Abstract
As shown by R5 antibody-based sandwich and competitive enzyme-linked immunosorbent assay (ELISA), selected sourdough lactobacilli, in combination with fungal proteases, hydrolyzed gluten (72 h at 37°C) of various cultivars of Triticum turgidum L. var. durum to less than 20 ppm. Complementary electrophoretic, chromatography, and mass spectrometry techniques were used to characterize the gluten and epitope hydrolysis. Nine peptidases were partially purified from the pooled cytoplasmic extract of the sourdough lactobacilli and used to hydrolyze the 33-mer epitope, the most immunogenic peptide generated during digestion of Triticum species. At least three peptidases (general aminopeptidase type N [PepN], X-prolyl dipeptidyl aminopeptidase [PepX], and endopeptidase PepO) were necessary to detoxify the 33-mer without generation of related immunogenic epitopes. After 14 h of incubation, the combination of all or at least six different peptidases totally hydrolyzed the 33-mer (200 mM) into free amino acids. The same results were found for other immunogenic epitopes, such as fragments 57-68 of α9-gliadin, 62-75 of A-gliadin, and 134-153 of γ-gliadin. When peptidases were used for fermentation of durum wheat semolina, they caused the hydrolysis of gluten to ca. 2 ppm. The in vivo digestion was simulated, and proteins/peptides extracted from pepsin-trypsin (PT) digestion of durum wheat semolina fermented with selected sourdough lactobacilli induced the expression of gamma interferon and interleukin 2 at levels comparable to those of the negative control. Durum wheat semolina fermented with sourdough lactobacilli was freeze-dried and used for making Italian-type pasta. The scores for cooking and sensory properties for this pasta were higher that those of conventional gluten-free pasta.
Celiac disease (CD) is an inflammatory disorder of the small intestine that affects genetically predisposed individuals when they ingest gluten from any Triticum species and similar proteins of barley and rye, and their crossbred varieties. Epidemiological studies in Europe and the United States indicate that CD is increasing; the prevalence of CD is approximately 1% of the general population (29). Pathogenesis of CD involves interactions between genetic, immunological, and environmental/technological factors. Human leukocyte antigen (HLA) DQ2 (or DQ8) molecules of antigen-presenting cells bind and submit gluten peptides to the lamina propria of CD4+ T cells. The latter trigger the T helper 1 (Th1)-based immune response, with the consequent synthesis of gamma interferon (IFN-γ). Among the main dietary proteins, gluten is unique in that it contains ca. 15% proline and 35% glutamine residues (40). The high concentration of glutamine and, especially, proline prevents the complete degradation by human gastric and pancreatic enzymes and results in the build up of oligopeptides in the small intestine that are resistant to further proteolysis and toxic to genetically predisposed CD patients. To date, in silico analyses revealed the presence of more than 60 immunogenic oligopeptides from gluten of the Triticum species (37). The length of these oligopeptides may vary from 7/9 to 91 amino acid residues, and the common feature is represented by the high number of glutamine and proline residues (Immune Epitope Database [IEDB]). The most important example of such oligopeptides is the 33-mer epitope. The 33-mer epitope was identified as one of the main stimulators of the inflammatory response to gluten. This epitope was shown to be a potent inducer of gut-derived human T-cell lines in all 14 CD patients (35).
A strict, lifelong gluten-free diet is the only accepted treatment for CD. Nevertheless, complete exclusion of dietary gluten is difficult due to the ubiquitous nature of this protein, cross-contamination of foods, inadequate food labeling regulations, and social constraints. Furthermore, gluten-free food provides inadequate supply of fibers, minerals, and vitamins and excess calories in the diet and exhibits poor sensory properties (9, 24). Alternative therapeutic options included the proposed oral supplementation with microbial oligopeptidases (8, 31, 42). Recently, a new combination of enzymes, consisting of glutamine-specific endoprotease (EP-B2 from barley) and prolyl-endopeptidase (SC PEP from Sphingomonas capsulata), was investigated for the capacity to digest gluten under gastrointestinal conditions (22). Another prolyl-endoprotease from Aspergillus niger (ANPEP) was selected for its stability at pH 2 and resistance to pepsin digestion (27, 39). Unfortunately, to date, the medium- and long-term safety of oral administration of proteases for gluten degradation has not yet been demonstrated in CD patients.
Recently, it was shown that selected sourdough lactobacilli (12, 16, 17), in combination with fungal proteases, decreased the residual concentration of gluten (Triticum aestivum flour) below 10 ppm during food fermentation (32). Despite the extensive research carried out on T. aestivum, less attention has been paid to durum wheat flour from Triticum turgidum L. var. durum (18). Durum wheat is an important food crop of the Mediterranean area, not only because of the large acreage but also for its importance in the human diet (20). Durum wheat is largely used for making pasta, especially in the European and North American countries. Bread, burghul, couscous and frekeh are also manufactured with durum wheat in several countries. Currently, dietary recommendations from the USDA-HHS (U.S. Department of Agriculture-Health and Human Services) promote the consumption of pasta as the optimal source of complex carbohydrates and carotenoids (30). Previously, it was shown that making pasta by T. durum wheat fermented by selected sourdough lactobacilli markedly decreased the toxicity of prolamin epitopes. Nevertheless, the concentration of gluten still remained above 1,000 ppm (18).
This paper aimed at exploiting the potential of the new selected pool of sourdough lactobacilli (32) for the manufacture of gluten-free pasta (GFP) made of durum wheat semolina (DWS). The mechanism of hydrolysis of various Pro-rich immunogenic epitopes (containing high level of proline residues) by peptidases of sourdough lactobacilli was highlighted. Hydrolysis of gliadins and glutenins was determined by complementary electrophoresis, chromatography, and immunology analyses. Assays of cytokines IFN-γ and interleukin 2 (IL-2) on duodenal biopsy specimens from CD patients were used to determine the toxicity of hydrolyzed gluten.
MATERIALS AND METHODS
Microorganisms and enzymes.
Lactobacillus sanfranciscensis 7A, LS3, LS10, LS19, LS23, LS38, and LS47, Lactobacillus alimentarius 15M, Lactobacillus brevis 14G, and Lactobacillus hilgardii 51B were selected on the basis of their peptidase activities, with particular reference to degradation of Pro-rich peptides (12, 16). Previous studies (17, 32) showed that all the above strains were indispensable to obtain degradation of gluten to ca. 10 ppm. Strains were propagated for 24 h at 30°C in modified MRS broth (mMRS) (Oxoid, Basingstoke, Hampshire, United Kingdom), with the addition of fresh yeast extract (5%, vol/vol) and 28 mM maltose at a final pH of 5.6. When used for fermentation of durum wheat semolina, lactobacilli were cultivated until the late exponential phase of growth was reached (ca. 12 h). Fungal proteases from Aspergillus oryzae (500,000 hemoglobin units on the tyrosine basis/g) and Aspergillus niger (3,000 spectrophotometric acid protease units/g), routinely used as improvers in the bakery industry, were purchased from BIO-CAT Inc. (Troy, VA).
Durum wheat cultivars and milling.
Eight Italian durum wheat cultivars (Arcangelo, Ciccio, Claudio, Colosseo, Duilio, Gargano, Simeto, and Svevo) were used. Samples were supplied by Divella S.p.A. (Rutigliano, Bari, Italy). Divella is the second largest producer of pasta in Italy. Cultivars were grown (year 2007) in the same location of the Apulia region (southern Italy) under the same agroecological conditions. The cultivar Ocotillo from Arizona (United States) was also used. Under industrial conditions (Divella S.p.A.), the grain was hand cleaned, conditioned to 15% moisture, and milled into DWS. The yield was ca. 55%. The particle size of DWS was determined according to Italian legislation methods (D.P.R. 09.02.2001). Protein analysis was carried out by the Kjeldhal method and expressed using the conversion factor N × 5.7 (2). All determinations were carried out on three different samples of DWS, and the values were averaged.
Fermentation of durum wheat semolina.
Eighty grams of DWS and 320 g of tap water, containing ca. 5 × 108 CFU/g (final cell density in the dough) of L. sanfranciscensis 7A, L. alimentarius 15M, L. brevis 14G, and L. hilgardii 51B and ca. 1 × 109 CFU/g of L. sanfranciscensis LS3, LS10, LS19, LS23, LS38, and LS47 and 200 ppm of both fungal proteases were used to produce the fermented durum wheat semolina 1 (FDWS1). Fermented DWS2 (FDWS2) was the same as the FDWS1 formula with the addition of 400 ppm of both fungal proteases. Fermentation was conducted for 48 or 72 h (DSW1 and DSW2, respectively) at 37°C while the mixture was stirred (ca. 200 rpm). After fermentation, FDWS2 was subjected to spray drying to remove water, and the resulting hydrolyzed flour was further milled and used.
Extraction of proteins from durum what semolina and 2-DE.
Before and after fermentation, DWS proteins were selectively extracted according to the method of Weiss et al. (46). For immunological analysis, protein extraction was carried out directly with 60% ethanol to include both proteins and peptides that were hydro-alcoholically soluble (39). An aliquot of dough (12.75 g) was diluted with 30 ml of ethanol (75%, vol/vol), stirred at 25°C for 2 h, and centrifuged at 20,000 × g for 20 min. The supernatant contained albumins, globulins, and alcohol-soluble polypeptides. The protein concentration was determined by the Bradford method (6). Two-dimensional electrophoresis (2-DE) of ca. 30 μg of proteins extracted from various fractions was carried out with the Immobiline-polyacrylamide system (16). Gels were silver stained, and spot intensities were normalized by the method of Bini et al. (5). Four gels from independent fermentations were analyzed.
Reversed-phase high-performance liquid chromatography (RP-HPLC) and amino acid analyses.
Aliquots of 1 ml of the 60% ethanol extracts were mixed with 0.05% (vol/vol) trifluoroacetic acid (TFA) and centrifuged at 10,000 × g for 10 min. The supernatant was filtered through a 0.22-μm pore size filter and used for HPLC analysis with an ÄKTA purifier system (GE Healthcare) equipped with a UV detector operating at 214 nm using a reverse-phase C18 XTerra column (Waters, Milford, MA). Total and individual free amino acids (FAA) were analyzed using a Biochrom 30 series amino acid analyzer (Biochrom Ltd., Cambridge Science Park, England) with a cation-exchange column (20- by 0.46-cm inner diameter) by the method of De Angelis et al. (12).
Immunological analysis.
Immunological analysis was carried out by using R5 antibody-based sandwich and competitive ELISA (R5-ELISA). The R5-ELISA (43) was carried out with the Transia plate detection kit according to the instructions of the manufacturer (Diffchamb, Västra Frölunda, Sweden).
Preparation of the cell cytoplasmic extract.
To assay the cytoplasm peptidase activities, cultures of each strain from the late exponential phase of growth (ca. 3 × 109 CFU/g) were used. This cell density corresponded to that found in sourdough during fermentation (17). Aliquots (0.3 g [dry weight]) of washed cell pellets were resuspended in 50 mM Tris-HCl (pH 7.0), incubated at 30°C for 30 min, and centrifuged at 13,000 × g for 10 min to remove loosely associated cell surface enzymes. The cytoplasmic extract was prepared by treatment with lysozyme in 50 mM Tris-HCl (pH 7.5) buffer containing 24% sucrose (10) at 37°C for 60 min while the mixture was stirred (ca. 160 rpm). Spheroplasts were resuspended in isotonic buffer and sonicated for 40 s at 16 A/s (Sony Prep model 150; Sanyo, United Kingdom). The cytoplasmic extract was concentrated 10-fold by freeze-drying (Edwards MOD E1PTB; Edwards, Milan, Italy), resuspended in 5 mM Tris-HCl (pH 7.0), and dialyzed for 24 h at 4°C.
Enzyme assays.
The cytoplasmic extracts of the 10 lactobacilli were pooled and assayed using synthetic substrates. All of the synthetic substrates used were from Sigma Chemical Co (Milan, Italy). General aminopeptidase type N (EC 3.4.11.11; PepN), proline iminopeptidase (EC 3.4.11.9; PepI), X-prolyl dipeptidyl aminopeptidase (EC 3.4.14.5; PepX), endopeptidase (EC 3.4.23; PepO), and prolyl endopeptidyl peptidase (EC 3.4.21.26; PEP) activities were determined by the methods of Gobbetti et al. (23) and Rizzello et al. (32), using Leu-p-nitroanilides (p-NA), Pro-p-NA, Gly-Pro-p-NA, and Gly-Pro-Ala, Z-Gly-Gly-Leu-p-NA, and Z-Gly-Pro-NH-trifluoromethylcoumarin (AMC) substrates, respectively. The assay mixture contained 900 μl of 2.0 mM substrate in 0.05 M potassium phosphate buffer, pH 7.0, and 100 μl of cytoplasmic extract. The mixture was incubated at 30°C for 1 h, and the absorbance was measured at 410 nm. The data were compared to standard curves set up by using p-nitroaniline (23). One unit of activity was defined as the amount of enzyme required to liberate 1 μmol of p-nitroaniline min−1 under the assay conditions. Peptidase activity on Z-Gly-Pro-AMC was determined by measuring the fluorescence at excitation and emission wavelengths of 400 and 505 nm, respectively. A unit of enzyme activity on Z-Gly-Pro-AMC substrate was the amount of enzyme that produced an increase in fluorescence of 0.1 min−1.
Tripeptidase (EC 3.4.11.4; PepT), dipeptidase (EC 3.4.13.11; PepV), prolidase (EC 3.4.13.9; PepQ), and prolinase (EC 3.4.13.8; PepR) activities were determined using Leu-Leu-Leu, Leu-Leu, Val-Pro, and Pro-Gly substrates, respectively. Activities on tri- and dipeptides were determined by the Cd-ninhydrin method (23). The assay conditions used were the same as those for p-nitroanilide substrates. One unit of activity was defined as the amount of enzyme required to liberate 1 μM amino acid per min under the assay conditions. The data obtained were compared to standard curves set up by using leucine (23).
Partial purification of peptidases.
After the pooled cytoplasmic extracts had been freeze-dried, they were resuspended in 0.05 M potassium phosphate buffer, pH 6.0, and applied for gel filtration into an FPLC (fast protein liquid chromatography) Superose 12 HR 10/30 column (Amersham Pharmacia Biotech, Milan, Italy). Elution with 0.05 M potassium phosphate buffer (pH 6.0) containing 0.15 M NaCl was at a flow rate of 18 ml/h. Active fractions were pooled, dialyzed, concentrated 10 times by freeze-drying, and subjected to further purification on an FPLC Mono-Q HR 5/5 column (Amersham Pharmacia Biotech). Elution with a linear NaCl gradient from 0 to 0.5 M in the same buffer was at a flow rate of 24 ml/h. Active fractions showing more than one peptidase activity were further purified using an FPLC Resource HIC column (Amersham Pharmacia Biotech). Proteins were eluted with a linear (NH4)2SO4 gradient (1.7 to 0 M) in 0.05 M potassium phosphate buffer, pH 7.5, at a flow rate of 60 ml/h. After purification, the concentration of proteins of each active fraction was estimated by the Bradford method (6).
Hydrolysis of Pro-rich synthetic epitopes.
Immunogenic epitopes corresponding to fragments 57-68 (Q-L-Q-P-F-P-Q-P-Q-L-P-Y) of α9-gliadin (3), 62-75 (P-Q-P-Q-L-P-Y-P-Q-P-Q-S-F-P) of A-gliadin (37), 134-153 (Q-Q-L-P-Q-P-Q-Q-P-Q-Q-S-F-P-Q-Q-Q-R-P-F) of γ-gliadin (1), and 57-89 (L-Q-L-Q-P-F-P-Q-P-Q-L-P-Y-P-Q-P-Q-L-P-Y-P-Q-P-Q-L-P-Y-P-Q-P-Q-P-F) (33-mer) of α2-gliadin (35) were chemically synthesized by PolyPeptide Laboratories (Strasbourg, France) and used in this study. The hydrolysis of peptides was carried out using the partially purified peptidases (alone or in combination) which derived from the initial cell density of ca. 3 × 109 CFU/g (protein concentration of ca. 1 μg/ml). The mixture, containing 100 μl of partially purified enzyme(s), 200 mM synthetic peptide, and 0.05% (wt/vol) NaN3 in 1 ml of 50 mM phosphate buffer (pH 7.5) was incubated for 24 h at 37°C while being stirred (150 rpm). Hydrolysis of peptides was monitored searching for liberated peptides and FAA through HPLC analysis and by using an amino acid analyzer, respectively. Multidimensional liquid chromatography (MDLC) coupled with electrospray ionization (ESI)-ion trap mass spectrometry (MS) was used to complete or confirm the analysis. The HPLC apparatus consisted of an Ettan MDLC (GE Healthcare, Milan, Italy) equipped with a Zorbax 300 SD C18 precolumn (5 by 0.3 mm) and a Thermo Electron BioBasic-8 column (150 by 0.18 mm). The MDLC was connected to a Finningan LCQ Deca XP Max ion trap mass spectrometer (ThermoElectron Co., San Jose, CA) through the nano-ESI interface. Aliquots of 10 μl of each sample were injected. HPLC separation was carried out at a flow rate of 75 μl/min by using a gradient elution with water (eluent A) and 84% acetonitrile (eluent B), both containing 0.1% (vol/vol) formic acid. The following program was used: 0% eluent B for 30 min; 0 to 100% (vol/vol) eluent B in 100 min, isocratic elution with 100% eluent B for 100 min, return to 0% eluent B in 5 min, and column reconditioning for 30 min. The flow rate at the nano-ESI source was 2.5 μl/min. The LCQ spectrometer, completely controlled by the Xcalibur software (ThermoElectron), operated in the positive ion mode; MS chromatograms in the total ion current (m/z range, 50 to 2,000) and selected ion monitoring modes were recorded for each sample.
Gluten digestion using partially purified peptidases.
The following reaction mixture was used: 80 g of DWS from Triticum turgidum L. var. durum cv. Arcangelo and 320 g of tap water, containing partially purified peptidases (protein concentration of ca. 1 μg/ml) and 400 ppm of both fungal proteases. To allow the hydrolysis of durum wheat gluten, the dough was fermented for 72 h at 37°C while being stirred (ca. 200 rpm).
Pepsin-trypsin (PT) digestion.
Sixty percent ethanol-soluble polypeptides and urea-dithiothreitol-soluble glutenins extracted from Triticum turgidum L. var. durum cv. Arcangelo DWS and related FDWS2 were subjected to sequential PT hydrolysis to simulate the in vivo digestion (11). One hundred grams of each freeze-dried protein fraction was digested in 1 liter of 0.2 N HCl (pH 1.8) with 2 g of purified porcine gastric mucosa pepsin (2,500 to 3,500 units/mg; Sigma Chemical Co.) at 37°C for 2 h while being stirred (160 rpm). The resultant peptic digestion product was further digested by the addition of 2 g of purified porcine pancreas trypsin (13,700 units/mg; Sigma Chemical Co.) after pH adjustment to pH 8.0 with 2 N NaOH. The reaction mixture was incubated at 37°C for 4 h under stirred conditions (160 rpm). After digestion, PT digestion products were heated at 100°C for 30 min to inactivate enzymes and freeze-dried for further analysis.
Mucosal biopsy specimens and organ culture.
Duodenal biopsy specimens were obtained from three CD patients on a gluten-free diet (age range, 19 to 30 years). All CD patients expressed the HLA-DQ2 phenotype. CD was diagnosed according to the criteria of the European Society for Pediatric Gastroenterology, Hepatology and Nutrition (19). Immediately after excision, all biopsy specimens were placed in ice-chilled culture medium (RPMI 1640; Gibco-Invitrogen, United Kingdom) and brought to the laboratory within 30 min.
Duodenal biopsy specimens were cultured for 4 h using the organ culture method of Browing and Trier (7). Briefly, biopsy specimens were orientated villous side up on a stainless steel mesh, positioned over the central well of an organ culture dish (Falcon). The well contained RPMI 1640 (Gibco-Invitrogen, United Kingdom) supplemented with 15% fetal calf serum (Gibco-Invitrogen, United Kingdom) and 1% penicillin/streptomycin solution (Gibco-Invitrogen, United Kingdom). Before the dishes were sealed and incubated at 37°C, they were placed in an anaerobic jar which was gassed with 95% O2 and 5% CO2. Four biopsy specimens (ca. 3 mm2) from each CD patient were cultured with the PT digestion product (5 mg/ml) of gliadin/glutenins from DWS of Triticum turgidum L. var. durum cv. Arcangelo, FDWS2 of cv. Arcangelo, or medium alone (negative control). The final volume of the assay was 1 ml.
RNA extraction and cDNA synthesis.
Tissue samples were taken and stored in RNA Later (Qiagen GmbH, Germany) to preserve RNA. Total RNA was extracted from tissue using the RNeasy minikit (Qiagen GmbH) according to the manufacturer's instructions. The concentration of mRNA was estimated by UV absorbance at 260 nm. Aliquots of total RNA (500 ng) were reverse transcribed using random hexamers and the TaqMan reverse transcription reagents (Applied Biosystems, Monza, Italy) with 3.125 U/μl of MultiScribe reverse transcriptase in a final volume of 50 μl. Samples of cDNA were stored at −20°C.
Quantitative real-time PCR (RT-PCR) for IFN-γ and IL-2 genes.
RT-PCR was carried out in 96-well plates of the ABI Prism 7500HT fast sequence detection system (Applied Biosystems). Data collection and analyses were carried out using the machine software. TaqMan gene expression assay for IFN-γ and IL-2 genes and Pre-Developed TaqMan assay for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (endogenous control) gene (Applied Biosystems) were used. Assays were carried out using 20× mix of PCR primers and TaqMan minor groove binder 6-carboxyfluorescein (6-FAM) dye-labeled probes with a nonfluorescent quencher at the 3′ end of the probe. Two-step reverse transcription-PCR was carried out using first-strand cDNA with a final concentration of 1× TaqMan gene expression assay and 1× TaqMan universal PCR master mix. The final reaction mixture volume was 25 μl. Each sample was analyzed in triplicate, and all experiments were repeated twice. A no-template control (RNase-free water) was included on each plate. The thermal cycler conditions were 2 min hold at 50°C (UNG activation), 10 min hold at 95°C, followed by 40 cycles, with 1 cycle consisting of 15 s at 95°C and 1 min at 60°C. Initially, a standard curve and a validation experiment were carried out for each primer/probe set. Six serial dilutions (20 to 0.1 ng/μl) of IFN-γ cDNA or IL-2 cDNA were used as the template for each primer/probe set. The standard curve was generated by plotting the threshold cycle (CT) values against the log of the amount of cDNA. The average value of the target gene was normalized using the GAPDH gene. A duodenal biopsy specimen from a healthy individual was used to calibrate all experiments.
IFN-γ and IL-2 proteins secreted in the supernatant were quantified by ELISA in round-bottom plates (Tema Ricerca, Milan, Italy) according to the manufacturer's recommendations.
Pasta making.
Pasta was manufactured at the pilot plant of the Department of Plant Protection and Applied Microbiology. One kilogram of FDWS2 from Triticum turgidum L. var. durum cv. Arcangelo containing less than 20 ppm of gluten, 1.75 kg of pregelatinized native maize (Zea mays) starch (Pregeflo C 25 G; Roquette Italia S.p.A., Alessandria, Italy), and 0.25 kg of pregelatinized rice (Oryza sativa) flour (Caremoli S.p.A., Milan, Italy) were blended together and mixed with tap water (ca. 1.4 liters). Two controls were also manufactured: (i) gluten-containing pasta (GCP), using unfermented DWS from cv. Arcangelo as the only ingredient; and (ii) gluten-free pasta (GFP) using pregelatinized native maize and pregelatinized rice. Doughs were extruded using a benchtop pasta maker (La Parmigiona s.r.l., Fidenza, Italy) to produce the Italian pasta type “sedani.” Sedani were placed into pasta frames and allowed to dry for 24 h at 25°C.
Sensory analysis of cooked pasta.
Sensory analysis of pasta was carried out after cooking sedani for 8 min, which was determined as the optimum cooking time. Sensory characteristics were evaluated by six trained panelists by the method of D'Egidio et al. (14) under test conditions of International Standard 7304 (25). Stickiness (material adhering to the surface of cooked pasta), firmness (resistance to bite through the cooked pasta with the incisors), odor, and flavor were determined. These characteristics were evaluated by a score from 20 to 100 (25), and each panelist analyzed the same sample twice. The scores for stickiness were assigned as follows: <20 (very high); 20 to 40 (high); 41 to 60 (average); 61 to 80 (almost absent); and 81 to 100 (absent). The scores for firmness were assigned as follows: <20 (very low); 20 to 40 (low); 41 to 60 (sufficient); 61 to 80 (good); and 81 to 100 (very good). The scores for odor and flavor were assigned as follows: <20 (very unpleasant); 20 to 40 (unpleasant); 41 to 60 (sufficient); 61 to 80 (good); and 81 to 100 (pleasant).
Statistical analysis.
All data were obtained from at least three replicates. The percentages were arcsine transformed for data analysis (47). Analysis of variance (ANOVA) was carried out on transformed data, and the means were separated by Tukey's honestly significant difference (HSD) test using the statistical software program Statistica for Windows (Statistica 6.0 per Windows 1998) (StatSoft, Vigonza, Italy). Peptide profiles obtained in this study were analyzed by UNICORN 5.0 from GE Healthcare, followed by statistical analysis of the peak areas using Statistica 6.0 per Windows 1998.
RESULTS
Fermentation of durum wheat semolina (DWS).
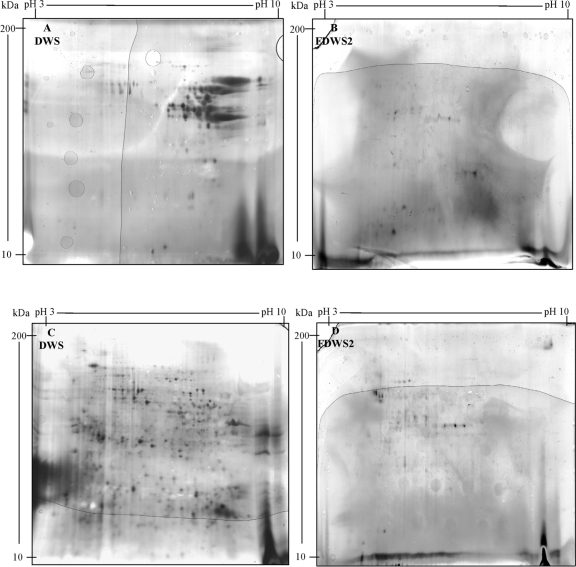
The concentration of proteins and gluten of DWS from the nine cultivars of Triticum turgidum L. var. durum differed (from 11.5% ± 0.07% to 14.8% ± 0.05% and from 10.2% ± 0.02% to 13.4% ± 0.08% [wt/wt], respectively). Almost all gliadins had pI and molecular mass values distributed in the range of 4.2 to 9.0 and 18 to 94 kDa, respectively. The highest number of gliadin spots was found for cv. Ocotillo (70 spots), Arcangelo (61 spots), Ciccio (59 spots), Colosseo (58 spots), and Gargano (51 spots). Glutenin polypeptides were widespread with pI and molecular mass values of 3.8 to 8.9 and 15 to 94 kDa, respectively. The highest number of glutenin spots was found for cv. Svevo (200 spots), Ocotillo (179 spots), Colosseo (109 spots), and cultivars Claudio, Arcangelo, and Simeto (97 spots). 2-DE analyses of gliadin and glutenin polypeptides of DWS from the cv. Arcangelo are shown in Fig. 1A and C.
FIG. 1.
Two-dimensional gel electrophoresis (2-DE) analysis of gliadin (A and B) and glutenin (C and D) polypeptides of durum wheat semolina (DWS) from Triticum turgidum L. var. durum cv. Arcangelo before (DWS) and after fermentation for 72 h at 37°C (FDWS2). The Arcangelo cultivar showed the highest resistance to hydrolysis. The formula of FDWS2 is described in Materials and Methods.
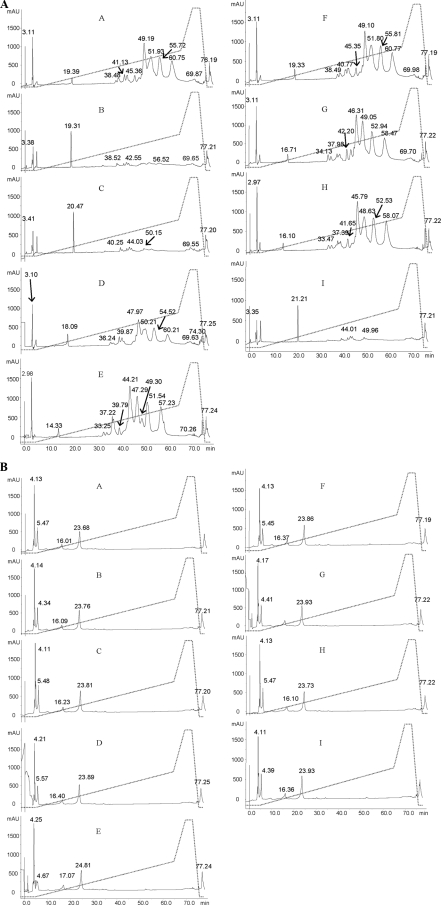
In a preliminary experiment, the fermentation of DSW was carried out according to the protocol of Rizzello et al. for Triticum aestivum flour (32). As shown by R5-ELISA, the residual gluten concentration (ppm) of FDWS1 (fermented durum wheat semolina 1) exceeded the threshold of 20 ppm for the following cultivars of Triticum turgidum L. var. durum: Gargano (581 ppm), Colosseo (506 ppm), Duilio (478 ppm), Ciccio (354 ppm), Simeto (294 ppm), Arcangelo (275 ppm), Claudio (262 ppm), Ocotillo (233 ppm), and Svevo (170 ppm). Consequently, the time of fermentation was extended to 72 h, and the concentration of both fungal proteases was increased to 400 ppm. Under these conditions (fermented durum wheat semolina 2 [FDWS2]), the residual concentration of gluten was always less than 20 ppm. Except for a few protein spots in the cv. Arcangelo (Fig. 1A and B), all the other FDWS2 did not contain gliadin polypeptides detectable through 2-DE analysis. A few glutenin polypeptides persisted in all FDWS2, including cv. Arcangelo (Fig. 1C and D). Ethanol-soluble peptides extracted from both DWS and FDWS2 were also quantified by RP-HPLC analysis (Fig. 2A and B). DWS of cv. Arcangelo, Colosseo and, especially, Duilio, Gargano, Ocotillo, and Simeto showed the highest number of peaks and largest areas of the peaks. Compared to DWS, the number and areas of peaks of FDWS2 markedly decreased, especially in the central zone of the acetonitrile gradient (30 to 60 min). The profiles of ethanol-soluble peptides of FDWS2 were almost similar between cultivars. Proteolysis during fermentation was also estimated by the determination of the concentration of FAA. DWS of cv. Ciccio, Claudio, Colosseo, Ocotillo, and Svevo had the highest concentration of FAA (901 to 1,284 mg/kg). The other cultivars contained 313 to 521 mg/kg of FAA. The concentration of FAA markedly increased in FDWS2. The highest values were found for cultivars Arcangelo and Claudio (24,601 to 24,618 mg/kg), followed by Ocotillo and Svevo (23,296 to 23,577 mg/kg) and Ciccio (22,304 mg/kg). The lowest values (16,344 to 16,059 mg/kg) were found for Simeto and Colosseo.
FIG. 2.
Reversed-phase high-performance liquid chromatography (RP-HPLC) analysis of 60% ethanol extract of durum wheat semolina (DWS) (A) and DWS fermented for 72 h at 37°C (FDWS2) (B). The formula of FDWS2 is described in Materials and Methods. DWS and FDWS2 from cultivars Arcangelo (A), Ciccio (B), Claudio (C), Colosseo (D), Duilio (E), Gargano (F), Ocotillo (G), Simeto (H), and Svevo (I) are shown. Time in minutes is shown on the x axis of each graph, and absorbance in milliabsorbance units (mAU) is shown on the y axis of each graph. The dotted line shows the concentration (as a percentage, vol/vol) of acetonitrile.
Since sourdough lactobacilli had the capacity to hydrolyze gluten from cultivars having proteomic differences and since the mechanism of hydrolysis of Pro-rich polypeptides is still unknown, the intracellular peptidase activities of sourdough lactobacilli were studied in depth.
Partial purification of peptidases.
In a preliminary experiment, the pooled cytoplasmic extracts of the 10 lactobacilli were assayed using synthetic substrates. The following peptidase activities were found: 7.94 ± 0.04 U for PepN, 2.80 ± 0.01 U for PepI, 3.55 ± 0.03 U for PepX, 5.37 ± 0.02 U for PepO, 6.05 ± 0.07 U for PEP, 16.74 ± 0.21 U for PepT, 20.54 ± 0.44 U for PepV, 23.10 ± 0.89 U for PepQ, and 18.52 ± 0.56 U for PepR. The peptidases contained in the pooled cytoplasmic extracts were partially purified using three chromatographic steps. After purification, active fractions contained only single enzyme activity.
Peptidase activities toward Pro-rich synthetic epitopes.
The initial cell densities of the lactobacilli used to ferment DWS varied from 5 × 108 to 1 × 109 CFU/g. During fermentation, the cell densities of the various lactobacilli normalized to values of ca. 3 × 109 to 5 × 109 CFU/g (22). Therefore, partially purified peptidases derived from the initial cell density of ca. 3 × 109 CFU/g (protein concentration of ca. 1 μg/ml) were used. PepN, PepI, PepX, PepO, PEP, PepT, PepV, PepQ, and PepR peptidases were singly incubated (24 h at 37°C) with the 33-mer epitope at a concentration of 200 mM. Hydrolysis was monitored by amino acid analyzer and RP-HPLC. Single peaks from RP-HPLC were analyzed by nano-ESI tandem mass spectrometry (nano-ESI-MS/MS). PepN liberated only leucine (L) from the NH2 terminus of the 33-mer. No traces of proline (P) or other FAA were found (Table 1). PepO cleaved the internal peptide bond Q↓L with the liberation of four different peptides. Three of these peptides were immunogenic epitopes. FAA were not found. PEP hydrolyzed the internal peptide bond P↓F and liberated two peptides. One of these two peptides was an immunogenic epitope. No hydrolysis of the 33-mer was found using the partially purified PepI, PepX, PepQ, PepT, PepV, PepQ, or PepR. Further, all combinations between purified peptidases were assayed for complementary hydrolysis (Table 1). The combinations of PepN and PepI or PepN and PepX liberated only leucine (L) from the NH2 terminus of the 33-mer. This was probably due to PepN activity. The combination of PepN and PepO hydrolyzed the 33-mer into four peptides (three of them were epitopes) and free leucine (L). PepN and PEP hydrolyzed the 33-mer into two peptides (both of them were epitopes) and free leucine (L). PepN together with PepT, PepV, PepQ, or PepR liberated only leucine (L). No complementary hydrolysis of the 33-mer was found when PepI was used together with PepO or PEP. Similar results were found using PepI with PepT, PepV, PepQ, or PepR. Complementary hydrolysis was found using PepX and PEP, PepX and PepO, PepO and PEP, or PepO and PepV. Nevertheless, only the combined activity of PepN, PepX and PepO or PepN, PepX, PepO and PEP resulted in the partial hydrolysis of the 33-mer without the generation of related immunogenic epitopes. To obtain the complete hydrolysis of the 33-mer into FAA, one of the following combination of peptidases had to be used: PepN, PepI, PepX, PepO, PEP, PepT, PepV, PepQ, and PepR or PepN, PepX, PepO, PEP, PepV, and PepQ (Table 1 and Fig. 3). When subjected to the combined activity of PepN, PepX, PepO, PEP, PepT, PepV, and PepQ peptidases, the hydrolysis of the 33-mer was completed in 14 h. Based on the complementary enzyme activities, peptides were found in the interval from 1 to 12 h of incubation and only FAA were detectable after 14 h of incubation. Other synthetic immunogenic epitopes (37), such as fragments 57-68 of α9-gliadin, 62-75 of A-gliadin, and 134-153 of γ-gliadin were completely hydrolyzed to FAA (12 to 14 h) by the same combination of peptidases (data not shown).
TABLE 1.
In vitro hydrolysis of the 33-mer epitope by partially purified peptidases from the pooled cytoplasmic extract of 10 sourdough lactobacillia
| Epitope or peptidaseb | Sequence(s) of epitope or peptide fragment(s)c | No. of epitopes | Free amino acid(s) |
|---|---|---|---|
| Epitopes | |||
| 33-mer epitope | L1QLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF33 | ||
| Presumptive immunogenic epitopesd | [2-33], [3-10], [4-10], [6-33], [25-33], [11-17], [18-24], [6-12], [10-33] | ||
| Peptidases | |||
| PepN | QLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF | 1 | L |
| PepI | LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF | 1 | |
| PepX | LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF | 1 | |
| PepO | LQ; LQPFPQPQ; 2 LPYPQPQ; LPYPQPQPF | 3 | |
| PEP | LQLQP; FPQPQLPYPQPQLPYPQPQLPYPQPQPF | 1 | |
| PepT | LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF | 1 | |
| PepV | LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF | 1 | |
| PepQ | LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF | 1 | |
| PepR | LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF | 1 | |
| PepN/PepI | QLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF | 1 | L |
| PepN/PepX | QLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF | 1 | L |
| PepN/PepO | LQ; QPFPQPQ; LPYPQPQ; LPYPQPQ; LPYPQPQPF | 3 | L |
| PepN/PEP | QLQP; FPQPQLPYPQPQLPYPQPQLPYPQPQPF | 1 | L |
| PepN/PepT/PepV/PepQ/PepR | QLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF | 1 | L |
| PepI/PepX | LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF | 1 | |
| PepI/PepO | LQ; LQPFPQPQ; LPYPQPQ; LPYPQPQ; LPYPQPQPF | 3 | |
| PepI/PEP | LQLQP; FPQPQLPYPQPQLPYPQPQLPYPQPQPF | 1 | |
| PepI/PepT/PepV/PepQ/PepR | LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF | 1 | |
| PepX/PepO | LQ; LQPFPQPQ; LP; YP; QPQ; LP; YP; QPQ; LP; YP; QP; QPF | 1 | |
| PepX/PEP | LQLQP; FP; QP; QLPYPQPQLPYPQPQLPYPQPQPF | 1 | L |
| PepX/PepT/PepV/PepQ/PepR | LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF | 1 | |
| PepO/PEP | LQ; LQLQP; FPQPQ; LPYPQPQ; LPYPQPQ; LPYPQPQPF | 3 | |
| PepO/PepV | LQPFPQPQ; LPYPQPQ; LPYPQPQ; LPYPQPQPF | 3 | L; Q |
| PepO/PepT/PepQ/PepR | LQ; LQPFPQPQ; 2 LPYPQPQ; LPYPQPQPF | 3 | |
| PEP/PepT/PepV/PepQ/PepR | LQLQP; LQLQ; FPQPQLPYPQPQLPYPQPQLPYPQPQPF | 1 | |
| PepN/PepI/PepX | QLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF | 1 | L |
| PepN/PepI/PepO | LQ; QPFPQPQ; 2 LPYPQPQ; LPYPQPQPF | 3 | L |
| PepN/PepI/PEP | QLQP; FPQPQLPYPQPQLPYPQPQLPYPQPQPF | 1 | L |
| PepN/PepI/PepT/PepV/PepQ/PepR | QLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF | 1 | L |
| PepN/PepX/PepO | LQ; QP; FP; QPQ; LP; YP; QPQ; LP; YP; QPQ; LP; YP; QP; QPF | L | |
| PepN/PepX/PEP | QLQP; FP; QP; QLPYPQPQLPYPQPQLPYPQPQPF | 1 | L |
| PepN/PepX/PepT/PepV/PepQ/PepR | QLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF | 1 | L |
| PepI/PepX/PepO | LQ; LQPFPQPQ; LP; YP; QPQ; LP; YP; QPQ; LP; YP; QP; QPF | 1 | |
| PepI/PepX/PEP | LQLQP; FP; QP; QLPYPQPQLPYPQPQLPYPQPQPF | 1 | |
| PepI/PepX/PepT/PepV/PepQ/PepR | QLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF | 1 | |
| PepX/PepO/PEP | LQPFPQPQ; LP; YP; QPQ; LP; YP; QPQ; LP; YP; QP; QPF | 1 | |
| PepX/PepO/PepT/PepV/PepQ/PepR | LQ; LQPFPQPQ; LP; YP; QPQ; LP; YP; QPQ; LP; YP; QP; QPF | 1 | |
| PepN/PepI/PepX/PepO | LQ; QP; FP; QPQ; LP; YP; QPQ; LP; YP; QPQ; LP; YP; QP; QPF | L | |
| PepN/PepI/PepX/PEP | QLQP; FP; QP; QLPYPQPQLPYPQPQLPYPQPQPF | 1 | L |
| PepN/PepI/PepX/PepT/PepV/PepQ/PepR | QLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF | 1 | |
| PepN/PepI/PepX/PepO/PEP | LQ; QP; FP; QPQ; LP; YP; QPQ; LP; YP; QPQ; LP; YP; QP; QPF | L | |
| PepN/PepI/PepX/PepO/PEP/PepT/PepV/PepQ/PepR | 13P; 10Q; 5L; 3Y; 2F | ||
| PepN/PepX/PepO/PEP/PepV/PepQ | 13P; 10Q; 5L; 3Y; 2F |
In vitro hydrolysis of the 33-mer epitope (LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF) by partially purified peptidases from the pooled cytoplasmic extracts of 10 sourdough lactobacilli. See Materials and Methods for details of the reaction mixture.
PepN, general aminopeptidase type N; PepI, proline iminopeptidase; PepX, X-prolyl dipeptidyl aminopeptidase; PepO, endopeptidase; PEP, prolyl endopeptidyl peptidase; PepT, tripeptidase; PepV, dipeptidase; PepQ, prolidase; PepR, prolinase.
Peptides derived from the 33-mer epitope. For instance, PepO cleaved the internal peptide bond Q↓L of the 33-mer epitope and liberated four different peptides, and three of these peptides were immunogenic epitopes. The combination of PepN and PepI or of PepN and PepX liberated only leucine (L) from the NH2 terminus of the 33-mer epitope.
The presumptive immunogenic epitope row shows the number position of the presumptive immunogenic epitopes that are contained in the native sequence of the 33-mer epitope (37). The fragments are indicated by their number positions (e.g., [2-33] is the fragment containing positions 2 to 33 of the native sequence of the 33-mer epitope). Presumptive immunogenic epitopes may have the lowest size of 7/9 amino acid residues. When the size is 7 amino acid residues, epitopes should be contained in a larger fragment to be immunogenic (37).
FIG. 3.
Schematic representation of the sequence of hydrolysis of the 33-mer immunogenic epitope by partially purified peptidases from the pooled cytoplasmic fraction of 10 sourdough lactobacilli. PepN, general aminopeptidase type N; PepO, endopeptidase; PEP, prolyl endopeptidyl peptidase; PepX, X-prolyl dipeptidyl aminopeptidase; PepQ, prolidase. Cleavage sites are indicated (↑).
Gluten digestion by partially purified peptidases.
To assay the in situ detoxification of gliadin/glutenin epitopes, all partially purified PepN, PepI, PepX, PepO, PEP, PepT, PepV, PepQ, and PepR were pooled and used, instead of sourdough lactobacilli cells, during 72 h of fermentation of DSW (Triticum turgidum L. var. durum cv. Arcangelo). As in the formula of FDWS2, fungal proteases were also added to the reaction mixture. Pooled partially purified peptidases were used at the protein concentration of ca. 1 μg/ml which approximately corresponded to that of ca. 3 × 109 cells of lactobacilli per gram. As estimated by R5-ELISA, the residual concentration of gluten was ca. 2 ppm. The RP-HPLC profile did not significantly (P > 0.05) differ from those found in FDWS2 (data not shown). No gliadin/glutenin epitopes were found at the lowest detectable concentration (20 ppm) by nano-ESI-MS/MS analysis. The lowest detectable concentration was estimated by injecting different concentrations (1 to 100 ppm) of the synthetic immunogenic peptide QLQPFPQPQLPY (fragment 57-68 of α9-gliadin) together with the sample containing gliadin/glutenin epitopes (32).
Cytokine expression in duodenal biopsy specimens of CD patients.
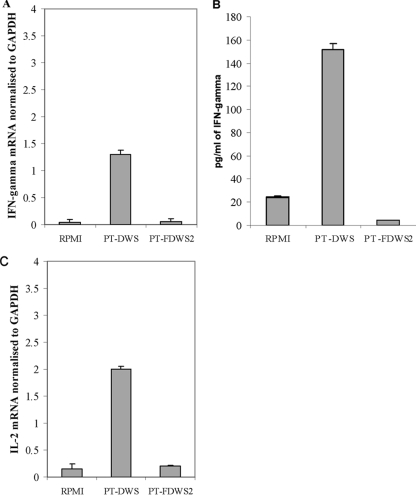
Sixty percent ethanol-soluble polypeptides and urea-dithiothreitol-soluble glutenins were extracted from DWS and FDWS2 of Triticum turgidum L. var. durum cv. Arcangelo, subjected to PT digestion, pooled, and used to treat duodenal biopsy specimens from CD patients. The exposure of biopsy specimens to PT digestion products from DWS resulted in a significant (P < 0.05) increase in the production of IFN-γ mRNA (Fig. 4A). The release of IFN-γ mRNA by biopsy specimens treated with the PT digestion product of FDWS2 did not significantly (P > 0.05) differ from that of the negative control (medium alone). Increased levels of IFN-γ protein were found in the culture supernatant of all biopsy specimens. A good agreement was found between the proteins and the level of mRNA. The highest production of IFN-γ proteins was found in biopsy specimens treated with PT digestion product from DWS (Fig. 4B).
FIG. 4.
Effect of the PT (pepsin-trypsin) digestion of polypeptides soluble in 60% ethanol and glutenins soluble in urea-dithiothreitol extracted from durum wheat semolina (DWS) (PT-DWS) and DWS fermented for 72 h at 37°C (FDWS2) (PT-FDWS2) on the expression of gamma interferon (IFN-γ) mRNA (A), IFN-γ protein (B), and interleukin 2 (IL-2) mRNA (C). RPMI, negative control (medium alone). GAPDH, glyceraldehyde-3-phosphate dehydrogenase (endogenous control). Data are the means plus standard deviations (error bars) from three independent fermentations using duodenal biopsy specimens from three CD patients.
IL-2 expression in the culture supernatants was measured in duodenal biopsy specimens from CD patients after 4 h of stimulation with PT digestion products. Stimulated biopsy specimens produced significant levels of IL-2 mRNA in response to PT digestion products of DWS of Triticum turgidum L. var. durum cv. Arcangelo (Fig. 4C). In contrast, no IL-2 mRNA overexpression was found following stimulation by the PT digestion products corresponding to FDWS2. Overexpression of IL-2 protein was found only in the supernatant of duodenal biopsy specimens from CD patients treated with PT digestion products of gliadins from DWS (data not shown).
Sensory analysis of the cooked pastas.
After the pastas were cooked for 8 min, fermented durum wheat pasta (FDWP), GCP, and GFP (sedani type) were subjected to sensory analysis by a trained panel (Table 2). The scores for stickiness and firmness of FDWP were significantly (P < 0.05) lower than those found for GCP (68.3 ± 6.7 [average] versus 80 ± 5.7 [almost absent] and 59.4 ± 8.6 [sufficient] versus 72.1 ± 12.2 [good]). The lowest scores for stickiness and firmness were found for GFP (44.3 ± 2.7 and 36.1 ± 5.8, respectively) (P < 0.05). Odor of FDWP and GCP did not significantly differ (P > 0.05). Compared to GFP, a more pronounced flavor was found for FDWP.
TABLE 2.
Sensory analysis of fermented durum wheat semolina from Triticum turgidum L. var. durum cv. Arcangelo (FDWP), gluten-containing (GCP) and gluten-free (GFP) pasta
| Pasta | Avg score ±SD for the following characteristic(s)a: |
||
|---|---|---|---|
| Stickiness | Firmness | Odor and flavor | |
| FDWP | 68.3 ± 6.7 B | 59.4 ± 8.6 B | 75.0 ± 5.0 A |
| GCP | 80.0 ± 5.7 A | 72.1 ± 12.2 A | 86.2 ± 4.7 A |
| GFP | 44.3 ± 2.7 B | 36.1 ± 5.8 C | 62.0 ± 8.1 B |
Average values (n = 8) ± standard deviations in the same column with different letters (A, B, and C) differ significantly (P < 0.05). The scores and scoring methods are explained in “Sensory analysis of cooked pasta” in Materials and Methods.
DISCUSSION
Food-grade sourdough lactobacilli, as cell factories for multiple and complementary proteolytic enzymes, supplemented with fungal proteases, were previously proven to efficiently degrade the gluten of Triticum aestivum wheat flour (32). The hydrolyzed wheat flour used for making sweet baked goods was tolerated by CD patients during daily administration (8 g of gluten equivalent) for 60 days (13). Nevertheless, the efficiency of this biotechnology approach was not validated on durum wheat semolina, and especially, the mechanism of hydrolysis of the gluten immunogenic epitopes was still unknown. Nine cultivars of Triticum turgidum L. var. durum having large proteomic differences were subjected to gluten hydrolysis according to Rizzello et al. (32) with some modifications. Cultivars of durum wheat were more resistant to hydrolysis than T. aestivum and the modifications regarded the higher concentration of fungal proteases (400 ppm) and the longer time of fermentation (72 h). Under these conditions, gluten in all cultivars of durum wheat semolina was degraded to less than 20 ppm.
During food processing, an effective enzyme treatment for gluten detoxification has to degrade all or the major part of the gluten-derived immunogenic peptides. Highly antigenic gluten epitopes share a number of properties (35). They are mainly located in Pro-rich regions of gliadins and occur within the sequence of long and multivalent peptides. Pro-rich peptides are protected from proteolysis by gastric, pancreatic, and intestinal brush border membrane enzymes. Most of the inflammatory epitopes are the preferred substrates for human transglutaminase 2 (TG2). DQ2 has the unique feature of enabling the molecules to bind and present Pro-rich peptides with glutamate residues to CD4+ T cells of the intestinal mucosa (6). Recently, it was shown that endopeptidases derived from bacteria are useful to abolish the antigenicity of the 33-mer and other Pro-rich putative immunotoxic epitopes (L. Greco, M. Gobbetti, R. Auricchio, R. Di Mase, F. Paparo, R. Di Cagno, M. De Angelis, C. G. Rizzello, A. Cassone, G. Terrone, L. Timpone, M. D'Aniello, R. Troncone, and S. Auricchio, submitted for publication). Nevertheless, there are still problems with the delivery of endopeptidases, especially with efficient mixing with gluten and with their stability under acidic gastric conditions (36). The use of microbial peptidases during food processing does not show the same problems. Lactobacilli possess a very complex peptidase system (for reviews, see references 4 and 33). Nevertheless, no unique strain may have the entire portfolio of peptidases needed for hydrolyzing all potential gluten polypeptides where Pro is located at the different positions (for a review, see reference 24). Notwithstanding that cell autolysis may occur at some extent during dough mixing and sourdough fermentation, peptidases of sourdough lactobacilli are located in the cytoplasm. Previously, it was shown that the size of immunogenic oligopeptides was suitable for the bacterial transport system (32). In vitro analysis revealed that the 33-mer was found in the cell cytoplasm of lactobacilli as soon as the incubation started. The cytoplasmic extracts of the 10 sourdough lactobacilli were pooled, and several peptidases were partially purified and used alone or in combination to hydrolyze the 33-mer epitope. At least three peptidases (PepN, PepX, and PepO) were needed to hydrolyze the 33-mer epitope without generation of derived immunogenic peptides. First, PepN, PepO, PEP, and PepX hydrolyzed the 33-mer to di- and tripeptides. Then, these peptides were further hydrolyzed by PepT, PepV, PepQ, and PepR, liberating only free amino acids as end products (4, 33). The activity of only one (e.g., PepO) or two (e.g., PepN and PepO) peptidases liberated two or more epitopes, thus maintaining/increasing the toxicity of the native 33-mer. Only the complementary activities of most of the intracellular peptidases of the 10 sourdough lactobacilli guaranteed the complete hydrolysis of the 33-mer within 12 to 14 h of incubation. The same results were found using other synthetic epitopes. The complete hydrolysis of gluten was confirmed in situ. Fungal proteases and partially purified peptidases were used to hydrolyze durum wheat semolina. Partially purified peptidases were used at the concentration which approximately coincided with that contained in the lactobacilli during sourdough fermentation. Fungal proteases, routinely used as bakery improvers, are indispensable to start the primary proteolysis of gluten (32). Polypeptides of intermediate dimensions (e.g., 4 to 40 amino acids), generated from the native proteins, are the substrates for secondary proteolysis by complementary peptidases of sourdough lactobacilli. According to this scheme, the hydrolyzed durum wheat semolina contained less than 20 ppm of residual gluten. The use of sourdough lactobacilli during food processing could be considered the easiest and noninvasive way to eliminate gluten from wheat (13, 32). In contrast, administration of peptidases with the meal actually enhanced the immunoresponse, exposing the small intestinal mucosa more rapidly to immunopeptides from fragmented gluten proteins (36).
After hydrolysis, some glutenin polypeptides persisted. Glutenins may contain sequences that activate T cells from the small intestine, thus resulting in the secretion of large amounts of IFN-γ (39, 44). High-molecular-mass glutenin subunits (HMM-GS) stimulate T-cell lines from some CD patients and exacerbate CD in vivo (15). On the basis of these considerations, the diagnostic value of R5 antibody to detect toxic epitopes might be limited. In vitro assays are imposed to confirm the absence of toxicity (26). The traditional approach consists of organ culture of treated CD intestinal biopsy specimens in the presence of gluten or gluten fractions (36). As expected, the PT digestion products from durum wheat semolina (DWS) caused a marked increase of IFN-γ (26). IFN-γ coordinates many aspects of the innate and adaptive immune responses, and it is the principal cytokine produced by α-β CD4+ reactive T cells upon gluten activation (28, 39, 45). The PT digestion products of fermented durum wheat semolina (FDWS2) induced IFN-γ as the negative control. Similar results were found for the synthesis of IL-2.
The findings of this study provide evidence that the immunogenic sequences of gliadins/glutenins were not present in fermented durum wheat semolina which might be safely used for pasta making. Compared to pasta containing gluten (GCP), the cooking properties (stickiness and firmness) of the pasta made from the hydrolyzed semolina (FDWP) were obviously lower but still appreciable and preferable with respect to the conventional gluten-free pasta (GFP). Given the encouraging results of this study, further research should include a long-term in vivo challenge on CD patients.
Acknowledgments
This work was supported by the Italian Ministry of University and Research, project “Pasta alimentare: Miglioramento della qualità tecnologica e riduzione dell'intolleranza alimentare al glutine-Qualitech-Pasta” 7134 (1 August 2005).
We thank Ilario Losito (Department of Chemistry, Centro Interdipartimentale di Ricerca SMART, University of Bari) for scientific assistance on nano-ESI-MS/MS analysis; Silvestro Borracci (Divella S.p.A.) for technical assistance; and Leonardo Caputo (CNR-ISPA, Bari, Italy), Francesco Divella, and Fabio Divella (both of Divella S.p.A.) for critical discussions of the results.
Footnotes
Published ahead of print on 30 November 2009.
REFERENCES
- 1.Aleanzi, M., A. M. Demonte, C. Esper, S. Garcilazo, and M. Waggener. 2001. Celiac disease: antibody recognition against native and selectively deaminated gliadin peptides. Clin. Chem. 47:2023-2028. [PubMed] [Google Scholar]
- 2.American Association of Cereal Chemists. 2000. Methods 55-10, 44-15, 56-81B, 76-13, and 76-21. In Approved methods of the American Association of Cereal Chemists, 10th ed. American Association of Cereal Chemists, St. Paul, MN.
- 3.Arentz-Hansen, H., R. Korner, O. Molberg, H. Quarsten, W. Vader, Y. M. Kooy, K. E. A Lundin, F. Koning, P. Roepstorff, L. M. Sollid, and S. N. McAdam. 2000. The intestinal T cell response to alpha-gliadin in adult coeliac disease is focused on a single deamidated glutamine targeted by tissue transglutaminase. J. Exp. Med. 191:603-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bethune, M. T., E. Ribka, C. Khosla, and K. Sestak. 2008. Transepithelial transport and enzymatic detoxification of gluten in gluten-sensitive rhesus macaques. PLoS One 1857:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bini, L., B. Magi, B. Marzocchi, F. Arcuri, S. Tripodi, M. Cintorino, J. C. Sanchez, S. Frutiger, and D. Hochstrasser. 1997. Protein expression profiles in human breast ductal carcinoma and histologically normal tissue. Electrophoresis 18:2832-2841. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Browning, T. H., and J. S. Trier. 1969. Organ culture of mucosal biopsies of small intestine. J. Clin. Invest. 48:1423-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catassi, C., E. Fabiani, G. Iacono, C. D'Agate, R. Francavilla, F. Biagi, U. Volta, S. Accomando, A. Picarelli, I. De Vitis, G. Pianelli, R. Gesuita, F. Carle, A. Mandolesi, I. Bearzi, and A. Fasano. 2007. A prospective, double-blind, placebo-controlled trial to establish a safe gluten threshold for patients with celiac disease. Am. J. Clin. Nutr. 85:160-166. [DOI] [PubMed] [Google Scholar]
- 9.Chand, N., and A. A. Mihas. 2006. Celiac disease: current concepts in diagnosis and treatment. J. Clin. Gastroenterol. 40:3-14. [DOI] [PubMed] [Google Scholar]
- 10.Crow, V. L., R. Holland, and T. Coolbear. 1993. Comparison of subcellular fractionation methods for Lactococcus lactis ssp. lactis and Lactococcus lactis ssp. cremoris. Int. Dairy J. 3:599-612. [Google Scholar]
- 11.De Angelis, M., C. G. Rizzello, A. Fasano, M. G. Clemente, C. De Simone, M. Silano, M. De Vincenzi, I. Losito, and M. Gobbetti. 2006. VSL#3 probiotic preparation has the capacity to hydrolyze gliadin polypeptides responsible for celiac sprue. Biochim. Biophys. Acta 1762:80-93. [DOI] [PubMed] [Google Scholar]
- 12.De Angelis, M., R. Di Cagno, G. Gallo, M. Curci, S. Siragusa, C. Crecchio, E. Parente, and M. Gobbetti. 2007. Molecular and functional characterization of Lactobacillus sanfranciscensis strains isolated from sourdoughs. Int. J. Food Microbiol. 114:69-82. [DOI] [PubMed] [Google Scholar]
- 13.De Angelis, M., F. Minervini, L. Caputo, A. Cassone, R. Coda, M. P. Calasso, F. Divella, F. Divella, and M. Gobbetti. 2008. Proteomic analysis by two-dimensional gel electrophoresis and starch characterization of Triticum turgidum L. var. durum cultivars for pasta making. J. Agric. Food Chem. 56:8619-8628. [DOI] [PubMed] [Google Scholar]
- 14.D'Egidio, M. G., B. M. Mariani, S. Nardi, and P. Novaro. 1993. Viscoelastograph measures and total organic matter test: suitability in evaluating textural characteristics of cooked pasta. Cereal Chem. 70:67-72. [Google Scholar]
- 15.Dewar, D. H., M. Amato, H. J. Ellis, E. L. Pollock, N. Gonzalez-Cinca, H. Wieser, and P. J. Ciclitira. 2006. The toxicity of high molecular weight glutenin subunits of wheat to patients with coeliac disease. Eur. J. Gastroenterol. Hepatol. 18:483-491. [DOI] [PubMed] [Google Scholar]
- 16.Di Cagno, R., M. De Angelis, P. Lavermicocca, M. De Vincenzi, C. Giovannini, M. Faccia, and M. Gobbetti. 2002. Proteolysis by sourdough lactic acid bacteria: effects on wheat flour protein fractions and gliadin peptides involved in human cereal intolerance. Appl. Environ. Microbiol. 68:623-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Cagno, R., M. De Angelis, S. Auricchio, L. Greco, C. Clarke, M. De Vincenzi, C. Giovannini, M. D'Archivio, F. Landolfo, G. Parrilli, F. Minervini, E. Arendt, and M. Gobbetti. 2004. Sourdough bread made from wheat and nontoxic flours and started with selected lactobacilli is tolerated in celiac sprue patients. Appl. Environ. Microbiol. 70:1088-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Cagno, R., M. De Angelis, G. Alfonsi, M. De Vincenzi, M. Silano, O. Vincentini, and M. Gobbetti. 2005. Pasta made from durum wheat semolina fermented with selected lactobacilli as a tool for a potential decrease of the gluten intolerance. J. Agric. Food Chem. 53:4393-4402. [DOI] [PubMed] [Google Scholar]
- 19.European Society of Paediatric Gastroenterology, Hepatology and Nutrition. 1990. Revised criteria for diagnosis of coeliac disease. Report of Working Group of European Society of Paediatric Gastroenterology and Nutrition. Arch. Dis. Child. 65:909-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flagella, Z. 2006. Qualità nutrizionale e tecnologica del frumento duro. Ital. J. Agron. 1:203-239. [Google Scholar]
- 21.Gass, J., M. T. Bethune, M. Siegel, A. Spencer, and C. Khosla. 2007. Combination enzyme therapy for gastric digestion of dietary gluten in patients with celiac sprue. Gastroenterology 133:472-480. [DOI] [PubMed] [Google Scholar]
- 22.Gobbetti, M. 1998. The sourdough microflora: interactions between lactic acid bacteria and yeasts. Trends Food Sci. Technol. 9:267-274. [Google Scholar]
- 23.Gobbetti, M., R. Lanciotti, M. De Angelis, M. R. Corbo, R. Massini, and P. Fox. 1999. Study of the effects of temperature, pH and NaCl on the peptidase activities of non-starter lactic acid bacteria (NSLAB) by quadratic response surface methodology. Int. Dairy J. 9:865-875. [Google Scholar]
- 24.Gobbetti, M., C. G. Rizzello, R. Di Cagno, and M. De Angelis. 2007. Sourdough lactobacilli and celiac disease. Food Microbiol. 24:187-196. [DOI] [PubMed] [Google Scholar]
- 25.International Organization for Standardization. 1985. Durum wheat semolinas and alimentary pasta-estimation of cooking quality of spaghetti by sensory analysis. International standard ISO 7304:1985. International Organization for Standardization, Geneva, Switzerland.
- 26.Kilmartin, C., S. Lynch, M. Abuzakouk, H. Wieser, and C. Feighery. 2003. Avenin fails to induce a Th1 response in coeliac tissue following in vitro culture. Gut 52:47-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitea, C., R. Havenaar, J. W. Drijfhout, L. Edens, L. Dekking, and F. Koning. 2008. Efficient degradation of gluten by a prolyl endoprotease in a gastrointestinal model: implications for coeliac disease. Gut 57:25-32. [DOI] [PubMed] [Google Scholar]
- 28.Monteleone, I. G., G. Del Vecchio Blanco, P. Vavassori, S. Cucchiara, T. T. MacDonald, and F. Pallone. 2004. Regulation of the T helper cell type 1 transcription factor T-bet in coeliac disease mucosa. Gut 53:1090-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niewinski, M. M. 2008. Advances in celiac disease and gluten-free diet. J. Am. Diet. Assoc. 108:661-672. [DOI] [PubMed] [Google Scholar]
- 30.Pagnotta, M. A., A. Blanco, A. Gadaleta, and C. Fares. 2005. Functional determinants of grain quality, p. 483-527. In C. Royo et al. (ed.), Durum wheat breeding. Current approaches and future strategies. Food Products Press, Binghamton, NY.
- 31.Pyle, G. G., B. Paaso, B. E. Anderson, D. D. Allen, T. Marti, Q. Li, M. Siegel, C. Koshla, and G. M. Gray. 2005. Effect of pretreatment of food gluten with prolyl endopeptidase on gluten-induced malabsorption in celiac spue. Clin. Gastroenterol. Hepatol. 3:687-694. [DOI] [PubMed] [Google Scholar]
- 32.Rizzello, C. G., M. De Angelis, R. Di Cagno, A. Camarca, M. Silano, I. Losito, M. De Vincenzi, M. D. De Bari, F. Palmisano, F. Maurano, C. Gianfrani, and M. Gobbetti. 2007. Highly efficient gluten degradation by lactobacilli and fungal proteases during food processing: new perspectives for celiac disease. Appl. Environ. Microbiol. 73:4499-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Savijoki, K., H. Ingmer, and P. Varmanen. 2006. Proteolytic systems of lactic acid bacteria. Appl. Microbiol. Biotechnol. 71:394-406. [DOI] [PubMed] [Google Scholar]
- 34.Seilmeier, W., I. Valdez, E. Mendez, and H. Wieser. 2001. Comparative investigations of gluten proteins from different wheat species. Eur. Food Res. Technol. 121:355-363. [Google Scholar]
- 35.Shan, L., O. Molberg, I. Parrot, F. Hausch, F. Filiz, G. M. Gray, L. M. Sollid, and C. Khosla. 2002. Structural basis for gluten intolerance in celiac sprue. Science 297:2275-2279. [DOI] [PubMed] [Google Scholar]
- 36.Shan, L., T. Marti, L. M. Sollid, G. M. Gray, and C. Khosla. 2004. Comparative biochemical analysis of three bacterial prolyl endopeptidases: implications for celiac sprue. Biochem. J. 383:311-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shan, L., S. W. Qiao, H. Arentz-Hansen, Ø. Molberg, G. M. Gray, L. M. Sollid, and C. Khosla. 2005. Identification and analysis of multivalent proteolytically resistant peptides from gluten: implications for celiac sprue. J. Proteome Res. 4:1732-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silano, M., and M. De Vincenzi. 1999. Bioactive antinutritional peptides derived from cereal prolamins: a review. Nahrung. 43:175-184. (In German.) [DOI] [PubMed] [Google Scholar]
- 39.Søndergaard, I., K. Jensen, and B. Krath. 1994. Classification of wheat varieties by isoelectric focusing patterns of gliadins and neural network. Electrophoresis 15:584-588. [DOI] [PubMed] [Google Scholar]
- 40.Stepniak, D., L. Spaenij-Dekking, C. Mitea, M. Moester, A. de Ru, R. Baak-Pablo, P. van Veelen, L. Edens, and F. Koning. 2006. Highly efficient gluten degradation with a newly identified prolyl endoprotease: implications for celiac disease. Am. J. Physiol. Gastrointest. Liver Physiol. 291:G621-G629. [DOI] [PubMed] [Google Scholar]
- 41.Stern, M., P. J. Ciclitira, R. van Eckert, C. Feighery, F. W. Janssen, E. Méndez, T. Mothes, R. Troncone, and H. Wieser. 2001. Analysis and clinical effects of gluten in coeliac disease. Eur. J. Gastroenterol. Hepatol. 13:741-747. [DOI] [PubMed] [Google Scholar]
- 42.Troncone, R., N. Caputo, M. Micillo, L. Maiuri, and V. Poggi. 1994. Immunologic and intestinal permeability tests as predictors of relapse during gluten challenge in childhood coeliac disease. Scand. J. Gastroenterol. 29:144-147. [DOI] [PubMed] [Google Scholar]
- 43.Valdés, I., E. Garcia, M. Lorente, and E. Méndez. 2003. Innovative approach to low-level gluten determination in foods using a novel sandwich enzyme-linked immunosorbent assay protocol. Eur. J. Gastroenterol. Hepatol. 15:465-474. [DOI] [PubMed] [Google Scholar]
- 44.van de Wal, Y., M. C. Kooy, P. van Veelen, W. Vader, S. A. August, J. W. Drijfhout, S. A. Peña, and F. Koning. 1999. Glutenin is involved in the gluten-driven mucosal T cell response. Eur. J. Immunol. 29:3133-3139. [DOI] [PubMed] [Google Scholar]
- 45.Wapenaar, M. C., M. J. van Belzen, J. H. Fransen, A. F. Sarasqueta, R. H. Houwen, J. W. Meijer, C. J. Mulder, and C. Wijmenga. 2004. The interferon γ gene in celiac disease: augmented expression correlates with tissue damage but no evidence for genetic susceptibility. J. Autoimmun. 23:183-190. [DOI] [PubMed] [Google Scholar]
- 46.Weiss, W., C. Volgelmeier, and A. Gorg. 1993. Electrophoretic characterization of wheat grain allergens from different cultivars involved in bakers' asthma. Electrophoresis 14:805-816. [DOI] [PubMed] [Google Scholar]
- 47.Winer, B. J., D. R. Brown, and K. M. Michels. 1991. Statistical principles in experimental design, 3rd ed. McGraw-Hill, New York, NY.