Abstract
We have developed the conversion of glycerol into thermoplastic poly(3-hydroxypropionate) [poly(3HP)]. For this, the genes for glycerol dehydratase (dhaB1) of Clostridium butyricum, propionaldehyde dehydrogenase (pduP) of Salmonella enterica serovar Typhimurium LT2, and polyhydroxyalkanoate (PHA) synthase (phaC1) of Ralstonia eutropha were expressed in recombinant Escherichia coli. Poly(3HP) was accumulated up to 11.98% (wt/wt [cell dry weight]) in a two-step, fed-batch fermentation. The present study shows an interesting application to engineer a poly(3HP) synthesis pathway in bacteria.
Glycerol has become an inexpensive and abundant carbon source due to its occurrence as a by-product of biodiesel production particularly in Europe. With every 100 tons of biodiesel produced via transesterification of vegetable oils or animal fats, about 10 tons of crude glycerol is produced. The tremendous growth of the biodiesel industry has generated a glycerol surplus, resulting in a sharp decrease in crude glycerol prices between 2005 and 2007 (16, 33). Therefore, the development of processes to convert crude glycerol into higher-value products is an urgent need. One possibility is the production of thermoplastics, like polyhydroxyalkanoates (PHAs).
PHAs are bacterial storage compounds for carbon and energy. They are synthesized and intracellularly deposited as granules in many bacteria when the cells are cultivated in the presence of an excess of a carbon source and if one other nutrient limits growth. PHAs exhibit a high degree of polymerization. Molecular masses of up to several million Da have been reported previously (14). They are biodegradable, insoluble in water, nontoxic, biocompatible, piezoelectric, thermoplastic, and/or elastomeric. These features make PHAs suitable for several applications in the packaging, medicine, pharmacy, agriculture, and food industries, as well as for raw material for production of enantiomerically pure chemicals and for production of paints (1, 28). One interesting polymer is poly(3-hydroxypropionate) [poly(3HP)]. It combines the properties of poly(3-hydroxybutyrate) [poly(3HB)] and poly(2-hydroxypropionate) [poly(2HP)], which is also referred to as polylactic acid. Poly(3HP) has a higher stability than poly(2HP) with regard to hydrolytic cleavage but is more easily enzymatically cleaved than poly(3HB) due to the lacking methyl groups at the polymer backbone (7). Until now, poly(3HP) could be synthesized only chemically via the ring-opening polymerization of β-propiolactone (2) or biotechnologically by a propionyl-coenzyme A (CoA) synthase, like PrpE from Salmonella enterica, and a PHA synthase, like PhaC1 from Ralstonia eutropha, with 3-hydroxypropionate (3HP) as a precursor molecule (27, 31). In addition, poly(3HB-co-3HP) copolyesters with a very low 3HP content not exceeding 2.1 mol% of the constituents were obtained from structurally unrelated carbon sources in recombinant strains of R. eutropha expressing enzymes of the 3HP cycle of Chloroflexus aurantiacus (5). Furthermore, poly(3HB-co-3HP) copolyesters were obtained from R. eutropha and Alcaligenes latus when the cells were cultivated on structurally related carbon sources, like 3-hydroxypropionate and various α,ω-alkanediols (7, 17).
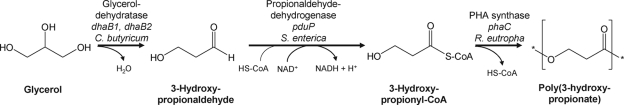
The aim of this study was to engineer a novel, non-naturally existing pathway for the biotechnological conversion of glycerol into poly(3HP) in Escherichia coli (Fig. 1). For this, we used (i) the glycerol dehydratase DhaB1 from Clostridium butyricum, which has already been used for the large-scale biotechnological production of 1,3-propanediol from glycerol as the sole carbon source by C. butyricum and E. coli (6, 18, 19, 24); (ii) the propionaldehyde dehydrogenase PduP, which is involved in 1,2-propanediol degradation in Salmonella enterica (3, 12); and (iii) the PHA synthase PhaC1 from Ralstonia eutropha (7, 20, 26, 27, 34), which is involved in the biosynthesis of poly(3-hydroxypropionate).
FIG. 1.
Pathway for conversion of glycerol into poly(3HP) in the engineered strain of E. coli.
Bacterial strains and plasmids.
All strains and plasmids used in this study are listed in the supplemental material. For routine cloning, plasmids were introduced into E. coli TOP10 or E. coli XL1-Blue. For poly(3HP) production, the plasmid was transferred into E. coli HMS174(DE3).
Media.
To produce poly(3HP) in fed-batch fermentation, we used Riesenberg medium (Rb) at 37°C, with an initial glycerol (Caldic) concentration of 300 mM (8). The feeding solution for the anaerobic production phase contained, in addition to the ingredients described by Korz et al. (8), 0.5 M di-Na+-fumarate (Carl Roth), 0.5 M K+-Na+-tartrate (Carl Roth), and 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The cultivation was performed with a 2-liter Biostat B plus (Sartorius) fermentor. All other cultivations of E. coli or Salmonella enterica serovar Typhimurium LT2 were done under aerobic conditions at 37°C in lysogeny broth (LB) medium (21). The cultivation of Clostridium butyricum was done at 30°C under anaerobic conditions in 2× YT medium containing 2% (wt/vol) glucose (19). Antibiotics were added at appropriate concentrations as follows: ampicillin, 75 μg/ml; chloramphenicol, 25 μg/ml; kanamycin, 50 μg/ml; gentamicin, 20 μg/ml; and rifampin, 20 μg/ml.
Plasmid construction.
The construction of pCOLADuet-1::dhaB1B2::pduP::phaC1 is described in the supplemental material.
Testing of the expression plasmid.
All in vitro tests were done with crude extracts obtained by sonification from cells of E. coli HMS174(DE3)/pCOLADuet-1::dhaB1B2::pduP::phaC1 and cells of E. coli HMS174(DE3)/pCOLADuet-1 as a negative control. Expression of the cloned genes dhaB1, dhaB2, pduP, and phaC1 was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) performed according to Laemmli (10) and by staining with Coomassie brilliant blue R-250 (32). The protein amount was determined by the method of Lowry et al. (13).
The glycerol dehydratase activity was measured as described by Raynaud et al. (19), and the propionaldehyde dehydrogenase activity was quantified according to Leal et al. (12). The basal activity was subtracted from the enzyme activity. Samples with recombinant E. coli harboring dhaB1, dhaB2, pduP, and phaC1 dehydrated glycerol to 3-hydroxypropionate with an average (±standard deviation) activity of 12.7 ± 1.3 μmol·min−1·mg−1 protein. The addition of coenzyme A initiated the coenzyme A acylation of the aldehyde group of 3HP by PduP (average [±standard deviation] of 6.5 ± 1.6 μmol·min−1·mg−1 protein). [Levels for E. coli HMS174(DE3)/pCOLADuet-1 were not detectable. All assays were done in triplicate with crude extracts from different cultures.] Former studies revealed that PduP catalyzes the CoA acylation of propionaldehyde (12). We show that PduP also has the ability to use 3-hydroxypropionaldehyde as a substrate for coenzyme A acylation.
In vivo PHA accumulation in fed-batch cultures.
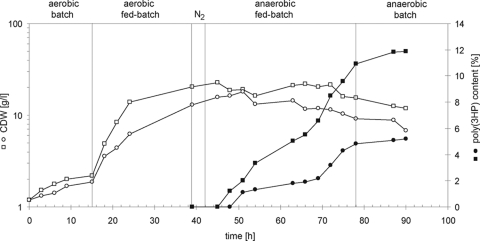
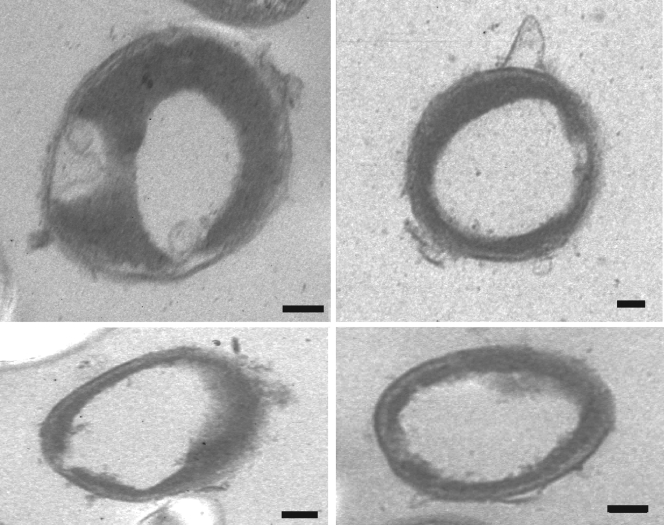
For poly(3HP) accumulation, E. coli HMS174(DE3)/pCOLADuet-1::dhaB1B2::pduP::phaC1 was cultivated in fed-batch fermentation for 92 h at 37°C in Rb (Table 1). Similarly to the microbial conversion of glycerol to 1,3-propanediol (24), the cultivation of our recombinant E. coli strain was performed in two different steps, with an aerobic growth phase and an anaerobic PHA production phase (Fig. 2). We used pure or crude glycerol, respectively, as the sole carbon source. In the first phase, an agitation of 400 rpm and an airflow of 2.5 ml/h were applied. The pH and the temperature were adjusted to 6.7 and 37°C, respectively. When the cells reached the early stationary growth phase, the aerobic fed batch was started and the Rb feeding solution, supplemented with 2 M glycerol, was added with a flow of 20 ml/h. After 45 h, the airflow was stopped, the culture was flushed with nitrogen, and the anaerobized system was closed. During the anaerobic production phase, di-Na+-fumarate, K+-Na+-tartrate, and IPTG were added to the feeding solution and applied with a flow of 20 ml/h for 33 h to the fermentor to induce poly(3HP) production. The main fermentation products were succinate and ethanol. Detection of growth-inhibiting acetic acid was only marginal (Fig. 3; Table 1). Accumulation of PHAs within the cells was monitored by fluorescence microscopy (22), transmission electron microscopy (TEM) (30), and gas chromatography-mass spectrometry (GC-MS) (4, 23). For cultures with pure glycerol, accumulation of PHAs started about 6 h after protein expression was induced with IPTG; for cultures with crude glycerol, it started at 9 h. After 92 h of incubation, the cells were harvested by centrifugation for PHA analysis and isolation. The appearance of the accumulated poly(3HP) within the cells is typical for PHA synthesis in recombinant strains without any phasins (15, 25). Often, one big granule was seen in the middle of the cell (Fig. 4).
TABLE 1.
Main fermentation products of the poly(3HP)-producing strain E. coli HMS174(DE3)/pCOLADuet-1::dhaB1B2::pduP::phaC1 after 92 h of incubation in Rb with pure or crude glycerol
| Carbon source | Glycerol consumption (g/liter) | Cell dry weight (g/liter) | poly(3HP) (g/liter) | Succinate (g/liter) | Ethanol (g/liter) | Acetate (g/liter) | Lactate (g/liter) | 3HP yield (mmol/mol glycerol) |
|---|---|---|---|---|---|---|---|---|
| Pure glycerol | 83.26 | 11.98 | 1.42 | 48.92 | 8.04 | 0.26 | 0 | 17.53 |
| Crude glycerol | 68.34 | 5.15 | 0.27 | 46.33 | 7.54 | 0.22 | 0 | 3.31 |
FIG. 2.
Two-step fermentation of E. coli HMS174(DE3)/pCOLADuet-1::dhaB1B2::pduP::phaC1, with pure (□) or crude (○) glycerol as the sole carbon source. The cultivation of the cells was divided into an aerobic growth phase and an anaerobic production phase and was carried out using Riesenberg medium (19). The content of poly(3HP) derived from pure (▪) or crude (•) glycerol was monitored by GC-MS. CDW, cell dry weight.
FIG. 3.
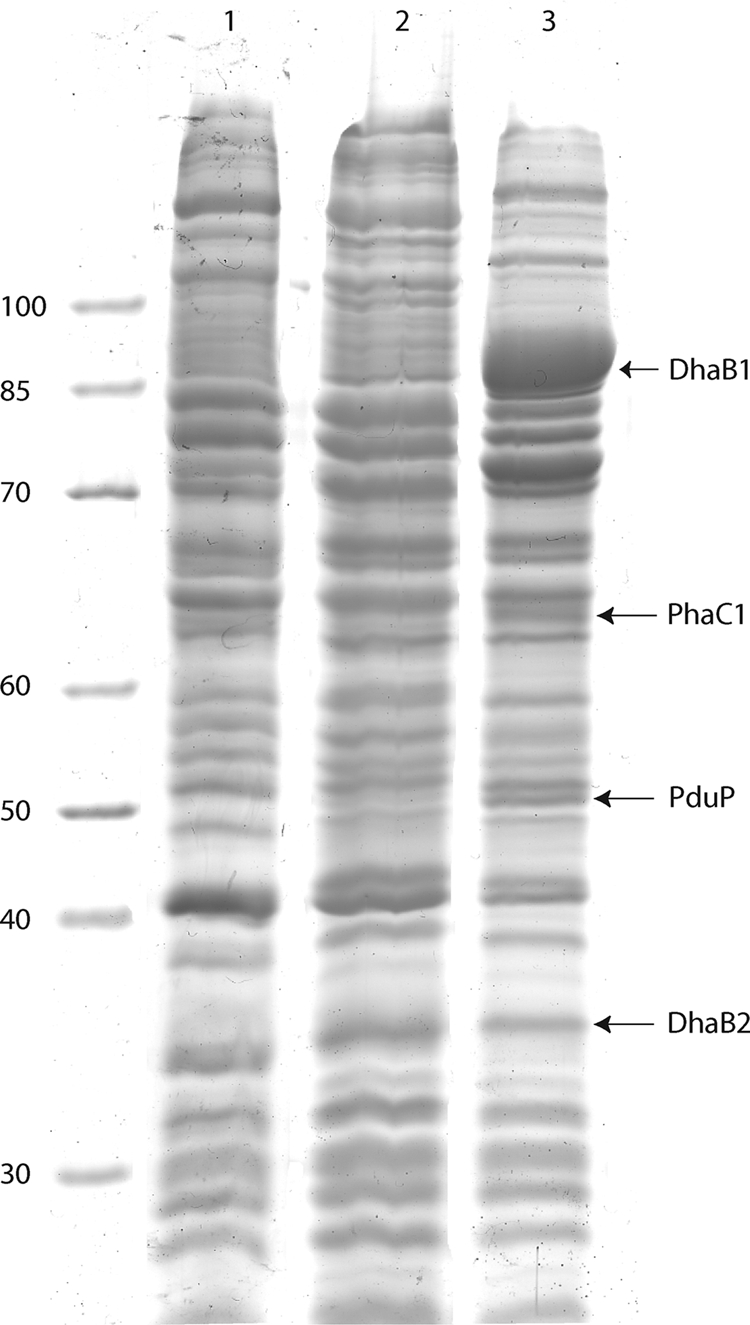
Proteins in crude extracts of cells of E. coli HMS174(DE3), E. coli HMS174(DE3)/pCOLADuet-1, and E. coli HMS174(DE3)/pCOLADuet-1::dhaB1B2::pduP::phaC1 grown in LB medium. Protein expression was induced with 1 mM IPTG at an optical density at 600 nm of 0.5, and the cells were then cultivated for an additional 8 h. The proteins were separated using 12.5% (wt/vol) SDS-polyacrylamide gels and stained with Coomassie brilliant blue. Lane 1, E. coli HMS174(DE3); lane 2, E. coli HMS174(DE3)/pCOLADuet-1; lane 3, E. coli HMS174(DE3)/pCOLADuet-1::dhaB1B2::pduP::phaC1. The values on the left indicate molecular mass (in kDa).
FIG. 4.
TEM of poly(3HP)-accumulating cells of E. coli HMS174(DE3)/pCOLADuet-1::dhaB1B2::pduP::phaC1. The fermentation was divided into an aerobic growth phase and an anaerobic production phase and was carried out using Riesenberg medium (19). The feeding solution for the anaerobic production phase was supplemented with 0.5 M di-Na+-fumarate, 0.5 M K+-Na+-tartrate, and 1 mM IPTG. Bars, 100 nm.
PHA isolation and identification.
PHAs were extracted from lyophilized cells with hot chloroform in a Soxhlet apparatus. The extract was concentrated with a rotary evaporator, and the PHAs were precipitated by pouring this solution into 10 vol of ice-cold ethanol (11). This procedure was repeated at least four times to obtain highly purified polymer samples. Dried precipitate appeared as a white powder. Identification of the polyester was done by GC-MS (4, 23) and electrospray ionization-MS (ESI-MS) (see the supplemental material). Methyl esters of the extracted polymer eluted at a retention time of 7.045 min, which was identical to the retention time of 3-hydroxypropionate methyl esters. This particular peak was fractionated by electron impact (EI) to record the GC-MS spectrum. Again, the extracted peak spectrum of the produced PHAs corresponded with the extracted peak spectrum of 3-hyroxypropionate. The ESI-MS analysis of the monomer, achieved by hydrolysis of the extracted polymer, showed a mass of 89 kDa, which corresponds to the mass of ionized 3-hydroxypropionate. The same peak occurred when commercially purchased 3-hydroxypropionate was injected. Collision-induced dissociation of the respective peaks led to nearly identical masses, i.e., 59 and 43 kDa (data not shown).
With pure glycerol as the sole carbon source, the final poly(3HP) content of the cells amounted to 11.98% (wt/wt [cell dry weight]), whereas cultures with crude glycerol accumulated the polymer only up to 5.2% (wt/wt [cell dry weight]) (Table 1). This could be due to growth- and/or enzyme activity-inhibiting residues like methanol, methyl ester, or ash from the transesterification of vegetable oils or animal fats.
Compared to that for other PHA-producing bacteria, this content is low. This is probably caused by a loss of pCOLADuet-1::dhaB1B2::pduP::phaC1. To verify plasmid stability, appropriately diluted samples of the fermentation broth were spread on LB agar plates in the presence or absence of kanamycin and incubated overnight at 37°C. The number of colonies on the plates without antibiotics was set to 100%. After 92 h of fermentation, only 56% of the cells possessed the expression plasmid. To achieve more stable production, a plasmid addiction system as recently described (9, 29) could be established. The advantages of such systems are numerous: (i) they provide antibiotic-independent selection, (ii) they result in higher plasmid stability, and (iii) due to the higher copy number, the expression level is higher than in the case of a chromosomal insertion. Addiction systems may therefore contribute to a reduction in production cost. Studies with such a system will be done during further strain optimization.
One further problem with poly(3HP) production is the redox balance (see the supplemental material) and the disposal of reducing equivalents. During polymer accumulation, large amounts of NADH were formed. A solution for that could be the coexpression of an NADH-consuming enzyme and coproduction of a reduced product. It is obvious to use the 1,3-propanediol dehydrogenase (DhaT) from C. butyricum (19). This enzyme has two major advantages: (i) the surplus of NADH will be oxidized to NAD+, and (ii) another valuable product will be generated from glycerol.
In summary, the recombinant E. coli strain HMS174(DE3)/pCOLADuet-1::dhaB1B2::pduP::phaC1 can accumulate poly(3HP) at up to 11.98% of the cell dry weight. For an industrially relevant large-scale production of poly(3HP), further developments are necessary to obtain a high polymer yield on glycerol.
Supplementary Material
Acknowledgments
Financial support from BMVEL/FNR (FKZ 22015806, 06NR158) is gratefully acknowledged.
We also thank K. Schlattmann, C. Köppler, and M. Opalka for skillful technical introduction, advice, and assistance during transmission electron microscopy.
Footnotes
Published ahead of print on 20 November 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Anderson, A. J., and E. A. Dawes. 1990. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54:450-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba, H., N. Tanahashi, Y. Kumagai, and Y. Doi. 1992. Effects of molecular-structure on enzymatic degradation of polyesters. J. Chem. Soc. Jpn. 5:527-533. [Google Scholar]
- 3.Bobik, T. A., G. D. Havemann, R. J. Busch, D. S. Williams, and H. C. Aldrich. 1999. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B(12)-dependent 1,2-propanediol degradation. J. Bacteriol. 181:5967-5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandl, H., R. A. Gross, R. W. Lenz, and R. C. Fuller. 1988. Pseudomonas oleovorans as a source of poly(β-hydroxyalkanoates) for potential applications as biodegradable polyesters. Appl. Environ. Microbiol. 54:1977-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukui, T., M. Suzuki, T. Tsuge, and S. Nakamura. 2009. Microbial synthesis of poly((R)-3-hydroxybutyrate-co-3-hydroxypropionate) from unrelated carbon sources by engineered Cupriavidus necator. Biomacromolecules doi: 10.1021/bm801391. [DOI] [PubMed]
- 6.Gonález-Pajuelo, M., J. C. Andrade, and I. Vasconcelos. 2004. Production of 1,3-propanediol by Clostridium butyricum VPI 3266 using a synthetic medium and raw glycerol. J. Ind. Microbiol. Biotechnol. 31:442-446. [DOI] [PubMed] [Google Scholar]
- 7.Hiramitsu, M., and Y. Doi. 1993. Microbial synthesis and characterization of poly(3-hydroxybutyrate-co-3-hydroxypropionate). Polymer 34:4782-4786. [Google Scholar]
- 8.Korz, D. J., U. Rinas, K. Hellmuth, E. A. Sanders, and W.-D. Deckwer. 1995. Simple fed-batch technique for high cell density cultivation of Escherichia coli. J. Biotechnol. 39:59-65. [DOI] [PubMed] [Google Scholar]
- 9.Kroll, J., A. Steinle, R. Reichelt, C. Ewering, and A. Steinbüchel. 2009. Establishment of a novel anabolism-based addiction system with an artificially introduced mevalonate pathway: complete stabilization of plasmids as universal application in white biotechnology. Metab. Eng. 11:168-177. [DOI] [PubMed] [Google Scholar]
- 10.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 11.Lageveen, R. G., G. Huisman, H. Preusting, P. Ketelaar, G. Eggink, and B. Witholt. 1988. Formation of polyesters by Pseudomonas oleovorans: effect of substrates on formation and composition of poly-(R)-3-hydroxyalkanoates and poly-(R)-3-hydroxyalkenoates. Appl. Environ. Microbiol. 54:2924-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leal, N. A., G. D. Havemann, and T. A. Bobik. 2003. PduP is a coenzyme-a-acylating propionaldehyde dehydrogenase associated with the polyhedral bodies involved in B12-dependent 1,2-propanediol degradation by Salmonella enterica serovar Typhimurium LT2. Arch. Microbiol. 180:353-361. [DOI] [PubMed] [Google Scholar]
- 13.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 14.Lütke-Eversloh, T., and A. Steinbüchel. 2003. Novel precursor substrates for polythioesters (PTE) and limits of PTE biosynthesis in Ralstonia eutropha. FEMS Microbiol. Lett. 221:191-196. [DOI] [PubMed] [Google Scholar]
- 15.Lütke-Eversloh, T., K. Bergander, H. Luftmann, and A. Steinbüchel. 2001. Identification of a new class of biopolymer: bacterial synthesis of a sulfur-containing polymer with thioester linkages. Microbiology 147:11-19. [DOI] [PubMed] [Google Scholar]
- 16.McCoy, M. 2006. Glycerin surplus. Chem. Eng. News 84:7-8. [Google Scholar]
- 17.Nakamura, S., M. Kunioka, and Y. Doi. 1991. Biosynthesis and characterization of bacterial poly(3-hydroxybutyrate-co-3-hydroxypropionate). J. Macromol. Sci. Chem. A 28:15-24. [Google Scholar]
- 18.O'Brien, J. R., C. Raynaud, C. Croux, L. Girbal, P. Soucaille, and W. N. Lanzilotta. 2004. Insight into the mechanism of the B12-independent glycerol dehydratase from Clostridium butyricum: preliminary biochemical and structural characterization. Biochemistry 43:4635-4645. [DOI] [PubMed] [Google Scholar]
- 19.Raynaud, C., P. Sarçabal, I. Meynial-Salles, C. Croux, and P. Soucaille. 2003. Molecular characterization of the 1,3-propanediol (1,3-PD) operon of Clostridium butyricum. Proc. Natl. Acad. Sci. U. S. A. 100:5010-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rehm, B. H. A. 2003. Polyester synthases: natural catalysts for plastics. Biochem. J. 376:15-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 22.Spiekermann, P., B. H. A. Rehm, R. Kalscheuer, D. Baumeister, and A. Steinbüchel. 1999. A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch. Microbiol. 171:73-80. [DOI] [PubMed] [Google Scholar]
- 23.Stein, S., A. Levitsky, O. Fateev, and G. Mallard. 1998. The NIST mass spectral search program. Windows software version 1.6d. National Institute of Standards and Technology, Gaithersburg, MD.
- 24.Tang, X., Y. Tan, H. Zhu, K. Zhao, and W. Shen. 2009. Microbial conversion of glycerol to 1,3-propanediol by an engineered strain of Escherichia coli. Appl. Microbiol. Biotechnol. 75:1628-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tessmer, N., S. König, U. Malkus, R. Reichelt, M. Pötter, and A. Steinbüchel. 2007. Heat-shock protein HspA mimics the function of phasins sensu stricto in recombinant strains of Escherichia coli accumulating polythioesters or polyhydroxyalkanoates. Microbiology 153:366-374. [DOI] [PubMed] [Google Scholar]
- 26.Valentin, H. E., and A. Steinbüchel. 1993. Cloning and characterization of the Methylobacterium extorquens polyhydroxyalkanoic-acid-synthase structural gene. Appl. Microbiol. Biotechnol. 39:309-317. [DOI] [PubMed] [Google Scholar]
- 27.Valentin, H. E., T. A. Mitsky, D. A. Mahadeo, M. Tran, and K. J. Gruys. 2000. Application of a propionyl coenzyme A synthetase for poly(3-hydroxypropionate-co-3-hydroxybutyrate) accumulation in recombinant Escherichia coli. Appl. Environ. Microbiol. 66:5253-5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Walle, G. A., G. J. de Koning, R. A. Weusthuis, and G. Eggink. 2001. Properties, modifications and applications of biopolyesters. Adv. Biochem. Eng. Biotechnol. 71:263-291. [DOI] [PubMed] [Google Scholar]
- 29.Voss, I., and A. Steinbüchel. 2006. Application of a KDPG-aldolase gene-dependent addiction system for enhanced production of cyanophycin in Ralstonia eutropha strain H16. Metab. Eng. 8:66-78. [DOI] [PubMed] [Google Scholar]
- 30.Wältermann M., A. Hinz, H. Robenek, D. Troyer, R. Reichelt, U. Malkus, H.-J. Galla, R. Kalscheuer, T. Stöveken, P. von Landenberg, and A. Steinbüchel. 2005. Mechanism of lipid-body formation in prokaryotes: how bacteria fatten up. Mol. Microbiol. 55:750-763. [DOI] [PubMed] [Google Scholar]
- 31.Wang, Y., and Y. Inoue. 2001. Effect of dissolved oxygen concentration in the fermentation medium on transformation of the carbon sources during the biosynthesis of poly(3-hydroxybutyrate-co-3-hydroxypropionate) by Alcaligenes latus. Int. J. Biol. Macromol. 28:235-243. [DOI] [PubMed] [Google Scholar]
- 32.Weber, K., and M. Osborn. 1969. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J. Biol. Chem. 244:4406-4412. [PubMed] [Google Scholar]
- 33.Yazdani, S. S., and R. Gonzalez. 2008. Engineering Escherichia coli for the efficient conversion of glycerol to ethanol and co-products. Metab. Eng. 10:340-351. [DOI] [PubMed] [Google Scholar]
- 34.Yuan, W., Y. Jia, J. Tian, K. D. Snell, U. Müh, A. J. Sinskey, R. H. Lambalot, C. T. Walsh, and J. Stubbe. 2001. Class I and III polyhydroxyalkanoate synthases from Ralstonia eutropha and Allochromatium vinosum: characterization and substrate specificity studies. Arch. Biochem. Biophys. 394:87-98. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.