Abstract
Streptococcus thermophilus is perceived as a recently emerged food bacterium that evolved from a commensal ancestor by loss and gain of functions. Here, we provide data allowing a better understanding of this evolutionary scheme. A multilocus sequence typing approach that we developed showed that S. thermophilus diverges significantly from its potential ancestors of the salivarius group and displays a low level of allelic variability, confirming its likely recent emergence. An analysis of the origin and dissemination of the prtS gene was carried out within this evolutionary scheme. This gene encodes a protease that allows better growth in milk by facilitating casein breakdown to supply amino acids. The S. thermophilus protease exhibits 95% identity to the animal Streptococcus suis protein PrtS. Genomic analysis showed that prtS is part of an island flanked by two tandem insertion sequence elements and containing three other genes which present the best identities and synteny with the S. suis genome. These data indicate a potential origin for this “ecological” island in a species closely related to S. suis. The analysis of the distribution of the prtS gene in S. thermophilus showed that the gene is infrequent in historical collections but frequent in recent industrial ones. Moreover, this “ecological” island conferring an important metabolic trait for milk adaptation appears to have disseminated by lateral transfer in the S. thermophilus population. Taken together, these data support an evolutionary scheme of S. thermophilus where gene acquisition and selection by food producers are determining factors. The source and impact of genes acquired by horizontal gene transfer on the physiology and safety of strains should be addressed.
Streptococcus thermophilus bacteria are lactic acid bacteria (LAB) with major economic importance originating from dairy products. This species is generally recognized as safe for food products and was granted qualified presumption of safety (QPS) status in Europe. It is historically widely used for the manufacture of yogurt and cheese with other LAB, such as Lactobacillus delbrueckii subsp. bulgaricus and Lactococcus lactis (18).
Comparative genomic analysis of two S. thermophilus genomes provides new insights in the evolution of this species (4). The high proportion of pseudogenes (10%) in the genome of this species suggests that S. thermophilus mainly evolved by loss of functions. Recombination and horizontal gene transfer (HGT) events were also proposed to contribute to the plasticity of the S. thermophilus genome (4, 23, 27). Although the mechanism of HGT remains largely uncharacterized, S. thermophilus harbors various mobile genetic elements such as insertion sequence (IS), phage-related integrase/recombinase, and integrative and potentially conjugative elements (ICEs) which could be involved in these events (4, 15, 29, 34). IS elements (15) and gene cassettes (34) were pointed out as probably having been acquired by S. thermophilus by lateral gene transfer from dairy bacteria such as Lactococcus lactis (34). Furthermore, the conjugative transfer of ICESt3 from Enterococcus faecalis to S. thermophilus was identified after a first gene exchange from S. thermophilus (2). A convincing example of HGT from LAB to S. thermophilus is a 17-kb region containing IS copies and a mosaic of fragments with more than 90% identity with the DNAs of Lb. bulgaricus, L. cremoris, and L. lactis (4). Genomic searches suggest that several regions encoding important industrial phenotypic traits such as bacteriocin production (blp, lab), restriction-modification systems, or oxygen tolerance (4, 18) were acquired by HGT. In most cases, the origin of genes acquired by S. thermophilus is suggested to be other LAB that are living in recurrent association with S. thermophilus in cheese or yogurt manufacture (4, 15). Lastly, the diversification of eps clusters encoding extracellular polysaccharide biosynthesis proteins and displaying a modular organization combined with high sequence diversity could be the result of recurrent HGT. The origin of these events was not pointed out in most cases (5, 6, 32), suggesting that the range of gene acquisition in S. thermophilus could be broader than that described previously. This hypothesis addresses more generally the question of the evolutionary scheme in food bacteria that may have an impact on their use. Indeed, and for example, in Europe, the QPS status of microorganisms depends not only on a long history of safe use but also on concerns provided by the knowledge associated with each species (9). In a QPS context, the nature of gene transfers (function, origin, etc.) is clearly highlighted as a possible concern.
Little is known about the diversity of the genetic population and the evolutionary scheme of this food bacterium. Recently, a phylogenetic relationship study of some 50 dairy S. thermophilus strains proposed that the patterns observed by genome hybridization could be explained by the high frequency of occurrence of gene transfer and recombination (32). S. thermophilus belongs to the salivarius group, which also includes Streptococcus salivarius and Streptococcus vestibularis, commensal bacteria commonly isolated from the human oral cavity (12). The three species of the salivarius group are genetically closely related, as has been shown by comparing their 16S rRNA and sodA genes (21, 31). For several years, S. thermophilus was classified as an S. salivarius subspecies (Streptococcus salivarius ssp. thermophilus) before regaining full species status based on DNA-DNA reassociation experiments (35). Recently, a population genetic analysis using multilocus sequence typing (MLST) was performed to better understand the phylogenetic relationship between S. salivarius and S. vestibularis and assess the genetic diversity within each species. Evidence of intraspecies recombination and HGT with other oral streptococci was reported, and it was suggested that such events may be an important factor driving the population evolution of S. salivarius and S. vestibularis (11).
In LAB, PrtS is a key enzyme of the proteolytic system, initiating the breakdown of caseins into oligopeptides before their uptake. prtS gene is harbored on a conjugative plasmid in Lactococcus lactis and on the chromosome in lactobacilli and S. thermophilus. Despite this localization and the relevant role of this enzyme in growth in milk, only 3 S. thermophilus strains among 97 screened from an INRA historical collection were found to express cell wall-associated proteinase activity (36). The enzyme was previously purified, and the prtS gene was sequenced (13). To our knowledge, the origin of the S. thermophilus prtS gene is not known yet. It could be questioned whether part of this gene is gradually lost from its commensal ancestor as the result of a genome restriction process or is a new function acquired by HGT.
To provide a better understanding of S. thermophilus genome evolution, we determined the population structure of S. thermophilus and the phylogenic relationship of S. thermophilus in the salivarius group by an MLST study. We complemented this study by genetic investigations of the origin and dissemination of the prtS gene in S. thermophilus. Our study reveals a clonal structure of S. thermophilus strains; supports the emergence of the species from the salivarius group in the recent past, concomitant with the development of the dairy practice; and suggests HGT from a distant streptococcal pathogen or commensal. Such transfer raises concerns about food safety that will have to be addressed in the future.
MATERIALS AND METHODS
Bacterial strains and chromosomal DNA.
The reference collection, made of 27 Streptococcus thermophilus strains used for MLST analysis, is listed in Table 1. The bacteria were isolated from different products (cheese, yogurt, fermented milk, and starter) in 11 countries over a 40-year period (1962 to 2002). Strain CCHSS9, isolated from human blood, was supplied by C. Poyart (INSERM E44, Institut Cochin, Centre National de Référence des Streptocoques, Service de Bactériologie Hôpital Cochin, Faculté de Médecine Paris 5, France). JIM, CNRZ, and LMG strains are available from our laboratory collections (Centre National de Recherche Zootechniques, INRA, Jouy-en-Josas, France, and the Belgium Coordinated Collections of Microorganisms, Ghent University, Ghent, Belgium). Additional strains were selected from the INRA historical collection; 21 were used for glcK locus analysis, and 135 were used for prtS distribution analysis. Strains were grown overnight at 42°C in M17 broth medium using lactose at a final concentration of 1% in an anaerobic atmosphere. Chromosomal DNAs of 20 S. suis strains, including isolates from humans (n = 6), clinically healthy pigs (n = 7), and pigs suffering from meningitis and septicemia (n = 7), were obtained from C. Marois [Agence Française de Sécurité Sanitaire des Aliments (AFSSA), Unité de Mycoplasmologie-Bactériologie, Ploufragan, France].
TABLE 1.
MLST and phenotypic data of the 27 S. thermophilus isolates used in this study
| Strain | ST | Allele no. |
Source | Location | Yr | Presence of prtSc | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| glcK | ddlA | pepO | ilvC | thrS | tkt | pyrE | dnaE | ||||||
| Lineage 1 | |||||||||||||
| JIM7444 | 3a | 1 | 1 | 1 | 1 | 2 | 3 | 6 | 1 | Starter | France | NAb | + |
| CNRZ1592 | 3a | 1 | 1 | 1 | 1 | 2 | 3 | 6 | 1 | Yogurt starter | Germany | 1987 | − |
| CNRZ703 | 3a | 1 | 1 | 1 | 1 | 2 | 3 | 6 | 1 | Fermented milk | Mongolia | 1974 | + |
| CNRZ1595 | 17 | 1 | 1 | 1 | 1 | 2 | 4 | 6 | 1 | Hard cheese | Austria | 1979 | − |
| JIM7446 | 4 | 1 | 1 | 1 | 1 | 1 | 3 | 6 | 1 | Starter | France | NA | − |
| CNRZ1691 | 20 | 4 | 1 | 1 | 1 | 1 | 3 | 6 | 1 | Cheese starter | Ireland | 1992 | − |
| CNRZ24B | 14 | 1 | 1 | 3 | 2 | 1 | 3 | 6 | 1 | Cheese starter | France | 1959 | − |
| JIM7442 | 1 | 1 | 1 | 1 | 1 | 2 | 7 | 11 | 1 | Starter | France | NA | + |
| JIM7443 | 1 | 1 | 1 | 1 | 1 | 2 | 7 | 11 | 1 | Starter | France | NA | + |
| JIM8232 | 2 | 1 | 1 | 1 | 1 | 2 | 7 | 12 | 1 | Milk | France | 2002 | + |
| LMD-9 | 8 | 1 | 1 | 2 | 1 | 2 | 7 | 5 | 1 | Cheese | United States | NA | + |
| CCHSS9 | 21 | 1 | 1 | 2 | 1 | 5 | 11 | 5 | 1 | Human blood | Hôpital Cochin, Paris, France | 2002 | + |
| Lineage 2 | |||||||||||||
| LMG18311 | 6 | 1 | 2 | 1 | 1 | 2 | 1 | 3 | 1 | Yogurt | United Kingdom | 1974 | − |
| CNRZ388 | 6 | 1 | 2 | 1 | 1 | 2 | 1 | 3 | 1 | Yogurt starter | France | 1971 | − |
| JIM7455 | 5 | 5 | 2 | 1 | 1 | 1 | 1 | 3 | 1 | Starter | France | NA | − |
| CNRZ1066 | 5 | 5 | 2 | 1 | 1 | 1 | 1 | 3 | 1 | Yogurt starter | France | 1986 | − |
| Lineage 3 | |||||||||||||
| CNRZ385 | 9 | 2 | 2 | 5 | 1 | 1 | 8 | 13 | 1 | Yogurt | Japan | 1971 | + |
| CNRZ781 | 11 | 3 | 2 | 5 | 1 | 1 | 8 | 13 | 1 | Starter | France | 1979 | − |
| CNRZ368 | 13 | 3 | 2 | 5 | 1 | 1 | 8 | 9 | 1 | Yogurt starter | The Netherlands | 1970 | − |
| Singletons | |||||||||||||
| CNRZ1202 | 10 | 2 | 2 | 1 | 1 | 1 | 3 | 1 | 1 | Greek yogurt | Heraklion Greece | 1988 | − |
| CNRZ1213 | 10 | 2 | 2 | 1 | 1 | 1 | 3 | 1 | 1 | Greek yogurt | Heraklion, Greece | 1988 | − |
| CNRZ758 | 18 | 2 | 2 | 3 | 1 | 1 | 6 | 1 | 5 | Yogurt starter | Bulgaria | 1978 | − |
| CNRZ759 | 7 | 1 | 2 | 1 | 1 | 1 | 10 | 10 | 3 | Yogurt starter | Bulgaria | 1978 | − |
| CNRZ1575 | 16 | 7 | 2 | 1 | 1 | 2 | 5 | 4 | 1 | Soft cheese | Italy | 1962 | − |
| CNRZ1120 | 19 | 1 | 2 | 3 | 2 | 2 | 3 | 8 | 2 | Cheese starter | France | 1987 | − |
| CNRZ702 | 12 | 1 | 3 | 6 | 1 | 4 | 2 | 2 | 4 | Greek sheep yogurt | Athens, Greece | 1973 | − |
| CNRZ1447 | 15 | 6 | 2 | 4 | 1 | 3 | 9 | 7 | 4 | Fermented milk | Delhi, India | 1990 | − |
Ancestral type in lineage 1.
NA, not available.
+, present; −, absent.
MLST.
The eight housekeeping gene loci used in this study are those designed for MLST analysis of S. salivarius and S. vestibularis (11). They are spread around the chromosome, except for pepO and ilvC, which are only 14 kb apart (4). The methods used for DNA extraction, purification, and sequencing and the PCR conditions and oligonucleotides used were described previously (11). Sequences from strains LMD-9 (25), LMG18311, and CNRZ1066 (4) were obtained from the NCBI database.
Analysis of prtS genes.
Sequencing of the prtS gene region was done by primer walking reactions for strain JIM8232 (Table 2). The sequence of the strain CNRZ385 prtS gene (accession no. AF243528) was verified, a few errors in the first hundred nucleotides were corrected, and it was found to be identical to that of strains JIM8232 and LMD-9 (accession no. YP_820283) in this region. Comparative genomic analysis of this region used S. thermophilus sequences from LMD-9 (CP000419), CNRZ1066 (CP000024), and LMG18311 (CP000023) and Streptococcus suis sequences from 05ZYH33 (NC 009442.1), 98HAH33 (NC_009443.1), P1/7 (Sanger Institute), and 89/1591 (DOE Joint Genomic Institute) strains.
TABLE 2.
Oligonucleotide primers used in this study
| Locus | Primer name | Sequence (5′-3′) |
|---|---|---|
| ciaH | bprt1 | GATGCGTCTTTTGACGAC |
| rpsT-coaA | bprt4 | TCGAACCCACACGAAATC |
| prtS | prt2 | ACAACAGCCTACACTTCC |
| prt3 | TCGTGGTCAACGTCAAGA | |
| prt4 | AGAGACTAAGGTGGAAGC | |
| eriC | aprt2 | TCTCCCAGCTCCGTATTA |
| aprt1 | GCCACCTAAGATGTCCAT | |
| potD | aprt2b | AATCATTCCTCTCCCAG |
| aprt-3 | TCCCCAGAAGTAGGGGATA | |
| aprt-3b | TGGGGAACCTTGGGCATT | |
| aprt-4b | TTGGCTGCAGGAATTGGT |
Data treatment.
For each locus, all of the sequences were compared and arbitrary allele numbers were assigned to each different sequence. The combination of alleles at each locus defined an allelic profile or sequence type (ST) for a strain. Strains with the same allelic profile were assigned to the same ST. The STs were identified by arbitrary numbers. The unweighted-pair group method using average linkages (UPGMA) tree of STs was drawn with the START version 1.1 program (K. Jolley; http://www.mlst.net/links/software.asp). The STs were grouped into lineages or clonal complexes with the BURST program developed by E. Feil and located in the START program. The members of a lineage were defined as groups of independent isolates that had identical alleles at six or more of eight loci.
The number of polymorphic nucleotide sites and the maximal and average percentages of nucleotide divergence of alleles at a given locus were calculated using the MEGA version 3 software (http://www.megasoftware.net) (22). Phylogenetic analysis of the nucleotide sequences of each housekeeping gene and the concatenated ddlA, thrS, pyrE, and dnaE sequences was performed using the neighbor-joining (NJ) method (33). The Kimura two-parameter distance model was used for estimating distances for nucleotide sequences. To determine the significance of the observed groupings in trees constructed by the NJ method, bootstrap analysis with 1,000 replicates was performed.
Nucleotide sequence accession numbers.
The sequences of all of the alleles included in this study have been deposited in the GenBank database under accession numbers FJ200327 to FJ200328 (ilvC fragment), FJ200311 to FJ200313 (ddlA fragment), FJ200319 to FJ200326 (glcK fragment), FJ200329 to FJ200334 (pepO fragment), FJ200348 to FJ200352 (thrS fragment), FJ200300 to FJ200310 (tkt fragment), FJ200335 to FJ200347 (pyrE fragment), and FJ200314 to FJ200318 (dnaE fragment). The sequence of prtS and its flanking region from strain JIM8232 has been deposited in the GenBank database under accession number FJ200299.
RESULTS
Development of an MLST scheme for S. thermophilus.
An S. thermophilus reference collection representative of product and geographical diversity has been established. For this purpose, 26 strains isolated from different dairy products (yogurt, fermented milk, cheese, different starters), from different countries (France, India, Italy, Mongolia, Greece, Bulgaria, etc.) over a 40-year period (1962 to 2002) were collected to obtain a broad representation of dairy strains. In addition, one strain was isolated from a patient with bacteremia at the Hôpital Cochin (Paris, France) (Table 1).
Since S. thermophilus is closely related to S. salivarius, we tested the same set of primers used recently in an MLST scheme of S. salivarius species to assess the phylogenetic relationship between the two species. It is based on the nucleotide sequence of an internal portion of nine housekeeping genes (11). Eight of these genes (glcK, ddlA, pepO, ilvC, thrS, tkt, pyrE, and dnaE) gave reliable amplification products and were used for MLST analysis. The nucleotide sequences of these loci in S. thermophilus isolates were determined; for the polymorphic sites among these alleles, see Fig. S1 in the supplemental material.
The sequence diversity within the S. thermophilus genes is low, with an average level of 0.2%, and allows distinguishing 2 to 13 alleles per locus (Table 3). The ilvC locus is the least divergent, with one variable site in 492 bp corresponding to only two alleles. The maximum percent nucleotide sequence divergence present in MLST alleles ranges from 0.2% at the ilvC locus to 1.7% at the glcK locus (Table 3). It is ≤1% for all of the loci except glcK (1.7%) and pyrE (1.2%). The frequency at which each allele occurred in the sample population is represented by one or two predominant alleles. The remaining alleles are observed in only one or two isolates for the glcK, thrS, ddlA, ilvC, and dnaE loci (Table 1). To demonstrate that the low genetic variability of S. thermophilus alleles is independent of an unexpected bias in the selected set of strains, the locus presenting the maximal divergence (1.7%), glcK, was analyzed in 21 supplementary S. thermophilus strains. To ensure maximal potential diversity, these strains were isolated from several dairy sources in different countries and between 1960 and 1987, as for the former set of strains. This 77% increase in the number of isolates revealed only one new allele (glk8th), which did not provide any new divergent nucleotide site (see Fig. S1 in the supplemental material).
TABLE 3.
Genetic diversity at S. thermophilus and S. salivarius MLST loci
| Locus | Sequence length (bp) | No. of alleles in S. thermophilus | No. of variable sites in S. thermophilus | Maximal % divergence |
||
|---|---|---|---|---|---|---|
| Within S. thermophilus | Within S. salivarius | Between S. thermophilus and S. salivarius | ||||
| ddlA | 459 | 3 | 2 | 0.4 | 8.1 | 9.8 |
| thrS | 497 | 5 | 4 | 0.6 | 6.1 | 8.6 |
| pyrE | 519 | 13 | 12 | 1.2 | 7.3 | 10.4 |
| dnaE | 480 | 5 | 4 | 0.6 | 7 | 18.8 |
| glcK | 470 | 7 | 12 | 1.7 | 11.9 | 15.3 |
| pepO | 460 | 6 | 5 | 0.4 | 45.3 | 44.1 |
| ilvC | 492 | 2 | 1 | 0.2 | 15.5 | 17.1 |
| tkta | 527 | 11 | 12 | 1 | ||
The tkt locus was amplified only in S. thermophilus isolates.
Relatedness of S. thermophilus isolates.
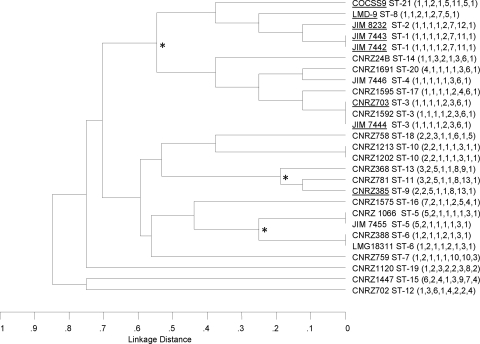
Table 1 summarizes the allelic profiles of the 27 S. thermophilus strains from the reference collection. Each unique combination of allele numbers represents one allelic profile or ST. Twenty-one different allelic profiles were found, corresponding to ST-1 to ST-21. Sixteen of the 21 STs are represented by only a single isolate, and ST-1, ST-3, ST-5, ST-6, and ST-10 contain two or three isolates (Table 1). The two strains belonging to ST-10 were isolated from the same product in the same isolation campaign and might correspond to multiple isolates of the same strain. ST-3 isolates were from several dairy products sampled in different countries over a 10-year period (Table 1).
A dendrogram was constructed by cluster analysis using UPGMA from the matrix of pairwise differences in the allelic profiles of the 27 S. thermophilus isolates (Fig. 1). No cluster correlates with the source, geographic origin, or isolation year of isolation of the isolates. Assignment of STs to clonal complexes by BURST analysis revealed that 7 STs are unrelated to any others while 14 STs are assigned to three lineages or clonal complexes (Table 1). Each lineage is composed of strains with identical STs or STs that vary at one or two loci (single- or double-locus variants) with at least one other member of the group. Lineage 1 is the largest and contains 12 isolates representing 9 STs (Table 1). Strain CCHSS9 (ST-21), isolated from human blood, belongs to this major group. A founder strain, whose ST differs at only one of eight loci from the highest number of other isolates in this lineage, was searched. ST-3 was identified as the ancestral type of lineage 1, which contains strains isolated from different products such as cheese, yogurt, and milk in six different countries. Lineages 2 and 3 consist of four and three strains, respectively, all isolated from yogurt or yogurt starters (Table 1).
FIG. 1.
UPGMA dendrogram showing the genetic relatedness of the 27 S. thermophilus strains used in this study. The dendrogram was constructed from a matrix of the pairwise distances between the allelic profiles of the 27 S. thermophilus strains. Three branch points showing the lineages obtained by BURST analysis are indicated by asterisks. Underlined strains contain the prtS gene.
Phylogenetic analysis of S. thermophilus within the salivarius group.
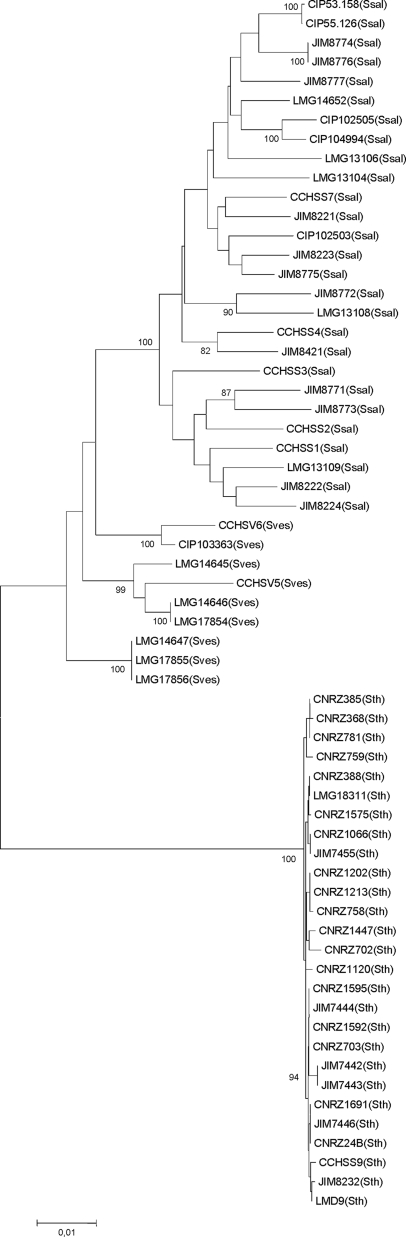
S. thermophilus species belongs to the salivarius group together with the two oral streptococci S. salivarius and S. vestibularis. The relationships among these three species were analyzed by combining results of this work and those obtained for 27 S. salivarius and 9 S. vestibularis isolates from humans (11). This analysis was performed with the ddlA, thrS, pyrE, and dnaE genes (total length of 1,955 bp) only because the glcK, pepO, ilvC, and tkt sequences were likely affected by HGT in S. salivarius (11). Phylogenetic analysis of the concatenated sequences yielded the tree shown in Fig. 2. It revealed a clear separation of the species S. thermophilus, S. salivarius, and S. vestibularis supported by significant bootstrap values. Phylogenetic trees were constructed for each of the eight housekeeping loci, and all of them resolved S. thermophilus alleles into clusters distinct from those containing the oral streptococcus alleles (data not shown). The identity between S. thermophilus and S. salivarius within the four loci is 89.4% at the nucleotide sequence level. The maximum percent nucleotide sequence divergence between S. thermophilus and S. salivarius loci ranges from 8.6% for the thrS locus to 18.8% for the dnaE locus (Table 3).
FIG. 2.
Phylogenetic tree of 63 strains (Ssal, S. salivarius; Sves, S. vestibularis; Sth, S. thermophilus) based on the concatenated sequences of the housekeeping genes ddlA, thrS, pyrE, and dnaE. The tree was based on the NJ method. Bootstrap values of >80% are indicated. The midpoint rooting method was used to root the tree. The scale bar indicates the number of nucleotide substitutions per site.
Analysis of the prtS region in S. thermophilus.
The gene encoding PrtS was initially characterized in strain CNRZ385 (13) and is present in newly sequenced strain LMD-9 (25). In addition, we sequenced this and the neighboring genes in JIM8232 by primer walking and with the help of the two existing sequences. A comparison of the amino acid sequences of the three S. thermophilus PrtS proteins showed that they are >98% identical (data not shown).
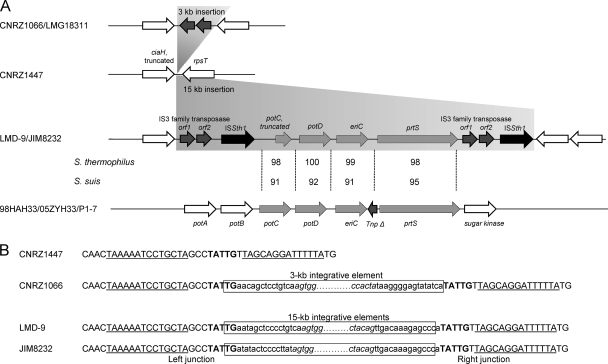
Comparative genomic analysis of prtS region of strains LMD-9 and JIM8232 indicates that the prtS gene is located in a 15-kb fragment in the intergenic region between the pseudogene ciaH and the gene rpsT (Fig. 3A). Three open reading frames, potC (truncated), potD, and eriC, are present upstream of the prtS gene, and they encode proteins that display identities with a polyamine (putrescine/spermidine) ABC uptake transporter membrane-spanning protein, a periplasmic protein, and chloride channel protein EriC, respectively. These three genes and prtS are flanked by two tandem sequences consisting of IS elements of the IS3 family transposase and ISSth1 (ISL3 family) (Fig. 3A). The tandem sequences are not identical, and their proteins are only 90 and 82% identical to the IS3 family transposase and ISSth1 elements, respectively. The IS3 family transposase is only found in S. thermophilus species, whereas ISSth1 is also widespread in S. suis species. The genetic organization of prtS and flanking genes is reminiscent of that of mobile elements.
FIG. 3.
Chromosomal region targeted by the prtS island in S. thermophilus. (A) Genetic organization of the region in strains CNRZ1447, CNRZ1066, LMG18311, LMD-9, and JIM8232. Synteny regions present in the S. suis and S. thermophilus genomes are light gray, IS3 family transposase and ISSth1 regions are dark gray and black, respectively. In S. suis, the intergenic region between the eriC and prtS genes contains a vestige of a transposase gene. Percentages of identity of prtS island proteins with proteins present in the species S. thermophilus (first line) and S. suis (second line) are indicated. (B) Sequences of the target and junctions of the prtS island and IS elements in different strains. The 5-bp target site, which is duplicated after element integration, is in bold, and the inverted repeats that flank the target site are underlined. Boxes represent 3- and 15-kb integrative elements with the IS ends. Italic sequences belong to the 22-bp (AGTGGACAATGAAAAAATAAAA/TTTTATTATTTCATTGTCCACT) and 14-bp (CTGTAGTGGGTAAA/TCTACCCACTACAG) terminal inverted repeats found at the ends of the IS3 family transposase and ISSth1, respectively.
Variability and distribution of the PrtS island in S. thermophilus.
PCR amplifications were carried out to amplify the internal region of the prtS gene in a large set of S. thermophilus historical collection strains. Twenty-one of 135 strains gave a band of the expected size (data not shown), including 8 that belong to the reference collection (Table 1). Strains containing the prtS gene are not related in the phylogenetic tree constructed on the basis of MLST data (Fig. 1). The identities of the PCR products were checked by sequencing strain CNRZ385, CNRZ703, and JIM8232 fragments.
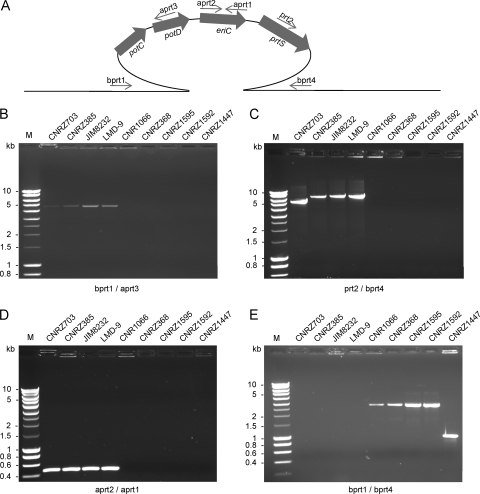
The organization of the island and its border regions was further analyzed by PCR amplification and sequence analysis in four prtS-positive and five prtS-negative strains (Fig. 4 and 3B). The internal fragment (Fig. 4D) and both right and left boundary fragments (Fig. 4B and C) of the prtS island were obtained in all of the prtS-positive S. thermophilus strains but failed to be amplified in the five prtS-negative S. thermophilus strains. This result indicates that the prtS island is present in the four prtS-positive S. thermophilus strains at the same chromosomal location. The only difference found is the size of the right border in strain CNRZ703, which is about 1 kb shorter than expected. This deletion may affect the IS element, since this strain was shown earlier to display strong protease activity and therefore should contain an intact prtS gene (36). PCR products were obtained with primers bprt-1 and bprt-4, flanking the prtS island, from the five prtS-negative S. thermophilus strains, giving fragments of 3 kb for strains CNRZ1066, CNRZ1595, CNRZ368, and CNRZ1592 and 1 kb for strain CNRZ1447 (Fig. 4E). Sequence analysis of these fragments revealed the presence of one copy of ISSth1 in the 3-kb fragment and the absence of any element in the 1-kb fragment. The copy of ISSth1 in Prt− strains is integrated at the same location as the prtS island but in the reverse orientation (Fig. 3A and B). The potential target site (TATTG) present in the chromosome of strain CNRZ1447 is duplicated by the insertion of the prtS island (strains JIM8232 and LMD-9) or IS3 family transposase (strain CNRZ1066) and is located in the loop of a stem-and-loop structure corresponding to a potential bidirectional terminator present in the intergenic region between the ciaH and rpsT genes (Fig. 3B).
FIG. 4.
Detection of the 15-kb prtS island and the 3-kb insertion in S. thermophilus strains. (A) Schematic representation of the prtS island and the primers used for amplification. (B) PCR assay of the left border of the prtS island. (C) PCR assay of the right border of the prtS island. (D) PCR assay of the internal eriC gene of the prtS island. (E) PCR assay of the complete region spanning the prtS island. The names of the strains are indicated above the electrophoresis gel lanes. M, marker.
Search for a potential origin of the S. thermophilus prtS island.
A search for PrtS homologues in different genomic databases showed that the protein, over its entire length, is 95% identical to the subtilisin-like serine protease of Streptococcus suis (97% identical at the nucleotide level) and, as a second best score, 48% identical to CspA of S. agalactiae. The main difference between the S. thermophilus and S. suis proteases is a duplication of 32 amino acids in the N-terminal part of the S. thermophilus protease (positions 63 to 90). Remarkably, the genes present in the S. thermophilus prtS island have homologues in the S. suis genome that are similarly clustered and ordered (Fig. 3A). However, in S. suis, this region is not flanked by IS elements. Moreover, potC appears to be intact and is clustered with two additional genes, potA and potB (Fig. 4). Taken together, these genes constitute the potABCD operon, the homologue of which encodes an ABC transporter for polyamines in Streptococcus pneumoniae (42).
To address the question of whether prtS and flanking genes are prevalent in S. suis populations, we tested the presence of this region in 20 S. suis strains isolated from humans and pigs by PCR amplification with the set of primers previously designed from S. thermophilus sequences (Table 2). In all of the strains, the amplification of 387-, 445-, and 491-bp PCR fragments confirmed the presence of the prtS, eriC, and potD genes, respectively (data not shown). The nucleotide sequences of these amplified fragments were determined to investigate the genetic diversity of the prtS and eriC genes among S. suis strains and compare them to those of S. thermophilus that were sequenced earlier. Available sequences of prtS and eriC from the S. suis genome were also included in the analysis. For prtS, three alleles were found in each species while two and six alleles were found for eriC in S. thermophilus and S. suis, respectively (see Fig. S2 in the supplemental material). The maximal levels of divergence of the prtS nucleotide sequences were 2.9% and 1.3% within S. thermophilus and S. suis, respectively, and for eriC, they were 0.3% and 1.5%, respectively (see Fig. S2 in the supplemental material). Between the S. thermophilus and S. suis alleles, the maximal level of nucleotide sequence divergence was 4.5% for the prtS allele and 9.1% for the eriC allele.
DISCUSSION
Comparative population study in the salivarius group.
In this study, we investigated the genetic diversity within the species S. thermophilus and its relationship with S. salivarius and S. vestibularis, the two commensal species of the salivarius group. MLST, a typing method involving the identification of nucleotide variations in housekeeping genes, could provide information about the “backbone” structure of strains and the phylogenetic lineage in a population. Recently, the population structure and genetic diversity of the two commensal species were studied by using this method (11). The use of the same primer set in this work allowed us to better assign the place of S. thermophilus in this group. Phylogenetic analysis of the concatenated sequences of four loci generated clusters that corresponded to each species (Fig. 2). Analysis of this phylogenetic tree shows that S. thermophilus is clearly separated from the two oral streptococci. Furthermore, examination of each locus separately showed that S. thermophilus and the two commensal streptococci have no alleles in common. These results confirm the status of S. thermophilus as a distinct species (35).
Because S. salivarius and S. thermophilus are phylogenetically closely related, these two warrant a more detailed comparison. The basic features of the population structure of S. thermophilus described here differ markedly from those of an equivalent number of S. salivarius isolates. A comparable MLST study of 27 S. salivarius strains using the same set of genes provided a very high level of allele sequence diversity (over 6%) (11). By contrast, the 27 S. thermophilus isolates recovered from various products at several geographic locations over a 40-year period displayed a small pool of unique alleles with few polymorphic nucleotide sites, indicating that the species is genetically rather uniform. These data are in agreement with the low polymorphism observed in the S. thermophilus genome (4, 18). This feature is evocative of a clonal S. thermophilus population structure, probably consequent to its recent emergence, preventing the possibility of substantial variation accumulation. Low sequence variations in the core genome, suggesting a recent species origin, were described for several pathogenic species, such as Streptococcus agalactiae (7, 20, 24, 40), Yersinia pestis, and Mycobacterium tuberculosis (1, 37). These observations support the suggestion that S. thermophilus emerged at the beginning of human dairy activity, about 7,000 years ago (4, 14).
prtS was likely acquired from a species close to S. suis.
Two hypotheses could be proposed to explain the scattered distribution of the prtS gene in the S. thermophilus population. The first supposes that prtS is lost in the genome restriction process occurring in S. thermophilus evolution, implying its presence in its commensal ancestor. The second assumes that prtS is gained by HGT. S. salivarius and S. vestibularis are not known to display strong extracellular proteolytic activity, and the analysis of the genomes of two S. salivarius strains revealed that they do not contain a prtS gene homolog (data not shown), which is at variance with the former hypothesis. In contrast, the present study indicates that the prtS gene was acquired by horizontal transfer from a Streptococcus species closely related to S. suis.
Genomic analysis of the prtS island showed that this region is flanked by two tandem sequences of IS elements, suggesting its potential acquisition by HGT. The inner region, containing one truncated (potC) and three complete (potD, eriC, and prtS) genes, presents synteny and shows high levels of identity at the nucleotide and protein sequence level with the S. suis chromosome. In S. suis, potC is complete and clustered in a four-gene operon, potABCD. Homologues of potABCD and eriC genes are present elsewhere and in different locations in S. thermophilus genomes. Phylogenic analysis of potD and eriC confirms that the island genes are closely related to those of S. suis genomes, whereas their S. thermophilus counterparts are closely related to those present in the S. salivarius genome (data not shown). Therefore, the structure of the prtS island and its synteny and high level of identity with an S. suis genome region support the hypothesis of the acquisition of a genomic fragment provided by HGT from a streptococcal strain closely related to the species S. suis.
Potential scheme of prtS gene dissemination in S. thermophilus.
In S. thermophilus, the protease activity performs the first step of protein breakdown, an important metabolic trait in milk. It is a key enzyme to increase bacterial fitness in this medium, which contains a limited amount of directly metabolizable nitrogen. Our study of 135 strains from the historical INRA collection collected at various times (1956 to 2008) from different products, including traditional ones and in different countries, suggests a low prevalence of the protease gene in this species (21 of 135 strains, corresponding to 15% of the strains). This is in agreement with previous studies of the historical INRA collection showing that only 3 of 97 strains were Prt+ (13, 36). In contrast, a recent study by whole-genome hybridization showed that 35 of 47 strains from a collection of industrial strains contain the prtS gene (32). A likely explanation for the high prevalence of prtS-positive strains in the industrial strain collection is bias in the method and the date of isolation. Among the selection criteria, the ability to “grow well in milk” will favor the selection of Prt+ strains (32). Furthermore, analysis of our collection data indicates that the ratio of prtS-positive strains increased drastically in strains isolated in the last 10 years. The first prtS-positive strains, CNRZ385 and CNRZ703, were isolated in Asia in 1971 and 1974, respectively (36). Most (80%) of the strains were isolated later than 1999, a few years after the characterization of strains displaying high proteolytic activity (36). These observations may reflect the expansion of the prtS island in the population by intraspecies exchange and further selection for its occurrence in commercial starters.
The prtS island is organized similarly in the four strains studied in detail here, strongly suggesting that protease acquisition in S. thermophilus occurred only from one source. The fact that several prtS-positive strains are not related in the phylogenic tree constructed on the basis of MLST alleles indicates that the prtS island is disseminated by gene transfer. However, although the prtS island is flanked by an IS element that may facilitate its integration into new genomes, it lacks any element for transfer. Its dissemination would therefore depend on other mobile elements (transposons or ICEs) or on processes like competence. The recent discovery that S. thermophilus is able to develop natural competence shows that such transfers are possible in this bacterium (3).
Remarkably, prtS alleles display a higher number of variable sites in S. thermophilus (2.9%) than in S. suis (1.3%). The opposite situation is observed for eriC, which is more conserved in prtS islands (0.3%) than in S. suis chromosomes (1.5%). Furthermore, a region of the S. thermophilus prtS propeptide domain is duplicated. This domain functions as an intramolecular chaperone in the subtilisin protease family (39), suggesting that this change is required to direct correct folding of PrtS in its new host. These modifications indicate that prtS could evolves rapidly in S. thermophilus, possibly under positive selection, contrary to the surrounding region that contains duplicates of genes that already exist in S. thermophilus. The PrtS island would therefore be a recently acquired “ecological island” carrying a new metabolic function that contributes to S. thermophilus competiveness in the milk environment (10, 16).
S. thermophilus probably has good potential to acquire new genetic material by HGT and recombination, similar to other streptococcal species (17, 19, 23, 30, 38, 43). In agreement with this suggestion, the large variability of several clusters, such as the eps locus, would be a consequence of a high frequency of gene transfer and recombination (5, 32). Until now, the origin of genes acquired by this bacterium was thought to be confined to bacteria largely associated in dairy products such as LAB (4) or a contaminant of milk (28, 29). The prtS island is the first example of the acquisition of a gene encoding a significant metabolic function from a species closely related to a commensal and/or pathogen. S. suis is commonly carried by pigs, and several strains are virulent (26). The exact way the two species were able to exchange genes is not known, since they do not share the same ecological niche in the farm environment. Cell envelope proteases are virulence factors in pathogenic streptococci, such as CspA in S. agalactiae (8) and possibly PrtS in S. suis (41). However, it is very likely that the protease has only a fitness function in S. thermophilus, aiding its growth in milk. The understanding of the role of PrtS in the physiology of S. thermophilus and its dissemination in food confirms that it has no effect on the safety of strains. Given the increasing amount of sequence data in the microbial world, new examples of potentially alarming transfer of genes will probably be discovered in food bacteria, and the ways this concern will be addressed to ensure food safety should be anticipated as well as possible.
Supplementary Material
Acknowledgments
We thank Claire Poyart for providing S. thermophilus strain CCHSS9 and Corinne Marois for S. suis DNA.
This work was supported by grant Flore-QPS 2.21P from the French Research Agency (ANR).
Footnotes
Published ahead of print on 13 November 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Achtman, M., K. Zurth, G. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U. S. A. 96:14043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellanger, X., A. P. Roberts, C. Morel, F. Choulet, G. Pavlovic, P. Mullany, B. Decaris, and G. Guédon. 2009. Conjugative transfer of the integrative conjugative elements ICESt1 and ICESt3 from Streptococcus thermophilus. J. Bacteriol. 191:2764-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blomqvist, T., H. Steinmoen, and L. S. Havarstein. 2006. Natural genetic transformation: a novel tool for efficient genetic engineering of the dairy bacterium Streptococcus thermophilus. Appl. Environ. Microbiol. 72:6751-6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolotin, A., B. Quinquis, P. Renault, A. Sorokin, S. D. Ehrlich, S. Kulakauskas, A. Lapidus, E. Goltsman, M. Mazur, G. D. Pusch, M. Fonstein, R. Overbeek, N. Kyprides, B. Purnelle, D. Prozzi, K. Ngui, D. Masuy, F. Hancy, S. Burteau, M. Boutry, J. Delcour, A. Goffeau, and P. Hols. 2004. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 22:1554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourgoin, F., A. Pluvinet, B. Gintz, B. Decaris, and G. Guédon. 1999. Are horizontal transfers involved in the evolution of the Streptococcus thermophilus exopolysaccharide synthesis loci? Gene 233:151-161. [DOI] [PubMed] [Google Scholar]
- 6.Broadbent, J. R., D. J. McMahon, D. L. Welker, C. J. Oberg, and S. Moineau. 2003. Biochemistry, genetics, and applications of exopolysaccharide production in Streptococcus thermophilus: a review. J. Dairy Sci. 86:407-423. [DOI] [PubMed] [Google Scholar]
- 7.Brochet, M., E. Couve, M. Zouine, T. Vallaeys, C. Rusniok, M. C. Lamy, C. Buchrieser, P. Trieu-Cuot, F. Kunst, C. Poyart, and P. Glaser. 2006. Genomic diversity and evolution within the species Streptococcus agalactiae. Microbes Infect. 8:1227-1243. [DOI] [PubMed] [Google Scholar]
- 8.Bryan, J. D., and D. W. Shelver. 2009. Streptococcus agalactiae CspA is a serine protease that inactivates chemokines. J. Bacteriol. 191:1847-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chamba, J. F., and E. Jamet. 2008. Contribution to the safety assessment of technological microflora found in fermented dairy products. Int. J. Food Microbiol. 126:263-266. [DOI] [PubMed] [Google Scholar]
- 10.Courtin, P., V. Monnet, and F. Rul. 2002. Cell wall proteinases PrtS and PrtB have a different role in Streptococcus thermophilus/Lactobacillus bulgaricus mixed cultures in milk. Microbiology 148:3413-3421. [DOI] [PubMed] [Google Scholar]
- 11.Delorme, C., C. Poyart, S. D. Ehrlich, and P. Renault. 2007. Extent of horizontal gene transfer in evolution of streptococci of the salivarius group. J. Bacteriol. 189:1330-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Facklam, R. 2002. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin. Microbiol. Rev. 15:613-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez-Espla, M. D., P. Garault, V. Monnet, and F. Rul. 2000. Streptococcus thermophilus cell wall-anchored proteinase: release, purification, and biochemical and genetic characterization. Appl. Environ. Microbiol. 66:4772-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox, P. F. 1993. Cheese: chemistry, physics and microbiology. Chapman & Hall, London, United Kingdom.
- 15.Guédon, G., F. Bourgoin, M. Pebay, Y. Roussel, C. Colmin, J. M. Simonet, and B. Decaris. 1995. Characterization and distribution of two insertion sequences, IS1191 and iso-IS981, in Streptococcus thermophilus: does intergeneric transfer of insertion sequences occur in lactic acid bacteria co-cultures? Mol. Microbiol. 16:69-78. [DOI] [PubMed] [Google Scholar]
- 16.Hacker, J., and E. Carniel. 2001. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2:376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hakenbeck, R., A. Konig, I. Kern, M. van der Linden, W. Keck, D. Billot-Klein, R. Legrand, B. Schoot, and L. Gutmann. 1998. Acquisition of five high-Mr penicillin-binding protein variants during transfer of high-level beta-lactam resistance from Streptococcus mitis to Streptococcus pneumoniae. J. Bacteriol. 180:1831-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hols, P., F. Hancy, L. Fontaine, B. Grossiord, D. Prozzi, N. Leblond-Bourget, B. Decaris, A. Bolotin, C. Delorme, S. Dusko Ehrlich, E. Guédon, V. Monnet, P. Renault, and M. Kleerebezem. 2005. New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol. Rev. 29:435-463. [DOI] [PubMed] [Google Scholar]
- 19.Hoshino, T., T. Fujiwara, and M. Kilian. 2005. Use of phylogenetic and phenotypic analyses to identify nonhemolytic streptococci isolated from bacteremic patients. J. Clin. Microbiol. 43:6073-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones, N., J. F. Bohnsack, S. Takahashi, K. A. Oliver, M. S. Chan, F. Kunst, P. Glaser, C. Rusniok, D. W. Crook, R. M. Harding, N. Bisharat, and B. G. Spratt. 2003. Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 41:2530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawamura, Y., X. G. Hou, F. Sultana, H. Miura, and T. Ezaki. 1995. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int. J. Syst. Bacteriol. 45:406-408. [DOI] [PubMed] [Google Scholar]
- 22.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 23.Lefébure, T., and M. J. Stanhope. 2007. Evolution of the core and pan-genome of Streptococcus: positive selection, recombination, and genome composition. Genome Biol. 8:R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luan, S. L., M. Granlund, M. Sellin, T. Lagergard, B. G. Spratt, and M. Norgren. 2005. Multilocus sequence typing of Swedish invasive group B Streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. J. Clin. Microbiol. 43:3727-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goodstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J. H. Lee, I. Diaz-Muniz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U. S. A. 103:15611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marois, C., S. Bougeard, M. Gottschalk, and M. Kobisch. 2004. Multiplex PCR assay for detection of Streptococcus suis species and serotypes 2 and 1/2 in tonsils of live and dead pigs. J. Clin. Microbiol. 42:3169-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marri, P. R., W. Hao, and G. B. Golding. 2006. Gene gain and gene loss in streptococcus: is it driven by habitat? Mol. Biol. Evol. 23:2379-2391. [DOI] [PubMed] [Google Scholar]
- 28.Mathur, S., and R. Singh. 2005. Antibiotic resistance in food lactic acid bacteria—a review. Int. J. Food Microbiol. 105:281-295. [DOI] [PubMed] [Google Scholar]
- 29.Pavlovic, G., V. Burrus, B. Gintz, B. Decaris, and G. Guédon. 2004. Evolution of genomic islands by deletion and tandem accretion by site-specific recombination: ICESt1-related elements from Streptococcus thermophilus. Microbiology 150:759-774. [DOI] [PubMed] [Google Scholar]
- 30.Poulsen, K., J. Reinholdt, C. Jespersgaard, K. Boye, T. A. Brown, M. Hauge, and M. Kilian. 1998. A comprehensive genetic study of streptococcal immunoglobulin A1 proteases: evidence for recombination within and between species. Infect. Immun. 66:181-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poyart, C., G. Quesne, S. Coulon, P. Berche, and P. Trieu-Cuot. 1998. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J. Clin. Microbiol. 36:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rasmussen, T. B., M. Danielsen, O. Valina, E. Johansen, and M. B. Pedersen. 2008. The Streptococcus thermophilus core genome: comparative genome hybridization study of 47 strains. Appl. Environ. Microbiol. 74:4703-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 34.Schirawski, J., W. Hagens, G. F. Fitzgerald, and D. Van Sinderen. 2002. Molecular characterization of cadmium resistance in Streptococcus thermophilus strain 4134: an example of lateral gene transfer. Appl. Environ. Microbiol. 68:5508-5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schleifer, K. H., M. Ehrmann, U. Krusch, and H. Neve. 1991. Revival of the species Streptococcus thermophilus (ex Orla-Jensen, 1919) nom. rev. Syst. Appl. Microbiol. 14:386-388. [Google Scholar]
- 36.Shahbal, S., D. Hemme, and P. Renault. 1993. Characterization of a cell envelope-associated proteinase activity from Streptococcus thermophilus H strains. Appl. Environ. Microbiol. 59:177-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. U. S. A. 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stadler, C., and M. Teuber. 2002. The macrolide efflux genetic assembly of Streptococcus pneumoniae is present in erythromycin-resistant Streptococcus salivarius. Antimicrob. Agents Chemother. 46:3690-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subbian, E., Y. Yabuta, and U. Shinde. 2004. Positive selection dictates the choice between kinetic and thermodynamic protein folding and stability in subtilases. Biochemistry 43:14348-14360. [DOI] [PubMed] [Google Scholar]
- 40.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. U. S. A. 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vanier, G., N. Fittipaldi, J. D. Slater, M. de la Cruz Dominguez-Punaro, A. N. Rycroft, M. Segura, D. J. Maskell, and M. Gottschalk. 2009. New putative virulence factors of Streptococcus suis involved in invasion of porcine brain microvascular endothelial cells. Microb. Pathog. 46:13-20. [DOI] [PubMed] [Google Scholar]
- 42.Ware, D., Y. Jiang, W. Lin, and E. Swiatlo. 2006. Involvement of potD in Streptococcus pneumoniae polyamine transport and pathogenesis. Infect. Immun. 74:352-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whatmore, A. M., V. A. Barcus, and C. G. Dowson. 1999. Genetic diversity of the streptococcal competence (com) gene locus. J. Bacteriol. 181:3144-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.