Abstract
To prevent transmission of mycobacterial pathogens, medical devices must be disinfected by germicides with proven mycobactericidal activity. The quantitative carrier test EN 14563 provides an international standard for evaluation of the mycobactericidal activity of disinfectants under practical conditions. However, tests according to the EN 14563 standard are based on cultivation, and results are available only after 21 days. The aim of this study was to accelerate assessment of dosage and contact times of mycobactericidal preparations based on the EN 14563 standard. To this end, a gfp gene was constructed with a codon usage adapted for Mycobacterium tuberculosis. Expression of the gfpm2+ gene in Mycobacterium terrae improved the detection sensitivity by 10-fold over that with a previously used reporter strain. Peracetic acid and a cation-active formulation were tested as commercially available disinfectants for medical devices. M. terrae expressing gfpm2+ was used to determine dosage and contact times for the two test germicides. Fluorescence measurements correlated well with growth of the reporter strain, demonstrating that the fluorescence reliably indicated the number of viable cells. The fluorescence enabled us to determine the mycobactericidal efficacy of the test disinfectants according to the quantitative carrier test EN 14563 standard within at least 15 days. In conclusion, this study establishes gfpm2+-expressing M. terrae as a new reporter strain for reliable evaluation of mycobactericidal activities of disinfectants with a superior sensitivity and in a significantly shorter time than previously possible.
Prevention of infections by mycobacteria is an important goal due to their paramount clinical relevance. This task presents a significant challenge, since mycobacteria are intrinsically resistant to many chemical agents (2). In order to avoid device-related transmission of mycobacterial pathogens, germicides developed for disinfection of medical devices must be proven to possess mycobactericidal activity. Conventional testing of disinfectants for their tuberculocidal activity is based on cultivation techniques. Mycobacterium terrae, which has been demonstrated to possess resistance to chemical disinfectants similar to that of Mycobacterium tuberculosis (5), is used as a surrogate for establishing tuberculocidal activity of germicides. However, such an assay takes at least 3 weeks because of the slow growth of M. terrae. In order to accelerate the process of screening chemicals for mycobactericidal activity, Zafer et al. has described the use of gfp-expressing M. terrae in quantitative suspension tests (18). Generally, suspension tests are used to determine the overall efficacy of disinfectants against mycobacteria. However, mycobacteria are used in cell suspensions in these experiments and thus are likely to be more accessible to disinfectants. Therefore, it is essential to demonstrate the mycobactericidal efficacy of disinfectants for medical devices under practical conditions. For these reasons, the European Committee for Standardization developed the quantitative carrier test EN 14563, which evaluates the mycobactericidal activity of chemical disinfectants for medical instruments under more practical conditions (4). The aim of this study was to establish an assay which allows rapid determination of the dosage and contact times in the presence of organic soiling, as demanded by the EN 14563 standard. In order to achieve this goal, an optimized variant of the green fluorescent protein (Gfp) for use in mycobacteria was constructed. By expression of gfpm2+ in M. terrae, a significant improvement in sensitivity was achieved, which enabled us to rapidly evaluate the performance of disinfectants under practical conditions.
MATERIALS AND METHODS
Test organisms and growth conditions.
Escherichia coli DH5α was used for cloning experiments and was routinely grown in Luria-Bertani broth at 37°C. Mycobacterium smegmatis mc2155 and Mycobacterium bovis BCG were grown in 7H9 medium (Difco) supplemented with 0.2% glycerol and 0.05% Tween 80 at 37°C and for M. bovis BCG additionally enriched with ADS (50 g/liter bovine serum albumin [BSA], 20 g/liter glucose, and 8.5 g/liter NaCl). Antibiotic concentrations were as follows: kanamycin, 10 (mycobacteria) or 30 (E. coli) μg/ml; hygromycin B, 50 (mycobacteria) or (E. coli) 20 μg/ml.
M. terrae ATCC 15755 was cultivated according to the EN 14563 standard (4). M. terrae transformed with the plasmid pBEN was cultivated in the presence of 30 μg/ml kanamycin; M. terrae transformed with the plasmid pMN437 was cultivated in the presence of 75 μg/ml hygromycin B.
Liquid cultures of all M. terrae strains were obtained by cultivation in Middelbrook 7H9 broth (Becton-Dickinson, Germany), supplemented with ADC enrichment (Becton-Dickinson, Germany) in the presence of the respective antibiotic for transformed M. terrae.
Synthesis of gfp genes with codon usage optimized for translation in mycobacteria.
The gfpm+ and gfpm2+ genes were synthesized with codon usage optimized for expression in M. tuberculosis using a table similar to that provided recently (12). The Gfp protein encoded by gfpm+ contains the same fluorescence- and folding-enhancing mutations (F64L, S65T, F99S, M153T, and V163A) as published for Gfp+ (9). The gfpm2+ gene encodes a protein with the two additional mutations I167T and S175G, which enhance the thermostability of Gfp (10). The GenBank accession number for the gfpm2+ gene is GU112756.
Construction of gfp expression plasmids.
All plasmids contained a transcriptional fusion of the gfp allele to the rather weak pwmyc promoter (6) for comparison of different Gfp variants in M. smegmatis and M. bovis BCG. Vectors and PCR fragments were digested with PacI and ClaI and cloned into the same sites of pMN012, a multicopy shuttle vector for E. coli and mycobacteria (6). Additionally, the gfpm2+ gene was also cloned into pMN014 (6) using the PacI and ClaI restriction sites to give vector pMN437, in which transcription of gfpm2+ is driven by the strong mycobacterial promoter psmyc. All plasmids were verified by DNA sequencing (Table 1).
TABLE 1.
Plasmids used in this worka
| Plasmid | Parent vector, relevant genotype, and properties | Source or reference |
|---|---|---|
| pMS2 | ColE1 origin; PAL5000 origin; Hygr; 5,229 bp | 6 |
| pMN012 | pwmyc-mspA; ColE1 origin; PAL5000 origin; Hygr; 6,000 bp | 6 |
| pMN016 | psmyc-mspA; ColE1 origin; PAL5000 origin; Hygr; 6,164 bp | 14 |
| pEGFP | egfp expression vector; Ampr; 3,355 bp | Clontech |
| pMN411 | pMN012 derivative, pwmyc-gfp+; pAL5000 origin; Hygr, 6,250 bp | This study |
| pMN412 | pMN012 derivative, pwmyc-gfpSDm+; pAL5000 origin; Hygr, 6,250 bp | This study |
| pMN413 | pMN012 derivative, pwmyc-gfpm+; pAL5000 origin; Hygr, 6,210 bp | This study |
| pMN414 | pMN012 derivative, pwmyc-gfpm2+; pAL5000 origin; Hygr, 6,210 bp | This study |
| pMN415 | pMN012 derivative, pwmyc-egfp; pAL5000 origin; Hygr, 6,220 bp | This study |
| pMN437 | pMN016 derivative, psmyc- gfpm2+; pAL5000 origin; Hygr, 6,250 bp | This study |
“Origin” denotes origin of replication. The annotation Hygr or Kanr indicates that the plasmid confers resistance to hygromycin or kanamycin, respectively.
In vivo fluorescence measurements.
The plasmids pMN411, pMN412, pMN413, pMN414, pMN415, and pUV25 (Table 1) were transformed into M. smegmatis and M. bovis BCG, respectively. A preculture was grown for 48 h at 37°C. Twenty ml medium was inoculated with 0.3 ml of the preculture and shaken at 37°C until the culture reached an optical density at 600 nm (OD600) of 0.5. Cells were harvested by centrifugation and resuspended in 2 ml phosphate-buffered saline (PBS) containing Tween 80 (4.3 mM Na2HPO4, 1.4 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, 0.5 g/liter Tween 80). The ratio of fluorescence intensity to cell density was taken as a measure for Gfp expression levels. Fluorescence measurements were carried out essentially as described previously (9). M. smegmatis and M. bovis BCG cells expressing Gfpuv, Gfpm+, enhanced green fluorescent protein (Egfp), and Gfpm2+ were excited at 395 nm or 488 nm, respectively. Fluorescence emission was detected at 510 nm. Background fluorescence of M. smegmatis and M. bovis BCG cells was determined at the same wavelengths using the vector pMS2, which does not contain a gfp gene (Table 1).
For the quantitative carrier test according to EN 14563, M. terrae ATCC 15755 was transformed with pMN437 (gfpm2+) and with pBEN (gfpmut3), a gift from S. A. Sattar (University of Ottawa, Ottawa, Ontario, Canada) (18). Expression of the Gfp variants was verified by fluorescence microscopy.
Quantitative carrier tests and subsequent Gfp-based determination of efficacy of tested germicides.
The quantitative carrier tests were carried out according to EN 14563 (4). As test germicides, two commercially available formulations for disinfection of medical devices were used, one being a peracetic acid-based germicide (PAA) (Gigasept once; Schülke & Mayr, United Kingdom) and the other being a product consisting of a combination of 14% (wt/wt) cocospropylenediamineguanidinediacetate, 35% (wt/wt) phenoxypropanols, and 2.5% (wt/wt) benzalkoniumchloride (“Gigasept Instru AF”; Schülke & Mayr, Germany). For neutralization Middlebrook 7H9 broth containing ADC-enrichment and β-cyclodextrine (1.135% [wt/vol]), sodium thiosulfate (0.5% [wt/vol]), L-histidine (0.1% [wt/vol]), and glycerol (0.2% [vol/vol]) was demonstrated to be effective in the neutralization experiments for both biocidal formulations tested and was therefore used throughout the experiments.
In the carrier test, 1 ml and 100 μl, respectively, of the neutralized test suspension were used for determination of the logarithmic reduction factor (R = log N0 − log Na, where N0 is CFU/ml of control and Na is CFU/ml in sample after exposure to disinfectant) as described in the EN 14563 standard (4) in parallel to the Gfp-based detection. For the Gfp-based determination of efficacy, the residual 8.9 ml of the neutralized test suspensions was filtered through a funnel containing sterile glass wool in order to retain the glass beads used in the carrier test setup. The filtered mycobacterial suspensions were washed twice in 10 ml Middlebrook 7H9 medium supplemented with ADC enrichment and hygromycin (75 μg/ml) and were subsequently resuspended in the same medium. One ml of the resuspended mycobacterial suspensions was used for initial determination (day 0) of the CFU by serial dilution in tryptone-NaCl solution (EN 14563) and subsequent plating on 7H10 agar supplemented with oleic acid-albumin-dextrose-catalase (OADC) enrichment and hygromycin (75 μg/ml). Plates were sealed and incubated for 21 days at 37°C.
The residual 9 ml of the mycobacterial suspension was aliquoted each into three screw-cap, clear glass vials (3 ml/vial). For determination of the relative fluorescence units (RFU), each glass vial was inserted in the cuvette holder of the fluorimeter (FP-6300; Jasco, Germany). Excitation and emission of Gfp were measured at 485 nm and 512 nm, respectively, with a bandwidth of 5 nm. Each glass vial was vortexed for 5 s before readings were taken, and reading was done in triplicate for each glass vial within 6 s. Parallel to the RFU readings, CFU determination was carried out as described above, using 100 μl of the corresponding sample. Experiments were carried out as indicated either in the presence or in the absence of organic load (3 g/liter bovine serum albumin [BSA] and 3 ml/liter sheep erythrocytes).
RESULTS
Optimization of Gfp fluorescence in mycobacteria.
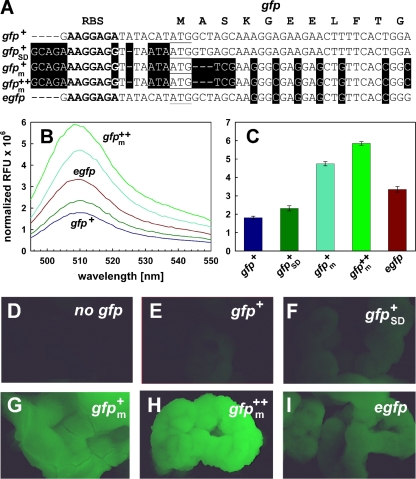
Previously, we had developed Gfp+, a variant with mutations which improved the brightness and the folding of the protein, resulting in a 130-fold-increased fluorescence compared to that of wild-type Gfp (9). However, the fluorescence of M. smegmatis and M. bovis BCG containing a gfp+ expression plasmid was low in suspended cells and barely visible in colonies on agar plates (Fig. 1; see also Fig. S1 in the supplemental material). We hypothesized that the expression levels of Gfp+ were lower in mycobacteria due to the much lower G+C content of gfp+ than of mycobacterial genes. In a first step to increase Gfp expression levels, we altered the ribosome binding site to a sequence perfectly complementary to the 16S rRNA of M. tuberculosis (7). The distance between this mycobacterial consensus Shine-Dalgarno sequence (SDm) and the start codon was reduced from eight to six nucleotides (Fig. 1A). These changes barely increased the fluorescence of Gfp+, indicating that binding of the ribosome and translation initiation to the original gfp+ mRNA did not limit expression. In a second step, the codon usage of gfp+ was adapted to that of M. tuberculosis as described previously (12). This change removed 28 rare codons (less than 5% frequency in M. tuberculosis H37Rv) and elevated the G+C content of the gfp+ gene from 42% to 61%. The protein sequence and the spectral characteristics of Gfp+ were not affected by these alterations (Fig. 1B). However, the fluorescence of M. bovis BCG cells transformed with the synthetic gfpm+ gene increased by 2.5-fold (Fig. 1C), demonstrating that the less-favorable codon usage of gfp+ indeed limited its expression in mycobacteria. In a last step, we added the mutations I167T and S175G to enhance the thermostability of Gfp+ (10). This increased the fluorescence intensity of M. bovis BCG by approximately 20% (Fig. 1C), indicating that the increased thermostability is indeed beneficial for Gfp fluorescence in mycobacteria. The fluorescence of M. bovis BCG containing the gfpm2+ gene was 2-fold higher than that with egfp, a gene which is frequently used in mycobacteria (Fig. 1C). The fluorescence properties of these Gfp variants are very similar (Fig. 1B). Microscopy of colonies visualized the bright green fluorescence of M. bovis BCG containing the gfpm2+ gene in contrast to that containing gfp+ or egfp (Fig. 1D to I). Very similar results for the gfp variants created in this study were obtained in M. smegmatis (see Fig. S1 in the supplemental material), indicating that expression signals, codon usage, and optical transmittance at the excitation and emission wavelengths are similar in both organisms. It also showed that the gfpm2+ gene is a superior gfp variant in both fast- and slow-growing mycobacteria.
FIG. 1.
Fluorescence of codon usage-adapted gfp variants in M. bovis BCG. (A) Sequence of the gfp alleles. The sequence of the ribosome binding site (RBS) is shown in bold. The start codon ATG is underlined. The amino acid sequences of the proteins encoded by these genes are identical except for the mutations introduced to enhance fluorescence and are indicated in one-letter code above the DNA sequence for the first 11 codons. (B and C) Fluorescence of M. bovis BCG containing different pwmyc-gfp transcriptional fusions. (B) Emission spectra. Fluorescence was excited at 488 nm for all Gfp variants. Fluorescence emission was detected at 510 nm. (C) All measurements were carried out in triplicate, corrected for background fluorescence, and normalized to the cell density and are shown in relative fluorescence units (RFU). Standard deviations are shown as error bars. (D to I) M. bovis BCG cells containing described plasmids were inoculated onto 7H10 agar plates (Difco) supplemented with 0.2% glycerol and additionally enriched with ADS at 37°C for 3 weeks. The colonies were visualized using a fluorescence stereomicroscope and a 20-fold magnification. (D) No gfp (pMS2); (E) gfp+ (pMN411); (F) gfp+SD (pMN412); (G) gfpm+ (pMN413); (H) gfpm2+ (pMN414); (I) egfp (pMN415). The fluorescence was recorded using excitation and emission filter sets for Gfp.
Expression of the optimized gfpm2+ gene in M. terrae.
For the reasons described above, we used the gfpm2+ gene in M. terrae to accelerate the testing of biocides for their efficacy against mycobacteria. First, we examined the performance of gfpm2+ compared to gfpmut3, which was previously used in M. terrae for a similar purpose (18). These experiments revealed that the fluorescence of M. terrae cells containing the gfpm2+ gene (pMN437) was 10-fold higher than that of cells transformed with the gfpmut3 expression vector pBEN (18), while titers of the respective cell suspensions were identical. This is reflected by a 10-fold-lower detection limit for M. terrae expressing gfpm2+ than for M. terrae expressing gfpmut3 (5 × 104 versus 4 × 105 CFU/ml) (data not shown). The superior performance of the new gfpm2+ gene should enable a faster evaluation of the mycobactericidal activity of chemical germicides.
Test of germicides for mycobactericidal efficacy by a quantitative carrier test without organic load.
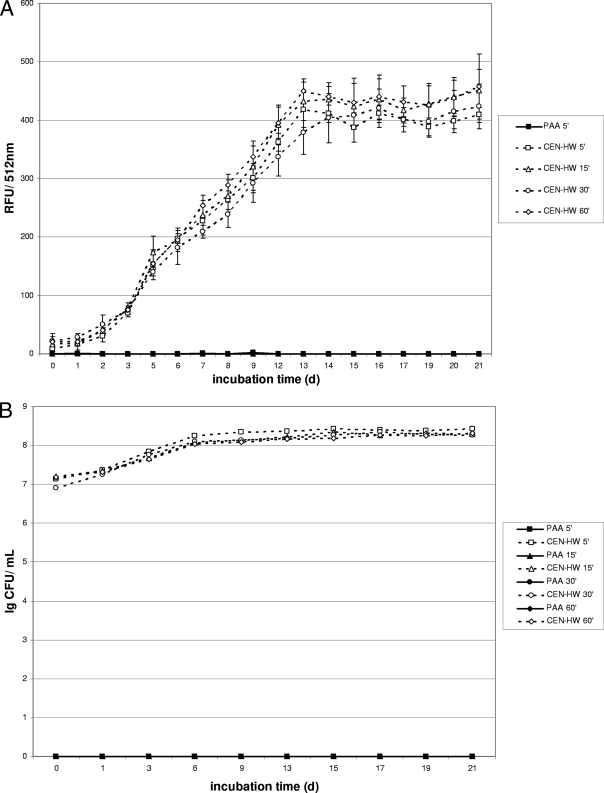
To examine the performance of gfpm2+ under standard testing conditions, formulations based on peracetic acid (PAA) and on cationic actives and phenoxypropanols (GIGA) were chosen. The efficacies of these biocides against M. terrae were examined using a quantitative carrier test as required by the European standard EN 14563 in the absence of any organic load (see Materials and Methods). For the peracetic acid-based formulation (PAA), no increase in fluorescence was detected for all samples treated with PAA even at the shortest contact time (5 min), indicating complete killing of mycobacteria by PAA at a concentration of 1.5% within 5 min in the absence of any organic load (Fig. 2A). For the controls treated with standardized hard water (CEN-HW) instead of PAA, a fluorescence increase was detected within the first 3 to 5 days (Fig. 2A). Comparison of the fluorescence intensity showed a good correlation with the growth of M. terrae (pMN437) throughout the experiment (Fig. 2B). No viable cells were detected in any samples treated with 1.5% PAA, consistent with the lack of fluorescence in these samples, thus enabling a rapid evaluation of the mycobactericidal potential of the concentrations and contact times for this biocidal formulation.
FIG. 2.
Fluorescence and growth of gfpm2+-expressing M. terrae in a quantitative carrier test without organic load. Kinetics were recorded for disinfectants at a concentration of 1.5% (vol/vol). (A and B) Fluorescence (A) is expressed as relative fluorescence units (RFU) and growth of M. terrae (B) is expressed in CFU after exposure to the peracetic acid based-disinfectant (PAA). Since the shortest contact time (5 min) was already effective, data for longer contact times are not shown. (C and D) Fluorescence (C) is expressed as relative fluorescence units (RFU) and growth of M. terrae (D) is expressed in CFU after exposure to the cation-based disinfectant (Gigasept Instru AF). Data are the means ± SD of three RFU determinations and one CFU determination for each disinfectant.
Similar experiments for a formulation based on cationic actives and phenoxypropanols (GIGA) revealed a fluorescence increase within 7 to 9 days for the samples treated for 5 min and within 15 to 17 days for the samples treated for 15 min with the germicide at a concentration of 1.5% (Fig. 2C). Samples treated for 30 or 60 min with this disinfectant did not show any fluorescence increase throughout experiment, thus suggesting 30 min as the effective contact time at the tested concentration of 1.5% for this formulation in the absence of any organic load. These data correlated well with growth experiments using liquid cultures (Fig. 2D), where no viable cells were detected in the samples treated for either 30 or 60 min with this formulation. Both growth and fluorescence of gfpm2+-expressing M. terrae increased after a lag period for contact times with only limited mycobactericidal efficacy. This is consistent with the results obtained for PAA and demonstrates that the Gfp fluorescence of the M. terrae reporter strain is a reliable indicator of its viability and can be used to rapidly evaluate the mycobactericidal efficacies of germicides.
Test of germicides for mycobactericidal efficacy by quantitative carrier test with high organic load.
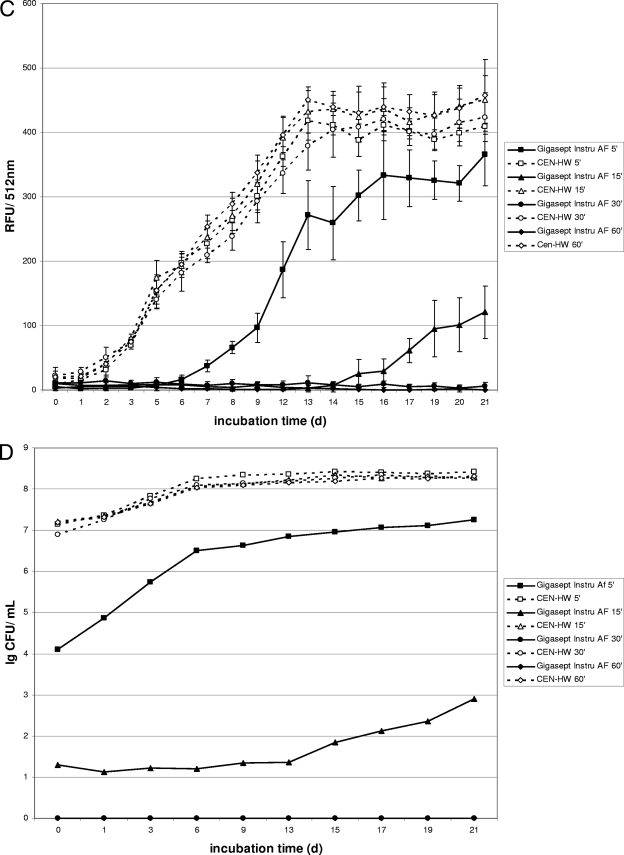
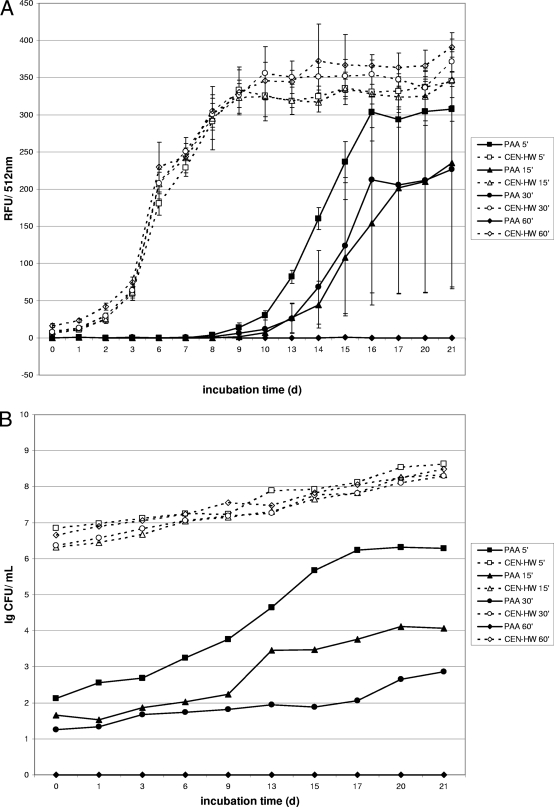
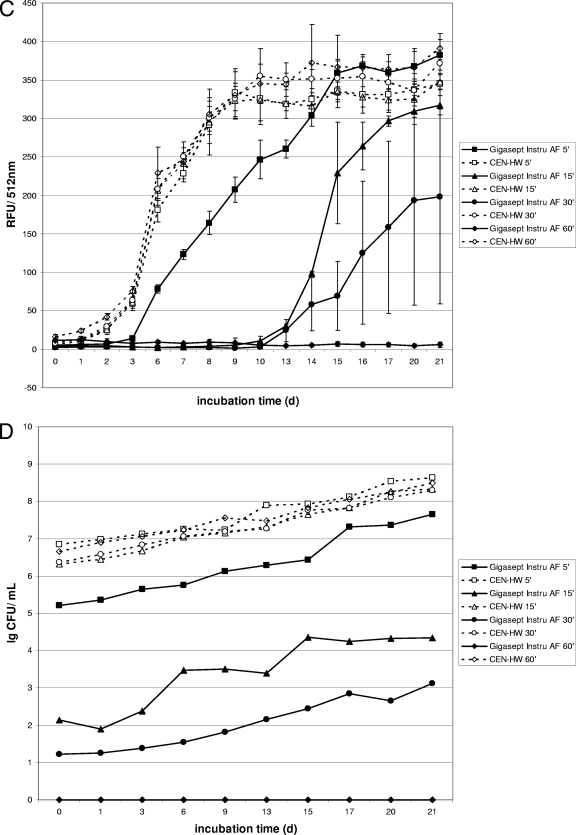
The mycobactericidal efficacy of germicidal formulations according to the EN 14563 standard was tested in the presence of 0.3% serum albumin and 0.3% sheep erythrocytes to simulate a high organic load (“dirty conditions”). Therefore, the carrier test with gfpm2+-expressing M. terrae was also carried out in the presence of a high organic load. Under “dirty conditions,” a fluorescence increase for the shorter contact times within the first 10 to 15 days was obtained for both test formulations (PAA and GIGA) (Fig. 3A and C). However, after a contact time of 60 min, no fluorescence increase was detected for either formulation, indicating complete killing of mycobacteria by PAA and GIGA at a concentration of 1.5% within 60 min in the presence of a high organic load. These data correlate well with results of the growth experiments for both test formulations (Fig. 3B and D), which showed no viable cells for either formulation after a contact time of 60 min.
FIG. 3.
Fluorescence and growth of gfpm2+-expressing M. terrae in a quantitative carrier test in the presence of a high organic load. Kinetics were recorded for disinfectants at a concentration of 1.5% (vol/vol). (A and B) Fluorescence (A) is expressed as relative fluorescence units (RFU) and growth is expressed in CFU for M. terrae after exposure to the peracetic acid based-disinfectant (PAA). (C and D) Fluorescence (C) is expressed in relative fluorescence units (RFU) and growth of M. terrae is expressed in CFU (D) after exposure to the cation-based disinfectant (Gigasept Instru AF). Data are the means ± standard deviations for three RFU determinations and one CFU determination for each disinfectant.
DISCUSSION
Repeated reports of device-related transmission of mycobacterial infections for more than a decade (8, 13) stress the importance of assessing the efficacy of disinfectants used for disinfection of medical devices under practical conditions. Especially for medical devices, organic contamination, in particular in the form of biofilms, is frequent (15, 16). Therefore, disinfectants need to be tested in the presence of organic soiling that mimics realistic conditions. An additional common problem in susceptibility assays is the presence of bovine serum albumin in liquid medium. Serum albumin quenches biocidal activities of disinfectants, thereby impairing the efficacy of biocidal formulations markedly (11). In our experiments, we also found this reported quenching of biocidal actives by organic soiling for both test formulations, indicated by the prolonged contact times necessary to achieve complete killing of mycobacteria in the presence of a high organic load.
Our results demonstrate that the assessment of mycobactericidal efficacy in the quantitative carrier test according to the EN 14563 standard (4) can be significantly accelerated by using an M. terrae reporter strain which contains the optimized gfpm2+ gene. Based on the good correlation of the fluorescence of the reporter strain with its growth throughout the experiments, effective mycobactericidal concentrations and contact times were determined within at least 15 days of incubation. Thus, the assay results can be obtained significantly faster using the fluorescence readout than with the conventional method, which requires cultivation of M. terrae for 21 days (4). For use of concentrations and contact times with only limited mycobactericidal activity, the Gfp-based assay is even more rapid, since the fluorescence increase is detectable within the first few days of incubation. Furthermore, the Gfp-based assay also helps to determine the adequate contact times of disinfectants under practical conditions, including testing under “dirty conditions.” We found the use of the Gfp-based assay in the carrier test even more beneficial, since the use of spread plates can be reduced or even omitted, thus resulting in a significant reduction in hands-on time.
Mycobacteria form clumps which artificially increase drug resistance and therefore represent a well-known problem in susceptibility experiments (3). It has been shown that the degree of clumping increases with time of storage (17). Therefore, storage and preparation of the test organisms prior to testing have an impact on the outcome of the susceptibility tests. In order to minimize the impact of clumping on the experiments, only freshly homogenized mycobacteria were used. However, clumping could not be prevented entirely and resulted in some variation in the readings of Gfp fluorescence of the samples, as is indicated by larger standard deviations. Clumping of M. terrae also could not be completely prevented by addition of Tween 80 (polysorbate 80) (data not shown). However, the use of Tween 80 in susceptibility experiments with mycobacteria cannot be recommended, since Tween 80 is known to interfere with many disinfectants (1). In the experiments with a high organic load, clumping was more pronounced than in the experiments without organic load. However, the differences in fluorescence of the reporter strain in response to effective or ineffective biocide treatments throughout the experiments were large enough to reliably interpret the results. In order to minimize the impact of clumping on the experimental data, only freshly homogenized mycobacteria should be used.
In conclusion, this study establishes gfpm2+-expressing M. terrae as a new reporter strain to reliably evaluate mycobactericidal activities of disinfectants with a superior sensitivity and in a significantly shorter time than previously possible. Many laboratories will benefit from these findings.
Supplementary Material
Acknowledgments
This work was funded by Schülke & Mayr GmbH, Norderstedt, Germany, and by a grant from the Deutsche Forschungsgemeinschaft to M.N. (NI 412).
We thank Mirjana Jevtić for excellent technical assistance and A. Zafer and S. A. Sattar for helpful discussions and for kindly providing us with M. terrae pBEN.
Supplemental material for this article may be found at http://aem.asm.org/.
Footnotes
Published ahead of print on 30 November 2009.
REFERENCES
- 1.Assadian, O., and A. Kramer. 2008. Desinfektion unbelebter Materialien, p. 161-184. In O. Assadian and A. Kramer (ed.), Wallhäußers Praxis der Sterilisation, Desinfektion, Antiseptik und Konservierung, 1st ed. Georg Thieme Verlag, Stuttgart, Germany.
- 2.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 64:29-63. [DOI] [PubMed] [Google Scholar]
- 3.Danilchanka, O., M. Pavlenok, and M. Niederweis. 2008. Role of porins for uptake of antibiotics by Mycobacterium smegmatis. Antimicrob. Agents Chemother. 52:3127-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.European Committee for Standardisation. 2009. European standard EN 14563: chemical disinfectants—quantitative carrier test for evaluation of mycobactericidal activity of chemical disinfectants for instruments used in medical area. Test method and requirements (phase2/step2). European Committee for Standardisation, Brussels, Belgium.
- 5.Griffiths, P. A., J. R. Babb, and A. P. Fraise. 1998. Mycobacterium terrae: a potential surrogate for Mycobacterium tuberculosis in a standard disinfectant test. J. Hosp. Infect. 38:183-192. [DOI] [PubMed] [Google Scholar]
- 6.Kaps, I., S. Ehrt, S. Seeber, D. Schnappinger, C. Martin, L. W. Riley, and M. Niederweis. 2001. Energy transfer between fluorescent proteins using a co-expression system in Mycobacterium smegmatis. Gene 278:115-124. [DOI] [PubMed] [Google Scholar]
- 7.Kempsell, K. E., Y. E. Ji, I. C. Estrada, M. J. Colston, and R. A. Cox. 1992. The nucleotide sequence of the promoter, 16S rRNA and spacer region of the ribosomal RNA operon of Mycobacterium tuberculosis and comparison with Mycobacterium leprae precursor rRNA. J. Gen. Microbiol. 138:1717-1727. [DOI] [PubMed] [Google Scholar]
- 8.Metchock, B. G., F. S. Nolte, and R. J. Wallace, Jr. 1999. Mycobacterium, p. 399-437. In P. R. Murray, E. J. Baron, and M. A. Pfaller (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, DC.
- 9.Scholz, O., A. Thiel, W. Hillen, and M. Niederweis. 2000. Quantitative analysis of gene expression with an improved green fluorescent protein. Eur. J. Biochem. 267:1565-1570. [DOI] [PubMed] [Google Scholar]
- 10.Siemering, K. R., R. Golbik, R. Sever, and J. Haseloff. 1996. Mutations that suppress the thermosensitivity of green fluorescent protein. Curr. Biol. 6:1653-1663. [DOI] [PubMed] [Google Scholar]
- 11.Simoes, M., M. O. Pereira, I. Machado, L. C. Simoes, and M. J. Vieira. 2006. Comparative antibacterial potential of selected aldehyde-based biocides and surfactants against planktonic Pseudomonas fluorescens. J. Ind. Microbiol. Biotechnol. 33:741-749. [DOI] [PubMed] [Google Scholar]
- 12.Song, H., and M. Niederweis. 2007. Functional expression of the Flp recombinase in Mycobacterium bovis BCG. Gene 399:112-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spach, D. H., F. E. Silverstein, and W. E. Stamm. 1993. Transmission of infection by gastrointestinal endoscopy and bronchoscopy. Ann. Intern. Med. 118:117-128. [DOI] [PubMed] [Google Scholar]
- 14.Stephan, J., J. Bender, F. Wolschendorf, C. Hoffmann, E. Roth, C. Mailänder, H. Engelhardt, and M. Niederweis. 2005. The growth rate of Mycobacterium smegmatis depends on sufficient porin-mediated influx of nutrients. Mol. Microbiol. 58:714-730. [DOI] [PubMed] [Google Scholar]
- 15.Talsma, S. S. 2007. Biofilms on medical devices. Home Healthc. Nurse 25:589-594. [DOI] [PubMed] [Google Scholar]
- 16.Vijayaraghavan, R., R. Chandrashekhar, Y. Sujatha, and C. S. Belagavi. 2006. Hospital outbreak of atypical mycobacterial infection of port sites after laparoscopic surgery. J. Hosp. Infect. 64:344-347. [DOI] [PubMed] [Google Scholar]
- 17.Woelk, E., P. Goroncy-Bermes, and W. Sand. 2003. Influence of storage on monodispersed cells of Mycobacterium terrae used for quantitative carrier test prEN 14563. Appl. Environ. Microbiol. 69:6932-6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zafer, A. A., Y. E. Taylor, and S. A. Sattar. 2001. Rapid screening method for mycobactericidal activity of chemical germicides that uses Mycobacterium terrae expressing a green fluorescent protein gene. Appl. Environ. Microbiol. 67:1239-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.