Abstract
The highly variable flagellin-encoding flaA gene has long been used for genotyping Campylobacter jejuni and Campylobacter coli. High-resolution melting (HRM) analysis is emerging as an efficient and robust method for discriminating DNA sequence variants. The objective of this study was to apply HRM analysis to flaA-based genotyping. The initial aim was to identify a suitable flaA fragment. It was found that the PCR primers commonly used to amplify the flaA short variable repeat (SVR) yielded a mixed PCR product unsuitable for HRM analysis. However, a PCR primer set composed of the upstream primer used to amplify the fragment used for flaA restriction fragment length polymorphism (RFLP) analysis and the downstream primer used for flaA SVR amplification generated a very pure PCR product, and this primer set was used for the remainder of the study. Eighty-seven C. jejuni and 15 C. coli isolates were analyzed by flaA HRM and also partial flaA sequencing. There were 47 flaA sequence variants, and all were resolved by HRM analysis. The isolates used had previously also been genotyped using single-nucleotide polymorphisms (SNPs), binary markers, CRISPR HRM, and flaA RFLP. flaA HRM analysis provided resolving power multiplicative to the SNPs, binary markers, and CRISPR HRM and largely concordant with the flaA RFLP. It was concluded that HRM analysis is a promising approach to genotyping based on highly variable genes.
Campylobacter jejuni and Campylobacter coli are the most common causes of human bacterial gastroenteritis in industrialized countries (21). The flagellin-encoding genes flaA and flaB share 95% sequence homology and are arranged in tandem (9, 20). While flaA gene expression appears critical for motility, colonization, and pathogenesis, this is not the case for flaB, which is thought to be a largely nonfunctional reservoir of genetic variation that can increase the diversity of flaA by recombination and so assist the cell in evading host immune responses (1, 7, 8, 28).
The flaA gene is commonly used for typing C. jejuni and C. coli. Two methods have gained wide acceptance: flaA restriction fragment length polymorphism (RFLP) (19) and flaA short variable region (SVR) sequencing (16). The flaA RFLP technique involves PCR amplification of the entire flaA gene followed by RFLP of the PCR product (19). Sequencing the SVR of the flaA gene was developed as a more streamlined and portable alternative to flaA RFLP protocols (16), and sequence variants are compiled at a central website (http://pubmlst.org/campylobacter/flaA/) (10). While both of these methods are very effective, they have several disadvantages: the RFLP approach is multistep, because the PCR product must be cleaved with a restriction enzyme, and the fragments must be subsequently resolved by electrophoresis (33). Also, there are many changes in the sequence that will not alter the sizes of the restriction fragments. SVR sequencing is also multistep; the targeted region is small, which limits resolving power; and DNA sequencing requires expensive equipment in specialized facilities (6).
High-resolution melting (HRM) analysis is an emerging method that has been applied to the interrogation of single-nucleotide polymorphisms (SNPs), hypervariable repeat regions in PCR products, and also the discovery of new SNPs (3, 24, 26, 27, 30). It is based upon the accurate monitoring of the reduction in fluorescence as a PCR product stained with a double-strand-specific fluorescent dye is heated through its melting temperature (Tm). In contrast to traditional melting analysis, the information in HRM analysis is contained in the shape of the melting curve, rather than just the calculated Tm, so HRM analysis may be considered a form of spectroscopy. HRM analysis is single step and closed tube, because the amplification and melting can be run as a single protocol on a real-time PCR device.
Our group has an ongoing interest in the use of HRM to interrogate loci with complex variation (23, 24, 27) and has previously reported an HRM-based method for interrogating the C. jejuni CRISPR locus (24). The purpose of this study was to develop a C. jejuni/C. coli typing method based on HRM analysis of flaA.
MATERIALS AND METHODS
Isolates and DNA extraction.
Eighty-seven C. jejuni and 15 C. coli isolates were used in this study. The C. jejuni isolates have previously been described (17). They were all obtained from chicken farms in South-East Queensland, Australia. The collection contained three groups of 10 C. jejuni isolates, which were termed L1, L2, and L3. The isolates within each group were obtained at the same time and place, and a variety of genotyping methods have shown a clonal relationship between the members of each group (17). The members of each group are therefore regarded as being epidemiologically linked. The flaA RFLP types of L1, L2, and L3 were FT-XXVI, FT-I, and FT-VII, respectively. The remainder of the collection consisted of 26 C. jejuni isolates that were obtained at different times and/or places and were selected on the basis of being FT-I and 46 isolates (31 C. jejuni and 15 C. coli) that were selected on the basis that each possessed a different flaA RFLP type. More detailed descriptions of these isolates are available in the supplemental material.
Genomic DNA was extracted using the DNeasy blood and tissue lysis kit per the manufacturer's instructions (Qiagen, Clifton Hill, Australia).
HRM protocol for flaA and CRISPR interrogation.
All the HRM analyses were performed on a Corbett Rotor-Gene 6000 (Corbett Research, Sydney, Australia). Due to a corporate acquisition, the Corbett Rotor-Gene 6000 instrument is no longer available, but the Qiagen Rotorgene Q with HRM capability is an essentially identical device.
The flaA HRM analysis method developed in the course of this study was as follows: DNA was amplified using primers flaA4f and flaA625RU (16). The 10-μl reaction mixtures were comprised of 5 μl of Platinum SYBR green qPCR SuperMix-UDG (Invitrogen Australia, Mulgrave, Victoria, Australia), 5 pmol of each primer, 1 μl of template, and 3.5 μl of H2O. The cycling conditions were 95°C for 2 min; 40 cycles of 95°C for 30 s, 59°C for 20 s, and 72°C for 45 s; and 72°C for 2 min and 50°C for 2 min. The amplified DNA was then subjected to HRM with 0.05°C increments in temperature ranging from 72°C to 84°C. The HRM curves were normalized using the sections of the raw fluorescence data from 72.6°C to 73.2°C and from 82.8°C to 83.2°C. The reference isolate NCTC11168 was included in each run as a control to monitor interrun variability. The reactions were carried out routinely in duplicate.
CRISPR interrogation was performed as previously described (24). The different melting profiles assigned were called CRISPR types (CTs).
Analysis of HRM data.
Three different strategies were used to estimate whether pairs of HRM curves were derived from the same or different sequences. First, in many cases HRM curves could be discriminated on the basis of obvious differences in curve shape and/or on the basis of Tm, with a difference of 0.2°C regarded as significant. Second, we have previously reported using difference graph analysis, with an amplitude of >5 normalized fluorescence units being indicative that the baseline curve and the “comparator” curve arose from different sequences (24). Finally, we used an approach similar to that described by Andersson et al. (2). This is a development of the difference graph-based method and involves deriving the 3rd and 97th centiles from the mean ± 1.96 standard deviations for the fluorescence at every temperature. This generates centile curves analogous to those used to monitor growth in children. Andersson et al. (2) termed these “confidence limits,” but we now believe that the term “centile curves” is more technically correct. In the current study, the 3rd and 97th centile curves were calculated from at least nine replicates, and in general, the data for the derivative of fluorescence with respect to temperature (dF/dT) rather than those for the normalized fluorescence were used.
Rotor-Gene 6000 software version 1.7.34 or 1.7.87 was used to analyze HRM data within a run, while exported data were analyzed using either Teechart Office 2.0 or Microsoft Excel 2003.
DNA sequencing.
PCR products ranging in amount from 25 to 35 ng with 6.4 pmol of primers were submitted for sequencing to the Australian Genome Research Facility (AGRF), Brisbane, Australia.
RESULTS
Identification of a flaA fragment suitable for HRM analysis.
Defining a flaA fragment suitable for HRM analysis was a compromise between minimizing the size of the fragment in order to simplify the discrimination of alleles and maximizing the size of the fragment so as to maximize the number of alleles and consequent resolving power. An additional factor was the presence of flaB, which is very similar to flaA. A mixed PCR fragment that is derived from both flaA and flaB would be essentially impossible to analyze meaningfully by HRM, so it was regarded as essential to ensure that the primer set was flaA specific.
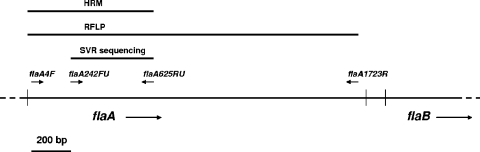
Initial experiments were carried out using the PCR fragment that is commonly used for flaA SVR sequence typing. This was amplified by primers flaA242FU and flaA625RU (Fig. 1). Sequence analysis revealed significant numbers of double peaks. This was probably caused by the presence of the flaB-derived sequence in the PCR product (data not shown). It was concluded that this primer set was unsuitable for flaA HRM analysis.
FIG. 1.
The flaA fragments used in RFLP, SVR sequencing, and HRM-based genotyping. The PCR primers are depicted by the small arrows.
The next primer set to be tested incorporated the upstream primer used for flaA RFLP analysis (flaA4F) and the downstream primer used for SVR sequence analysis (flaA625RU) (Fig. 1). Amplification and sequencing of this fragment from three isolates (one isolate from each of the three epidemiologically linked groups L1, L2, and L3) yielded sequence traces with no double peaks, indicating that there was no significant contamination with flaB-derived sequences. It was concluded that this 620-bp fragment was promising for HRM-based genotyping.
HRM analyses.
HRM analysis of the 620-bp fragment was carried out for all isolates. The analyses were performed prior to sequence determination for all isolates, except the three used for the validation of the 620-bp fragment that was described in the previous section.
The HRM curves were compared on the basis of shape and Tm, and in some instances, difference graphs were used (see Materials and Methods). Based on these analyses, it was estimated that there were likely to be 47 sequence variants. A sample of HRM curves is shown in Fig. 2, and all curves are provided in the supplemental material. The different melting curves were named flaA HRM type (FHT)-1 to FHT-47.
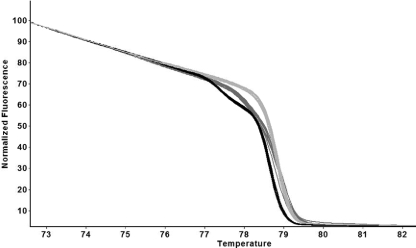
FIG. 2.
Examples of normalized HRM curves and their reproducibility. These are derived from 10 isolates of each of the three epidemiologically linked groups L1 (light gray), L2 (black), L3 and (dark gray). One DNA preparation from each isolate was assayed in duplicate. The “replicate view” option was used, so each curve is the mean of the data from the duplicates.
After sequence analysis was carried out, it was found that the HRM analyses had been completely successful in resolving all the sequence variants, and there were no occasions when HRM curves derived from identical sequences had been classified as different. In other words, the HRM analysis was 100% in accord with the sequence-based gold standard. The sequences also confirmed that the flaA HRM amplification primers generated product not contaminated with flaB or any other sequence. The 10 novel alleles have been deposited in the flaA SVR sequence database under numbers 1066 to 1068, 1093 to 1098, and 1160. All sequences have been deposited in GenBank, and the accession numbers are in the supplemental material file that contains the HRM data.
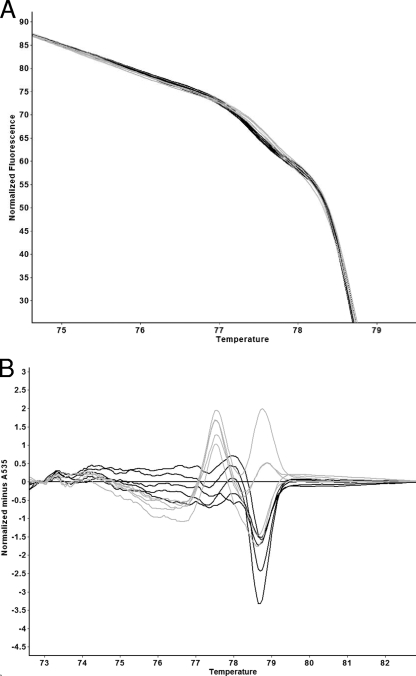
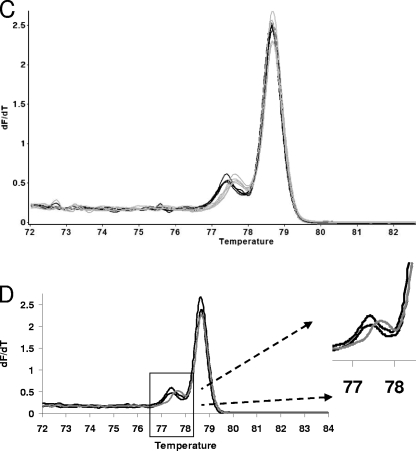
In order to increase the sophistication of our HRM data analysis methods, the FHT-1 and FHT-2 HRM curves were further analyzed. The data for this were obtained by analyzing in duplicate 10 isolates with these genotypes. FHT-1 and FHT-2 were chosen because the HRM curves are very similar. The results are shown in Fig. 3. In Fig. 3A, it can be seen that there is a visible, albeit subtle, difference between the normalized FHT-1 and FHT-2 curves. Figure 3B shows that difference graph analysis normalized using an FHT-1 HRM curve does appear to divide the FHT-1 and FHT-2 curves into two groups. However, there is some variation within the two genotypes. Also, the FHT-2 difference graphs do not reach ±5. An amplitude of ±5 is one of the criteria previously used to classify a difference graph as being derived from a sequence that is different from the one that defines the baseline (24). In other words, the difference graph was not clearly indicating sequence variation. However, Fig. 3C shows that the dF/dT curves for both FHTs consist of two peaks and that there appears to be a consistent difference between the FHT-1 and FHT-2 curves that is manifested as the position of the small peak. Figure 3D depicts the 3rd and 97th centile curves (see Materials and Methods) for the dF/dT of the FHT-1 HRM curve. These were calculated from the 10 isolates that belong to the epidemiologically linked group L2. A typical FHT-2 dF/dT curve is shown, and as expected, a substantial portion of the small peak is outside the 3rd and 97th centile curves. Interestingly, the amplitude of the FHT-2 large peak is also outside the FHT-1 3rd and 97th centile curves, suggesting a subtle but consistent difference in the slope of the normalized curves at temperatures close to the Tm.
FIG. 3.
Detailed comparison of the FHT-1 and FHT-2 HRM curves. (A) Magnified portion of replicate FHT-1 (black) and FHT-2 (gray) normalized HRM curves. (B) Difference graph depicting FHT-1 (black) and FHT-2 (gray). Data from a randomly chosen FHT-1 isolate were used as the x axis. (C) Comparison of the dF/dT curves for replicates of FHT-1 (black) and FHT-2 (gray). (D) The 95% 3rd and 97th centile curves for the dF/dT curve for FHT-1 (black), and a typical FHT-2 curve (gray).
Correlation of flaA HRM with flaA SVR sequence analysis and flaA RFLP.
The fragment of the flaA gene interrogated by HRM includes the SVR and 238 bp upstream of the SVR (Fig. 1). HRM, therefore, has the potential to split single SVR sequence types. We found one such example in our data set; two isolates with the same flaA SVR allele (allele 117) were resolved by HRM into different HRM types (FHT-10 and FHT-44), which are discriminated from each other by two SNPs upstream of the SVR.
The flaA RFLP method involves the amplification and restriction profiling of the entire flaA gene (1.7 kb) (Fig. 1). The flaA RFLP fragment is thus larger than the HRM fragment and will contain more polymorphisms. However, not all polymorphisms may occur within restriction sites, so an identical restriction profile may be observed across sequence variants. Therefore, the question as to whether flaA RFLP or flaA HRM will provide the higher resolution is interesting. Our results showed that FT-I, comprising 36 isolates, was resolved into five HRM types (FHT-1, 2, 3, 13, and 47). However, there were six instances (FHT-3, 5, 13, 21, 23, and 42) in which HRM was unable to resolve the different FTs (FT-I and XXV, FT-XLVI and XV, FT-I and IV, FT-LXI and XI, FT-VIII and LIX, and FT-LV and LXII).
The resolving power of flaA HRM analysis is multiplicative to the resolving power of CRISPR HRM analysis.
Our research group has previously reported the application of HRM to the interrogation of the highly variable C. jejuni CRISPR locus (24). It would be expected that the combinatorial resolving power of CRISPR HRM and fla HRM would be greater than that of either method on its own, particularly given the high recombination rate of C. jejuni (5, 28, 34). Isolates were selected for this study on the basis of flaA RFLP type and included a substantial collection of epidemiologically unlinked flaA RFLP type I isolates and another set that were all chosen to be different, with respect to flaA RFLP. This precluded direct comparisons of the resolving powers of the two HRM methods alone or in combination. However, we were able to observe that FHT-2, which represents the majority of the epidemiologically unlinked FT-I isolates, was resolved into nine CRISPR HRM types, thus demonstrating the potential of CRISPR HRM to subdivide FHTs.
The C. jejuni isolates included in this study have previously been subjected to typing based on resolution-optimized SNPs derived from the C. jejuni/C. coli multilocus sequence typing (MLST) database and resolution-optimized binary markers derived from microarray data (17) The results of all the typing methods from this and previous studies have been collated and are supplied in the supplemental material. As expected, none of the typing methods divide the isolates into exactly the same groups, so the resolving powers of all are multiplicative, with the qualification that in this study, SVR sequencing did not provide discrimination within any flaA HRM types.
DISCUSSION
The flagellin gene has been a well-accepted marker for Campylobacter genotyping for over 2 decades (4, 16, 18, 19, 22, 25, 32). In this study we have developed a genotyping method based on flaA interrogation using HRM analysis. The procedure is single step and closed tube, takes approximately 3 h to perform, and costs approximately $1.00.
One unusual aspect of this study is the large size of the fragment (620 bp) that we were able to successfully analyze using HRM. HRM is usually applied to the analysis of single SNPs in much smaller fragments (13-15, 29). The ability of HRM to correctly resolve the 47 flaA 620-bp fragment alleles included in this study was somewhat unexpected. It would not be appropriate to extrapolate from this to conclude that HRM can detect any change in this sequence. However, it does suggest that this method could effectively replace either flaA RFLP or SVR sequencing as a first-pass technique for testing hypotheses regarding epidemiological linkage.
It is desirable that any bacterial typing method yield data that is easily portable between laboratories. The successful comparison of HRM data between runs and between different devices has recently been reported (27, 31). Our experience with the Corbett Rotor-Gene 6000 is that the relative temperature calibration is extremely accurate but that the absolute calibration can vary by up to 0.5°C between different instruments. This means that the shape of the HRM curve is completely consistent from device to device, but temperature normalization may sometimes be necessary. This could easily be achieved by including a control sequence with a known Tm in any batch of HRM analyses. The portability of the HRM data raises the possibility of online libraries of HRM curves, which ideally would include centile curves or analogous descriptors of variation, and applications for carrying out temperature normalization and searches for similar curves.
In this study, the FHT-1 and FHT-2 curves were used as a model system to further explore HRM curve comparison strategies. In the case of FHT-1 and FHT-2, the presence of a second peak assisted greatly in discrimination. We were concerned that this peak may have arisen from a secondary PCR product, but this was never visible on any electrophoresis gel (data not shown), and the relative amplitudes of the large and small peaks were highly consistent. This suggests that the presence of two peaks is a function of different melting domains within the same sequence. In a sequence of 620 bp, this is plausible. For these curves, inspection of the dF/dT curves was the most effective means of discrimination, and a consistent difference between FHT-1 and FHT-2 was confirmed by calculating the FHT-1 dF/dT 3rd and 97th centile curves.
The isolates included in this study have been subjected to typing based on variation in resolution-optimized MLST database-derived SNPs and resolution-optimized binary markers (17) as well as to the methods addressed in the present study. The SNP-, binary marker-, and HRM-based methods can all be performed on the real-time PCR platform, and they all yield digitizable results (17, 23-25), thus providing a readily accessible choice of methods or combinations of methods that can be adapted to different tasks and questions. It has previously been shown that the resolution-optimized SNP-defined genotypes correlate well with the MLST-defined population structure; i.e., there was a strong tendency for isolates with identical SNP types to have identical or very similar MLSTs (25). It was also found that addition of flaA SVR sequencing resulted in the correlation between genotype and population structure becoming essentially perfect (25). The present study suggests that SVR sequencing could be replaced with flaA HRM analysis and that a combination of the SNP-based typing and flaA HRM analysis, both of which can be carried out on a real-time PCR machine, would provide both an indication of the evolutionary position of the genome backbone and good resolution. CRISPR HRM and binary marker-based typing would be expected to add yet more resolving power. The strategy of using selected SNPs in combination with markers that may evolve more rapidly is reminiscent of the phylogenetic hierarchical assays using nucleic acids strategy articulated by Keim and coworkers (11, 12), who have applied it to the large-scale reconstruction of anthrax epidemiology. The performance of these real-time PCR-based typing methods will not equal the performance of whole-genome sequence analysis, high-density array-based methods, or the interrogation of large numbers of SNPs. However, real-time PCR is ideal for immediate closed-tube analysis of samples at locations away from large laboratories.
Supplementary Material
Acknowledgments
This study was funded by the Australian Rural Industries Research and Development Corporation and Queensland University of Technology (QUT). S.M.-P. is in receipt of a QUT capacity-building postgraduate scholarship.
We thank the curators and data contributors for the Campylobacter flaA database (http://pubmlst.org/campylobacter/flaA/) hosted at the University of Oxford and funded by DEFRA grant 0Z0615.
Footnotes
Published ahead of print on 20 November 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alm, R. A., P. Guerry, and T. J. Trust. 1993. Significance of duplicated flagellin genes in Campylobacter. J. Mol. Biol. 230:359-363. [DOI] [PubMed] [Google Scholar]
- 2.Andersson, P., T. Harris, S. Y. Tong, and P. M. Giffard. 2009. Analysis of blaSHV codon 238 and 240 allele mixtures using Sybr green high-resolution melting analysis. Antimicrob. Agents Chemother. 53:2679-2683. doi: 10.1128/AAC.00063-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erali, M., K. V. Voelkerding, and C. T. Wittwer. 2008. High resolution melting applications for clinical laboratory medicine. Exp. Mol. Pathol. 85:50-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ertas, H. B., B. Cetinkaya, A. Muz, and H. Ongor. 2004. Genotyping of broiler-originated Campylobacter jejuni and Campylobacter coli isolates using fla typing and random amplified polymorphic DNA methods. Int. J. Food Microbiol. 94:203-209. [DOI] [PubMed] [Google Scholar]
- 5.Fearnhead, P., N. G. Smith, M. Barrigas, A. Fox, and N. French. 2005. Analysis of recombination in Campylobacter jejuni from MLST population data. J. Mol. Evol. 61:333-340. [DOI] [PubMed] [Google Scholar]
- 6.Fitzgerald, C., L. O. Helsel, M. A. Nicholson, S. J. Olsen, D. L. Swerdlow, R. Flahart, J. Sexton, and P. I. Fields. 2001. Evaluation of methods for subtyping Campylobacter jejuni during an outbreak involving a food handler. J. Clin. Microbiol. 39:2386-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerry, P. 2007. Campylobacter flagella: not just for motility. Trends Microbiol. 15:456-461. [DOI] [PubMed] [Google Scholar]
- 8.Guerry, P. 2007. The role of flagella in Campylobacter virulence, p. 197-207. In K. A. Brogden, F. C. Minion, N. Cornick, T. B. Stanton, Q. Zhang, L. K. Nolan, and M. J. Wannemuehler (ed.), Virulence mechanisms of bacterial pathogens, 4th ed. ASM Press, Washington, DC.
- 9.Guerry, P., S. M. Logan, S. Thornton, and T. J. Trust. 1990. Genomic organization and expression of Campylobacter flagellin genes. J. Bacteriol. 172:1853-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jolley, K. A., and M. C. Maiden. 2006. AgdbNet—antigen sequence database software for bacterial typing. BMC Bioinformatics 7:314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keim, P., T. Pearson, and R. Okinaka. 2008. Microbial forensics: DNA fingerprinting of Bacillus anthracis (anthrax). Anal. Chem. 80:4791-4799. [DOI] [PubMed] [Google Scholar]
- 12.Keim, P., M. N. Van Ert, T. Pearson, A. J. Vogler, L. Y. Huynh, and D. M. Wagner. 2004. Anthrax molecular epidemiology and forensics: using the appropriate marker for different evolutionary scales. Infect. Genet. Evol. 4:205-213. [DOI] [PubMed] [Google Scholar]
- 13.Krypuy, M., G. M. Newnham, D. M. Thomas, M. Conron, and A. Dobrovic. 2006. High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-small cell lung cancer. BMC Cancer 6:295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehmensiek, A., M. W. Sutherland, and R. B. McNamara. 2008. The use of high resolution melting (HRM) to map single nucleotide polymorphism markers linked to a covered smut resistance gene in barley. Theor. Appl. Genet. 117:721-728. [DOI] [PubMed] [Google Scholar]
- 15.Maltese, P., E. Canestrari, L. Palma, A. Ruzzo, F. Corini, M. Menotta, F. Andreoni, A. Latiano, V. Annese, and M. Magnani. 2009. High resolution melting (HRM) analysis for the detection of ER22/23EK, BclI, and N363S polymorphisms of the glucocorticoid receptor gene. J. Steroid Biochem. Mol. Biol. 113:269-274. [DOI] [PubMed] [Google Scholar]
- 16.Meinersmann, R. J., L. O. Helsel, P. I. Fields, and K. L. Hiett. 1997. Discrimination of Campylobacter jejuni isolates by fla gene sequencing. J. Clin. Microbiol. 35:2810-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merchant-Patel, S., P. J. Blackall, J. Templeton, E. P. Price, J. K. Miflin, F. Huygens, and P. M. Giffard. 2008. Characterisation of chicken Campylobacter jejuni isolates using resolution optimised single nucleotide polymorphisms and binary gene markers. Int. J. Food Microbiol. 128:304-308. [DOI] [PubMed] [Google Scholar]
- 18.Miflin, J. K., J. M. Templeton, and P. J. Blackall. 2007. Antibiotic resistance in Campylobacter jejuni and Campylobacter coli isolated from poultry in the South-East Queensland region. J. Antimicrob. Chemother. 59:775-778. [DOI] [PubMed] [Google Scholar]
- 19.Nachamkin, I., K. Bohachick, and C. M. Patton. 1993. Flagellin gene typing of Campylobacter jejuni by restriction fragment length polymorphism analysis. J. Clin. Microbiol. 31:1531-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nuijten, P. J., F. J. van Asten, W. Gaastra, and B. A. van der Zeijst. 1990. Structural and functional analysis of two Campylobacter jejuni flagellin genes. J. Biol. Chem. 265:17798-17804. [PubMed] [Google Scholar]
- 21.Park, S. F. 2002. The physiology of Campylobacter species and its relevance to their role as foodborne pathogens. Int. J. Food Microbiol. 74:177-188. [DOI] [PubMed] [Google Scholar]
- 22.Pittenger, L. G., M. D. Englen, C. T. Parker, J. G. Frye, B. Quinones, S. T. Horn, I. Son, P. J. Fedorka-Cray, and M. A. Harrison. 2009. Genotyping Campylobacter jejuni by comparative genome indexing: an evaluation with pulsed-field gel electrophoresis and flaA SVR sequencing. Foodborne Pathog. Dis. 6:337-649. [DOI] [PubMed] [Google Scholar]
- 23.Price, E. P., F. Huygens, and P. M. Giffard. 2006. Fingerprinting of Campylobacter jejuni by using resolution-optimized binary gene targets derived from comparative genome hybridization studies. Appl. Environ. Microbiol. 72:7793-7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price, E. P., H. Smith, F. Huygens, and P. M. Giffard. 2007. High-resolution DNA melt curve analysis of the clustered, regularly interspaced short-palindromic-repeat locus of Campylobacter jejuni. Appl. Environ. Microbiol. 73:3431-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price, E. P., V. Thiruvenkataswamy, L. Mickan, L. Unicomb, R. E. Rios, F. Huygens, and P. M. Giffard. 2006. Genotyping of Campylobacter jejuni using seven single-nucleotide polymorphisms in combination with flaA short variable region sequencing. J. Med. Microbiol. 55:1061-1070. [DOI] [PubMed] [Google Scholar]
- 26.Reed, G. H., J. O. Kent, and C. T. Wittwer. 2007. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics 8:597-608. [DOI] [PubMed] [Google Scholar]
- 27.Stephens, A. J., J. Inman-Bamber, P. M. Giffard, and F. Huygens. 2008. High-resolution melting analysis of the spa repeat region of Staphylococcus aureus. Clin. Chem. 54:432-436. [DOI] [PubMed] [Google Scholar]
- 28.Suerbaum, S., M. Lohrengel, A. Sonnevend, F. Ruberg, and M. Kist. 2001. Allelic diversity and recombination in Campylobacter jejuni. J. Bacteriol. 183:2553-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szvetko, A. L., A. Jones, J. Mackenzie, L. Tajouri, P. A. Csurhes, J. M. Greer, M. P. Pender, and L. R. Griffiths. 2009. An investigation of the C77G and C772T variations within the human protein tyrosine phosphatase receptor type C gene for association with multiple sclerosis in an Australian population. Brain Res. 1255:148-152. [DOI] [PubMed] [Google Scholar]
- 30.Taylor, C. F. 2009. Mutation scanning using high-resolution melting. Biochem. Soc. Trans. 37:433-437. [DOI] [PubMed] [Google Scholar]
- 31.Tong, S. Y., R. A. Lilliebridge, D. C. Holt, M. I. McDonald, B. J. Currie, and P. M. Giffard. 2009. High-resolution melting analysis of the spa locus reveals significant diversity within sequence type 93 methicillin-resistant Staphylococcus aureus from northern Australia. Clin. Microbiol. Infect. [Epub ahead of print.] doi: 10.1111/j.1469-0691.2009.02732.x. [DOI] [PubMed]
- 32.Unicomb, L. E., L. C. O'Reilly, M. D. Kirk, R. J. Stafford, H. V. Smith, N. G. Becker, M. S. Patel, and G. L. Gilbert. 2008. Risk factors for infection with Campylobacter jejuni flaA genotypes. Epidemiol. Infect. 136:1480-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wassenaar, T. M., and D. G. Newell. 2000. Genotyping of Campylobacter spp. Appl. Environ. Microbiol. 66:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson, D. J., E. Gabriel, A. J. Leatherbarrow, J. Cheesbrough, S. Gee, E. Bolton, A. Fox, C. A. Hart, P. J. Diggle, and P. Fearnhead. 2009. Rapid evolution and the importance of recombination to the gastroenteric pathogen Campylobacter jejuni. Mol. Biol. Evol. 26:385-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.