Abstract
Ubiquitous isethionate (2-hydroxyethanesulfonate) is dissimilated by diverse bacteria. Growth of Cupriavidus necator H16 with isethionate was observed, as was inducible membrane-bound isethionate dehydrogenase (IseJ) and inducible transcription of the genes predicted to encode IseJ and a transporter (IseU). Biodiversity in isethionate transport genes was observed and investigated by transcription experiments.
The importance of isethionate (2-hydroxyethanesulfonate) was realized when the compound was shown to be the major anion in the squid giant axon (18); it was then found to be a normal component in mammalian tissue (20) and in human urine (15). The source of this isethionate was considered to be the gut flora (11), and a pathway of isethionate formation from taurine (2-aminoethanesulfonate) in bacteria was established (28). The compound is found in large amounts (at about 250 mM) in red algae (for an example, see reference 13) and on orb spiders’ webs (∼2 M) (30). Isethionate is widely used in commerce as a counter-ion in the formulation of cationic pharmaceuticals and, in derivatized form, in shampoos and soap replacements (for examples, see references 6 and 29).
We presume that the ubiquity of isethionate explains the widespread bacterial dissimilation of the compound (5, 21). The degradative pathway of isethionate requires an unknown uptake system (see reference 5) and, in Paracoccus denitrificans NKNIS, it involves an inducible, 62-kDa, cytochrome c-dependent, membrane-bound isethionate dehydrogenase (EC 1.1.2.-), whose product, sulfoacetaldehyde, is desulfonated to acetyl phosphate and sulfite by sulfoacetaldehyde acetyltransferase (Xsc; EC 2.3.3.15) (3, 5, 23, 31). The genes for isethionate dissimilation were unknown (3); only one xsc gene was known in strain NKNIS, and it was located in a gene cluster encoding taurine degradation.
Dissimilation of C2 sulfonates converges at a single Xsc in many organisms (5, 31). Some organisms, however, have several xsc paralogues which might represent different complete pathways for different C2 sulfonates. Ruegeria pomeroyi DSS-3 contains the xsc gene (SPO3561), which is expressed during growth with taurine (12). Additionally, SPO2360 is annotated as an xsc pseudogene (the gene product lacks key amino acids in the active site). The presence of this second xsc, coupled with isethionate utilization (8), led us to hypothesize that the flanking genes could encode isethionate uptake and isethionate dehydrogenase genes (see below). Orthologues of putative isethionate dehydrogenase (IseJ) were found in other organisms, several of them known isethionate degraders, whereas iseJ was absent in the genomes of isethionate nonutilizers. The hypothesis was tested in a terrestrial isethionate degrader (Cupriavidus necator H16) and a marine isethionate degrader (R. pomeroyi DSS-3), where data from growth kinetics, enzymology, and reverse transcription-PCR (RT-PCR) largely confirmed the initial hypothesis.
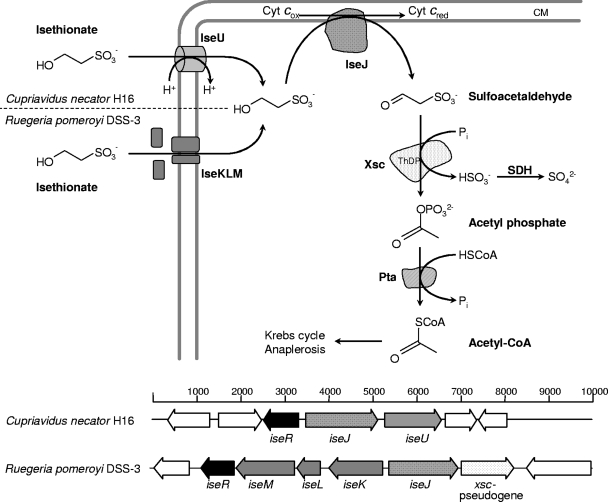
Gene candidates for isethionate biodegradation.
Sequence data were analyzed with Lasergene, SignalP, TMHMM (http://www.cbs.dtu.dk./services/), and PROSITE (http://www.expasy.org/) software. We concentrated on the flanking regions of the xsc pseudogene in R. pomeroyi DSS-3 (genome accession numbers NC_003911 and NC_006569), especially the five genes in the upstream region. They could encode a dehydrogenase (SPO2359) of the BetA family (glucose-methanol-choline [GMC] oxidoreductases; COG2303), particulate enzymes of 60 to 64 kDa (http://www.brenda-enzymes.info/php/result_flat.php4?ecno=1.1.99.1&organism=), and, divergently transcribed, a tripartite ATP-independent (TRAP) transporter (SPO2358 to SPO2356; TC 2.A.56.-.-) and an IclR-type transcriptional regulator (SPO2355). This allowed the following hypothesis (Fig. 1): that isethionate was transported into the cell via a TRAP transporter, that an orthologue of the membrane-associated, 62-kDa isethionate dehydrogenase found earlier (3) was encoded by the BetA-type gene, and that both moieties were under the control of an IclR-type transcriptional regulator. Degradation of sulfoacetaldehyde would then proceed via Xsc (SPO3561) and Pta (SPO3560) as described elsewhere (Fig. 1) (12).
FIG. 1.
Initial steps of isethionate degradation in Cupriavidus necator H16 and Ruegeria pomeroyi DSS-3 with the corresponding gene clusters in C. necator H16 and R. pomeroyi DSS-3. CM, cytoplasmic membrane; IseJ, isethionate dehydrogenase; Xsc, sulfoacetaldehyde acetyltransferase; Pta, phosphate acetyltransferase; SDH, sulfite dehydrogenase; ThDP, thiamine diphosphate. In strain C. necator H16, where data are available, SDH is periplasmic (9).
The putative gene products were compared individually with protein sequences in the NCBI database (http://www.ncbi.nlm.nih.gov/) using the BLAST algorithm. Many orthologues of IseJ were found, and a dendrogram showed four major clusters, two of which presumably represent IseJ in three groups (see Fig. S1 in the supplemental material).
Some 35 organisms, shown in Fig. S1 in the supplemental material, contained at least one additional ise gene candidate (see Table S3 in the supplemental material), as well as xsc, and these organisms were thus candidates to dissimilate isethionate. The terrestrial isolates listed in Table S3 in the supplemental material, e.g., C. necator H16 (genome accession numbers NC_008313, NC_008314, and NC_005241), were predicted to use a major facilitator superfamily (MFS) isethionate transporter (IseU) instead of the TRAP system (Fig. 1). Our rationale for this is that we predict a higher dilution of isethionate in marine environments than in terrestrial environments. Thus, a facilitator suffices in terrestrial organisms, whereas a powerful pump is needed by marine bacteria, which is consistent with a favored use of TRAP transporters in saline environments (22).
Growth physiology of organisms utilizing isethionate.
All organisms (see Table S1 in the supplemental material) except Rhodobacterales sp. HTCC2150 were grown aerobically at 30°C in batch culture (media are listed in Table S1 in the supplemental material). Taurine (>99%; Fluka), isethionate (>98%; Fluka), or acetate (>99%; Merck) were sterilized by autoclaving and used as sole added sources of carbon and energy for growth (10 to 20 mM). Growth experiments were done as described elsewhere (31). Growth was followed turbidimetrically at 580 nm or determined as protein in whole cells (16). Isethionate was determined by ion chromatography (IC) (28). Sulfate was quantified turbidimetrically as an insoluble suspension of BaSO4 (26), and sulfite was quantified as the fuchsin adduct (7).
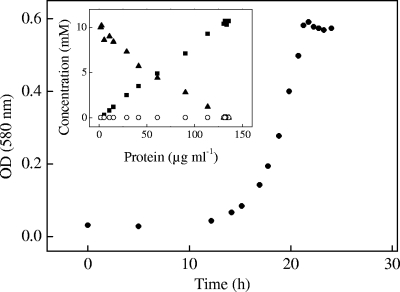
In growth experiments with C. necator H16, ammonium chloride was replaced by ammonium nitrate, which allowed the first determination of isethionate concentration during its degradation by a microorganism (chloride ions interfere during IC detection) (28). Growth with isethionate as the sole source of carbon and energy was rapid (μ = 0.35 h−1) and concomitant with both isethionate utilization and sulfate release, which was quantitative (Fig. 2). The growth yield was 6.6 g protein (mol C)−1, confirming quantitative utilization of isethionate (see reference 4). R. pomeroyi DSS-3 utilized isethionate (μ = 0.06 h−1) with quantitative recovery of sulfate.
FIG. 2.
Growth of Cupriavidus necator H16 with 10 mM isethionate as sole source of carbon and energy. Shown are concentrations of substrate and products during growth of C. necator H16, plotted as a function of the protein concentration (inlay), isethionate (filled triangles), sulfate (filled squares), and sulfite (open circles). An optical density at 580 nm [OD (580 nm)] of 0.5 corresponded to 113 μg protein ml−1.
Rhodobacterales sp. HTCC2150 was cultivated at 16°C in sterilized supplemented seawater medium (see Table S1 in the supplemental material) because the organism is not cultivable in defined media. Isethionate (2, 5, 10, and 20 μM) or glucose (1 and 10 μM) was added, and the increase in cell densities and final cell yields were determined by flow cytometry on a Guava EasyCyte cell counter (Guava Technologies, Hayward, CA) (27). Growth was carbon limited as shown by comparing counts of cells in outgrown medium which had contained 0.0, 1.0, and 10.0 μM glucose; replicate flasks without the addition of glucose yielded 1.1 × 106 cells ml−1, while additions of glucose resulted in a linear increase of about 5.5 × 104 cells (nmol glucose-carbon)−1. Growth with isethionate (2 to 20 μM) resulted in a linear increase in cell yield of about 1.3 × 105 cells (nmol isethionate-carbon)−1, showing that the organism utilized isethionate and growth was proportional to the substrate concentration.
Enzyme activities involved in isethionate degradation in C. necator H16.
Preparation of cell extracts and separation of membrane and soluble fractions were done as described elsewhere (3), and protein content was assayed by protein-dye binding (2). IseJ was assayed spectrophotometrically as the isethionate-dependent reduction of beef heart cytochrome c at 550 nm. The reaction mixture contained 50 μmol potassium phosphate buffer, pH 7.2 (with 5 mM MgCl2), 12.5 μmol isethionate, 50 nmol cytochrome c, and 0.1 to 0.5 mg protein. The reaction was linear for at least 2 min. Xsc was assayed by the colorimetric determination of acetyl phosphate (23).
Cell extracts of isethionate-grown cells showed activities of both IseJ (0.3 mkat kg−1) and Xsc (2.8 mkat kg−1), whereas extracts of acetate-grown cells did not. After separation of soluble and membrane fractions, Xsc activity (3.4 mkat kg−1) was found solely in the soluble fraction, whereas IseJ (0.1 mkat kg−1) was active in the membrane fraction, as predicted (3). An inducible membrane protein of 60 to 62 kDa was detected in the membrane fraction by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). These enzyme activities confirmed the inducible presence of the predicted pathway (Fig. 1) (3, 17, 19).
RT-PCR of ise gene candidates.
Total RNA was isolated using the E.Z.N.A. bacterial kit (Omega Bio-Tek, Doraville, GA). Primers (see Table S2 in the supplemental material) were synthesized by Microsynth (Balgach, Switzerland), and reverse transcription and PCR were performed as described elsewhere (14, 31) using materials from Fermentas GmbH (St Leon-Rot, Germany). PCR products were visualized on 1.5% agarose gels (25). The hypothesis that gene candidates in C. necator H16 would encode uptake (iseU) and oxidation (iseJ) of isethionate was tested by RT-PCR. No transcript was detected in acetate-grown cells, whereas the transcript of each gene (monocistronic transcription) was detected in isethionate-grown cells (see Fig. S2 in the supplemental material). The gene candidate iseJKLM of R. pomeroyi DSS-3 was hypothesized to encode a TRAP transporter and IseJ (Fig. 1). No transcript was detected in acetate-grown cells, whereas a transcript of each gene was detected in isethionate-grown cells (not shown). We interpret these data as evidence that IseJ and IseKLM in R. pomeroyi DSS-3, or IseJ and IseU in C. necator H16, are involved in isethionate utilization.
Data from genome sequencing projects.
The gene cluster identified in C. necator H16 (iseRJU) (sometimes iseRUJ) was found in 16 other terrestrial isolates whose genomes have been sequenced (see Table S3 in the supplemental material). This allowed us to predict utilization of isethionate by these organisms, which we confirmed by growth experiments with seven organisms that were available to us (Table S3) (1, 3, 8, 21). In some organisms, e.g., P. denitrificans PD1222, Sinorhizobium medicae WSM419, and Dinoroseobacter shibae DFL 12, the ise genes are contiguous with the xsc and pta genes, presumably encoding a complete and independent isethionate degradative pathway.
The gene cluster identified in R. pomeroyi DSS-3, iseJKLMR, was found in 13 other marine genome-sequenced organisms (see Table S3 in the supplemental material), which allowed us to predict isethionate dissimilation in those isolates. The hypothesis was confirmed by growth experiments with two strains available to us (Table S3). We regard this as confirmation of the function of IseJKLM.
Two more marine isolates (Alphaproteobacterium sp. HTCC2255 and Rhodobacterales sp. HTCC2150) (see Table S3 in the supplemental material) contain iseRJ (as well as xsc and pta), but neither iseKLM nor iseU. Rhodobacterales sp. HTCC2150 did, however, utilize isethionate (see above). A TerC family (TC 9.A.30.-.-) hypothetical protein is encoded adjacent to iseJ, with a 1-bp overlap. This protein, with six predicted transmembrane helices, may represent a third type of isethionate transporter (RB2150_10781), possibly involving the neighboring genes (RB2150_10776 to RB2150_10771). These three genes are conserved upstream of iseJ in Alphaproteobacterium sp. HTCC2255 and in two sequences from a metagenome project (24).
Brüggemann et al. (3) predicted the presence of a regulator of isethionate dissimilation. We now presume that IseR, an IclR-type protein, fulfils this function, because it is almost always colocalized with the genes encoding the degradative pathway (Fig. 1; see Table S3 in the supplemental material). The biodiversity of the isethionate pathway, with at least three types of transporters, concurs with the widespread occurrence of isethionate in the environment.
Sequence prediction in P. denitrificans NKNIS.
The biochemical background to the present paper is the presence of an inducible, 62-kDa, membrane-bound protein in P. denitrificans NKNIS, assumed to be IseJ (3). The genome of a different strain of P. denitrificans, PD1222, has now been sequenced (NC_010355), and its nine-gene taurine cluster is essentially identical to the corresponding sequence in strain NKNIS (not shown). We predicted that the ise gene cluster in strain NKNIS would also show high similarity to that in strain PD1222. We amplified and sequenced overlapping fragments of the cluster in NKNIS. Chromosomal DNA was isolated as described elsewhere (10), and the PCR primers listed in Table S2 in the supplemental material were used to amplify the fragments. PCR products were purified with the E.Z.N.A. Cycle-Pure kit (Omega Bio-Tek). Sequencing was done at MWG-Biotech AG (Ebersberg, Germany). We confirmed our hypothesis by finding a 3.5-kb fragment which shared 99% identity with the corresponding region (iseRUJ-xsc) in strain PD1222. Thus, the isethionate-positive phenotype of P. denitrificans NKNIS was confirmed to be accompanied by the iseRUJ-xsc genotype.
The power of prediction—that the presence of iseJ indicates isethionate utilization in an organism on the one hand, and that membrane-bound isethionate dehydrogenase indicates the presence of iseJ in the organism's genome on the other hand—is surely sufficient to establish that iseJ encodes isethionate dehydrogenase (EC 1.1.2.-). Nevertheless, in the future, it will hopefully be possible to purify IseJ and identify any cofactors. As an alternative, one could construct a deletion mutant defective in iseJ, but these experiments were unsuccessful, despite being able to delete other genes in the same organism (S. Weinitschke, unpublished data).
Nucleotide sequence accession number.
The gene sequence reported in this paper has been deposited in the GenBank database (accession no. EU025134).
Supplementary Material
Acknowledgments
We are grateful to S. J. Giovannoni for making available both lab space and apparatus, to S. Vuilleumier for providing sequence data of Methylobacterium spp. prior to publishing, to B. Bowien, M. A. Moran, and J. M. Tiedje for providing bacteria, and to K. Denger for discussions.
S.W. was supported by a grant (Co 206/6) from the Deutsche Forschungsgemeinschaft to A.M.C. and T.H.M.S. U.S. (and S. J. Giovannoni) were supported by the Marine Microbiology Initiative of the Gordon and Betty Moore Foundation. P.I.S. was supported by funds from the University of Konstanz.
Footnotes
Published ahead of print on 20 November 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Baldock, M. I., K. Denger, T. H. M. Smits, and A. M. Cook. 2007. Roseovarius sp. strain 217: aerobic taurine dissimilation via acetate kinase and acetate-CoA ligase. FEMS Microbiol. Lett. 271:202-206. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Brüggemann, C., K. Denger, A. M. Cook, and J. Ruff. 2004. Enzymes and genes of taurine and isethionate dissimilation in Paracoccus denitrificans. Microbiology 150:805-816. [DOI] [PubMed] [Google Scholar]
- 4.Cook, A. M. 1987. Biodegradation of s-triazine xenobiotics. FEMS Microbiol. Rev. 46:93-116. [Google Scholar]
- 5.Cook, A. M., and K. Denger. 2002. Dissimilation of the C2 sulfonates. Arch. Microbiol. 179:1-6. [DOI] [PubMed] [Google Scholar]
- 6.Delobel, P., and R. Pradinaud. 2003. Rhabdomyolysis associated with pentamidine isethionate therapy for American cutaneous leishmaniasis. J. Antimicrob. Chemother. 51:1319-1320. [DOI] [PubMed] [Google Scholar]
- 7.Denger, K., J. Ruff, U. Rein, and A. M. Cook. 2001. Sulphoacetaldehyde sulpho-lyase (EC 4.4.1.12) from Desulfonispora thiosulfatigenes: purification, properties and primary sequence. Biochem. J. 357:581-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denger, K., T. H. M. Smits, and A. M. Cook. 2006. L-Cysteate sulpho-lyase, a widespread pyridoxal 5′-phosphate-coupled desulphonative enzyme purified from Silicibacter pomeroyi DSS-3T. Biochem. J. 394:657-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denger, K., S. Weinitschke, T. H. M. Smits, D. Schleheck, and A. M. Cook. 2008. Bacterial sulfite dehydrogenases in organotrophic metabolism: separation and identification in Cupriavidus necator H16 and in Delftia acidovorans SPH-1. Microbiology 154:256-263. [DOI] [PubMed] [Google Scholar]
- 10.Desomer, J., M. Crespi, and M. Van Montagu. 1991. Illegitimate integration of non-replicative vectors in the genome of Rhodococcus fascians upon electrotransformation as an insertional mutagenesis system. Mol. Microbiol. 5:2115-2124. [DOI] [PubMed] [Google Scholar]
- 11.Fellman, J. H., E. S. Roth, N. A. Avedovech, and K. D. McCarthy. 1980. The metabolism of taurine to isethionate. Arch. Biochem. Biophys. 204:560-567. [DOI] [PubMed] [Google Scholar]
- 12.Gorzynska, A. K., K. Denger, A. M. Cook, and T. H. M. Smits. 2006. Inducible transcription of genes involved in taurine uptake and dissimilation by Silicibacter pomeroyi DSS-3T. Arch. Microbiol. 185:402-406. [DOI] [PubMed] [Google Scholar]
- 13.Holst, P. B., S. E. Nielsen, U. Anthoni, K. S. Bisht, C. Christophersen, S. Gupta, V. S. Parmar, P. H. Nielsen, D. B. Sahoo, and A. Singh. 1994. Isethionate in certain red algae. J. Appl. Phycol. 6:443-446. [Google Scholar]
- 14.Innis, M. A., D. H. Gelfand, J. J. Sninsky, and T. J. White. 1990. PCR protocols. A guide to methods and applications. Academic Press, San Diego, CA.
- 15.Jacobsen, J. G., L. L. Collins, and L. H. Smith, Jr. 1967. Urinary excretion of isethionic acid in man. Nature 214:1247-1248. [DOI] [PubMed] [Google Scholar]
- 16.Kennedy, S. I. T., and C. A. Fewson. 1968. Enzymes of the mandelate pathway in bacterium N.C.I.B. 8250. Biochem. J. 107:497-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King, J. E., R. Jaouhari, and J. P. Quinn. 1997. The role of sulfoacetaldehyde sulfo-lyase in the mineralization of isethionate by an environmental Acinetobacter isolate. Microbiology 143:2339-2343. [DOI] [PubMed] [Google Scholar]
- 18.Koechlin, B. A. 1954. The isolation and identification of the major anion fraction of the axoplasm of squid giant nerve fibers. Proc. Natl. Acad. Sci. U. S. A. 40:60-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondo, H., H. Niki, S. Takahashi, and M. Ishimoto. 1977. Enzymatic oxidation of isethionate to sulfoacetaldehyde in bacterial extract. J. Biochem. 81:1911-1916. [DOI] [PubMed] [Google Scholar]
- 20.Kumpulainen, E., I. Pesonen, and P. Lahdesmaki. 1982. Exchange of isethionate between blood and tissues in adult and 7-day-old mice. Acta Physiol. Scand. 114:419-423. [DOI] [PubMed] [Google Scholar]
- 21.Lie, T. L., J. R. Leadbetter, and E. R. Leadbetter. 1998. Metabolism of sulfonic acids and other organosulfur compounds by sulfate-reducing bacteria. Geomicrobiol. J. 15:135-149. [Google Scholar]
- 22.Mulligan, C., D. J. Kelly, and G. H. Thomas. 2007. Tripartite ATP-independent periplasmic transporters: application of a relational database for genome-wide analysis of transporter gene frequency and organization. J. Mol. Microbiol. Biotechnol. 12:218-226. [DOI] [PubMed] [Google Scholar]
- 23.Ruff, J., K. Denger, and A. M. Cook. 2003. Sulphoacetaldehyde acetyltransferase yields acetyl phosphate: purification from Alcaligenes defragrans and gene clusters in taurine degradation. Biochem. J. 369:275-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sabehi, G., A. Loy, K. H. Jung, R. Partha, J. L. Spudich, T. Isaacson, J. Hirschberg, M. Wagner, and O. Béjà. 2005. New insights into metabolic properties of marine bacteria encoding proteorhodopsins. PLoS Biol. 3:e273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Laboratory Press, Cold Spring Harbor, NY.
- 26.Sörbo, B. 1987. Sulfate: turbidimetric and nephelometric methods. Methods Enzymol. 143:3-6. [DOI] [PubMed] [Google Scholar]
- 27.Stingl, U., H. J. Tripp, and S. J. Giovannoni. 2007. Improvements of high-throughput culturing yielded novel SAR11 strains and other abundant marine bacteria from the Oregon coast and the Bermuda Atlantic Time Series study site. ISME J. 1:361-371. [DOI] [PubMed] [Google Scholar]
- 28.Styp von Rekowski, K., K. Denger, and A. M. Cook. 2005. Isethionate as a product from taurine during nitrogen-limited growth of Klebsiella oxytoca TauN1. Arch. Microbiol. 183:325-330. [DOI] [PubMed] [Google Scholar]
- 29.Sun, J. Z., J. W. Parr, and M. C. Erickson. 2003. Solubilization of sodium cocoyl isethionate. J. Cosmet. Sci. 54:559-568. [PubMed] [Google Scholar]
- 30.Townley, M. A., E. K. Tillinghast, and C. D. Neefus. 2006. Changes in composition of spider orb web sticky droplets with starvation and web removal, and synthesis of sticky droplet compounds. J. Exp. Biol. 209:1463-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinitschke, S., K. Denger, A. M. Cook, and T. H. M. Smits. 2007. The DUF81 protein TauE in Cupriavidus necator H16, a sulfite exporter in the metabolism of C2 sulfonates. Microbiology 193:3055-3060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.