B), GST-Yrb1p (C and D) or as a control, GST alone was trapped on the sensor chip through the mAb to GST. The purified GMPPNP–Gsp1p (A and C) or GDP–Gsp1p (B and D) was then injected at a concentration of 0.005 (trace 1), 0.01 (trace 2), 0.025 (trace 3), 0.05 (trace 4), or 0.1 (trace 5) μM. The relative response units from which the relative response units of GST alone had been subtracted are shown.
MOG1 Functionally Interacts with NTF2.
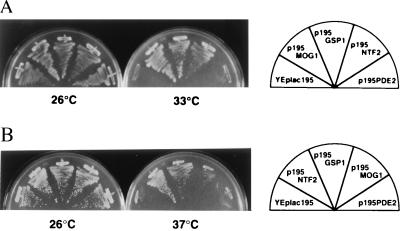
To examine whether MOG1 is essential for survival, we disrupted the gene by using pHIS3MOG1. The plasmid DNA of pHIS3MOG1 digested with the XbaI and EcoRI enzymes was introduced into the haploid YPH499 and YPH500 strains. Appropriate gene replacements in the resulting strains, designated as MOY1 and MOY2, were confirmed by Southern blot analysis. Both MOY1 and MOY2 strains showed temperature sensitivity for cell proliferation. Taking advantage of the ts character of Δmog1, we then examined whether overexpression of NTF2 can suppress Δmog1. NTF2 and (as controls) MOG1, GSP1, and PDE2, all of which were carried on the YEplac195 vector, and the vector alone were introduced into the MOY1 strain. Ura+ transformants were incubated on a synthetic-medium (ura−) plate at 26°C (the permissive temperature) or 33°C (the nonpermissive temperature; Fig. 4A). NTF2 conferred the ts+ phenotype to Δmog1, similar to MOG1 and GSP1. Conversely, we addressed the question of whether overexpression of MOG1 suppresses the temperature-sensitive mutations of NTF2 (27). MOG1 and (as controls) NTF2, GSP1, and PDE2 carried on the YEplac195 vector and the vector alone were introduced into the PSY852–1 strain possessing pPS919 (ntf2–1) or the PSY852–2 strain possessing pPS920 (ntf2–2). Ura+Leu+ transformants were incubated on a synthetic medium (ura−leu−) plate at 37°C. Both NTF2 and GSP1 suppressed ntf2–1 and ntf2–2 as reported (37), but MOG1 and PDE2 did not. Representative results of the PSY852–1 strain are shown in Fig. 4B.
Figure 4.
Functional interaction between Ntf2p and Mog1p. (A) The indicated plasmids were introduced into the MOY1 strain. Ura+ transformants incubated in synthetic medium lacking uracil at 26°C for 3 days were streaked on a synthetic-medium (ura−) plate and incubated at 26°C or 33°C for 3 days. (B) The indicated plasmids were introduced into the PSY852–1 (Δntf2∷HIS3 [pPS919 (CEN LEU2 ntf2–1)]) strain. Ura+Leu+ transformants were streaked on synthetic medium (ura−leu−) at 26°C or at 37°C for 3 days.
Mog1p Is Required for Nuclear-Protein Import.
The above results revealed that Mog1p has some functional interaction with Ntf2p, which is essential for nuclear pore transport function (26–28). To investigate whether the Δmog1 strain has a defect in nuclear pore transport function, the plasmid pFB1–33C (GAL10∷H2B-lacZ; ref. 38) was introduced into the MOY1 strain and, as a control, the wild-type YPH499 strain. Resultant Ura+ transformants were cultured to early logarithmic phase at 26°C in synthetic medium containing raffinose but lacking uracil. After induction with galactose for 1 hr at 26°C, half of the cells were then incubated at 36°C, the nonpermissive temperature, in synthetic medium containing galactose but lacking uracil. Two hours later, cells were fixed and stained with anti-β-galactosidase antibodies. In comparison to wild-type YPH499 cells, the nuclear import of β-galactosidase that was fused to the H2B-NLS was retarded in the MOY1 strain (Fig. 5A). Subsequently, pPS751 (URA3 NPL3-myc; ref. 39) was introduced into the MOY1 strain and, as a control, the wild-type YPH499 strain. Resulting Ura+ transformants were incubated at 26°C or 36°C in synthetic medium lacking uracil. Three hours later, cells were stained with the mAb to the Myc tag. At 36°C, the nonpermissive temperature for the MOY1 strain, Npl3-myc accumulated in the nucleus of wild-type cells, but not in the nucleus of MOY1 cells (Fig. 5B). Npl3p is an mRNA-binding protein and enters the nucleus in a manner independent of the classical NLS (40, 41).
Figure 5.
Nuclear-protein import and mRNA export in the wild-type and Δmog1 strains. (A) Exponentially growing cultures of the YPH499 (Upper) and MOY1 (Lower) stains containing the plasmid pFB1–33C were cultivated at 26°C for 5 hr in synthetic medium (2% raffinose, ura−) and induced for 1 hr with 2% galactose. After that, half of the cultures were incubated at 36°C. Two hours later, cells were fixed and doubly stained by anti-β-galactosidase antibodies and DAPI as indicated. The same field of cells is shown in each panel. (B) Exponentially growing cultures of the YPH499 (Upper) and MOY1 (Lower) strains containing pPS751 were cultivated at 26°C for 5 hr in synthetic medium (ura−). After that, half of the cultures were incubated at 36°C. Three hours later, cells were fixed and doubly stained by the mAb to the Myc-tag and DAPI, as indicated. The same field of cells is shown in each panel. (C) The YPH499 (Upper) and MOY1 (Lower) strains were cultured in YPD medium at 26°C until early logarithmic phase, and half of the cultures were incubated at 37°C. Two hours later, cells were fixed and hybridized with biotin-labeled oligo(dT)50 followed by incubation with FITC–avidin. Cellular DNA was stained with DAPI. The same field of cells is shown in each panel.
In contrast with the defect in nuclear accumulation of NLS proteins, mRNA that was in situ-hybridized with biotin-labeled oligo(dT) was distributed in the cytoplasm of MOY1 cells that had been cultured for 2 hr at 37°C, the nonpermissive temperature, similar to the case of wild-type cells (Fig. 5C). Even after a 6-hr incubation at 37°C, there was no accumulation of mRNA in the nucleus of MOY1 cells.
Mog1p Localized in the Nucleus.
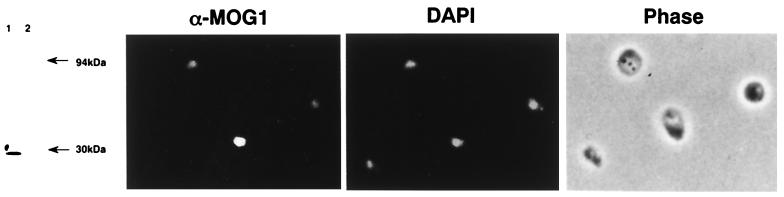
To determine the localization of Mog1p, anti-Mog1p antibodies were prepared and affinity-purified (Fig. 6, Left). By using the affinity-purified anti-Mog1p antibodies, the wild-type YPH499 cells were stained but there was no clear localization (data not shown). Therefore, p195MOG1 was introduced into the MOY1 strain to increase the protein level. Resulting Ura+ transformants that normally grew at 26°C were doubly stained with the anti-Mog1p antibodies and DAPI (Fig. 6, Center). Compared with the DAPI staining pattern, the majority of Mog1p seems to be localized in the nucleus.
Figure 6.
Nuclear localization of Mog1p. (Left) Total lysates of the YPH499 (lane 1) and MOY1 (lane 2) strains were electrophoresed in 11.25% SDS/PAGE gels and analyzed by immunoblotting using the affinity-purified 1:1,000 diluted anti-Mog1p antibodies. (Center) p195MOG1 was introduced into the MOY1 strain. Ura+ transformants were cultured in synthetic medium lacking uracil until early logarithmic phase and fixed to be doubly stained with the anti-Mog1p antibodies (α-MOG1 diluted 1:5,000) and DAPI. (Right) Phase-contrast microscopy. The same field of cells is shown in the Center and Right panels.
DISCUSSION
In this report, we found that PDE2 is a multicopy suppressor of gsp1. It has previously been reported that rna1–1 can be suppressed by either the reg1 mutation of REG1 (srn1; ref. 42) or by overexpression of PDE2 (31). Reg1p is required for glucose repression (43). In this context, it is notable that the expression of GSP2 is enhanced in glycerol medium (25), suggesting the possibility that gsp1 was suppressed via the increase in the expression of Gsp2p by PDE2. However, we found that even in the absence of the GSP2 gene, overexpression of Pde2p suppressed gsp1. This is also true for the other gsp1 suppressors NTF2 and MOG1. Therefore, the Ran network may be regulated by the Ras-cAMP pathway (44). However, only 6 of 25 gsp1 alleles were fully suppressed by PDE2. Although a total of 19 gsp1 alleles were fully or partially suppressed by PDE2, the relationship between the Ran network and the Ras-cAMP pathway remains to be further investigated.
In contrast to PDE2, more than half of the gsp1 alleles were fully suppressed by NTF2 and MOG1. Whereas Ntf2p is essential for survival, Mog1p is not. However, the disruption of the MOG1 gene makes yeast temperature-sensitive for growth. In Δmog1 cells, both classic and the nonclassic NLS-dependent nuclear-protein import seem to be defective, whereas mRNA export is apparently normal at the nonpermissive temperature. Thus, Mog1p is required for nuclear-protein import. A database search shows no known protein homologous to Mog1p. The findings that the efficiency of MOG1 suppression of gsp1 is comparable to that of NTF2 and that both NTF2 and MOG1 can suppress gsp1–479, even at single copy, suggest a functional interaction between Mog1p and Ntf2p. Consistent with this possibility, Δmog1 can be suppressed by overexpression of NTF2, although ntf2 cannot be suppressed by overexpression of MOG1. These results indicate that the function of Mog1p can be bypassed by Ntf2p. Although Ntf2p is concentrated at the nuclear pore (24), Mog1p is localized in the nucleus when overexpressed. This result reveals that Mog1p exists in the nucleus, but it remains to be investigated where in the nucleus Mog1p is localized.
Mog1p specifically binds to GTP–Gsp1p, whereas Ntf2p is reported to bind to GDP–Gsp1p (28, 29). This finding may suggest that Mog1p functions at a site upstream of the Gsp1p GTPase-activating protein, Rna1p, or downstream of the GDP/GTP exchange factor of Gsp1p, Prp20. Ntf2p is well conserved through evolution (27). The functional interaction with Ntf2p suggests that Mog1p may also be conserved, although no homology has yet been reported. Recently, the ability of Ntf2p to bind to GDP-Ran was shown to be important for efficient nuclear-protein import (46). However, the biological function of Ntf2p in nuclear-protein import is still obscure. Mog1p may be a missing link between Ntf2p and the nuclear pore transport function.
Acknowledgments
We thank N. Nakashima and Drs. P. Silver, E. Noguchi, A. Toh-e, M. Sakaguchi, and A. Tartakoff for the kind gifts of antibodies, yeast strains, and plasmids. We thank Dr. S. Sazer for critical reading and review of our manuscript. This work was supported by Grants-in-Aid for Specially Promoted Research and by the Human Frontier Science Program.
ABBREVIATIONS
- ts
temperature-sensitive
- GEF
gaunine nucleotide exchange factor
- RanGAP
RAN GTPase activator
- NLS
nuclear localization signal
- GST
glutathione S-transferase, DAPI, 4′,6-diamidino-2-phenylindole
References
- 1.Drivas G T, Shih A, Coutavas E, Rush M G, D’Eustachio P. Mol Cell Biol. 1990;10:1793–1798. doi: 10.1128/mcb.10.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore M, Blobel G. Trends Biochem Sci. 1994;19:211–216. doi: 10.1016/0968-0004(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 3.Gorlich D, Mattaj I W. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 4.Avis J M, Clarke P. J Cell Sci. 1996;109:2423–2427. doi: 10.1242/jcs.109.10.2423. [DOI] [PubMed] [Google Scholar]
- 5.Nigg E A. Nature (London) 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 6.Gorlich D. EMBO J. 1998;17:2721–2727. doi: 10.1093/emboj/17.10.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melchior F, Gerace L. Trends Cell Biol. 1998;8:175–179. doi: 10.1016/s0962-8924(98)01252-5. [DOI] [PubMed] [Google Scholar]
- 8.Wozniak R A, Rout M P, Aitchison J D. Trends Cell Biol. 1998;8:184–187. doi: 10.1016/s0962-8924(98)01248-3. [DOI] [PubMed] [Google Scholar]
- 9.Weis K. Trends Biochem Sci. 1998;23:185–189. doi: 10.1016/s0968-0004(98)01204-3. [DOI] [PubMed] [Google Scholar]
- 10.Wong D H, Corbett A H, Kent H M, Stewart M, Silver P A. Mol Cell Biol. 1997;17:3755–3767. doi: 10.1128/mcb.17.7.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oki M, Noguchi E, Hayashi N, Nishimoto T. Mol Gen Genet. 1998;257:624–634. doi: 10.1007/s004380050690. [DOI] [PubMed] [Google Scholar]
- 12.Bischoff F R, Ponstingl H. Nature (London) 1991;354:80–82. doi: 10.1038/354080a0. [DOI] [PubMed] [Google Scholar]
- 13.Clark K L, Sprague G F., Jr Mol Cell Biol. 1989;9:2682–2694. doi: 10.1128/mcb.9.6.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aebi M, Clark M W, Vijayraghavan U, Abelson J. Mol Gen Genet. 1990;224:72–80. doi: 10.1007/BF00259453. [DOI] [PubMed] [Google Scholar]
- 15.Nishitani H, Ohtsubo M, Yamashita K, Iida H, Pines J, Yasuda H, Shibata Y, Hunter T, Nishimoto T. EMBO J. 1991;10:1555–1564. doi: 10.1002/j.1460-2075.1991.tb07675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsumoto T, Beach D. Cell. 1991;66:347–360. doi: 10.1016/0092-8674(91)90624-8. [DOI] [PubMed] [Google Scholar]
- 17.Kadowaki T, Goldfarb D, Spitz L M, Tartakoff A M, Ohno M. EMBO J. 1993;12:2929–2937. doi: 10.1002/j.1460-2075.1993.tb05955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sazer S, Nurse P. EMBO J. 1994;13:606–615. doi: 10.1002/j.1460-2075.1994.tb06298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bischoff F R, Krebber H, Kempf T, Hermes I, Ponstingl H. Proc Natl Acad Sci USA. 1995;92:1749–1753. doi: 10.1073/pnas.92.5.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartwell L. J Bacteriol. 1967;93:1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hopper A K, Banks F, Evangelidis V. Cell. 1978;19:211–219. doi: 10.1016/0092-8674(78)90108-3. [DOI] [PubMed] [Google Scholar]
- 22.Amberg D C, Goldstein A L, Cole C N. Genes Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- 23.Forrester W, Stutz F, Rosbash M, Wickens M. Genes Dev. 1992;6:1914–1926. doi: 10.1101/gad.6.10.1914. [DOI] [PubMed] [Google Scholar]
- 24.Corbett A H, Koepp D M, Schlenstedt G, Lee M S, Hopper A K, Silver P A. J Cell Biol. 1995;130:1017–1026. doi: 10.1083/jcb.130.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belhumeur P, Lee A, Tam R, DiPaolo T, Fortin N, Clark M W. Mol Cell Biol. 1993;13:2152–2161. doi: 10.1128/mcb.13.4.2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paschal B M, Gerace L. J Cell Biol. 1995;129:925–937. doi: 10.1083/jcb.129.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corbett A H, Silver P A. J Biol Chem. 1996;271:18477–18484. doi: 10.1074/jbc.271.31.18477. [DOI] [PubMed] [Google Scholar]
- 28.Paschal B M, Delphin C, Gerace L. Proc Natl Acad Sci USA. 1996;93:7679–7683. doi: 10.1073/pnas.93.15.7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nehrbass U, Blobel G. Science. 1996;272:120–122. doi: 10.1126/science.272.5258.120. [DOI] [PubMed] [Google Scholar]
- 30.Sass P, Field J, Nikawa J, Toda T, Wigler M. Proc Natl Acad Sci USA. 1986;83:9303–9307. doi: 10.1073/pnas.83.24.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tung K-S, Hopper A K. Mol Gen Genet. 1995;247:48–54. doi: 10.1007/BF00425820. [DOI] [PubMed] [Google Scholar]
- 32.Carlson M, Botstein D. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- 33.Hill J, Donald K, Griffiths D E. Nucleic Acids Res. 1991;19:5791. doi: 10.1093/nar/19.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noguchi E, Hayashi N, Nakashima N, Nishimoto T. Mol Cell Biol. 1997;17:2235–2246. doi: 10.1128/mcb.17.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiomi T, Fukushima K, Suzuki N, Nakashima N, Noguchi E, Nishimoto T. J Biochem (Tokyo) 1998;123:883–890. doi: 10.1093/oxfordjournals.jbchem.a022020. [DOI] [PubMed] [Google Scholar]
- 36.End P, Gout I, Fry M J, Panayotou G, Dhand R, Yonezawa K, Kasuga M, Waterfield M D. J Biol Chem. 1993;268:10066–10075. [PubMed] [Google Scholar]
- 37.Paschal B M, Fritze C, Guan T, Gerace L. J Biol Chem. 1997;272:21534–21539. doi: 10.1074/jbc.272.34.21534. [DOI] [PubMed] [Google Scholar]
- 38.Moreland R B, Langevin G L, Singer R H, Garcea R L, Hereford L M. Mol Cell Biol. 1987;7:4048–4057. doi: 10.1128/mcb.7.11.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen E C, Henry M F, Weiss V H, Valentini S R, Silver P A, Lee M S. Genes Dev. 1998;12:679–691. doi: 10.1101/gad.12.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee M S, Henry M, Silver P A. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- 41.Senger B, Simos G, Bischoff F R, Podtelejnikov A, Mann M, Hurt E. EMBO J. 1998;17:2196–2207. doi: 10.1093/emboj/17.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tung K-S, Norbeck L L, Nolan S L, Atkinson N S, Hopper A K. Mol Cell Biol. 1992;12:2673–2680. doi: 10.1128/mcb.12.6.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tu J, Carlson M. EMBO J. 1995;14:5939–5946. doi: 10.1002/j.1460-2075.1995.tb00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Broach J R, Deschenes R J. Adv Cancer Res. 1990;54:79–139. doi: 10.1016/s0065-230x(08)60809-x. [DOI] [PubMed] [Google Scholar]
- 45.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clarkson W D, Corbett A H, Paschal B M, Kent H M, McCoy A J, Gerace L, Silver P A, Stewart M. J Mol Biol. 1997;272:716–730. doi: 10.1006/jmbi.1997.1255. [DOI] [PubMed] [Google Scholar]