Abstract
A method was developed for oriented immobilization of bacteriophage T4 through introduction of specific binding ligands into the phage head using a phage display technique. Fusion of the biotin carboxyl carrier protein gene (bccp) or the cellulose binding module gene (cbm) with the small outer capsid protein gene (soc) of T4 resulted in expression of the respective ligand on the phage head. Recombinant bacteriophages were characterized in terms of infectivity. It was shown that both recombinant phages retain their lytic activity and host range. However, phage head modification resulted in a decreased burst size and an increased latent period. The efficiency of bacteriophage immobilization with streptavidin-coated magnetic beads and cellulose-based materials was investigated. It was shown that recombinant bacteriophages form specific and strong bonds with their respective solid support and are able to specifically capture and infect the host bacterium. Thus, the use of immobilized BCCP-T4 bacteriophage for an Escherichia coli B assay using a phage multiplication approach and real-time PCR allowed detection of as few as 800 cells within 2 h.
Bacteriophages are the most diversified group of organisms known; they are ubiquitous in the environment and are used widely for typing bacteria due to their specific and fast interaction with target cells. The interaction between the bacteriophage and the bacterial cell is multivalent, very strong, and specific, making bacteriophages a promising alternative to antibodies for use as the recognition element in a sensor to capture, detect, and identify bacterial species. Phage-based methods for bacterial detection that utilize the following approaches have been developed: (i) monitoring the multiplication of phage particles in the presence of host cells (12, 28), (ii) detection of intracellular components released during phage-mediated cell lysis (3, 15, 21), (iii) detection of growth inhibition due to the presence of a specific bacteriophage (23), (iv) use of phages as specific bacterial staining agents (10, 14, 16), and (v) monitoring the expression of a reporter gene cloned into the phage genome that occurs after bacterial infection (6, 15, 32). The reported detection techniques based on these approaches were shown to be faster than conventional methods but still did not have sensitivities sufficient to meet the requirements for food and environmental applications.
The use of bacteriophage as a biosorbent has been proposed previously (1). A phage-based biosorbent consisting of a Salmonella-specific phage passively immobilized on polystyrene has been used to specifically separate Salmonella from food; however, the efficiency of cell capture was poor (1). Chemical biotinylation of the phage head proteins and construction of biosorbents by coating magnetic beads with phage via the biotin-streptavidin interaction increased capture efficiency and showed a significant improvement to the system described earlier (29). Sun et al. (29) showed that up to 20% of target cells could be concentrated on the surfaces of these streptavidin-coated magnetic beads covered with biotinylated bacteriophage within 30 min. However, the capture efficiency was still lower than that reported for immunosorbents. The capture efficiency may be improved by orientated immobilization of the phage via its head and thus lead to better performance of phages in biosensor applications.
Methods for targeted introduction of affinity tags in vivo circumvent several problems that arise with chemical methods. First, the label is inserted in the same position for all protein molecules, thus providing immobilization in the same orientation. Additionally, as the chemistry is performed in vivo, there are no hazardous reagents to handle and discard. The specificity of the reaction allays any concerns about the reproducibility or heterogeneity of the reaction products.
Specific biotinylation of fusion proteins in vivo has previously been reported by several researchers as a method to ensure oriented immobilization of the protein of interest (7, 8, 19, 30, 34). These authors describe the construction of fusion genes containing biotin acceptor domains derived from portions of biotin acceptor-type proteins (e.g., biotin carboxyl carrier protein, or BCCP). The approach has been successful in yielding in vivo biotinylated fusion proteins through enzyme-mediated coupling of BCCP with biotin (34). A similar approach was used to introduce cellulose binding modules (CBM) of xylanase 10A from Thermotoga maritima as a novel affinity tag for protein immobilization (4, 17, 26).
The goal of this study was to introduce affinity tags onto bacteriophage heads to ensure oriented immobilization that could be coupled with biosensor applications. Phage T4 and Escherichia coli B were used as a model system in this study. As an affinity tag, either BCCP or CBM9 was displayed on the phage head by use of a phage display technique, and the properties of the immobilized phage were investigated.
MATERIALS AND METHODS
Bacterial strains and bacteriophages.
Escherichia coli strain BL21(DE3) Nova blue competent cells were obtained from Novagen (Madison, WI). Escherichia coli B and bacteriophage T4 (D), as well as bacterial cultures used for testing the host ranges of the phages, were from the culture collection of the Canadian Research Institute for Food Safety, University of Guelph. T4-Z bacteriophage was a gift from Lindsay Black (24).
Bacterial strains were cultured at 37°C for 14 to 16 h in Luria-Bertani (LB; Difco) broth/agar that was supplemented with 100 μg/ml of ampicillin when necessary (LB-Amp). Enumeration of bacterial cells was done by spread plating aliquots of serial dilutions onto LB agar.
Propagation and enumeration of bacteriophages were performed using a soft agar overlay technique (26). Obtained stocks of bacteriophages were purified by dialysis.
Reagents.
Expression vector pET22(b) was obtained from Novagen (Madison, WI). Integration plasmid pRH was kindly provided by Lindsay Black (24). pET28-cbm-gfp was kindly provided by Charles A. Haynes (17). Restriction endonucleases were obtained from New England Biolabs (Beverly, MA). T4-DNA ligase was obtained from Roche Molecular Biochemicals (Laval, Quebec, Canada). Dynabeads (MyOne streptavidin; Dynal, Lake Success, NY) were used according to the manufacturer's instructions. Microcrystalline cellulose beads (CP-02) were obtained from Asthi Kasei Chem. Corp. (Japan). All other chemicals were from Sigma-Aldrich (Oakville, Ontario, Canada).
Cloning and expression of SOC-BCCP and SOC-CBM-GFP.
Isolation of plasmid DNA and transformation of competent cells were performed using standard methods (27). Restriction digestions and ligations were performed according to manufacturers' recommendations.
The bccp DNA fragment was generated by PCR using E. coli DNA as a template and the coding primer GCGGATCCACGGAACCCACTCATGGAT tailed with BamHI and the noncoding primer GCGAATTCAGCATGTTCGCCTCGTTACT tailed with EcoRI (the underlined sequences refer to the restriction sites). The soc fragment was obtained using T4 DNA as a template and the coding primer 5′-ATATCATATGGCTAGTACTCGCGTTATG-3′ tailed with NdeI and primer 5′-AGAAGGATCCAAACCAGTTACTTTCCACAAATC-3′ tailed with BamHI. The CBM-GFP fragment (where GFP is green fluorescent protein) was amplified using forward primer cbm-BamHI (CGCGCGGATCCCATGACTAGCGGAA TAATGGTAGC) and reverse primer cbm-EcoRI (CGCGCGAATTCTCAGTGGTGGTGGTGGTG), with pET28-cbm-gfp as a template.
Both soc and bccp or cbm-gfp PCR products were excised with BamHI and ligated together using T4 DNA ligase. The ligation products (for soc-bccp, ∼800 bp, and for soc-cbm-gfp, ∼1,100 bp) were PCR amplified using the forward primer for soc (NdeI) and the reverse primer for bccp or cbm-gfp (EcoRI), respectively. soc-bccp or soc-cbm fragments were cloned into the expression vector pET22(b). E. coli BL21(DE3) cells were transformed by electroporation with the constructed plasmids, and expression of SOC-BCCP and SOC-CBM-GFP fusions was confirmed by SDS-PAGE, Western blotting, and reverse transcriptase PCR (RT-PCR).
Integration of the soc-bccp or soc-cbm-gfp genes into the phage T4 genome.
soc-bccp and soc-cbm-gfp fragments were ligated into recombination plasmid pRH (2) and used for homologous recombination according to the method described by Ren et al. (25). According to this method, integration of the gene(s) of interest in the phage genome is accompanied by restoration of the lysozyme gene. Thus, the resulting recombinant phage is able to perform a full lytic propagation cycle. Briefly, the recombinant plasmid containing the fused genes was electroporated into E. coli DH10B competent cells. The cells containing the integration vector were grown in 100 ml of citrate medium (1 liter water; 10 g [1%, wt/vol] tryptone; 5 g [0.5%, wt/vol] NaCl; 50 ml Tris, pH 8.0, 1 M; and 10 ml 25% sodium citrate) to a concentration of ∼108 bacteria/ml. To 100 ml of this bacterial suspension, T4-Z bacteriophage (lysozyme-deficient phage) was added at a multiplicity of infection (MOI) of 0.3. After 2 h of incubation at 37°C, the bacterial culture was centrifuged at 5,000 × g for 10 min to remove bacterial debris. To harvest and concentrate progeny phage particles, the sample was centrifuged at 13,000 × g for 60 min (20). The recombinant phages containing fusion genes and full-length lysozyme were selected by plating the pellet on regular medium, and phage single-plaque DNA was used for the PCR to confirm integrants by use of the forward primer for soc and the reverse primer for bccp or cbm-gfp. Single phage plaques displaying BCCP (BCCP-T4) and CBM-GFP (CBM-T4) were grown, run on 15% SDS-PAGE gels, and electrotransferred onto ImmunoBlot polyvinylidene difluoride (PVDF) membranes. The membranes were blocked for 1 h at room temperature in 3% bovine serum albumin (BSA) with gentle agitation, followed by incubation for 30 min with streptavidin-horseradish peroxidase (HRP) or anti-GFP antibody for BCCP-T4 or CBM-T4, respectively. The membranes were washed twice and stained using an Opti-4CN substrate kit (Bio-Rad).
Scanning electron microscopy (SEM) images of immobilized BCCP-T4 bacteriophage were taken using a Hitachi S-4800 scanning electronic microscope. Bacteriophage was immobilized on the surface of gold modified with streptavidin, and pictures were taken as described previously (13).
Fluorescence spectra were obtained using a model RF-540 spectrofluorometer (Shimadzu Corporation, Kyoto, Japan).
One-step growth curve experiments were performed in order to determine the burst sizes and latent periods of phages T4, BCCP-T4, and CBM-T4. The procedure was a modification of the original method (11). An overnight culture of E. coli B was diluted (1:103) in LB broth and incubated with shaking at 37°C for 3 h. One milliliter of the culture was removed and placed into a separate tube that contained 100 μl of the respective phage suspension (109 PFU/ml) to give an MOI of 0.1. The remaining bacterial culture was left at room temperature to serve as indicator bacteria. The bacterium-phage suspension was diluted 1:104 in lambda buffer, and the diluted suspension was incubated with shaking at 37°C. Beginning at 0 min and at 10-min intervals for 120 min, the diluted suspension was plated by removing 100 μl of the suspension and adding 100 μl of indicator bacteria and 3 ml of top agar. The mixture was briefly vortexed and poured onto LB agar plates. The number of plaques was determined after 24 h of incubation at 37°C.
Immobilization of bacteriophages was performed by overnight incubation of BCCP-T4, and CBM-T4 (1 ml, 109 PFU/ml) with 100 μl of magnetic beads (109 beads/ml) or 100 μg of microcrystalline cellulose beads, respectively, with gentle shaking at room temperature on an Orbitron rotator II (Boekel Industries, Inc., Feasterville, PA). Wild-type T4 bacteriophage was used as a control in both cases. Unbound phage was removed by washing three times with phosphate-buffered saline (PBS)-Tween 20 solution (PBST) (NaCl, 8 g liter−1; KH2PO4, 0.2 g liter−1; Na2HPO4·12H2O, 2.9 g liter−1; KCl, 0.2 g liter−1; Tween 20, 0.5 g liter−1) and then three times in PBS. Immobilized phages were collected, resuspended in 100 μl of λ buffer, and used for further investigation. Phages were enumerated in the initial solution and supernatant/washing solutions. The number of attached phage particles was estimated by subtraction.
The infectivity of immobilized phages was tested by placing them on an LB agar plate and overlaying with molten semisolid LB agar containing 100 μl of a 12- to 14-h E. coli B culture lawn. Plates were examined for the presence of lysis around beads after 24 h of incubation at 37°C.
Assessment of bacterial capture by immobilized phages.
Tenfold serial dilutions of overnight E. coli B culture were prepared to give cell concentrations in the range of 10 to 105 CFU/ml, as determined by plate count. The constructed phage-based biosorbent (20 μl of suspension) was added to 1 ml of each dilution and rocked for 10 min. The supernatant was separated from the beads, and 0.1 ml of supernatant was placed on LB agar plates. Bacterial cells were enumerated in the initial suspension and the supernatant by plate count, and the number of captured cells was estimated by subtraction.
Detection of E. coli cells captured by immobilized phages using a phage multiplication assay coupled with real-time PCR.
Biosorbents with the attached bacteria were washed twice using λ buffer, resuspended in LB broth, and incubated at 37°C for 1 h to initiate phage-mediated lysis. Beads were removed, and the progeny phages were detected using real-time PCR. Phage lysate (30 μl) was subjected to heating for 10 min at 85°C. Real-time PCR standards were prepared by using 10-fold serial dilutions containing approximately 100 to 108 PFU/ml of phage stocks (BCCP-T4) to determine the sensitivity limit of the method. SYBR green I real-time PCR was performed with borosilicate glass capillaries (Roche Diagnostics) using a LightCycler-FastStart DNA Master SYBR green I kit (Roche Diagnostics) as described by the manufacturer. For detection of BCCP-T4 phage, the forward primer for soc (TATGGCTAGTACTCGCGGTTATG) and the reverse primer for bccp (TTCAGCATGTTCGCCTCGTTACT) were used. The PCR mixture was added to a capillary tube (Roche) and amplified using a Light Cycler system 1.2 (Roche Diagnostics). Samples were heated to 95°C for 10 min, followed by 50 cycles of denaturation at 95°C for 15 s, annealing and extension at 60°C for 1 min, and cooling to 4°C. Fluorescence was monitored during DNA amplification, and the data were analyzed using Roche System software version 3.0 (Roche Diagnostics).
Data analysis.
All experiments were performed at least in duplicate; the means and standard deviations were calculated using Microsoft Excel. The results were then analyzed statistically using a t test at a 95% level of confidence.
RESULTS
Engineering of bacteriophages with affinity tags displayed on their heads.
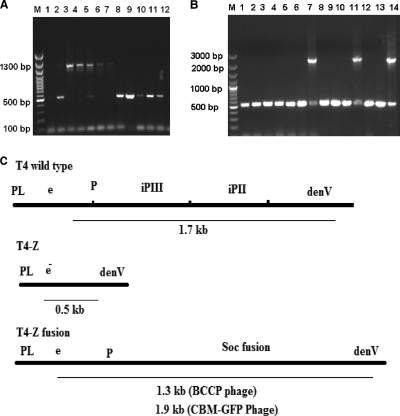
In order to construct a T4 phage that displays a biotin- or cellulose-binding domain, fusions of soc genes with added bccp or cbm-gfp coding sequences were inserted in a pRH recombination plasmid as described in Materials and Methods. In these constructs, the soc gene is flanked on its 5′ side by a 3′ portion of the e (lysozyme) gene of phage T4 and by the strong IPIII promoter to drive soc expression (25). At the 3′ end of soc, a portion of a downstream T4 gene, denV, allows homologous recombination between the phage and the plasmid on either side of the soc gene. T4-Z phage is deleted of 9.8 kb (alt, IPIII, and soc) and is also a partial deletion mutant for the lysozyme (e) gene. This construct allows 6 kb more DNA to be packed into the T4 head, with easy selection of positive mutants by plating on regular medium not supplemented with lysozyme (23, 33). The E. coli cells transformed with the pRH soc-bccp or the pRH soc-cbm-gfp plasmid were infected with T4-Z. During the phage infection, recombination occurred and soc fusions were incorporated into the lysozyme- and soc-deficient T4-Z phage. This was confirmed by PCR amplification of the region flanking gene e and denV. When the soc-bccp and soc-cbm-gfp fusions were incorporated into the defective phage genome, the corresponding ∼1.3-kb and 2.0-kb fragments, respectively, were detected, while the defective lysozyme-dependent phage produced a 0.5-kb fragment (Fig. 1).
FIG. 1.
Detection of soc insertion into the T4 genome by use of PCR. The primers at the 3′ end of gene e and the 3′ end of gene denV were used in the amplification reactions. (A) Lanes 3 to 7, 1.3-kb PCR products indicating the insertion of soc-bccp; lanes 8 to 12, fragment obtained using T4-Z phage; lane M, molecular size standard. (B) Lanes 7, 11, and 14, 2-kb PCR product indicating the presence of CBM-GFP fusions; lanes 1 to 6, 8 to 10, and 12 and 13, 0.5-kb DNA fragment obtained using T4-Z phage; lane M, molecular size standard. (C) Schematic presentation of the constructs used in the study.
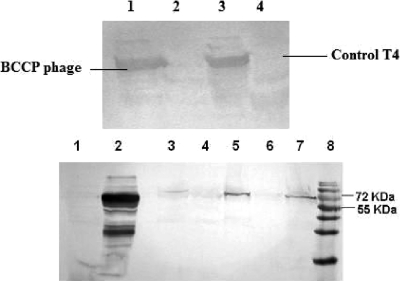
Progeny phage produced from infecting E. coli B with BCCP-T4 have been biotinylated by the host cell's biotin ligase protein (BLP, or BirA). Biotin, which is present in all living cells, is attached posttranslationally by BLP to a specific lysine residue in the BCCP peptide (18). It was reported previously that recombinant proteins carrying a biotinylation peptide expressed in small and large amounts are biotinylated at approximately 30% and 6% efficiencies, respectively, by endogenous levels of the BirA enzyme in E. coli (9). In the produced BCCP-T4 phage, the biotin-binding domain is highly expressed, as determined by RT-PCR. However, the level of biotinylation as detected by Western blotting appeared to be low, as only a faint band was observed upon application of the streptavidin-HRP conjugate (Fig. 2A). The HABA (4′-hydroxyazobenzene-2-carboxylic acid) reagent assay, usually employed for biotin detection, was not sensitive enough to detect the levels present. The low level of biotinylation may be attributed to the short latent period of BCCP-T4 phage (∼30 to 35 min) and to the very high expression level of the SOC protein, which may exceed the capacity of BirA. Despite the low level of biotinylation, it was assumed to be sufficient for oriented immobilization, as the presence of at least one biotin molecule per head is required.
FIG. 2.
Expression of the affinity tags on the phage head as detected by Western blotting. (A) Verification of biotinylation. Lanes 1 and 3, BCCP-T4 phage propagated in E. coli AVB-100 overexpressing BirA; lanes 2 and 4, T4 phage. (B) Verification of expression of green fluorescent protein on CBM-T4 phage by use of anti-GFP antibodies. Lane 1, E. coli B; lane 2, E. coli cells containing the cbm-gfp plasmid; lanes 3 to 5, and 7, CBM-T4 phage; lane 6, T4-Z phage (control); lane 8, molecular size standard.
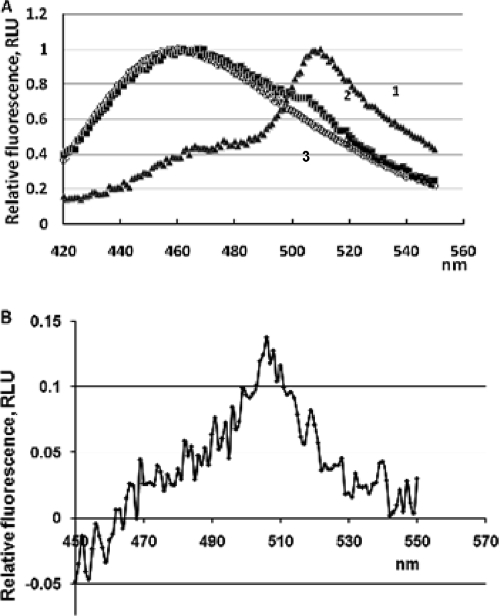
Expression of CBM-GFP in E. coli BL21 and on recombinant CBM-T4 phage was detected by Western blotting (Fig. 2), where a band corresponding to 62 kDa (53 kDa of CBM9-GFP and 9 kDa of SOC) was observed. Fluorescent spectra obtained for E. coli cells expressing CBM-GFP and for the CBM-T4 phage are presented in Fig. 3. For bacterial cells, a fluorescent spectrum characteristic of GFP was observed, with an emission maximum at 510 nm, which corresponds to the published data for the GFPUV variant used in this work (31). Despite the intensive band on the Western blot corresponding to the expression of GFP for the CBM-T4 bacteriophage, the observed fluorescent spectrum was only slightly different from the spectrum obtained for wild-type T4. However, the differential spectrum revealed the maximum at 510 nm as well, indicating that GFP is present in the CBM-T4 bacteriophage, although in small quantities. It is known that the formation of the fluorophore within GFP is a posttranslational autocatalytic oxidation process that could take up to several hours to complete (31). The relatively short latent period of the phage as well as some conformational restriction of the GFP expressed on the phage head may compromise the process of GFP maturation, which could account for the low level of the detected fluorescence.
FIG. 3.
(A) Normalized fluorescent spectra of E. coli expressing CBM-GFP (1), CBM-T4 phage (2), and T4 phage (3) (excitation at 395 nm). (B) Differential fluorescence spectra for CBM-T4 and T4 bacteriophages (excitation at 395 nm).
Determination of the host ranges of the bacteriophages was performed using a limited set of bacterial cultures including different strains of E. coli as well as other Gram-negative bacteria. There was no observed change in the host ranges of the constructed recombinant T4 bacteriophages, which confirmed that receptor elements were not affected by the genetic modifications.
One-step growth curves of the recombinant phages.
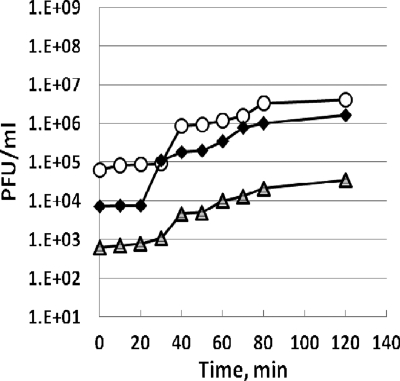
It was observed that the plaque sizes of recombinant phages were reduced by ∼75% compared to the plaque size of wild-type T4; therefore, one-step growth experiments for the recombinant phages were performed to investigate the burst sizes and latent periods of the constructed phages. The results are presented in Fig. 4. The latent period for the T4 phage was 25 min, with a burst size of 137 PFU; the BCCP-T4 latent period was ∼30 to 35 min, with a burst size of 35 PFU; and the CBM-T4 latent period was ∼35 min, with a burst size of 19. The burst size observed for the wild-type T4 phage was similar to previously reported values. Camara and Dealmeido (5) reported a latent period of 50 to 60 min and a burst size of 134 PFU/cell for T4 phage propagated in different E. coli strains (C600, C620, DV1211, and AD651) at 37°C. The burst size was 10 times lower in the same strain grown at 42°C. Matthews (22) reported a relatively large burst size of 200 phage particles for T4, with more phage per cell being produced within the first 15 to 30 min of infection. Thus, the expression of SOC::BCCP or SOC::CBM::GFP on the head of the T4 phage resulted in a significantly reduced burst size as well as a prolonged latent period.
FIG. 4.
One-step growth curves for bacteriophages T4 (⧫), BCCP-T4 (○), and CBM-T4 (▵).
The T4-Z phage used in the study is deleted of 9.8 kb (alt, IPIII, and soc), and it is a partial deletion mutant for the lysozyme (e) gene, so that it requires egg white lysozyme for propagation. Incomplete or inefficient lysozyme gene complementation in BCCP-T4 and CBM-T4 phages in addition to the insertion of bccp or cbm-gfp as a fusion with the soc gene may contribute to the increased latent period and the lower burst size of both of the recombinant phages.
Immobilization of bacteriophages.
BCCP-T4 and CBM-T4 bacteriophages were immobilized on streptavidin-coated magnetic beads and microcrystalline cellulose beads, respectively. Wild-type T4 phage served as a control in both cases. Phage particles were enumerated in initial samples and in supernatants after removal of the beads with captured phage. The efficiency of capture was assessed by subtraction.
For streptavidin-coated magnetic beads, the percentage of capture by BCCP-T4 phage increased from 37 to 84% when the number of added beads was increased (Table 1). Under the same conditions, <16% of wild-type T4 phage particles were immobilized on the magnetic beads. This indicated specificity of BCCP-T4 binding to the beads mediated by biotin-streptavidin interactions.
TABLE 1.
Efficiency of bacteriophage immobilization on magnetic and cellulose beadsa
| Type of support | No. of BCCP-T4 or CBM-T4 particles (log PFU/ml) |
% Particles bound | No. of T4 particles (log PFU/ml) |
% Particles bound | ||
|---|---|---|---|---|---|---|
| Initial | Supernatant | Initial | Supernatant | |||
| Magnetic beads (no.) | ||||||
| 2 × 109 | 9.09 ± 0.02 | 8.67 ± 0.07 | 62 ± 6 | 9.00 ± 0.05 | 8.98 ± 0.02 | 5 ± 6 |
| 7.31 ± 0.02 | 6.62 ± 0.06 | 79 ± 3 | ||||
| 1 × 109 | 9.30 ± 0.00 | 8.50 ± 0.11 | 84 ± 4 | 8.99 ± 0.02 | 8.98 ± 0.04 | 3 ± 8 |
| 7.30 ± 0.00 | 6.75 ± 0.05 | 72 ± 3 | ||||
| 5.30 ± 0.00 | 4.90 ± 0.03 | 60 ± 2 | ||||
| 2 × 108 | 9.21 ± 0.02 | 8.91 ± 0.07 | 49 ± 8 | |||
| 8.21 ± 0.02 | 7.93 ± 0.05 | 50 ± 5 | 8.07 ± 0.01 | 8.01 ± 0.01 | 13 ± 3 | |
| 7.21 ± 0.02 | 7.00 ± 0.01 | 37 ± 2 | 7.07 ± 0.01 | 7.01 ± 0.02 | 16 ± 4 | |
| Cellulose beads (mg) | ||||||
| 100 | 4.25 ± 0.04 | 2.55 ± 0.08 | 98 ± 0 | 7.01 ± 0.09 | 5.84 ± 0.33 | 92 ± 5 |
| 10 | 4.52 ± 0.16 | 4.15 ± 0.10 | 69 ± 9 | 4.45 ± 0.08 | 4.43 ± 0.04 | 4 ± 10 |
| 4.24 ± 0.16 | 3.61 ± 0.14 | 76 ± 8 | 4.15 ± 0.08 | 4.16 ± 0.04 | −3 ± 10 | |
| 3.94 ± 0.16 | 3.39 ± 0.08 | 71 ± 6 | 3.85 ± 0.08 | 3.91 ± 0.06 | −15 ± 16 | |
| 3.64 ± 0.16 | 3.17 ± 0.10 | 65 ± 8 | 3.55 ± 0.08 | 3.66 ± 0.05 | −30 ± 15 | |
Data (means ± standard deviations for 2 to 5 experiments) represent numbers of phage particles in the initial sample and in the supernatant (after bead removal). Results shown for magnetic beads were determined using BCCP-T4 phage, and results shown for cellulose beads were determined using CBM-T4 phage.
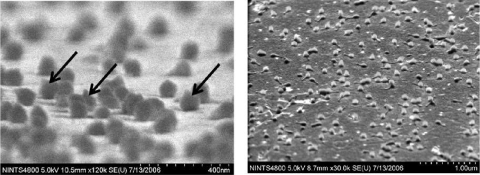
To assess the orientation of immobilized BCCP-T4 phage, SEM images of the phage on a streptavidin-modified gold surface were obtained (Fig. 5). The majority of the phage particles are attached to the surface through the head, thus confirming the oriented immobilization mediated by the display of biotin on the head of the phage.
FIG. 5.
Scanning electron micrographs of BCCP-T4 phage immobilized on the streptavidin-coated Au surface. Arrows point to the phage particles oriented with tails up.
In contrast, for immobilization on microcrystalline cellulose beads, there was significant attachment of both CBM-T4 and wild-type T4 phages to the beads, ranging from 92 to 98% when 100 mg of cellulose beads was used for immobilization (Table 1). When fewer cellulose beads were used, a significant difference in attachment of CBM-T4 and T4 was observed. To assess the strength and specificity of attachment of the recombinant and wild-type phages to cellulose, extensive washing of immobilized phages was performed and phage particles were enumerated in the washing solution. For wild-type T4 immobilized onto cellulose, up to 71% of bound phage was removed by repeatedly washing the beads six times every 24 h with buffer. However, only 12% of the CBM-T4 phage was removed by the washing, indicating the specificity and high affinity of the recombinant-phage binding to cellulose.
Bacterial capture by the immobilized phages.
To investigate the ability of immobilized bacteriophages to capture and infect bacterial cells, phage-based biosorbents were constructed using immobilization of BCCP-T4 or CBM-T4 phage on the respective support at different bead/phage ratios. The main strategy was to maximize the activity of immobilized phages by decreasing the extent of nonspecific binding that resulted in significant inactivation of the phage, as was shown previously (1). The other consideration was to avoid the possible presence of unbound phage particles in the samples, as they probably would interact with host cells much faster due to fewer diffusion restrictions.
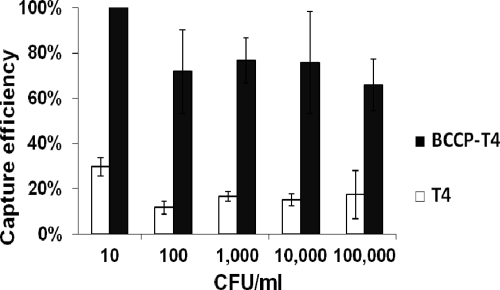
BCCP-T4-based biosorbents were produced using an excess of phage over beads (10:1), and extensive washing was used to remove the unbound and nonspecifically bound phage from the sample. The extent of capture of E. coli by immobilized bacteriophage was evaluated by assessing the depletion of the number of bacteria in the supernatant (Fig. 6). Biotinylated bacteriophage were able to capture E. coli cells with high efficiency after 10 min of contact time. At E. coli concentrations in the range of 10 to 105 CFU/ml, about 70% of cells were captured. The beads coated with nonbiotinylated phage captured a significantly lower percentage of cells (10 to 30%), thus showing the specificity of the constructed biosorbent.
FIG. 6.
Percentages of E. coli B cells captured by the immobilized phage. A portion (20 μl) of the constructed biosorbent (109 beads/ml) was used to capture the cells in 1 ml of LB broth. The time of contact was −10 min. Error bars show standard deviations.
CBM-T4-based biosorbents were produced by incubating microcrystalline cellulose beads (100 μg) with 109 phage particles overnight, followed by extensive washing to remove nonspecifically bound phage. However, the level of bacterial capture by immobilized T4-CBM phage was undetectable by the plate count technique.
Infectivity of immobilized bacteriophages was tested by a cultural method and by using real-time PCR to detect progeny phages originating from the bacteria captured and lysed by immobilized phages. Both BCCP-T4 and CBM-T4 immobilized on their respective beads produced a clear zone of lysis when applied to the lawn of E. coli cells. A significantly larger zone of lysis was observed for the BCCP-T4-based biosorbent than for the CBM-T4-based biosorbent. Wild-type T4 immobilized by nonspecific sorption also produced some clear zones around the beads, but these zones were much smaller than those produced by the recombinant phages. This indicates that despite the lower burst size of recombinant phages, the immobilized recombinant phages retain infectivity sufficient to produce effective lysis, while wild-type T4 was significantly inactivated by nonspecific sorption on the beads.
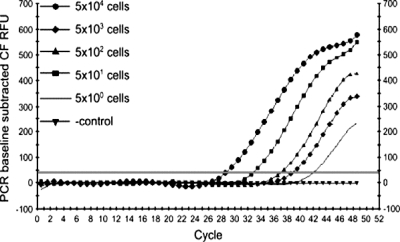
To quantitatively assess the infectivity of immobilized BCCP-T4 bacteriophage, the number of progeny phage in bacterial samples was determined by real-time PCR after treatment of the samples with immobilized bacteriophage for 10 min to allow attachment. Beads with attached bacteria were washed twice, resuspended in LB broth, and incubated for 1 h for the completion of the phage life cycle and production of progeny phage. Progeny phage were detected using real-time PCR after incubation and bead removal. The designed insert-specific primers produced a 400-bp product with a melting temperature of 90 ± 0.5°C originating from the fused soc-bccp gene of the recombinant phage. In the preliminary experiments, it was shown that for pure phage stocks, approximately 102 PFU of phage particles was needed to obtain a reliable PCR signal. One-milliliter samples containing E. coli cells at concentrations from 8 × 102 to 4 × 107 CFU/ml were treated with 20 μl of BCCP-T4-based biosorbent, and real-time PCR was performed. Nutrient medium served as a control. There was a correlation between the number of cycles (time) necessary for fluorescence detection and the number of bacteria present in the initial sample, i.e., the more cells present the less time needed for the fluorescence signal to reach the threshold (Fig. 7).
FIG. 7.
Real-time PCR detection of progeny phage produced in E. coli. The number of cells refers to the sample volume (6 μl). CF RFU, curve fit relative fluorescence units.
DISCUSSION
Bacteriophages are used widely for identification and typing of bacteria. Their interactions with host cells are fast, specific, and quite strong. The multivalent binding of bacteriophages to the host cell surface ensures a high affinity (avidity) that could potentially be used as a recognition event and employed in biosensors. Another feature of bacteriophages as biosensor elements is that, unlike antibodies, phages can both capture and detect the presence of live bacteria during the natural course of infection. If in antibody-based assays amplification of the signal is achieved through coupling with an enzyme, bacteriophages have a built-in amplification system derived through propagation during the infection cycle. Immobilization of bacteriophage would provide an additional advantage for using bacteriophage in biosensors, as it allows the easy separation of the bacteriophage with captured bacteria after infection is complete and eliminates problems due to the presence of exogenous phage. This would be especially advantageous in assays based on the detection of phage multiplication. To overcome problems associated with exogenous phage in other assays, the addition of a virucidal agent is required prior to lysis to be able to accurately enumerate the progeny phage particles (28). Although the use of immobilized phage is more convenient, there has been little research on this topic. Physiosorption of the phage on a plastic surface resulted in substantial inactivation, which is characteristic for this method of immobilization (1). Chemical biotinylation of amino groups available on bacteriophages resulted in partial phage inactivation, probably due to the modification of groups involved in bacteriophage interaction with the host cells (29). Immobilization of biotinylated phage on streptavidin-coated magnetic beads produced an effective biosorbent capable of capturing up to 25 to 30% of host cells. However, it was still below the capture efficiency reported for antibodies.
Here we present a new method for bacteriophage immobilization based on genetically modified phages expressing affinity tags on their heads. This provided a way for uniform and oriented immobilization of phage particles on the solid support carrying the respective receptors. Biotin carboxyl carrier protein (BCCP) and cellulose binding module (CBM) were chosen as affinity elements since they produce a very strong, almost irreversible interaction with their receptors streptavidin and cellulose, respectively. The phage display method used for introduction of affinity tags on the heads of T4 bacteriophage resulted in the construction of fully active recombinant bacteriophages expressing biotinylated BCCP or CBM that were recognized by the respective receptors. The host range of T4 phage did not change because of the modification. Though the host range of T4 bacteriophage does not allow the detection of all naturally occurring E. coli strains, the fact that the host range did not change as a result of genetic manipulations indicates that this phage display technique can be used more widely for the introduction of affinity tags onto phage heads. However, the burst size of the recombinant bacteriophages was affected by the modification. A substantially reduced burst size and a slightly longer latent period were observed for both BCCP-T4 and CBM-T4 compared with the results for wild-type T4. This could potentially compromise the application of the constructed phage in phage amplification assays.
Immobilization of the constructed phages on the surfaces of streptavidin-coated magnetic beads and microcrystalline cellulose beads resulted in the formation of strong and specific bonds between the phage and the support. Immobilized phages retained infectivity, as was shown by the observed lysis when immobilized phages were placed onto the lawn of the host bacterium. The efficiencies of bacterial capture by the immobilized phages were significantly different for the constructed BCCP-T4- and CBM-T4-based biosorbents. Biotinylated T4 immobilized onto streptavidin-coated magnetic beads was able to capture 72 to 99% of E. coli B cells present at levels between 10 and 105 CFU/ml. The capture of bacteria by immobilized CBM-T4, on the other hand, was undetectable by the plate counting technique. This may be explained by the fact that despite the availability of cellulose affinity tags on the phage head, there is still a possibility for phage tails to interact with sugar moieties on the surface of microcrystalline cellulose similarly to the interaction with sugars on the surface of the bacterial cell wall.
The high infectivity of immobilized BCCP-T4 bacteriophage was further confirmed by quantitative real-time PCR targeted to unique DNA sequences in the phage. It was shown that the more bacterial cells present in the sample the more progeny phage particles are detected. The experimental format included 10 min for attachment of target bacteria to the phage-based biosorbent, removal of the bisorbent with captured bacteria, and then incubation for 1 h for the completion of lytic cycle and real-time PCR for detection of progeny phages in the supernatant after removal of the biosorbent. The lowest bacterial cell concentration tested (800 cell/ml) gave a positive signal after approximately 42 cycles of DNA amplification. The signal from control media without bacteria did not reach the threshold during the length of the PCR (50 cycles). Taking into consideration the fact that approximately 70% of bacteria are captured by the BCCP-T4 biosorbent and that the burst size of this phage is ∼35, the expected number of progeny phage would be around 2 × 104 PFU/ml. The sample volume taken for PCR was 6 μl, which is equivalent to 1.2 × 102 PFU per sample and is in the range of sensitivity determined in preliminary tests with pure phage (100 PFU per sample). Thus, the use of immobilized T4 bacteriophage for the E. coli B assay using a phage multiplication approach and real-time PCR allowed detection of as few as 800 cells within 2 h. This time compares favorably with that for a traditional phage amplification assay format where the detection of progeny phage is performed with a plaque assay, which takes up to 48 h to complete. The assay is easy to perform and has a low background and a high signal-to-noise ratio.
The results of the present study confirm our hypothesis that the oriented immobilization of bacteriophage resulted in a highly active biosorbent that can simultaneously capture and detect a target bacterium with high sensitivity and in close to real-time format. The proposed approach could be extended further to other phages provided that their genetic structure is characterized sufficiently. This could open new avenues in the development of biosensors for rapid and user-friendly bacterial detection.
Acknowledgments
We thank OMAF, Sentinel Bioactive Paper Network (NSERC), for financial support.
We thank Lindsay W. Black (University of Maryland) and James Ren (Vencogen Laboratories) for generously supplying the pRH integration vector and phage T-Z, respectively.
Footnotes
Published ahead of print on 30 November 2009.
REFERENCES
- 1.Bennett, A. R., G. G. C. Davis, S. Vlahodimous, J. C. Banks, and P. R. Bell. 1997. Use of bacteriophages-based system for separation and concentration of Salmonella. J. Appl. Microbiol. 83:259-265. [DOI] [PubMed] [Google Scholar]
- 2.Black, L. W., M. K. Showe, and A. C. Steven. 1994. Morphogenesis of the T4 head, p. 218-258. In J. Karam (ed.), Molecular biology of bacteriophage T4. ASM Press, Washington, DC.
- 3.Blasco, R., M. J. Murphy, M. F. Sanders, and D. J. Squirrell. 1998. Specific assays for bacteria using phage mediated release of adenylate kinase. J. Appl. Microbiol. 84:661-666. [DOI] [PubMed] [Google Scholar]
- 4.Boraston, A. B., A. L. Creagh, M. M. Alam, J. M. Kormos, P. Tomme, C. A. Haynes, and D. G. Kilburn. 2001. Binding specificity and thermodynamics of a family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A. Biochemistry 40:6240-6247. [DOI] [PubMed] [Google Scholar]
- 5.Camara, F. P., and D. F. Dealmeido. 1991. Studies on the growth of the phage T4 in cell division mutants DV1211 and AD651 of Escherichia coli at normal and high temperature. Rev. Bras. Genet. 14:233-238. [Google Scholar]
- 6.Chen, J., and M. W. Griffiths. 1996. Salmonella detection in eggs using Lux+ bacteriophages. J. Food Prot. 59:908-914. [DOI] [PubMed] [Google Scholar]
- 7.Choi-Rhee, E., and J. E. Cronan. 2003. The biotin carboxylase-biotin carboxyl carrier protein complex of Escherichia coli acetyl-Co-A carboxylase. J. Biol. Chem. 278:30806-30812. [DOI] [PubMed] [Google Scholar]
- 8.Cronan, J. E., Jr. 1990. Biotinylation of proteins in vivo. A post-translational modification to label, purify, and study proteins. J. Biol. Chem. 265:10327-10333. [PubMed] [Google Scholar]
- 9.Cull, M. G., and P. J. Schatz. 2000. Biotinylation of proteins in vivo and in vitro using small peptide tags. Methods Enzymol. 326:430-440. [DOI] [PubMed] [Google Scholar]
- 10.Edgar, R., M. McKinstry, J. Hwang, A. B. Oppenheim, R. A. Fekete, G. Giulian, C. Merril, K. Nagashima, and S. Adhya. 2006. High-sensitivity bacterial detection using biotin-tagged phage and quantum-dot nanocomplexes. Proc. Natl. Acad. Sci. U. S. A. 103:4841-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellis, E. L., and M. Delbruck. 1939. The growth of bacteriophage. J. Gen. Physiol. 22:365-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Favrin, S. J., S. A. Jassim, and M. W. Griffiths. 2001. Development and optimization of a novel immunomagnetic separation-bacteriophage assay for detection of Salmonella enterica serovar Enteritidis in broth. Appl. Environ. Microbiol. 67:217-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gervais, L., M. Gel, B. Allain, M. Tolba, L. Brovko, M. Zourob, R. Mandeville, M. Griffiths, and S. Evoy. 2007. Immobilization of biotinylated bacteriophage on biosensor surfaces. Sens. Actuators B Chem. 125:615-621. [Google Scholar]
- 14.Goodridge, L. D. 1997. A fluorescent bacteriophage assay for detection of Escherichia coli O157:H7 in ground beef and raw milk. Ph.D. thesis. University of Guelph, Guelph, Ontario, Canada.
- 15.Goodridge, L., and M. W. Griffiths. 2002. Reporter bacteriophage assays as a means to detect food borne pathogen bacteria. Food Res. Int. 35:853-870. [Google Scholar]
- 16.Hennes, K. P., C. A. Suttle, and A. M. Chan. 1995. Fluorescently labeled virus probes show that natural virus populations can control the structure of marine microbial communities. Appl. Environ. Microbiol. 61:3623-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kavoosi, M., J. Meijer, E. Kwan, A. L. Creagh, D. G. Kilburn, and C. A. Haynes. 2004. Inexpensive one-step purification of polypeptide expressed in Escherichia coli as fusions with the family 9 carbohydrate-binding module of xylanase 10A from T. maritima. J. Chromotogr. B Analyt. Technol. Biomed. Life Sci. 807:87-94. [DOI] [PubMed] [Google Scholar]
- 18.Kwon, K., and D. Beckett. 2000. Function of a conserved sequence motif in biotin holoenzyme synthetases. Protein Sci. 9:1530-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, S., and J. E. Cronan. 1992. The gene encoding the biotin carboxlase subunit of Escherichia coli acetyl-CoA carboxylase. J. Biol. Chem. 267:855-863. [PubMed] [Google Scholar]
- 20.Malys, N., D. Y. Chang, R. G. Baumann, D. Xie, and L. W. Black. 2002. A bipartite bacteriophage T4 SOC, and HOC randomized peptide display library: detection and analysis of phage T4 terminase (gp17) and late σ factor (gp55) interaction. J. Mol. Biol. 319:289-304. [DOI] [PubMed] [Google Scholar]
- 21.Mandeville, R., M. W. Griffiths, L. Goodridge, L. McIntyre, and T. T. Iienchuk. 2003. Diagnostic and therapeutic application of lytic phages. Anal. Lett. 36:3241-3259. [Google Scholar]
- 22.Mathews, C. K. (ed.). 1971. Bacteriophage biochemistry. Van Nostrand Reinhold, New York, NY.
- 23.McIntyre, L. 1998. Application and evaluation of bacterial viruses in rapid methodologies for the detection of food-borne pathogens. Ph.D. thesis. University of Guelph, Guelph, Ontario, Canada.
- 24.Ren, Z. J., and L. W. Black. 1998. Phage T4 SOC and HOC display of biologically active, full length protein on the viral capsid. Gene 215:439-444. [DOI] [PubMed] [Google Scholar]
- 25.Ren, Z. J., G. K. Lewis, P. T. Wingfield, E. G. Locke, A. C. Steven, and L. W. Black. 1996. Phage display of intact domains at high copy number: a system based on SOC, the small outer capsid protein of bacteriophage T4. Protein Sci. 5:1833-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richins, R. D., A. Mulchandai, and W. Chen. 2000. Expression, immobilization, and enzymatic characterization of cellulose binding domains-organophosphorous hydrolase fusion proteins. Biotechnol. Bioeng. 68:591-596. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., vol. 1, p. 5.4-5.13. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 28.Stewart G. S., S. A. Jassim, S. P. Denyer, P. Newby, K. Linley, and V. K. Dhir. 1998. The specific and sensitive detection of bacterial pathogens within 4 h using bacteriophage amplification. J. Appl. Microbiol. 84:777-783. [DOI] [PubMed] [Google Scholar]
- 29.Sun, W., L. Brovko, and M. Griffiths. 2001. Use of bioluminescent Salmonella for assessing the efficiency of the constructed phage-based biosorbent. J. Ind. Microbiol. 25:273-275. [DOI] [PubMed] [Google Scholar]
- 30.Tatsumi, H., S. Fukda, M. Kikuchi, and Y. Koyama. 1996. Construction of biotinylated firefly luciferases using biotin acceptor peptides. Anal. Biochem. 243:176-180. [DOI] [PubMed] [Google Scholar]
- 31.Tsien, R. 1998. The green fluorescent protein. Annu. Rev. Biochem. 67:509-544. [DOI] [PubMed] [Google Scholar]
- 32.Ultzur, S., and J. Kuhn. 1987. Introduction of lux genes into bacteria, a new approach for specific determination of bacteria and their antibiotic susceptibility, p. 463-472. In J. Scholmerich, R. Anderson, A. Kapp, M. Ernst, and W. G. Woods (ed.), Bioluminescence and chemiluminescence: new perspectives. John Wiley and Sons, Inc., New York, NY.
- 33.Wu, D. G., C. H. Wu, and L. W. Black. 1991. Reiterated gene amplifications at specific short homology sequence in phage T4 produce Hp17 mutants. J. Mol. Biol. 218:705-721. [DOI] [PubMed] [Google Scholar]
- 34.Yamano, N., Y. Kawata, H. Kojimo, K. Yoda, and M. Yamasaki. 1992. In vivo biotinylation of fusion proteins expressed in Escherichia coli with a sequence of Propionibacterium freudenreichii transcarboxylase 1.3S biotin subunit. Biosci. Biotechnol. Biochem. 56:1017-1026. [DOI] [PubMed] [Google Scholar]