Abstract
Contaminated food is a significant vehicle for human norovirus transmission. The present study determined the effect of physicochemical treatments on the tenacity of infective human norovirus genogroup II in selected foods. Artificially contaminated produce was subjected to a number of processes used by the food industry for preservation and by the consumer for storage and preparation. Virus recovery was carried out by using ultrafiltration and was monitored by using bacteriophage MS2 as an internal process control. Norovirus was quantified by using monoplex one-step TaqMan real-time reverse transcription (RT)-PCR and an external standard curve based on recombinant RNA standards. An RNase pretreatment step was used to avoid false-positive PCR results caused by accessible RNA, which allowed detection of intact virus particles. Significant reductions in titers were obtained with heat treatments usually applied by consumers for food preparation (baking, cooking, roasting). Generally, processes used for preservation and storage, such as cooling, freezing, acidification (≥pH 4.5), and moderate heat treatments (pasteurization), appear to be insufficient to inactivate norovirus within a food matrix or on the surface of food. Besides data for persistence in processed food, comparable data for individual matrix-specific protective effects, recovery rates, and inhibitory effects on the PCRs were obtained in this study. The established procedure might be used for other noncultivable enteric RNA viruses that are connected to food-borne diseases. The data obtained in this study may also help optimize the process for inactivation of norovirus in food by adjusting food processing technologies and may promote the development of risk assessment systems in order to improve consumer protection.
Norovirus (NV) (formerly Norwalk-like virus) is a member of the family Caliciviridae and is a nonenveloped virus with a single-stranded RNA (ssRNA) genome. Genetically, the human noroviruses are subdivided into three distinct genogroups (genogroup I [GGI], GGII, and GGIV) and into at least 31 genetic clusters or genotypes (53). Noroviruses have emerged as the most common cause of food-borne outbreaks and sporadic cases of acute nonbacterial gastroenteritis worldwide (62, 87).
In addition to direct person-to-person infection, transmission via environmental contamination (70) and transmission via foods and drinking water (primary and secondary contamination) are known. Food can be contaminated by contact with sewage or sewage water before harvest; e.g., shellfish (3, 52, 58, 60, 81) and raspberries (33, 72) have been reported to be vehicles of NV infection. Food is often directly contaminated during production, storage, distribution, and preparation by infected persons (22); e.g., ill or asymptomatic food handlers have been identified as sources of virus contamination of fresh produce and ready-to-eat foods (25, 69). The percentage of cases that can be attributed to food- or waterborne transmission is estimated to be between 16% (56) and 57% (34). Human enteric viruses, such as NV, are environmentally stable; they are able to persist for long periods of time in contaminated food and appear to withstand various food processing and storage conditions (5, 22; for a review, see reference 76).
Comprehensive studies of the tenacity and inactivation of infective human norovirus in food, especially food with complex matrices, are still rare. Such studies are limited mainly to shellfish (42, 43) and berries (18, 19) or to cultivable enteric viruses, such as hepatitis A virus (HAV) (15, 24, 42, 66, 78), poliovirus (PV) (26, 84), or rotavirus (12). Due to the lack of a mammalian cell culture or animal model for norovirus, various studies of persistence and inactivation have been performed with genetically related surrogates of norovirus, including feline calicivirus (FCV) (17, 28, 32, 35, 68, 82, 86) and murine norovirus (MNV) (7, 8, 10, 11, 21, 51), which are cultivable nonenveloped viruses belonging to the family Caliciviridae. However, the validity of data obtained with these surrogates is limited because these viruses appear to be generally less stable than human NV when they are subjected to physicochemical treatments (21, 57). For detection and quantification of NV, real-time reverse transcription (RT)-PCR has been established as a rapid, sensitive, and reliable molecular biological method (63).
It is still unclear whether measures used in industrial food processing for preservation and processes usually used by the consumer for storage and preparation are sufficient to inactivate NV in contaminated foods. As a risk assessment approach, in this study representative foods belonging to various product groups, such as convenience foods, delicatessen foods, meat, fruits, and vegetables, were experimentally contaminated with a defined number of infective human NV GGII and subjected to physicochemical treatments (freezing, cooling, acidification, heating) to determine the inactivation potential of these treatments. Additionally, bacteriophage MS2 was utilized as an internal process control (29) to monitor the efficiency and reproducibility of extraction processes. After corecovery of NV and MS2 with an adapted ultrafiltration-based procedure, pretreatment with RNase was carried out to eliminate the problem of false-positive results caused by free NV RNA molecules (68) originating from destroyed virus particles. Because of the lack of an appropriate cell culture, detection and quantification were carried out by performing real-time RT-PCR with use of an external standard curve based on a recombinant NV RNA standard. Furthermore, individual food matrix-specific protective effects, rates of recovery from a product, and inhibitory effects on the PCRs were ascertained.
MATERIALS AND METHODS
Viruses and viral stock preparation.
Norovirus material was obtained from a stool sample kindly provided by the Federal Institute for Risk Assessment (Berlin, Germany). The norovirus in this stool sample was classified as GII cluster 3 based on nucleotide sequence analyses. For storage, the stool sample was diluted 1:5 with phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 10.1 mM Na2HPO4, 1.8 mM KH2PO4; pH 7.4) and clarified by centrifugation at 3,000 × g for 15 min. The supernatant was mixed with 15% glycerol in PBS (final glycerol concentration, 10%) and filtered through a 0.45-μm syringe filter with a polyethersulfone (PES) membrane (VWR International, Darmstadt, Germany). The NV inoculation standard was determined to contain 4 × 109 copies/ml using real-time quantitative RT-PCR (qRT-PCR) as described below and was stored in aliquots at −150°C until it was used.
Bacteriophage MS2 strain DSM13767 was kindly provided by the Institut fuer Laboratoriums und Transfusionsmedizin, Herz und Diabeteszentrum Nordrhein-Westfalen, Universitätsklinik der Ruhr-Universität Bochum (Bad Oeynhausen, Germany). MS2 was propagated by the double-agar-layer plaque technique and was quantified by plating assays as previously described (29). Aliquoted phage lysates were stored at −150°C until they were used.
Artificial inoculation of selected foods and sample processing.
Selected foods utilized in inoculation studies (Table 1) were purchased from local commercial sources and temporarily stored according to the producers’ recommendations.
TABLE 1.
Foodstuffs, physicochemical treatments, and modes of inoculation used in this study
| Food product | Inoculation | Process/treatment | Process conduct parametersc |
|---|---|---|---|
| Frozen pizza | Surface | Heating | Baking in table oven for 12 min at 200°Cd |
| Frozen pizza | Surface | Cryoconservation | Storage in freezer for 7 days at −18°Cd |
| Frozen pizza | Surface | Cryoconservation | Storage in freezer for 14 days at −18°Cd |
| Pizza baguette | Surface | Heating | Baking in table oven for 15 min at 220°Cd |
| Tomato ketchup | Internal | Acidification/cooling | Storage in refrigerator for 58 days at 6°C,d pH 4.5 |
| Spiced tomato sauce | Internal | Heating | Heating in water bath for 1 min at 72 to 74°Ce |
| Potato salad | Internal | Acidification/cooling | Storage in refrigerator for 24 days at 6°C,d pH 5.0 to 5.5 |
| Noodle salad | Internal | Acidification/cooling | Storage in refrigerator for 24 days at 6°C,d pH 5.0 to 5.5 |
| Mincemeata,b | Internal | Heating | Roasting in table ovenf for 30 min at 200°C |
| Mincemeata,b | Internal | Heating | Cooking in boiling liquidg for 30 min at 100°Ch |
| Mincemeata | Internal | Cryoconservation | Storage in freezer for 8 days at −18°C |
| Mincemeata | Internal | Cooling | Storage in refrigerator for 2 days at 6°Cd |
| Apple | Surface | Cooling | Storage in refrigerator for 7 days at 11°C |
| Iceberg lettuce | Surface | Cooling | Storage in refrigerator for 5 days at 11°C |
Consisted of 60% pork and 40% beef, and the fat content was <18%.
Meat was formed into meatballs (ca. 3 by 3 by 2.5 cm).
The temperature was measured in the direct environment of the product or was the setting of the device used.
According to the manufacturer's instructions.
Simulated pasteurization process (heating to 72 to 74°C, hold for 1 min, and immediate chilling on ice).
In a customary oven bag.
PBS was used as a substitute for water. For virus recovery the entire sample (including the PBS) was used.
Time measurement started when buffer reached 100°C.
Composite or hackled foods, such as convenience foods, delicatessen salads, and mincemeat, were artificially inoculated internally with a defined number of NV GGII. For this purpose, the diverse food samples (7 g of sauces, 15 g of all other samples) were transferred to stomacher bags with a mesh filter (spiced tomato sauce, mincemeat) or to 50-ml centrifuge tubes (Greiner Bio One, Frickenhausen, Germany). The samples were inoculated by adding 1 ml of the standard NV inoculum containing 4 × 109 copies/ml. Delicatessen salad samples were carefully inverted, and the convenience food and mincemeat samples were repeatedly mixed thoroughly over a 25-min time period. For heat experiments mincemeat was manually formed into meatballs, and the virus inoculum was mixed with the mincemeat evenly. Solid foods (fruits, salad, frozen pizza, and frozen pizza baguette) were inoculated by dispensing 1 ml of the standard NV inoculum evenly across the surface. The inoculum was air dried for 20 min at room temperature. Fruits and salad (15 g) were not chopped or disrupted as much as possible to prevent the release of potential inhibitory substances into the samples. Frozen food (15 g) was briefly thawed for 15 min at room temperature before inoculation.
Product treatments performed in this study (Table 1) were intended to simulate processes used in industrial food processing for preservation and during food storage and preparation by the consumer. Subsequent to processing, the samples were immediately brought to room temperature by chilling on ice or thawing.
The reference samples consisted of 1 ml of the standard NV inoculum and, instead of a food matrix, corresponding volumes of PBS. Reference samples were treated just like the food samples, being subjected to the same process.
Two replicates of each experimental sample were used, and each experiment was repeated under the same conditions.
Extraction of virus from experimentally inoculated food samples.
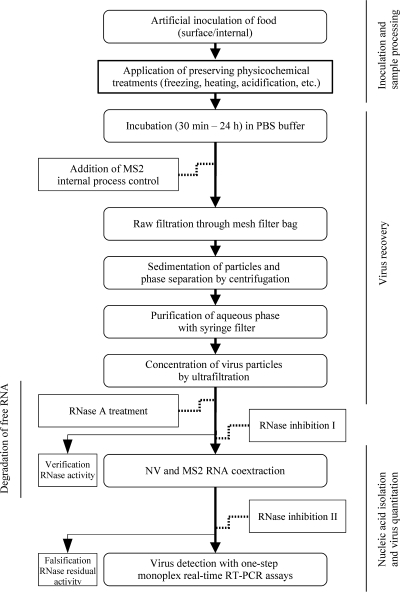
The virus recovery procedure was adapted from procedures described previously (30, 64), with minor modifications (Fig. 1). The processed samples were transferred into stomacher bags with a mesh filter. Depending on the dryness and viscosity of the food matrix, 7 to 30 ml PBS was added to the samples, together with 5 μl MS2 phage lysate containing 1.5 × 1011 PFU/ml (final concentration, 7.5 × 108 PFU/ml of ultrafiltration concentrate) as an internal process control (see below). After this, the samples were incubated for 30 min at room temperature or for 24 h at 4°C (sauces); during this period the samples were mixed several times in the stomacher bags. Delicatessen salads were gently shaken in PBS to avoid disruption of the components. Solid surface foods (fruits and salad) were rinsed in 17 ml PBS including an MS2 process control, with agitation for 30 min at room temperature. Subsequently, each liquid phase was transferred into a 50-ml centrifuge tube. Sedimentation of particulate debris and phase separation were achieved by centrifugation at room temperature for 10 min at 1,000 × g (fruits and salad) or for 30 min at 4,000 × g (all other samples). The aqueous phase was purified by filtration through 1.2- to 0.45-μm syringe filters with PES membranes. Each filtrate was then transferred to a Vivaspin 20 concentrator (molecular weight cutoff, 50,000) with a PES membrane (Vivascience AG, Hannover, Germany). Ultrafiltration devices were centrifuged at room temperature and 4,000 × g for 2 to 30 min. The centrifugation time was extended until the final retention volume was ca. 1 ml.
FIG. 1.
Flow chart of the experimental procedure.
Reference samples (without a food matrix) were diluted with 15 ml PBS inoculated with the MS2 internal process control (final concentration, 7.5 × 108 PFU/ml of ultrafiltration concentrate), analogous to the food samples (see above). After 30 min of incubation, each suspension was directly transferred into an ultrafiltration device and concentrated to a final volume of 1 ml by centrifugation at room temperature and 4,000 × g for approximately 2 min.
A total of 140 μl of each sample concentrate was used for further experimental procedures.
Controls.
Precautions were taken to prevent false-positive and false-negative results. In addition to spatial separation of workspaces at crucial experimental points (e.g., during RNase treatment and RNA extraction), each experiment included several overall control samples.
The internal process control with phage MS2 (29) was used to monitor the efficiency and reproducibility of recovery of virus from foods and to rule out false-negative results. Each sample except the amplification control samples (see below) was spiked with 5 μl MS2 phage lysate containing 1.5 × 1011 PFU/ml (final concentration, 7.5 × 108 PFU/ml of ultrafiltration concentrate) in an appropriate amount of PBS at the beginning of the virus recovery procedure (as described above). Thus, MS2 and NV were corecovered from the same food sample and concentrated. RNA was also coextracted from both NV and MS2, but the viruses were detected by performing individual monoplex real-time RT-PCRs (see below) in separate tubes.
An extraction-negative control was performed for both NV detection and MS2 detection by extracting food samples like the other samples but without any experimental virus contamination. This control was carried out to uncover potential false-positive results caused by cross contamination during the virus and RNA extraction or by contaminated kits, reagents, and foods.
External amplification controls were carried out to monitor the intensity of inhibitory effects on the PCRs mediated by the food matrices. Furthermore, amplification controls could reveal any suboptimal composition of the PCR mixture or the presence of residual RNase activity. An RNA eluate from a sample was spiked with defined concentrations of purchased genomic MS2 RNA (Roche Diagnostics, Mannheim, Germany); 1 μl of a 10−7 dilution of the MS2 stock solution was mixed with 9 μl RNA eluate from a corresponding sample. The solution was subsequently used as a template in real-time RT-PCR MS2 detection assays, as described below. The amplification control was included for each of the sample replicates which were without an MS2 process control.
Amplification-positive controls were applied for both NV detection and MS2 detection. The NV amplification control consisted of 10 μl RNA eluate isolated from 140 μl of an NV inoculation standard (see above). The MS2 control template comprised 1 μl of a 10−7 dilution of the MS2 stock solution mixed with 9 μl buffer AVE (Qiagen, Hilden, Germany).
Amplification-negative controls were also carried out for the NV and MS2 detection assays. These controls revealed potential contamination of the PCR mixture that would lead to false-positive results. The negative control PCR samples contained 10 μl RNase-free water instead of the RNA template.
Degradation of free RNA and RNase inhibition.
Digestion of free RNA was carried out by adding 35 μg RNase A (Qiagen) per sample and incubating the mixture at 37°C for 1 h. The RNase activity in the samples containing a food matrix was verified using the RNaseAlert lab test kit (Applied Biosystems, Darmstadt, Germany) according to the manufacturer's instructions. Fluorescence was excited using a transilluminator. After this, the RNase activity was inhibited by adding 140 U/sample of Qiagen RNase inhibitor (Qiagen) and incubating the mixture for 30 min at room temperature.
Nucleic acid isolation.
Virus RNA was extracted by the column centrifugation method, using the commercially available QIAamp viral RNA mini kit (Qiagen) according to the manufacturer's protocol. In brief, 140 μl of sample was denatured, adsorbed to a silica gel column, and washed twice. The RNA was finally eluted with 60 μl buffer AVE after 2 min of incubation at room temperature. To avoid residual RNase activity, 50 U/sample of Protector RNase inhibitor (Roche Diagnostics) was added, and the mixture was incubated for 30 min at room temperature. Suspensions were tested for residual RNase activity by using the RNaseAlert lab test kit as described above. The nucleic acid was immediately used in downstream PCR application or stored at −19°C until it was used.
Primer and probe design.
Primer and probe sequences for norovirus detection were selected based on a consensus sequence resulting from a multiple sequence alignment. The sequences aligned were obtained by performing sequence homology searches with nucleotide databases (GenBank, RefSeq Nucleotides, EMBL, and DDBJ) using a 450-bp sequence of a highly conserved region (ORF1-ORF2 junction) of the norovirus genome (49) as a reference sequence.
Sequence similarity database searches were performed with the BLAST algorithm (1) and the multiple alignment using CLUSTAL W (85). The oligonucleotides were designed using CLC Combined Workspace (version 3.01; CLC bio, Aarhus, Denmark) and Clone Manager Suite (version 6; Scientific & Educational Software, Cary, NC). The oligonucleotides were purchased from MWG Biotech AG (Ebersberg, Germany).
TaqMan real-time RT-PCR assays.
The real-time RT-PCRs were performed as one-step monoplex assays with TaqMan hydrolysis probes, using the QuantiTect Virus +ROX vial kit (Qiagen). Each PCR mixture (final volume, 15 μl) consisted of 5 μl QuantiTect Virus NR Master Mix, 0.25 μl QuantiTect Virus RT-Mix, 500 nM (final concentration) of each primer, 250 nM (final concentration) of a probe, and nuclease-free water. The primers and probes used for NV and MS2 detection are listed in Table 2. The final volume of each reaction mixture was adjusted to 25 μl by adding 10 μl of RNA solution (template) or RNase-free distilled water (control).
TABLE 2.
Primers and probes used for real-time RT-PCR in this study
| Primer or probe | Typee | Sequencef | Position |
|---|---|---|---|
| NLV-RT-FWDaa | NV, sense primer | TGT TYA GRT GGA TGA GRT TCT C | 5013-5034g |
| COG2Rb | NV, antisense primer | TCG ACG CCA TCT TCA TTC ACA | 5100-5079h |
| QNIFS-BHQc | NV, probe | FAM-AGC ACG TGG GAG GGC GAT CG-BHQ1 | 5042-5061h |
| MS2-TM2-Fd | MS2, sense primer | TGC TCG CGG ATA CCC G | 3169-3184i |
| MS2-TM2-Rd | MS2, antisense primer | AAC TTG CGT TCT CGA GCG AT | 3229-3210i |
| MS2-TM2JOEd | MS2, probe | JOE-ACC TCG GGT TTC CGT CTT GCT CGT-BHQ1 | 3186-3209i |
Modified sequence QNIF2 as described by Loisy et al. (61).
Data from reference 49.
Sequence QNIFS as described by Loisy et al. (61) modified with quencher dye BHQ1 at the 3′ end.
Data from reference 29.
NV, norovirus detection; MS2, MS2 detection.
Y = C or T; R = A or G; FAM, 6-carboxyfluorescein; BHQ1, black hole quencher 1; JOE, 6-carboxy-4′,5′ -dichloro-2′,7′-dimethoxyfluorescein.
Position in the nucleotide sequence of Lordsdale virus (GenBank accession no. X86557).
Position in the nucleotide sequence of Camberwell virus (accession no. AF145896).
Position in the nucleotide sequence of phage MS2 (accession no. V00642).
Amplification and detection were performed with the Rotor-Gene 3000 system (Corbett Research, Sydney, Australia). After the thermocycler rotor was loaded with the sample tubes, the run was initiated immediately. The thermal cycling conditions were reverse transcription at 50°C for 30 min, denaturation at 95°C for 5 min, and amplification for 65 cycles with denaturation at 95°C for 15 s, annealing, and fluorescence detection at 60°C for 45 s. Raw data were obtained during the annealing step of each cycle in the 510-nm channel (NV) and the 555-nm channel (MS2).
Data analysis.
The cycle threshold (CT) is defined as the fractional cycle at which the fluorescence rises above a given threshold value (41). CT values were calculated from the raw fluorescence data by using CAmpER (“calculation of amplification efficiencies for real-time RT-PCR experiments”), an open web-based application for automatic analysis of real-time PCR experiments (http://www.cebitec.uni-bielefeld.de/groups/brf/software/camper_info/index.html), allowing the standardized determination for single samples. After the Rotor-Gene output file was uploaded to the CAmpER web server, the CT value of each sample was calculated by using the implemented DART algorithm (71). The threshold is calculated as a multiple of the mean standard deviation of the first 10 cycles of the automated background-corrected fluorescence curve of all samples of the experiment.
Samples with CT values below 43 and with a sigmoidal amplification plot were considered suitable for analysis.
For determination of recovery rate, protective effect, and inactivation, the quantity of NV was calculated as described above using the average CT value derived from four replicates from two independent experiments. NV recovery, expressed as percent recovery, was calculated by using the following formula: % recovery rate = (virus quantity in untreated food sample)/(virus quantity in untreated reference sample) × 100. The food-specific protective effect, expressed as percent survival rate, was calculated by using the following formula: % survival rate = [(virus quantity in processed food sample) − (virus quantity in processed reference sample)]/(virus quantity in untreated food sample) × 100. Virus inactivation, expressed as logarithmic titer reduction, was calculated as follows: log10 (Nt/N0), where N0 is the quantity of virus in the untreated food sample and Nt is the quantity of virus in the processed food sample.
Mean values, standard deviations, and coefficients of variation were calculated by using the office application suite OpenOffice (version 3.01; Sun Microsystems free software community). Statistical analyses regarding significant differences between the mean numbers of viruses found in the treated and untreated samples were performed with Student's t test, and a P value of ≤0.05 was considered significant.
Quantification of norovirus.
Absolute quantification of NV RNA was performed by means of an external standard curve which displays the relationship between a given CT value and the corresponding number of genome equivalents per PCR assay. The external standard curve was derived from real-time RT-PCRs performed with known quantities of the NV target, a recombinant RNA (recRNA) standard.
Construction of the norovirus RNA standard was performed by using a previously described method (30), with some modifications. In brief, a recombinant plasmid (kindly provided by the Institut fuer Laboratoriums und Transfusionsmedizin, Herz und Diabeteszentrum Nordrhein-Westfalen, Universitätsklinik der Ruhr-Universität Bochum, Bad Oeynhausen, Germany), which harbors a cloned norovirus GGII sequence from the ORF1-ORF2 junction, was subjected to a PCR assay with the universal M13 primers to amplify the inserted DNA fragment, including phage promoter T7. The PCR product was purified with the QIAquick PCR purification kit (Qiagen) and subsequently used for runoff transcription using a T7 transcription kit (Fermentas, St. Leon-Rot, Germany) according to the manufacturer's protocols. The remaining DNA was digested with DNase I (RNase-Free DNase set; Qiagen) before the RNA transcripts were purified with a QIAamp viral mini kit (Qiagen) used according to the manufacturer's instructions, except for the addition of carrier RNA.
To generate the standard curve, a 10-fold dilution series of the recombinant RNA standard was prepared. RNA concentrations were obtained by spectroscopy at a wavelength of 260 nm. Quantification of RNA standard dilutions was carried out by using a formula that considers the RNA concentration, the fragment size, and the Avogadro constant (37). RNA dilutions from 10−1 to 10−6 were used as templates in real-time RT-PCR assays under the conditions described above. The standard curve was finally obtained by plotting the calculated quantities of the RNA standard dilutions (in genome equivalents per PCR assay) against the corresponding CT value.
RESULTS
Quantification of NV and limit of detection of PCR assays.
The quantities of NV were determined as genome equivalents with real-time qRT-PCR. For appropriate quantification, an external standard curve was constructed based on 10-fold serial dilutions of a recombinant RNA standard. With a slope of −3.43 and a coefficient of determination (R2) of >0.99, adequate correlation between the amount of target template and the CT value was obtained. The coefficient of variation (interassay) was ≤7.5%. The sensitivity of the TaqMan real-time RT-PCR system was calculated to be <19 viral genome copies per PCR assay.
Degradation of free RNA and RNase inhibition.
To properly estimate the effect of physicochemical treatments on the NV titer, our intention was to detect RNA molecules isolated from intact virus particles. Free RNA from inactivated virus particles in the sample could be coextracted and would therefore lead to false-positive results in the real-time RT-PCR. Preliminary experiments revealed that the native RNase activity in the food samples is generally not sufficient to digest amounts of added NV RNA corresponding to the number of artificially inoculated NV (data not shown). Thus, the samples were pretreated with RNase A prior to nucleic acid extraction to digest free RNA (68). For each food product tested in this study, the RNase activity was verified using a commercial kit, which indicated that the added RNase was functional in the sample and was not inhibited by the food matrix. An exception was tomato ketchup, for which RNase activity could not clearly ascertained. Because of the intense color of the sample it was not possible to distinguish between fluorescent and nonfluorescent signals.
Subsequent to the RNase digestion procedure, the RNase activity in the samples was deactivated in order to avoid degradation of the RNA extracted from intact virus particles, leading to false-negative PCR results. Preliminary experiments (data not shown) revealed that the combined application of RNase inhibitors immediately prior to and subsequent to the nucleic acid isolation step is sufficient to completely deactivate the artificially introduced RNase activity. Using a commercial kit, residual RNase activity was shown to be absent in each sample tested.
Inhibitory effects of food matrix on PCRs.
Amplification controls were carried out to uncover individual inhibitory effects on the PCRs caused by substances derived from the food matrix. In some cases inhibitory substances could not be completely removed by extraction and purification of virus RNA using a commercial kit. The inhibition of NV detection would lead to false-negative results and underestimation of the NV counts in the samples. Therefore, MS2 RNA was applied together with the NV RNA extracts in MS2 detection real-time RT-PCR assays. Comparison of the results of these assays with MS2 amplification-positive controls (without any food-derived RNA extract) revealed such inhibitory effects on the PCR. The food matrices tested had variable distinct inhibitory effects on the PCR (Table 3). Frozen pizza, pizza baguette, potato salad, noodle salad, apple, and lettuce showed partial inhibition in the range from 1 to 2.6 CT values compared to the amplification-positive controls, whereas for tomato ketchup, spiced tomato sauce, and mincemeat the differences were negligible (≤1 CT value) compared to the reference samples.
TABLE 3.
Food matrix-specific recovery rate for norovirus in different artificially contaminated foodstuff, protective effect during product treatment, and inhibitory effect on real-time RT-PCRs
| Food product | Process/treatment | Recovery rate, % (±SEM)a | Survival rate (protective effect), % (±SEM)b | Inhibition of real-time RT-PCRc |
|---|---|---|---|---|
| Frozen pizza | Heating (baking) | 0.5 (±0.25) | + | |
| Frozen pizza | Cryoconservation (7 days) | 2.4 (±0.44) | + | |
| Frozen pizza | Cryoconservation (14 days) | 1.5 (±1.42) | + | |
| Pizza baguette | Heating (baking) | 10.1 (±0.30) | 0.1 (±0.60) | + |
| Tomato ketchup | Acidification/cooling | 2.9 (±0.80) | − | |
| Spiced tomato sauce | Heating (pasteurization) | 21.7 (±0.37) | 44.6 (±0.61) | − |
| Potato salad | Acidification/cooling | 5.3 (±0.43) | + | |
| Noodle salad | Acidification/cooling | 9.4 (±1.11) | + | |
| Mincemeat | Heating (roasting) | 0.2 (±1.13) | 2.3 (±1.31) | − |
| Mincemeat | Heating (cooking) | 3.1 (±0.53) | − | |
| Mincemeat | Cryoconservation | 0.3 (±1.75) | − | |
| Mincemeat | Cooling | 1.2 (±0.71) | − | |
| Apple | Cooling | 43.3 (±0.28) | + | |
| Iceberg lettuce | Cooling | 64.0 (±0.37) | + |
Rate of recovery of NV from foodstuff based on comparison of untreated food samples with untreated reference samples (without food).
Food-specific protective effect (expressed as percent survival rate) based on comparison of treated food samples with treated reference samples (without food).
−, no or negligible inhibition (≤1 CT); +, partial inhibition (1 to 2.6 CT). Values were determined using an MS2 amplification control.
Norovirus recovery from food samples.
The norovirus recovery rates were obtained by comparing the numbers of NV detected in untreated food samples to the numbers of NV detected in untreated reference samples, in which the food was replaced by PBS. The percentages of NV recovered (Table 3) were determined to estimate the loss of virus due to the food matrix independent of the virus extraction process and the product treatments. The recovery rates (± standard errors of the means) for convenience products were determined to be 0.5% (±0.25) to 2.4% (±0.44) for frozen pizza, 10.1% (±0.30) for pizza baguette, 2.9% (±0.80) for tomato ketchup, and 21.7% (±0.37) for spiced tomato sauce. For the delicatessen products potato salad and noodle salad, the recovery rates were 5.3% (±0.43) and 9.4% (±1.11), respectively. And the NV recovery rates were between 0.2% (±1.13) and 3.1% (±0.53) for mincemeat, 43.3% (±0.28) for apples, and 64% (±0.37) for lettuce.
Food-specific protective effects during product treatment.
Protective effects, in which the food matrix protects microorganisms from physicochemical treatments (e.g., by preventing them from being inactivated), are well known for bacteria (45, 48) and have been described for some bacteriophages (67, 73). In this study, the food-specific protective effects were investigated by comparing treated food samples with treated reference samples, in which the food was replaced by PBS. The protective effect was expressed as the percent survival rate, which describes the amount of NV in the food sample that was not inactivated during product treatment. A clear protective effect of food was observed for some thermal treatments (Table 3). In pizza baguettes heated by baking, the survival rate (± standard error of the mean) was 0.1% (±0.60), in spiced tomato sauce that was pasteurized it was 44.6% (±0.61), and in mincemeat that was roasted it was 2.3% (±1.31).
Effects of physicochemical processes on the persistence and inactivation of NV in selected foods.
The reductions in the numbers of NV caused by product treatments usually used for food preservation in a variety of experimentally contaminated foods are shown in Table 4. Products with solid surfaces (frozen pizza, fruits, lettuce) were inoculated superficially, and the other foods (meat, sauces, delicatessen salads) were inoculated internally. The quantities of norovirus in the unprocessed foods were compared to the quantities in the processed samples (before and after the treatment) to assess the scale of the potential virus inactivation. The mean log10 reductions in norovirus titer and the corresponding P values are shown in Table 4; a P value of ≤0.05 was considerd significant.
TABLE 4.
Tenacity and inactivation of norovirus in artificially inoculated foodstuffs during physicochemical treatments
| Process/treatment | Food product | Log10 reduction in NV titera (±SEM) | P valueb |
|---|---|---|---|
| Heating (baking) | Frozen pizza | >−4 (±0.36) | 0.0001 |
| Heating (baking) | Pizza baguette | −3 (±0.60) | 0.001 |
| Heating (pasteurization) | Spiced tomato sauce | −0.4 (±0.51) | 0.148 |
| Heating (roasting) | Mincemeat | −1.6 (±1.31) | 0.05 |
| Heating (cooking) | Mincemeat | >−7 (±0.29) | 0.021 |
| Acidification/cooling | Tomato ketchup | −0.5 (±1.32) | 0.394 |
| Acidification/cooling | Potato salad | −1.7 (±0.83) | 0.01 |
| Acidification/cooling | Noodle salad | −0.5 (±1.07) | 0.412 |
| Cooling | Mincemeat | 0.0 (±0.61) | 0.740 |
| Cooling | Apple | −0.2 (±0.76) | 0.650 |
| Cooling | Iceberg lettuce | 0.1 (±0.15) | 0.272 |
| Cryoconservation (7 days) | Frozen pizza | −0.4 (±0.41) | 0.100 |
| Cryoconservation (14 days) | Frozen pizza | −1.1 (±0.92) | 0.174 |
| Cryoconservation | Mincemeat | 0.0 (±3.06) | 0.856 |
Reduction in the norovirus titer based on comparison of untreated food samples with treated food samples.
Significance calculation using Student's t test, based on the difference between the samples compared.
The effects of heating on the inactivation of NV in food were found to be divergent. Baking frozen pizza (200°C, 12 min) resulted in at least a 4-log10 reduction in the number of NV (P = 0.0001); no virus was detected in the treated samples. Also, inactivation so that the level was below the detection limit was found when mincemeat was cooked in boiling liquid (30 min; product core temperature, 94°C for 15 min), and the reduction in the number of NV was at least 7 log10 (P = 0.021). The reason for the greater magnitude of inactivation was the higher recovery rate for NV from the meat product than from the pizza. Baking pizza baguette (220°C, 15 min) resulted in a significant reduction (−3 log10; P = 0.001), as did roasting mincemeat (200°C, 30 min; core temperature, 96°C for 15 min) (−1.6 log10; P = 0.05). Based on the P value (0.148), pasteurization of spiced tomato sauce (72 to 74°C, 1 min) resulted in no significant decrease in the NV titer (−0.4 log10).
The effect of acidification on the tenacity of NV was tested by examining two delicatessen salads and one convenience product that were internally inoculated with NV. No significant reduction in the NV titer was found after storage (58 days, 6°C) at pH 4.5 for tomato ketchup (−0.5 log10, P = 0.394) and after storage (24 days, 6°C) at pH 5.0 to 5.5 for noodle salad (−0.5 log10, P = 0.412). However, the NV titer in potato salad was found to be significantly decreased (−1.7 log10, P = 0.01) under the same storage conditions that were used for the noodle salad (24 days, 6°C, pH 5.0 to 5.5).
The effect of cooling on the tenacity of NV in food was relatively consistent. No significant reduction in the NV titer was found after storage of mincemeat for 2 days at 6°C, nor was any reduction observed after storage of apples (−0.2 log10, P = 0.650) or iceberg lettuce (0.1 log10, P = 0.272).
The effect of freezing was tested with frozen pizza and mincemeat. There was no significant reduction in the NV titer after cryoconservation (7 and 14 days, −18°C) of frozen pizzas (−0.4 log10 [P = 0.100] and −1.1 log10 [P = 0.174]). Likewise, no reduction was observed for mincemeat (8 days, −18°C).
DISCUSSION
Some critical factors for reliable detection of NV in food are extraction from the food matrix or surface, the concentration of the virus, and RNA extraction and subsequent detection (e.g., using real-time RT-PCR). In this study selected foods were artificially inoculated with a defined number of NV GGII extracted from stool samples from an infected patient. The high virus concentration used (4 × 109 copies/ml) should have compensated for the expected loss of particles during virus extraction and ensured proper monitoring of the magnitude of inactivation.
Currently, there is no international standardized method for detection of NV in food. In fact, diverse methods have been developed, but they have been optimized for a limited number of food commodities, including fruits and vegetables (9, 20, 31, 39, 50), shellfish (6, 46, 59), deli ham (16), turkey and roast beef (80), hamburger (77), cheese (38), and water (40). The majority of these methods also include sample elution with a pH 9.5 glycine-buffered solution and subsequent polyethylene glycol (PEG) precipitation. Ausar et al. (4) observed significant Norwalk virus capsid disruption at alkaline pH, which agreed with the finding of White et al. (88) that native NV particles disassemble when they are suspended at pH 8.5. To circumvent the problem of capsid disruption at this point and to concentrate intact virus particles, we adapted an ultrafiltration-based extraction and concentration method for NV recovery described previously (30, 64). NV RNA can also be released by some physicochemical treatments (e.g., high temperature) with destruction of the virus capsid. Preliminary experiments indicated that free RNA in food samples is more stable than is generally assumed; in particular, the capability of native RNase seems to be limited in the digestion of huge amounts of NV RNA that was probably occurring under the experimental conditions used in this study. Released NV RNA could be coextracted with the RNA from intact virus particles, and this would lead to false-positive results in real-time RT-PCR assays and to overestimation of the amount of NV in the sample tested. One aim of this persistence study was detection of only intact virus particles, so that the effect of a given treatment on NV structural integrity could be monitored by comparing the numbers of intact particles before and after treatment. Studies with hepatitis A virus (HAV), poliovirus type 1 (PV-1), and feline calicivirus (FCV) showed that combined enzymatic treatment with RNase and proteinase K (resulting in complete removal of the coat protein) facilitated efficient prevention of false-positive RT-PCR results (47, 68). Therefore, in this study pretreatment with RNase was integrated into the virus extraction procedure to digest free NV RNA molecules; a proteinase K treatment would have affected the intact capsid. The RNase activity was tested with a sensitive commercial kit after addition of RNase to the food samples and prior to the PCR assays to monitor the functionality in the food matrices tested and to avoid false-negative results caused by degradation of the PCR template (residual activity).
It is generally accepted that RT-PCR is unable to discriminate between virus that is infectious and virus that has been inactivated (74). The question of whether NV is not only intact but still infective and therefore a threat to human health can be answered only by using a tissue culture or animal model, which is not available for NV at present (83). In studies with the cultivable HAV (42) quantitative real-time PCR results did not correlate with infectivity. Duizer et al. (32) observed in studies of FCV and canine calicivirus that a reduction in infectivity was not always correlated with diminished detectability of the viral genome. With the real-time RT-PCR strategy described here, viral genome copies were quantified, and the measured reduction in the titer was the actual reduction in the number of intact NV particles. However, it remains unclear if a technological process is able to abolish the infectivity of NV even more efficiently than disrupt its capsid.
Inhibitory food matrix effects.
The presence of PCR inhibitors, which may cause false-negative results, is a common problem with real-time RT-PCR detection methods (89). Such factors derived from food matrices (including organic and phenolic compounds, glycogen, fat, and Ca2+) can inhibit enzymatic reactions of the RT-PCR partially or even totally (75). A few foods are known to contain inhibitory substances (2, 75, 79). Detection of virus on berries is especially problematic due to the presence of both inhibitory chemicals and a low pH (20). In contrast to PEG virus precipitation, with which the PCR inhibitors can be diluted out, the use of an ultrafiltration-based virus extraction method leaves the possibility of coconcentration of inhibitory factors from food samples. Therefore, in this study the magnitude of PCR inhibition was monitored by using an external amplification control consisting of MS2 RNA. With the foods tested, the inhibition was found to be only partial and relatively small. The inhibition ranged from none or negligible for convenience products with complex matrices (tomato ketchup, spiced tomato sauce) and mincemeat (<1 log10) to partial (up to 2.6-CT value difference compared to the reference sample) for all other foodstuffs. In summary, the RNA extraction method based on a silica gel column (QIAamp viral RNA mini kit; Qiagen, Hilden, Germany) is able to remove the majority of inhibitors during the RNA purification process, and the foodstuffs tested are apparently less complicated with respect to inhibitory substances, as has been reported for, e.g., raspberries (20). A study determined that more than 1,000 HAV particles can be transferred from fecally contaminated fingers to food (14). Based on this information, a PCR inhibition of less 1 log10 still leaves the possibility of positive NV real-time PCR detection. Therefore, the process described here might even be applicable to detection of low levels of NV introduced by primary and secondary food contamination.
Recovery rates.
Recovery rates determined in different studies and for different foods are comparable only to a limited extent. Depending on what virus extraction and RNA extraction methods are used, the rates may differ significantly. For example, Kim et al. (50) reported that when a QIAamp viral RNA mini kit was used, the level of NV recovery from grapes was 80% and that when the Trizol reagent was used, the level of NV recovery was 0.4%. However, the rates of recovery determined in this study for solid surface foods, including lettuce (64%) and fruits (43.3%), are consistent with the findings of Fumian et al. (38) (up to 73.3% for lettuce and 56.3% for cheese). Further studies reported average rates of NV recovery from fresh vegetables of 46.3 to 9.5% and from different types of berries of 19.6 to 0.5% (20) and 13% (31). In composite foods with complex matrices and mincemeat products, the recovery rates are commonly lower than those for solid foods. This confirms the assumption that virus particles are more efficiently washed from a surface than eluted out of a complex food matrix. Nevertheless, the average rates of NV recovery obtained for convenience products (pizza, 1.5%; baguette, 10.1%; ketchup, 2.9%; spiced tomato sauce, 21.7%), delicatessen salads (potato salad, 5.3%; noodle salad, 9.4%), and meat (1.2%) indicate that the extraction protocol used enables efficient and reproducible recovery of NV also from complex food matrices. This was verified by using MS2 internal process controls for each food sample tested.
Inactivation studies.
Studies of inactivation of infectious human NV in food are rare. Various inactivation studies have been performed with FCV (17, 28, 32, 35, 68, 82, 86) and murine norovirus (7, 8, 10, 11, 21, 51), which are cultivable viruses belonging to the family Caliciviridae with ssRNA, as models for NV. The findings of these studies cannot be directly applied to NV because several results indicated that the surrogates are generally more sensitive to physicochemical treatments (21, 57). More stable enteric RNA viruses have been proposed as models for NV to assess the safety of a process (54). Moreover, the differences in the experimental conditions and the methods used complicate drawing general conclusions.
In this study, a significant reduction (−1.7 log10) in the NV titer was observed after storage at 6°C of NV under acid pH conditions in potato salad (24 days, pH 5.0 to 5.5); no reduction in the virus titer was observed for storage in noodle salad (24 days, pH 5.0 to 5.5) or tomato ketchup (58 days, pH 4.5). The latter findings are more consistent with findings which indicated that NV was stable for 3 h in pH 3 medium (27), at pH 3 to 7 (4), at approximately pH 3 to 4 on raspberries (72), and in orange juice (36). The NV surrogate murine norovirus (MNV) was observed to be still infective at pH 2 (21). Therefore, it is plausible to assume that NV is resistant in food with a low pH and might still be infective since enteric viruses overcome the conditions in the gastrointestinal tract. The reason for inactivation in delicatessen salad is unlikely to be the pH and might be related to the composition of the product; it was reported that some types of fresh produce showed antiviral activity (5). It was beyond the scope of this study to identify the facilitator, which may be an interesting subject for further studies.
No change in the NV titer during cooling was observed for apples (7 days, 11°C), lettuce (5 days, 11°C), or internally inoculated mincemeat (2 days, 6°C). These results are consistent with the results of studies in which survival of FCV on different food samples for 7 days at refrigerated temperatures was demonstrated (65), persistence of NV and FCV for 4 weeks at 4°C in marinated mussels was demonstrated (43), and tenacity of NV for at least 10 days on refrigerated lettuce was demonstrated (57). It is plausible that NV persists during refrigerated storage and might be infective under these conditions beyond the typical shelf life of the food.
Likewise, after cryoconservation of a superficially inoculated frozen pizza product (7 and 14 days, −18°C) and internally inoculated mincemeat (8 days, −18°C), no significant reductions in the NV titer were observed. It has been reported that freezing increased the titers of NV, FCV, HAV, and rotavirus in berries and herbs (19) and the titers of HAV, PV, and FCV in ice cream and frozen desserts (44) less than 1 log10. The infectivity of FCV was not affected by repeated cycles of freezing and thawing (32). Thus, given the stability of cryoconserved NV, it must be assumed that NV remains infective under freezing conditions. The results of epidemiological studies corroborate this conclusion, since outbreaks of NV infection were associated with consumption of frozen raspberries (23, 55, 72).
Thermal processes are used for production, preparation, and preservation of foods. To estimate the risk of infection, several studies were carried out in which the thermal stability of food-borne enteric and cultivable model viruses was investigated. Comparable data for NV in food are not available. An early study (27) with volunteers revealed that after heating (60°C, 30 min) there was incomplete inactivation and persistence of infective NV. Under the pasteurization conditions utilized in this study (72 to 74°C, 1 min), no significant reduction in the NV titer was observed. Conditions currently used for pasteurization processes appear to be insufficient to inactivate NV in the foodstuffs tested. This conclusion coincides with the conclusion of a study in which NV GGII.4 RNA in diluted clinical specimens was not exposed maximally until the temperature was 76.6°C (86). However, moderate heat treatments may eliminate NV infectivity by altering but not destroying the viral capsid and exposing the RNA to RNases (68). Therefore, active but noninfective particles are detected. This will be a problem until a culture model to monitor NV infectivity is established. These results are in agreement with those reported for HAV, which showed that pasteurization was insufficient for complete inactivation (13, 24, 42, 66). In addition to the temperature, the duration of a process must be taken into account, as reported in a study (32) in which FCV was found to be not completely inactivated by short-time pasteurization (72°C, 15 s). FCV appears to be generally more heat sensitive than NV since temperatures of >70°C caused clear FCV inactivation (21, 28, 32, 68, 82). Heat treatment of mincemeat (meatballs) (roasting for 30 min at 200°C in a table oven) resulted in a 1.6-log10 reduction, and the titer of mixed-in NV was reduced to a level below the detection limit (>7 log10) by boiling in liquid for 30 min. In other studies, HAV and PV were efficiently inactivated (>4 log10) by boiling contaminated solid food in water (44), and HAV was completely inactivated by a near-boiling temperature (95°C, 5 min) (13). However, Hewitt and Greening (42) obtained a 3.5-log10 reduction in the HAV titer but no reduction in the real-time RT-PCR titer of NV in heated mussels boiled for 180 s. Heat treatments used by the consumer for preparation of frozen pizza (200°C, 12 min) and frozen pizza baguette (220°C, 15 min) reduced the titer of NV inoculated on the surface prior to the process efficiently by at least 4 log10 and 3 log10, respectively.
Protective effect.
Matrix-derived protective effects have been described in connection with heat treatments. An increase in the fat content of milk appeared to have a protective effect on HAV, increasing its heat stability (15, 78). In this study, clear protective effects on NV were observed for some heat treatments with various intensities. In contrast to baked pizza (inactivation to levels below the detection limit), for baked pizza baguettes a survival rate of 0.1% was obtained, although the heat treatment was even longer and the temperature was higher than that used for pizza (3 min, 20°C). It was beyond the scope of this study to identify the factors responsible for this effect. It can only be speculated that the thicker base (subsidence of viruses), the divergent composition of toppings, or the higher content of fat (cheese) might have contributed to the protective effect. Likewise, for heated spiced tomato sauce (pasteurized), in which the survival rate was 44.6%, the facilitators of protection remain unclear. A noteworthy observation is that in roasted mincemeat the rate of survival of the mixed-in viruses was 2.3%, although boiling mincemeat completely inactivated NV. This might have been due to different heat distribution during the processes, but the reason is unclear at this point. The experiments revealed that the stability of NV in one food during heat treatment cannot be generalized to other products, since there may be individual food-specific protective effects that prevent virus inactivation.
In summary, it was shown in this study that food technology processes for preservation and storage, such as cooling (until the limit of product storability), freezing, acidification, and moderate heat treatments (pasteurization), appear to be insufficient to inactivate NV within a food matrix or on a food surface. Effective inactivation of NV can be obtained only with heat treatments used for food preparation (baking, cooking, roasting). Koopmans and Duizer (54) classified the risk of infection for the consumer as negligible and low if the reduction in the virus titer is at least 4 log10 and 3 log10, respectively. Using this classification, safety for the consumer could be determined only for the heat processes. It must be noted that in addition to the parameters of a technological process, matrix-specific protective effects (e.g., heated pizza or pizza baguette) and the process conduct (baking or boiling of mincemeat) can affect the inactivation of NV in food and should be considered when evaluating risk assessment. This approach might be applied to other enteric RNA viruses which are food borne and noncultivable. The methods established and data obtained in this study could also benefit process optimization for NV inactivation in foods and promote the development of risk assessment systems in order to improve consumer protection.
Acknowledgments
This study was funded by the German AiF (Arbeitsgemeinschaft industrieller Forschungsvereinigungen “Otto von Guericke” e.V.) with review and supervision by FEI (Forschungskreis der Ernährungsindustrie e.V.).
We thank Jens Dreier (Institut fuer Laboratoriums- und Transfusionsmedizin, Bad Oeynhausen, Germany) for providing the RNA standard plasmid and bacteriophage MS2 and for technical advice. We also thank Reimar Johne (Federal Institute for Risk Assessment, Berlin, Germany) for providing the NV stool samples and Jochen Blom (CeBiTec, University of Bielefeld, Bielefeld, Germany) for implementing Rotor-Gene analysis in the CAmpER framework. We gratefully acknowledge Dean O. Cliver (Department of Population Health and Reproduction, School of Veterinary Medicine, University of California, Davis, CA) for critical reading of the manuscript.
Footnotes
Published ahead of print on 20 November 2009.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atmar, R. L., T. G. Metcalf, F. H. Neill, and M. K. Estes. 1993. Detection of enteric viruses in oysters by using the polymerase chain reaction. Appl. Environ. Microbiol. 59:631-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atmar, R. L., F. H. Neill, J. L. Romalde, F. Le Guyader, C. M. Woodley, T. G. Metcalf, and M. K. Estes. 1995. Detection of Norwalk virus and hepatitis A virus in shellfish tissues with the PCR. Appl. Environ. Microbiol. 61:3014-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausar, S. F., T. R. Foubert, M. H. Hudson, T. S. Vedvick, and C. R. Middaugh. 2006. Conformational stability and disassembly of Norwalk virus-like particles. Effect of pH and temperature. J. Biol. Chem. 281:19478-19488. [DOI] [PubMed] [Google Scholar]
- 5.Baert, L., J. Debevere, and M. Uyttendaele. 2009. The efficacy of preservation methods to inactivate foodborne viruses. Int. J. Food Microbiol. 131:83-94. [DOI] [PubMed] [Google Scholar]
- 6.Baert, L., M. Uyttendaele, and J. Debevere. 2007. Evaluation of two viral extraction methods for the detection of human noroviruses in shellfish with conventional and real-time reverse transcriptase PCR. Lett. Appl. Microbiol. 44:106-111. [DOI] [PubMed] [Google Scholar]
- 7.Baert, L., M. Uyttendaele, E. Van Coillie, and J. Debevere. 2008. The reduction of murine norovirus 1, B. fragilis HSP40 infecting phage B40-8 and E. coli after a mild thermal pasteurization process of raspberry puree. Food Microbiol. 25:871-874. [DOI] [PubMed] [Google Scholar]
- 8.Baert, L., I. Vandekinderen, F. Devlieghere, E. Van Coillie, J. Debevere, and M. Uyttendaele. 2008. Inactivation of murine norovirus 1 and Bacteroides fragilis infecting phage B40-8 by the use of sodium hypochlorite and peroxyacetic acid as decontaminating agents for shredded iceberg lettuce. Commun. Agric. Appl. Biol. Sci. 73:97-101. [PubMed] [Google Scholar]
- 9.Baert, L., M. Uyttendaele, and J. Debevere. 2008. Evaluation of viral extraction methods on a broad range of ready-to-eat foods with conventional and real-time RT-PCR for norovirus GII detection. Int. J. Food Microbiol. 123:101-108. [DOI] [PubMed] [Google Scholar]
- 10.Baert, L., M. Uyttendaele, M. Vermeersch, E. Van Coillie, and J. Debevere. 2008. Survival and transfer of murine norovirus 1, a surrogate for human noroviruses, during the production process of deep-frozen onions and spinach. J. Food Prot. 71:1590-1597. [DOI] [PubMed] [Google Scholar]
- 11.Baert, L., I. Vandekinderen, F. Devlieghere, E. Van Coillie, J. Debevere, and M. Uyttendaele. 2009. Efficacy of sodium hypochlorite and peroxyacetic acid to reduce murine norovirus 1, B40-8, Listeria monocytogenes, and Escherichia coli O157:H7 on shredded iceberg lettuce and in residual wash water. J. Food Prot. 72:1047-1054. [DOI] [PubMed] [Google Scholar]
- 12.Benkaddour, M., S. Tache, C. Labie, G. Bodin, and M. Eeckhoutte. 1993. Role of temperature, acidic pH and lactic bacteria on the stability of rotavirus and coronavirus in milk. Rev. Med. Vet. 144:8-9. [Google Scholar]
- 13.Bhattacharya, S. S., M. Kulka, K. A. Lampel, T. A. Cebula, and B. B. Goswami. 2004. Use of reverse transcription and PCR to discriminate between infectious and non-infectious hepatitis A virus. J. Virol. Methods 116:181-187. [DOI] [PubMed] [Google Scholar]
- 14.Bidawid, S., J. M. Farber, and S. A. Sattar. 2000. Contamination of foods by food handlers: experiments on hepatitis A virus transfer to food and its interruption. Appl. Environ. Microbiol. 66:2759-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bidawid, S., J. M. Farber, S. A. Sattar, and S. Hayward. 2000. Heat inactivation of hepatitis A virus in dairy foods. J. Food Prot. 63:522-528. [DOI] [PubMed] [Google Scholar]
- 16.Boxman I. L. A., J. J. H. C. Tilburg, N. A. J. M. te Loeke, H. Vennema, E. de Boer, and M. Koopmans. 2007. An efficient and rapid method for recovery of norovirus from food associated with outbreaks of gastroenteritis. J. Food Prot. 70:504-508. [DOI] [PubMed] [Google Scholar]
- 17.Buckow, R., S. Isbarn, D. Knorr, V. Heinz, and A. Lehmacher. 2008. Predictive model for inactivation of feline calicivirus, a norovirus surrogate, by heat and high hydrostatic pressure. Appl. Environ. Microbiol. 74:1030-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butot, S., T. Putallaz, R. Amoroso, and G. Sánchez. 2009. Inactivation of enteric viruses in minimally processed berries and herbs. Appl. Environ. Microbiol. 75:4155-4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butot, S., T. Putallaz, and G. Sánchez. 2008. Effects of sanitation, freezing and frozen storage on enteric viruses in berries and herbs. Int. J. Food Microbiol. 126:30-35. [DOI] [PubMed] [Google Scholar]
- 20.Butot, S., T. Putallaz, and G. Sánchez. 2007. Procedure for rapid concentration and detection of enteric viruses from berries and vegetables. Appl. Environ. Microbiol. 73:186-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cannon, J. L., E. Papafragkou, G. W. Park, J. Osborne, L. Jaykus, and J. Vinjé. 2006. Surrogates for the study of norovirus stability and inactivation in the environment: a comparison of murine norovirus and feline calicivirus. J. Food Prot. 69:2761-2765. [DOI] [PubMed] [Google Scholar]
- 22.Cliver, D. O. 1997. Virus transmission via food. World Health Stat. Q. 50:90-101. [PubMed] [Google Scholar]
- 23.Cotterelle, B., C. Drougard, J. Rolland, M. Becamel, M. Boudon, S. Pinede, O. Traoré, K. Balay, P. Pothier, and E. Espié. 2005. Outbreak of norovirus infection associated with the consumption of frozen raspberries, France, March 2005. Euro Surveill. 10:E050428.1. [DOI] [PubMed] [Google Scholar]
- 24.Croci, L., M. Ciccozzi, D. De Medici, S. Di Pasquale, A. Fiore, A. Mele, and L. Toti. 1999. Inactivation of hepatitis A virus in heat-treated mussels. J. Appl. Microbiol. 87:884-888. [DOI] [PubMed] [Google Scholar]
- 25.Daniels, N. A., D. A. Bergmire-Sweat, K. J. Schwab, K. A. Hendricks, S. Reddy, S. M. Rowe, R. L. Fankhauser, S. S. Monroe, R. L. Atmar, R. I. Glass, and P. Mead. 2000. A foodborne outbreak of gastroenteritis associated with Norwalk-like viruses: first molecular traceback to deli sandwiches contaminated during preparation. J. Infect. Dis. 181:1467-1470. [DOI] [PubMed] [Google Scholar]
- 26.DiGirolamo, R., J. Liston, and J. R. Matches. 1970. Survival of virus in chilled, frozen, and processed oysters. Appl. Microbiol. 20:58-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dolin, R., N. R. Blacklow, H. DuPont, R. F. Buscho, R. G. Wyatt, J. A. Kasel, R. Hornick, and R. M. Chanock. 1972. Biological properties of Norwalk agent of acute infectious nonbacterial gastroenteritis. Proc. Soc. Exp. Biol. Med. 140:578-583. [DOI] [PubMed] [Google Scholar]
- 28.Doultree, J. C., J. D. Druce, C. J. Birch, D. S. Bowden, and J. A. Marshall. 1999. Inactivation of feline calicivirus, a Norwalk virus surrogate. J. Hosp. Infect. 41:51-57. [DOI] [PubMed] [Google Scholar]
- 29.Dreier, J., M. Störmer, and K. Kleesiek. 2005. Use of bacteriophage MS2 as an internal control in viral reverse transcription-PCR assays. J. Clin. Microbiol. 43:4551-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dreier, J., M. Störmer, D. Mäde, S. Burkhardt, and K. Kleesiek. 2006. Enhanced reverse transcription-PCR assay for detection of norovirus genogroup I. J. Clin. Microbiol. 44:2714-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dubois, E., C. Agier, O. Traoré, C. Hennechart, G. Merle, C. Crucière, and H. Laveran. 2002. Modified concentration method for the detection of enteric viruses on fruits and vegetables by reverse transcriptase-polymerase chain reaction or cell culture. J. Food Prot. 65:1962-1969. [DOI] [PubMed] [Google Scholar]
- 32.Duizer, E., P. Bijkerk, B. Rockx, A. De Groot, F. Twisk, and M. Koopmans. 2004. Inactivation of caliciviruses. Appl. Environ. Microbiol. 70:4538-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Falkenhorst, G., L. Krusell, M. Lisby, S. B. Madsen, B. Böttiger, and K. Mølbak. 2005. Imported frozen raspberries cause a series of norovirus outbreaks in Denmark, 2005. Euro Surveill. 10:E050922.2. [DOI] [PubMed] [Google Scholar]
- 34.Fankhauser, R. L., S. S. Monroe, J. S. Noel, C. D. Humphrey, J. S. Bresee, U. D. Parashar, T. Ando, and R. I. Glass. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 186:1-7. [DOI] [PubMed] [Google Scholar]
- 35.Fino, V. R., and K. E. Kniel. 2008. UV light inactivation of hepatitis A virus, Aichi virus, and feline calicivirus on strawberries, green onions, and lettuce. J. Food Prot. 71:908-913. [DOI] [PubMed] [Google Scholar]
- 36.Fleet, G. H., P. Heiskanen, I. Reid, and K. A. Buckle. 2000. Foodborne viral illness—status in Australia. Int. J. Food Microbiol. 59:127-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fronhoffs, S., G. Totzke, S. Stier, N. Wernert, M. Rothe, T. Brüning, B. Koch, A. Sachinidis, H. Vetter, and Y. Ko. 2002. A method for the rapid construction of cRNA standard curves in quantitative real-time reverse transcription polymerase chain reaction. Mol. Cell. Probes 16:99-110. [DOI] [PubMed] [Google Scholar]
- 38.Fumian, T. M., J. P. G. Leite, V. A. Marin, and M. P. Miagostovich. 2009. A rapid procedure for detecting noroviruses from cheese and fresh lettuce. J. Virol. Methods 155:39-43. [DOI] [PubMed] [Google Scholar]
- 39.Guévremont, E., J. Brassard, A. Houde, C. Simard, and Y. Trottier. 2006. Development of an extraction and concentration procedure and comparison of RT-PCR primer systems for the detection of hepatitis A virus and norovirus GII in green onions. J. Virol. Methods 134:130-135. [DOI] [PubMed] [Google Scholar]
- 40.Haramoto, E., H. Katayama, E. Utagawa, and S. Ohgaki. 2009. Recovery of human norovirus from water by virus concentration methods. J. Virol. Methods 160:206-209. [DOI] [PubMed] [Google Scholar]
- 41.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 42.Hewitt, J., and G. E. Greening. 2006. Effect of heat treatment on hepatitis A virus and norovirus in New Zealand greenshell mussels (Perna canaliculus) by quantitative real-time reverse transcription PCR and cell culture. J. Food Prot. 69:2217-2223. [DOI] [PubMed] [Google Scholar]
- 43.Hewitt, J., and G. E. Greening. 2004. Survival and persistence of norovirus, hepatitis A virus, and feline calicivirus in marinated mussels. J. Food Prot. 67:1743-1750. [DOI] [PubMed] [Google Scholar]
- 44.Hollinger, F., and J. Ticehurst. 1996. Hepatitis A virus, p. 735-782. In Field virology, 3rd ed. Lippincott-Raven, Philadelphia, PA.
- 45.Hurst, A., and A. Hughes. 1983. The protective effect of some food ingredients on Staphylococcus aureus MF31. J. Appl. Bacteriol. 55:81-88. [DOI] [PubMed] [Google Scholar]
- 46.Jaykus, L. A., R. De Leon, and M. D. Sobsey. 1996. A virion concentration method for detection of human enteric viruses in oysters by PCR and oligoprobe hybridization. Appl. Environ. Microbiol. 62:2074-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jothikumar, N., D. O. Cliver, and T. W. Mariam. 1998. Immunomagnetic capture PCR for rapid concentration and detection of hepatitis A virus from environmental samples. Appl. Environ. Microbiol. 64:504-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kadan, R. S., W. H. Martin, and R. Mickelsen. 1963. Effect of ingredients used in condensed and frozen dairy products on thermal resistance of potentially pathogenic staphylococci. Appl. Microbiol. 11:45-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kageyama, T., S. Kojima, M. Shinohara, K. Uchida, S. Fukushi, F. B. Hoshino, N. Takeda, and K. Katayama. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41:1548-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim, H., I. Kwak, I. Hwang, and G. Ko. 2008. Optimization of methods for detecting norovirus on various fruit. J. Virol. Methods 153:104-110. [DOI] [PubMed] [Google Scholar]
- 51.Kingsley, D. H., D. R. Holliman, K. R. Calci, H. Chen, and G. J. Flick. 2007. Inactivation of a norovirus by high-pressure processing. Appl. Environ. Microbiol. 73:581-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kingsley, D. H., G. K. Meade, and G. P. Richards. 2002. Detection of both hepatitis A virus and Norwalk-like virus in imported clams associated with food-borne illness. Appl. Environ. Microbiol. 68:3914-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koopmans M., C. H. von Bonsdorff, J. Vinjé, D. de Medici, and S. Monroe. 2002. Foodborne viruses. FEMS Microbiol. Rev. 26:187-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koopmans, M., and E. Duizer. 2004. Foodborne viruses: an emerging problem. Int. J. Food Microbiol. 90:23-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Korsager, B., S. Hede, H. Bøggild, B. E. Böttiger, and K. Mølbak. 2005. Two outbreaks of norovirus infections associated with the consumption of imported frozen raspberries, Denmark, May-June 2005. Euro Surveill. 10:E050623.1. [DOI] [PubMed] [Google Scholar]
- 56.Kruse, H., D. Brown, D. Lees, S. Le Guyader, S. Lindgren, M. Koopmans, A. Macri, and C. von Bonsdorff. 2002. Opinion of the Scientific Committee on Veterinary Measures Relating to Public Health on Norwalk-like Viruses. European Commission Health & Consumer Protection Directorate-General, Brussels, Belgium.
- 57.Lamhoujeb, S., I. Fliss, S. E. Ngazoa, and J. Jean. 2008. Evaluation of the persistence of infectious human noroviruses on food surfaces by using real-time nucleic acid sequence-based amplification. Appl. Environ. Microbiol. 74:3349-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lees, D. 2000. Viruses and bivalve shellfish. Int. J. Food Microbiol. 59:81-116. [DOI] [PubMed] [Google Scholar]
- 59.Lees, D. N., K. Henshilwood, J. Green, C. I. Gallimore, and D. W. Brown. 1995. Detection of small round structured viruses in shellfish by reverse transcription-PCR. Appl. Environ. Microbiol. 61:4418-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Le Guyader, F., F. H. Neill, M. K. Estes, S. S. Monroe, T. Ando, and R. L. Atmar. 1996. Detection and analysis of a small round-structured virus strain in oysters implicated in an outbreak of acute gastroenteritis. Appl. Environ. Microbiol. 62:4268-4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Loisy, F., R. L. Atmar, P. Guillon, P. Le Cann, M. Pommepuy, and F. S. Le Guyader. 2005. Real-time RT-PCR for norovirus screening in shellfish. J. Virol. Methods 123:1-7. [DOI] [PubMed] [Google Scholar]
- 62.Lopman, B. A., M. H. Reacher, Y. Van Duijnhoven, F. Hanon, D. Brown, and M. Koopmans. 2003. Viral gastroenteritis outbreaks in Europe, 1995-2000. Emerg. Infect. Dis. 9:90-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mackay, I. M., K. E. Arden, and A. Nitsche. 2002. Real-time PCR in virology. Nucleic Acids Res. 30:1292-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mäde, D., S. Kahle, K. Trübner, and R. Stark. 2005. Detection of norovirus in food and environmental samples by RT-PCR. Application in routine diagnostics. Arch. Lebensmittelhyg. 56:1-24. [Google Scholar]
- 65.Mattison, K., K. Karthikeyan, M. Abebe, N. Malik, S. A. Sattar, J. M. Farber, and S. Bidawid. 2007. Survival of calicivirus in foods and on surfaces: experiments with feline calicivirus as a surrogate for norovirus. J. Food Prot. 70:500-503. [DOI] [PubMed] [Google Scholar]
- 66.Millard, J., H. Appleton, and J. V. Parry. 1987. Studies on heat inactivation of hepatitis A virus with special reference to shellfish. Part 1. Procedures for infection and recovery of virus from laboratory-maintained cockles. Epidemiol. Infect. 98:397-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Müller-Merbach, M., H. Neve, and J. Hinrichs. 2005. Kinetics of the thermal inactivation of the Lactococcus lactis bacteriophage P008. J. Dairy Res. 72:281-286. [DOI] [PubMed] [Google Scholar]
- 68.Nuanualsuwan, S., and D. O. Cliver. 2002. Pretreatment to avoid positive RT-PCR results with inactivated viruses. J. Virol. Methods 104:217-225. [DOI] [PubMed] [Google Scholar]
- 69.Ozawa, K., T. Oka, N. Takeda, and G. S. Hansman. 2007. Norovirus infections in symptomatic and asymptomatic food handlers in Japan. J. Clin. Microbiol. 45:3996-4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Patterson, W., P. Haswell, P. T. Fryers, and J. Green. 1997. Outbreak of small round structured virus gastroenteritis arose after kitchen assistant vomited. Commun. Dis. Rep. CDR Rev. 7:R101- R 103. [PubMed] [Google Scholar]
- 71.Peirson, S. N., J. N. Butler, and R. G. Foster. 2003. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 31:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pönkä A., L. Maunula, C. H. von Bonsdorff, and O. Lyytikäinen. 1999. An outbreak of calicivirus associated with consumption of frozen raspberries. Epidemiol. Infect. 123:469-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quiberoni, A., D. M. Guglielmotti, and J. A. Reinheimer. 2003. Inactivation of Lactobacillus delbrueckii bacteriophages by heat and biocides. Int. J. Food Microbiol. 84:51-62. [DOI] [PubMed] [Google Scholar]
- 74.Richards, G. P. 1999. Limitations of molecular biological techniques for assessing the virological safety of foods. J. Food Prot. 62:691-697. [DOI] [PubMed] [Google Scholar]
- 75.Rossen, L., P. Nørskov, K. Holmstrøm, and O. F. Rasmussen. 1992. Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA-extraction solutions. Int. J. Food Microbiol. 17:37-45. [DOI] [PubMed] [Google Scholar]
- 76.Rzezutka, A., and N. Cook. 2004. Survival of human enteric viruses in the environment and food. FEMS Microbiol. Rev. 28:441-453. [DOI] [PubMed] [Google Scholar]
- 77.Sair, A. I., D. H. D'Souza, C. L. Moe, and L. Jaykus. 2002. Improved detection of human enteric viruses in foods by RT-PCR. J. Virol. Methods 100:57-69. [DOI] [PubMed] [Google Scholar]
- 78.Sattar, S. A., T. Jason, S. Bidawid, and J. Farber. 2000. Foodborne spread of hepatitis A: recent studies on virus survival, transfer and inactivation. Can. J. Infect. Dis. 11:159-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schwab, K. J., R. De Leon, and M. D. Sobsey. 1996. Immunoaffinity concentration and purification of waterborne enteric viruses for detection by reverse transcriptase PCR. Appl. Environ. Microbiol. 62:2086-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schwab, K. J., F. H. Neill, R. L. Fankhauser, N. A. Daniels, S. S. Monroe, D. A. Bergmire-Sweat, M. K. Estes, and R. L. Atmar. 2000. Development of methods to detect “Norwalk-like viruses” (NLVs) and hepatitis A virus in delicatessen foods: application to a food-borne NLV outbreak. Appl. Environ. Microbiol. 66:213-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schwab, K. J., F. H. Neill, F. Le Guyader, M. K. Estes, and R. L. Atmar. 2001. Development of a reverse transcription-PCR-DNA enzyme immunoassay for detection of “Norwalk-like” viruses and hepatitis A virus in stool and shellfish. Appl. Environ. Microbiol. 67:742-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Slomka, M. J., and H. Appleton. 1998. Feline calicivirus as a model system for heat inactivation studies of small round structured viruses in shellfish. Epidemiol. Infect. 121:401-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Straub, T. M., K. Höner zu Bentrup, P. Orosz-Coghlan, A. Dohnalkova, B. K. Mayer, R. A. Bartholomew, C. O. Valdez, C. J. Bruckner-Lea, C. P. Gerba, M. Abbaszadegan, and C. A. Nickerson. 2007. In vitro cell culture infectivity assay for human noroviruses. Emerg. Infect. Dis. 13:396-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Strazynski, M., J. Krämer, and B. Becker. 2002. Thermal inactivation of poliovirus type 1 in water, milk and yoghurt. Int. J. Food Microbiol. 74:73-78. [DOI] [PubMed] [Google Scholar]
- 85.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Topping, J. R., H. Schnerr, J. Haines, M. Scott, M. J. Carter, M. M. Willcocks, K. Bellamy, D. W. Brown, J. J. Gray, C. I. Gallimore, and A. I. Knight. 2009. Temperature inactivation of feline calicivirus vaccine strain FCV F-9 in comparison with human noroviruses using an RNA exposure assay and reverse transcribed quantitative real-time polymerase chain reaction—a novel method for predicting virus infectivity. J. Virol. Methods 156:89-95. [DOI] [PubMed] [Google Scholar]
- 87.Vinjé J., S. A. Altena, and M. P. Koopmans. 1997. The incidence and genetic variability of small round-structured viruses in outbreaks of gastroenteritis in The Netherlands. J. Infect. Dis. 176:1374-1378. [DOI] [PubMed] [Google Scholar]
- 88.White, L. J., M. E. Hardy, and M. K. Estes. 1997. Biochemical characterization of a smaller form of recombinant Norwalk virus capsids assembled in insect cells. J. Virol. 71:8066-8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wilson, I. G. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]