Abstract
The genus Burkholderia includes strains pathogenic to animals and plants, bioremediators, or plant growth promoters. Genome sequence analyses of representative Burkholderia cepacia complex (Bcc) and non-Bcc strains for the presence of the bce-I gene cluster, directing the biosynthesis of the exopolysaccharide (EPS) cepacian, further extended this previously described cluster by another 9 genes. The genes in the bce-II cluster were named bceM to bceU and encode products putatively involved in nucleotide sugar precursor biosynthesis and repeat unit assembly, modification, and translocation across the cytoplasmic membrane. Disruption of the B. cepacia IST408 bceQ and bceR genes, encoding a putative repeat unit flippase and a glycosyltransferase, respectively, resulted in the abolishment of cepacian biosynthesis. A mutation in the bceS gene, encoding a putative acyltransferase, did not affect EPS production yield significantly but decreased its acetylation content by approximately 20%. Quantitative real-time reverse transcription-PCR experiments confirmed the induction of genes in the bce-I and bce-II clusters in a Burkholderia multivorans EPS producer clinical isolate in comparison to the level for its isogenic EPS-defective strain. Fourier Transform infrared spectroscopy analysis confirmed that the exopolysaccharide produced by 10 Burkholderia isolates tested was cepacian. The ability of Burkholderia strains to withstand desiccation and metal ion stress was higher when bacteria were incubated in the presence of 2.5 g/liter of cepacian, suggesting that this EPS plays a role in the survival of these bacteria by contributing to their ability to thrive in different environments.
Many bacteria produce exopolysaccharides (EPSs), which play a wide range of roles in their biology. Besides their contribution to the fitness of the producing microorganism to their ecological niche (14, 36), EPSs are often important virulence determinants produced by pathogens of plants, animals, and humans. Cepacian is the major EPS produced by a large percentage of clinical isolates of the Burkholderia cepacia complex (Bcc) (11, 20, 37, 52). The Bcc comprises at least 17 distinct bacterial species, including soil and water saprophytes, rhizosphere parasites, bioremediators, plant growth promoters, and plant and animal pathogens (49). Bcc members are receiving particular attention due to their increasingly recognized importance as opportunistic pathogens in immunocompromised patients and in patients suffering from cystic fibrosis (CF) or chronic granulomatous disease (CGD) (27).
Several studies have pointed out cepacian as a virulence factor contributing to the overall pathogenicity of Bcc members and thus to their success as pathogens. For instance, Conway et al. (10) have shown that the EPS produced by a mucoid Burkholderia cenocepacia clinical isolate interfered with phagocytosis of bacteria by human neutrophils and facilitated bacterial persistence in the BALB/c mice model of infection. In a study performed using the gp91phox−/− CGD mouse model of infection, Sousa et al. (43) have shown that mutants defective in cepacian production were less virulent than the wild-type cepacian-producing strain or completely avirulent. Cepacian was also found to inhibit neutrophil chemotaxis and the production of reactive oxygen species, both essential components of the innate host defenses (5). The persistence of infections has been correlated with the ability of bacterial pathogens to form biofilms. Several studies have demonstrated the ability of the Bcc to form biofilms alone or together with other bacteria (11, 23). Studies performed with cepacian-defective mutants have demonstrated that, although not required for the initiation of biofilm formation, cepacian is required to the formation of thick and mature biofilms (11).
Cepacian is composed of a branched acetylated heptasaccharide repeat unit with d-glucose, d-rhamnose, d-mannose, d-galactose, and d-glucuronic acid in the ratio 1:1:1:3:1 (6). The biochemical pathway leading to the activated sugar nucleotides necessary for repeat unit formation was postulated, and the predicted enzyme activities were detected in crude extracts prepared from a cepacian-producing Bcc clinical isolate (38). A strategy based on random plasposon mutagenesis of the cepacian producer clinical isolate B. cepacia IST408 allowed the identification of the 16.2-kb bce cluster of genes involved in cepacian biosynthesis (31).
Although the bce clustered genes encode several proteins and enzymes required for the biosynthesis of the EPS (31), not all the proteins required for cepacian biosynthesis are encoded within this cluster. In the present work, we report the identification and partial functional analysis of a second cluster of genes, here named bce-II. The bce-II cluster contains genes encoding enzymes putatively involved in the synthesis of the d-rhamnose and d-glucose moieties of cepacian, a glycosyltransferase, a repeat unit flippase, and acyltransferases presumably required for the acetylation of cepacian. We also report results showing that the bce-I and bce-II clusters are widespread within all the sequenced Burkholderia strains, with the exception of Burkholderia mallei strains. In agreement with the results of these in silico studies, the abilities of several Bcc and non-Bcc strains to produce cepacian were confirmed. The ability of cepacian to confer resistance against desiccation and metal ion stress is also reported.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The strains and plasmids used in this study are listed in Table 1. Burkholderia strains were maintained in pseudomonas isolation agar (Difco) plates at 30 or 37°C. Mannitol medium (MM) (7) or S medium (37) supplemented with mannitol instead of glucose (SM) were used to quantify EPS production by Burkholderia strains at 30°C. Escherichia coli strains were grown in Lennox broth (LB) at 37°C. Growth media were supplemented with antibiotics when required, to maintain selective pressure, at the following final concentrations: for Burkholderia strains, 300 μg/ml chloramphenicol, 600 μg/ml kanamycin, and 100 μg/ml trimethoprim, and for E. coli, 100 μg/ml ampicillin, 50 μg/ml kanamycin, 25 μg/ml chloramphenicol, and 50 μg/ml trimethoprim.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| B. cepacia IST408 | Cystic fibrosis clinical isolate; cepacian producer | 37 |
| B. multivorans ATCC 17616 | Soil isolate | 48 |
| B. cenocepacia J2315 | Cystic fibrosis clinical isolate | 19 |
| B. vietnamiensis G4 | Isolated from a water treatment facility | 32 |
| B. dolosa AUO158 | Cystic fibrosis clinical isolate | 8 |
| B. ambifaria AMMD | Root-colonizing bacterium | 9 |
| B. lata 383 | Soil isolate | 44 |
| B. phytofirmans PsJN | Soil isolate; plant growth-promoting bacterium | 40 |
| B. phymatum STM815 | Soil isolate; nitrogen fixation | 47 |
| B. xenovorans LB400 | Soil isolate; degradation of polychlorinated biphenyl compounds | 18 |
| B. cepacia IST408 bceQ::pIS58-1 | pIS58-1 integrated into the bceQ gene region | This work |
| B. cepacia IST408 bceR::pIS58-2 | pIS58-2 integrated into the bceR gene region | This work |
| B. multivorans ATCC 17616 bceS::pSF71-8 | pSF71-8 integrated into the bceS gene region | This work |
| B. multivorans D2095 | Mucoid cystic fibrosis clinical isolate | D. P. Speert |
| B. multivorans D2214 | Nonmucoid cystic fibrosis clinical isolate | D. P. Speert |
| Escherichia coli XL1-Blue | recA1 lac (F′proAB lacIq ZαM15 Tn10 [Tcr]) thi | 4 |
| Plasmids | ||
| pDrive | 3.85-kb vector; lacZ α-peptide; Apr Kmr | Qiagen |
| pBCKS | 3.4-kb phagemid derived from pUC19; lac promoter; Cmr | Stratagene |
| pMLBAD | pBBR1 ori; araC-PBAD; Tpr; mob+ | 25 |
| pSF71-8 | pBCKS derivative carrying a 579-bp EcoRI/XbaI fragment with an internal fragment from the bceS gene | This work |
| pIS58-1 | pDrive derivative carrying a 1,133-bp HindIII fragment with an internal fragment from the bceQ gene | This work |
| pIS58-2 | pDrive derivative carrying a 1,028-bp HindIII/XbaI fragment with an internal fragment from the bceR gene | This work |
| pIS94-1 | pMLBAD derivative carrying a 2,509-bp KpnI/HindIII fragment with the coding region of the bceR gene | This work |
Ap, ampicillin; Km, kanamycin; Tp, trimethoprim; Cm, chloramphenicol.
DNA manipulation techniques.
Total DNA and plasmid DNA isolation, DNA restriction, agarose gel electrophoresis, Southern blot experiments, and E. coli transformation were carried out using standard procedures (39). Burkholderia multivorans ATCC 17616 or B. cepacia IST408 electrocompetent cells, prepared as described previously (15), were transformed by electroporation using a Bio-Rad Gene Pulser II system (200 Ω, 25 μF, 2.5 kV) and grown overnight before being plated in selective medium. Triparental conjugation was performed as described previously (15).
Construction of insertion mutants and phenotype complementation.
The primers bceQ-Fw/bceQ-Rev (Table 2) were used to amplify the 1,133-bp region encoding the putative flippase BceQ of B. cepacia IST408. The fragment obtained was digested with HindIII and ligated into the pDrive vector, originating plasmid pIS58-1. To amplify the glycosyltransferase-encoding gene bceR, primers bceR-Fw/bceR-Rev were used (Table 2). The 1,028-bp fragment amplified from B. cepacia IST408 was cloned into the XbaI/HindIII sites of the previously digested pDrive vector. The resulting plasmid was named pIS58-2. An internal 579-bp fragment from the B. multivorans ATCC 17616 bceS gene was amplified by PCR with primers bceS-Fw/bceS-Rev, and the amplified product was cloned into the XbaI/EcoRI sites of pBCKS, generating pSF71-8. The nucleotide sequences of the cloned internal gene regions of pIS58-1, pIS58-2, and pSF71-8 were confirmed by sequencing. Insertion mutations in each of the bceQ, bceR, and bceS genes were prepared from B. cepacia IST408 or B. multivorans ATCC 17616 by electroporation. Candidate mutants were further characterized by PCR amplification or Southern hybridization.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′)a |
|---|---|
| bceQ-Fw | CCTAAGCTTGGACGCTGATCGGCTAT |
| bceQ-Rev | CCTAAGCTTCGGATCGCCGACAGG |
| bceR-Fw | GCTTCTAGAGATCGTCGCGTGCT |
| bceR-Rev | CGAAAGCTTGCCGCCGGAACGTG |
| bceS-Fw | CATTCTAGACGGTCGTTCGAACA |
| bceS-Rev | CTCGAATTCGAAGTGCAGTTCTA |
| CbceR-Fw | CATGGTACCCGATCTGGCGGAAC |
| CbceR-Rev | GCAAAGCTTCCGATGCGAAAGGC |
| bceB_RT_Fw | TTCGTGAACATCCGCTTCATT |
| bceB_RT_Rev | CCGAGCACCTCGACCACTT |
| bceE_RT_Fw | CCGAGACCTATCCGGTTCATT |
| bceE_RT_Rev | CTTTCTGCAGCTGGTCCATCA |
| bceG_RT_Fw | ACGCTGTCCGGCAAGATC |
| bceG_RT_Rev | TAGCTCATGTTCGCGCCTTT |
| bceM_RT_Fw | GGCGAAGCGCATGAAGTC |
| bceM_RT_Rev | CAGTGTGCCGGTCGTATACG |
| bceR_RT_Fw | TTCGGCGAGGACGACTATG |
| bceR_RT_Rev | TGGAACCCGAGGAAATGC |
| bceS_RT_Fw | AACGGCCTCGTCCATCAC |
| bceS_RT_Rev | GCGTCCAGAACCAGACGAAA |
| gyrB_RT-Fw | GCGGACTGCCAGGAGAAAG |
| gyrB_RT-Rev | GACCCACCTGCCGAGTCA |
Restriction sites are in italic.
Plasmid pIS94-1 was constructed to complement the EPS-deficient phenotype of the B. cepacia bceR mutant as follows. The bceR coding region was amplified by PCR using primers CbceR-Fw/CbceR-Rev and B. cepacia IST408 genomic DNA as a template. The amplified fragment was restricted with KpnI/HindIII and inserted into the same restriction sites of the pMLBAD vector. The nucleotide sequence of the cloned gene was confirmed by sequencing.
Quantitative real-time RT-PCR experiments.
Cells of the B. multivorans strains D2095 and D2214 and the B. cepacia strains IST408 and bceQ::pIS58-1 were harvested at the late exponential phase of growth, and total RNA was extracted using an RNeasy minikit (Qiagen) with DNase treatment by following the recommendation of the manufacturer. RNA integrity was checked with an Agilent 2100 Bioanalyzer coupled with an RNA Nano-Assay (Agilent Technologies). For the reverse transcription step, 1 μg of total RNA from Burkholderia strains, derived from three independent samples, was used. cDNA was synthesized using TaqManR reverse transcription reagents (Applied Biosystems) in accordance with the manufacturer's instructions. The primers used to amplify the bce genes (Table 2) were designed using Primer Express 3.0 software (Applied Biosystems). Reverse transcription (RT) mixtures were properly diluted to use 400 ng of template cDNA, 2× SYBR green PCR master mix, and 0.4 mM reverse and forward primers for each gene in a total volume of 25 μl. Reaction mixtures containing nuclease-free water (Eppendorf) instead of the reverse transcriptase were included as a negative control. Reactions were performed with a model 7500 instrument from Applied Biosystems. The expression ratio of the target genes relative to the reference gene gyrB, which showed no variation in transcription abundance under the conditions tested, was determined. Relative quantification of gene expression by real time RT-PCR was determined using the ΔΔCT method (34).
Production and characterization of bacterial exopolysaccharides.
EPS production was assessed based on the dry weight of the ethanol-precipitated polysaccharide recovered from 100-ml culture samples of the different strains grown in liquid MM over 3 or 6 days at 30°C with orbital agitation, based on methods described before (37). For Fourier transform infrared (FTIR) spectroscopy analysis, the ethanol-precipitated EPSs were air dried and redissolved in distilled water prior to dialysis (molecular mass cutoff, 12 kDa) against water for 3 days at 4°C, followed by centrifugation at 10,000 × g for 30 min and freeze drying. The acetyl content of the EPSs produced was determined as described by McComb and McCready (30), using glucose penta-acetate as standard. The results shown are the mean values for at least 3 independent determinations.
In vivo complementation of the EPS-deficient phenotype of the IST408 bceR mutant.
Plasmid pIS94-1, carrying the parental bceR gene, was mobilized into the B. cepacia IST408 bceR mutant strain by triparental conjugation. The complemented IST408 bceR::pIS58-2/pIS94-1 strain was grown in solid MM supplemented with 1% (wt/vol) of l-arabinose, in order to induce the expression of the cloned gene from the PBAD promoter of E. coli present in this vector, at 30°C for 5 days, and the mucoidy of the corresponding colonies was assessed.
FTIR analysis.
The EPSs produced by the Burkholderia strains under study were analyzed by condensed-phase infrared spectroscopy in the wave number range of 400 to 4,000 cm−1 at a spectral resolution of 2 cm−1. The data were obtained with an FTIR spectrometer (Jasco FTIR 4100) in transmission mode, with at least 16 scans per sample. At least two independently prepared samples of each EPS with the same massic concentration (0.5 mg/50 mg), prepared with spectrometric-grade potassium bromide (Merck), were analyzed.
Metal ion stress assays and desiccation sensitivity.
Overnight-grown cultures of Burkholderia strains in SM were incubated at 30°C in the presence or absence of 2.5 g/liter of purified cepacian and of 50 mM FeSO4 or 50 mM ZnCl2. Viable cell counts (CFU) in LB solid medium were determined. For the quantification of bacterial survival to desiccation, 10 μl of overnight stationary-phase Burkholderia cultures was mixed with or without cepacian (final concentration, 2.5 g/liter) and aliquoted into the wells of a microtiter plate. After drying, the plate was incubated at 30°C for several days. Each viable count was performed by adding 100 μl of saline buffer to rehydrate, followed by serial dilutions and plating on solid LB medium. Experiments were performed at least three times.
Computational analysis of nucleotide and protein sequences.
Nucleotide and amino acid sequences were retrieved from Integrated Microbial Genomes (28) for the following strains: B. multivorans ATCC 17616; B. cenocepacia strains J2315, AU10546, HI2424, MC0-3, and PC184; Burkholderia ambifaria strains MC40-6 and AMMD; Burkholderia dolosa AUO158; Burkholderia vietnamiensis G4; B. mallei strains 2002721280, ATCC 23344, FMH, GB8 horse 4, JHU, NCTC 10229, NCTC 10247, SAVP1, and PRL-20; B. pseudomallei strains 1106a, 1106b, 1655, 1710a, 1710b, 305, 406e, 668, K96243, Pasteur, S13, 112, 14, 7894, 9, 91, B7210, BCC215, DM98, and NCTC 13177; Burkholderia lata sp. 383; Burkholderia thailandensis E264, BT4, MSMB43, ATCC 700388, and TXDOH; Burkholderia phymatum STM815; Burkholderia phytofirmans PsJN; Burkholderia oklahomensis strains EO147 and C6786; Burkholderia ubonensis Bu; Burkholderia graminis C4D1M; and Burkholderia xenovorans LB400. The BLAST algorithm (1) was used to compare the deduced amino acid sequences to database sequences available at the NCBI. Alignments were performed using the program CLUSTAL W (46). Transmembrane regions were predicted by TMHMM server v2.0. Signal sequences and subcellular localization were predicted by PSORTb v2.0. The B. vietnamiensis G4 genome sequence was used as a reference in the bioinformatic analysis.
RESULTS
Identification of a second gene cluster involved in cepacian biosynthesis.
In order to identify the cepacian biosynthesis genes missing from the bce cluster, the genome sequences of 7 representative Bcc strains and 8 non-Bcc strains were examined by bioinformatic tools. A genomic region containing 11 genes homologous to the bce cluster (bceA to bceK) previously identified by Moreira et al. (31), followed by 8 genes encoding proteins putatively involved in polysaccharide biosynthesis were identified in the genome sequences of the non-Bcc strains B. xenovorans LB400, B. phymatum STM815, B. phytofirmans PsJN, and B. graminis C4D1M (Fig. 1a). The search for homologues to these newly identified genes within the genomes of Bcc strains (B. cenocepacia J2315, B. ambifaria AMMD, B. dolosa AU0158, B. vietnamiensis G4, B. lata sp. 383, B. ubonensis Bu, and B. multivorans ATCC 17616) and other non-Bcc strains (B. pseudomallei 1106a, B. oklahomensis C6786, and B. thailandensis Bt4) indicated that they were present in all these genomes, approximately 155 to 314 kb downstream of the bceA-K genes, depending on the strain. The minimal distance between the two chromosomal regions was observed for B. cenocepacia J2315 (155 kb), with the maximum distance being observed for B. multivorans ATCC 17616 (314 kb). These size distances depend on the number/size of genes encoding phage-related proteins and insertion sequences present between them. B. mallei strains have this second region, but the previously identified bce genes are absent from their genomes. The second region was named the bce-II cluster, and the genes therein were named bceM through bceU (Fig. 1a and b).
FIG. 1.
Genetic organization of the bce gene cluster directing the biosynthesis of cepacian by Burkholderia bacteria. In representative strains of the species B. xenovorans, B. phymatum, B. phytofirmans, and B. graminis, the bce genes are clustered together in the same genomic region (a), while, in representative strains of the Burkholderia cepacia complex, comprising B. pseudomallei, B. oklahomensis, and B. thailandensis, the bce genes are split into two regions 155 to 314 kb apart (b). Strains from B. mallei have the bce-II cluster only. The locus tags for each gene in the B. vietnamiensis G4 genome are as follows: for bceA, Bcep1808_4200; for bceB, Bcep1808_4201; for bceC, Bcep1808_4202; for bceD, Bcep1808_4203; for bceE, Bcep1808_4204; for bceF, Bcep1808_4205; for bceG, Bcep1808_4206; for bceH, Bcep1808_4207; for bceI, Bcep1808_4208; for bceJ, Bcep1808_4209; for bceK, Bcep1808_4210; for bceM, Bcep1808_4471; for bceN, Bcep1808_4472; for bceO, Bcep1808_4473; for bceP, Bcep1808_4474; for bceQ, Bcep1808_4475; for bceR, Bcep1808_4476; for bceS, Bcep1808_4477; for bceT, Bcep1808_4479; and for bceU, Bcep1808_4480.
A thorough analysis revealed that strains with the two clusters together have the bceV gene encoding a putative acylesterase/lipolytic protein (Fig. 1a). Strains with the bce-II cluster separated possess the bceM gene encoding a putative NAD-dependent epimerase/dehydratase (Fig. 1b). Since the putative acyltransferase-encoding gene bceU was found in the genome sequences of all the Bcc strains as well as in B. thailandensis, B. oklahomensis, and B. pseudomallei strains, it was considered to belong to this genetic cluster (Fig. 1b).
Assignment of putative functions to the bce-II gene products.
The deduced amino acid sequences of the bce-II genes of B. vietnamiensis G4 were compared with protein sequences deposited in GenBank. BceT showed identity with putative or confirmed bacterial UDP-glucose pyrophosphorylases (Table 3) catalyzing the reversible formation of UDP-glucose from UTP and glucose-1-phosphate. The bceM and bceN genes were predicted to encode a GDP-6-deoxy-d-lyxo-4-hexulose reductase (RMD) and a GDP-d-mannose 6,4-dehydratase (GMD), respectively (Table 3). These two enzyme activities use the precursor GDP-d-mannose to synthesize GDP-d-rhamnose, the donor of the d-rhamnose moiety of cepacian. The B. vietnamiensis G4 bceR gene encodes a putative 817-amino-acid protein that, based on sequence similarity, shows two domains of the glycosyltransferase family GT4, which comprises retaining glycosyltransferases with a wide variety of donor and acceptor specificities (Table 3). With the exception of Burkholderia strains, no other bifunctional glycosyltransferase homologous to BceR was identified. The B. vietnamiensis G4 bceO, bceS, and bceU genes encode proteins homologous to functionally uncharacterized putative proteins (Table 3). In spite of the weak conservation, BceO, -S, and -U are also homologous to proteins that define a family of integral membrane proteins involved in the acylation of carbohydrate moieties of extracytoplasmic molecules. These proteins include OafA from Salmonella enterica serovar Typhimurium, which acetylates O antigen (41); NodX from Rhizobium leguminosarum biovar viciae, responsible for acetylation of the Nod factor and for conferring host range specificity (16); and ExoZ from Sinorhizobium meliloti, responsible for succinoglycan acetylation (3). The B. vietnamiensis G4 bceP gene encodes a predicted protein homologous to uncharacterized putative proteins from other bacteria (Table 3). Gene bceQ encodes a putative protein homologous to flippase or translocase proteins of the Wzx family, postulated to be involved in the transport of oligosaccharide repeat units of the lipopolysaccharide (LPS) O antigen, capsular polysaccharides, and EPSs across the inner membrane (29). The closest characterized homologues of B. vietnamiensis G4 BceQ were AceE from Gluconacetobacter xylinus, Wzx from E. coli, and RfbE from Shigella flexneri (Table 3). In a previous work (31), the putative Bce flippase was incorrectly assigned to the bceL gene in the bce-I cluster. However, the analysis of other Burkholderia genomes, such as those of B. phytofirmans or B. xenovorans, indicated that this protein is absent from the bce cluster.
TABLE 3.
Features of the Bce proteins from Burkholderia
| Protein | Predicted function | Homologue | % Identity/ % similaritya | Organism | Conserved domainsb | GenBank accession no. |
|---|---|---|---|---|---|---|
| BceM | GDP-6-deoxy-d-lyxo-4-hexulose reductase (RMD) | 54/67 | Pseudomonas syringae | NADP(H) binding Wierenga motif G(X)2G(X)2G; catalytic domain Y(X)3K and S/T | YP_273224 | |
| Rmd | 53/68 | Xanthomonas campestris | CAP53110 | |||
| Rmd | 32/48 | Pseudomonas aeruginosa | ABJ14840 | |||
| BceN | GDP-d-mannose 4,6-dehydratase (GMD) | Gmd | 58/72 | Pseudomonas aeruginosa | NADP(H) binding Wierenga motif G(X)2G(X)2G; catalytic domain Y(X)3K and S/T | NP_254140 |
| Gmd | 52/66 | Klebsiella pneumoniae | YP_002920358 | |||
| Gmd | 51/67 | Escherichia coli | NP_288559 | |||
| BceO | Acyltransferase | 46/65 | Acidobacteria bacterium | 9 transmembrane domains | ABF40784 | |
| 32/61 | Rhizobium etli | ZP_03501439 | ||||
| 32/45 | Solibacter usitatus | ABJ83241 | ||||
| BceP | Unknown | 38/55 | Nodularia spumigena | Six-bladed beta propeller TolB-like domain | EAW43145 | |
| 38/56 | Nostoc punctiforme | ACC81483 | ||||
| 38/54 | Anabaena variabilis | ABA20764 | ||||
| BceQ | Repeat unit flippase | Wzx | 26/48 | Escherichia coli | 12 transmembrane domains | AAT85651 |
| RfbE | 26/49 | Shigella flexneri | CAA50771 | |||
| AceE | 35/52 | Gluconacetobacter xylinus | CAA64437 | |||
| BceR-I | Glycosyltransferase | WbaZ | 66/81 | Bacillus cereus | Amino acids 46 to 424; two β-α-β | ZP_04230623 |
| RfaG | 64/81 | Desulfotomaculum reducens | Rossman-like domains | YP_001114454 | ||
| WbaZ-like | 62/78 | Salmonella enterica | characteristic of GT-B fold proteins | EDZ22349 | ||
| BceR-II | Glycosyltransferase | YqgM_like | 42/60 | Nostoc punctiforme | Amino acids 428 to 817; two β-α-β Rossman-like domains characteristic of GT-B fold proteins | ACC81485 |
| YqgM_like | 44/60 | Nodularia spumigena | EAW42619 | |||
| RfaG | 44/60 | Rhodothermus marinus | YP_003290427 | |||
| BceS | Acyltransferase | 27/43 | Cytophaga hutchinsonii | 8 transmembrane domains | ABG58185 | |
| 29/44 | Methylobacterium sp. | ACA19306 | ||||
| 26/43 | Lentisphaera araneosa | EDM29042 | ||||
| BceT | UDP-glucose pyrophosphorylase (UGP) | GalU | 53/68 | Xanthomonas campestris | Nucleotide binding domain: 12GXGTRXLPXTK (X)3KEXLP(X)4P35; glucose-1-P binding domain: 191EKP(X)4APSXL (X)3GRY208 | Q8P8Q1 |
| GalU | 51/65 | Escherichia coli | AP_001862 | |||
| ExoN | 46/62 | Sinorhizobium meliloti | AAA16043 | |||
| BceU | Acyltransferase | 35/50 31/45 32/46 | Bradyrhizobium japonicum Agrobacterium radiobacter Sinorhizobium meliloti | 9 transmembrane domains | BAC47625AC ACM30377 CAC49254 |
All the hits with proteins from Burkholderia strains were excluded from this table due to the high identity scores obtained by BLAST analysis. Instead, shown is the level of identity between Bce proteins and experimentally characterized ones or, in the absence of biochemical data, the best hits with uncharacterized proteins.
Domains refer to B. vietnamiensis G4 protein sequences.
Cepacian production is widespread among Burkholderia strains.
To determine whether Bcc and non-Bcc strains were able to produce EPS, 10 strains were grown in liquid MM for 3 to 6 days at 30°C. The results indicate that after 3 days of growth, the non-Bcc soil isolates B. xenovorans LB400, B. phytofirmans PsJN, and B. phymatum STM815 and the Bcc strains B. cepacia IST408, B. multivorans ATCC 17616, and B. ambifaria AMMD produced more than 2.6 g/liter of EPS (Fig. 2a). B. vietnamiensis G4, B. dolosa AU0158, and B. lata sp. 383 produced less than 0.9 g/liter of EPS after 3 days of incubation (Fig. 2a), but after 6 days of growth, 1.1, 0.6, and 0.3 g/liter of EPS were obtained, respectively. The B. cenocepacia J2315 isolate from the ET12 lineage was unable to produce EPS. Although the genome of B. cepacia IST408 is not sequenced, cepacian is the sole EPS produced by this strain (6), being used as a reference strain in this work.
FIG. 2.
EPS production by Burkholderia strains. Cells from different Burkholderia species (a) and from B. cepacia IST408 (•), B. cepacia IST408 bceR::pIS58-2 (▪), B. multivorans ATCC 17616 (▴), and B. multivorans ATCC 17616 bceS::pSF71-8 (▵) (b) were grown in MM for 3 days at 30°C and EPSs quantified by dry weight after ethanol precipitation. Data represent the means of results from at least three independent experiments. Error bars show standard deviations. Phenotype of B. cepacia IST408 bceR::pIS58-2 harboring pMLBAD (c) or pIS94-1 containing the bceR gene (d) grown in solid MM supplemented with 1% arabinose.
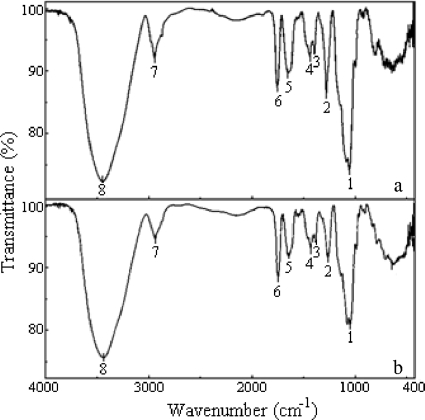
The EPSs obtained were analyzed by Fourier transform infrared (FTIR) spectroscopy using, as a reference, B. cepacia IST408 cepacian. All the EPSs originated spectra indistinguishable from the spectra obtained for B. cepacia IST408 cepacian, indicating that all strains analyzed produced cepacian. As an example, the FTIR spectra obtained for the EPSs of B. cepacia IST408 and B. phytofirmans PsJN are shown in Fig. 3.
FIG. 3.
FTIR analysis of the purified EPS produced by Burkholderia cepacia IST408 (a) and B. phytofirmans PsJN (b), showing similar spectra. Peaks: 1 and 2, carboxylic and/or hydroxyl C-O; 3 and 4, C-H; 5, water; 6, carbonyl; 7, C-C bonds; 8, water and hydroxyl groups of the EPS.
Altogether, our results suggest that cepacian production may be a common feature within the genus Burkholderia, independently of whether the source of the strain is environmental (B. xenovorans LB400, B. phytofirmans PsJN, B. phymatum STM815, B. multivorans ATCC 17616, B. vietnamiensis G4, B. lata sp. 383, and B. ambifaria AMMD) or clinical (B. cepacia IST408 and B. dolosa AU0158).
Characterization of the EPS phenotype of Burkholderia bceQ, bceR, and bceS mutants.
To clearly demonstrate the involvement of bce-II cluster genes in cepacian biosynthesis, the insertional inactivation of the genes bceQ, bceR, and bceS, encoding putative flippase, glycosyltransferase, and acyltransferase, respectively, was performed. The mutagenized strains were B. cepacia IST408 for the bceQ and bceR genes and B. multivorans ATCC 17616 for the bceS gene. The resulting mutants were named B. cepacia IST408 bceQ::pIS58-1, B. cepacia IST408 bceR::pIS58-2, and B. multivorans ATCC 17616 bceS::pSF71-8 (Table 1). The bceQ and bceR mutants were grown in liquid MM to assess EPS production. Under these conditions, no EPS production was detected over the 72 h of cultivation of the bceQ and bceR mutants, while the parental strain produced EPS (Fig. 2b). This result allowed for the conclusion that the bceQ and bceR genes are required for cepacian biosynthesis. In trans complementation using the bceR gene cloned in the replicative plasmid pMLBAD was performed. For this purpose, solid MM supplemented with 1% of arabinose was used. Under these conditions, the introduction of pIS94-1 into the bceR mutant led to the recovery of cepacian biosynthesis, associated with the mucoid phenotype of the colonies in solid medium (Fig. 2d).
The disruption of the B. multivorans ATCC 17616 bceS gene had no significant effect on cepacian biosynthesis, since the mutant and the parental strain produced similar quantities of EPS (Fig. 2b). In order to evaluate whether the mutation of bceS could have an impact in cepacian acetylation, the acetyl content of the EPSs produced by B. multivorans ATCC 17616 and the bceS mutant was quantified. The results obtained indicated 3.9 ± 0.1 and 3.1 ± 0.1 acetyl groups per repeat unit for the EPSs produced by the parental and the mutant strains, respectively. This reduction of approximately 20% in the acetyl content of the EPS produced by the bceS mutant is consistent with the predicted acetyltransferase activity of BceS.
Expression of bce genes in mucoid and nonmucoid strains.
To assess whether the genes from clusters bce-I and bce-II have similar expression patterns, quantitative real-time RT-PCR was performed with the two Bcc isogenic B. multivorans strains D2095 and D2214, isolated from a chronically infected cystic fibrosis patient (52), for the bceB, bceE, and bceG transcripts from the bce-I cluster and for the bceM, bceR, and bceS transcripts from the bce-II cluster. The expression values obtained indicate a 2-fold induction of the transcription of all the genes under study in the mucoid D2095 strain, compared to the level for the D2214 nonmucoid strain (Fig. 4a).
FIG. 4.
Quantitative real-time RT-PCR analysis of the relative transcript abundances in B. multivorans D2095 with respect to B. multivorans D2214 after 17 h of growth (a) and B. cepacia IST408 with respect to bceQ::pIS58-1 after 24 h of growth (b). For each gene, the data were standardized to values obtained for the internal control gene gyrB. The results were obtained from three independent experiments. Error bars represent standard deviations.
The expression of bceB, bceE, bceM, and bceS genes in the B. cepacia IST408 parental strain and in the bceQ::pIS58-1 mutant strain was also determined after 24 h of growth in SM. As is shown in Fig. 4b, the disruption of the bceQ gene had no significant influence on the expression levels of the bce-I region genes bceB and bceE. Concerning the two genes from bce-II region, the expression value of bceS was not found to change significantly when the parental and the mutant strain were compared, although a 2.5-fold induction was observed for the bceM gene in the parental strain B. cepacia IST408 (Fig. 4b).
Effect of the polysaccharide on the desiccation sensitivity of Burkholderia strains.
To determine whether the EPS produced by Burkholderia could contribute to their resistance to desiccation, the soil isolates B. xenovorans LB400 and B. multivorans ATCC 17616, the clinical isolate B. cepacia IST408, and the bceR::pIS58-2 mutant derivative were incubated in the presence or absence of 2.5 g/liter of EPS and kept dry for 7 days, and the numbers of CFU were determined (Fig. 5a). The results showed drastic reductions of CFU for B. xenovorans and B. multivorans incubated in the absence of the EPS in the first 24 h, and after 3 days, no viable cells were recovered (Fig. 5a). Contrastingly, when cells were dried in the presence of cepacian, viable cells could be recovered even after 7 days of incubation (Fig. 5a). B. cepacia IST408 and its mutant derivative bceR::pIS58-2 were highly susceptible to desiccation conditions, since no viable cells could be obtained after 24 h of exposure to desiccation either in the presence or in the absence of cepacian.
FIG. 5.
Protective role of EPS against desiccation and iron ion stress. Cells from overnight grown cultures of B. xenovorans LB400 (▪), B. multivorans ATCC 17616 (▴), and B. cepacia IST408 (•) were harvested by centrifugation and exposed to desiccation (a) or 50 mM ferrous sulfate (b) at 30°C in the presence (closed symbols) or absence (open symbols) of 2.5 g/liter of cepacian. The remaining viable bacterial counts were determined at different time points by determining the numbers of CFU. The data represents the means of results from three independent experiments. Error bars show standard deviations.
Effect of the exopolysaccharide on protection from metal ion stress.
The protective effects of cepacian against toxic levels of Fe2+ and Zn2+ were investigated by challenging B. xenovorans LB400, B. multivorans ATCC 17616, and B. cepacia IST408 with iron or zinc ions in the presence or absence of EPS. When these strains were challenged with 50 mM ferrous sulfate, the numbers of CFU of the strains incubated with 2.5 g/liter of EPS were 1 to 2 logs higher than those observed with the cells incubated with Fe2+ in the absence of EPS after 1 h of incubation (Fig. 5b). In the presence of EPS, only a slight increase in the survival rates was observed after treatment with zinc (data not shown). B. xenovorans and B. multivorans strains incubated with EPS seem to be more resistant to iron stress than B. cepacia under the same tested conditions. Indeed, we observed in 1 h a 1.5-log reduction when IST408 was incubated with EPS, while the two other strains exhibited no significant reduction in the number of viable cells. The B. cepacia IST408 bceR::IS58-2 mutant exhibited behavior similar to that observed for the parental strain (data not shown).
DISCUSSION
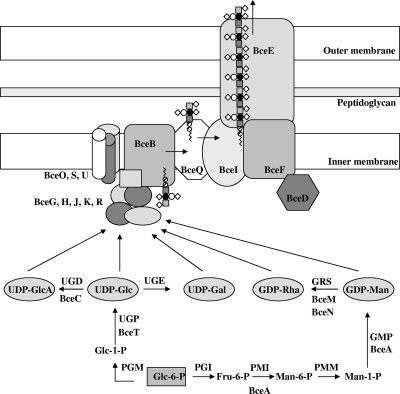
In this study, we report the identification of a second cluster of genes required for cepacian biosynthesis and compared the abilities of environmental and clinical Burkholderia strains to produce this EPS. In a previous work, Moreira et al. (31) identified 11 bce genes from B. cepacia IST408 as being involved in cepacian biosynthesis. Since the heptasaccharide repeat unit of cepacian is composed of 5 different sugars (6), it was evident that some protein activities were missing. A thorough analysis of the available Burkholderia genome sequences revealed that the genome sequences of four rhizosphere strains (B. graminis C4D1M, B. xenovorans LB400, B. phymatum STM815, and B. phytofirmans C6786) possess 8 additional genes immediately following the previously identified bce genes. These extra genes were also found in all other Burkholderia strains with a sequenced genome available, but, most probably due to genomic rearrangements caused by phages and insertion sequences, they are located several hundred kilobases apart from the initially identified bce-I cluster. The two clusters, bce-I and bce-II, account for most of the genes needed for cepacian biosynthesis, as depicted in Fig. 6. Most of the enzymes required for nucleotide sugar precursor synthesis are encoded by bce genes, with the exception of the genes encoding phosphoglucomutase (PGM), phosphoglucose isomerase (PGI), phosphomannomutase (PMM), and UDP-glucose epimerase (UGE). These enzyme activities are also involved in metabolic processes, such as the biosynthesis of lipopolysaccharide and other cell polysaccharides. The search for homologues to these proteins in Burkholderia genome sequences revealed that they are present in more than one copy and distributed in different locations of chromosomes 1 and 2 (data not shown). Two Bce proteins involved in nucleotide sugar precursor biosynthesis have been characterized: BceA, a bifunctional protein with phosphomannose isomerase and GDP-d-mannose pyrophosphorylase activities required for GDP-mannose biosynthesis (42), and BceC, a UDP-glucose dehydrogenase involved in UDP-glucuronic acid synthesis (26).
FIG. 6.
Pathway leading to the nucleotide-sugar precursors for cepacian biosynthesis by Burkholderia and model for the assembly and export of the EPS. With the exception of BceP, all the Bce proteins have confirmed or putative roles in EPS biosynthesis, as described in the text. Abbreviations: Glc, glucose; GlcA, glucuronic acid; Gal, galactose; Rha, rhamnose; Man, mannose; GDP, guanosine-5′-diphosphate; UDP, uridine-5′-diphosphate; PGM, phosphoglucomutase; UGE, UDP-glucose epimerase; PMM, phosphomannomutase; UGP, UDP-glucose pyrophosphorylase; PGI, phosphoglucose isomerase; GMP, GDP-d-mannose pyrophosphorylase; UGD, UDP-glucose dehydrogenase; PMI, phosphomannose isomerase; GRS, GDP-rhamnose synthetase.
The assembly of the heptasaccharide repeat unit of cepacian requires the priming glycosyltransferase BceB, catalyzing the addition of the first sugar, glucose, to the lipid carrier (50). The sequential addition of the six remaining sugars must be performed by BceG, -H, -J, and -K and the presumably bifunctional protein BceR, although the protein sequences do not give clues as to their specific glycosyltransferase activity. The presumably bifunctional protein BceR seems to have a unique organization, probably due to a process of fusion of two glycosyltransferase domains. Except for the orthologues found in the genome sequences of Burkholderia strains, no similar proteins could be found in databases.
The EPS produced by B. cepacia IST408, as well as from other strains, is acetylated. The exact number and position of the acetyl groups are still unknown (6). Three putative acyltransferases were found within the bce-II cluster, encoded by the bceO, bceS, and bceU genes. We show results indicating that the disruption of bceS caused a decrease in the acetylation content of cepacian, suggesting that BceS is involved in the repeat unit acetylation. Hydrophobicity analysis of the BceO, BceS, and BceU amino acid sequences suggests that they are probably located in the inner membrane. This is consistent with their possible role in the acetylation of cepacian since the repeat unit is synthesized on the cytoplasmic face of the inner membrane. O-acetylated extracellular and cell surface polysaccharides are synthesized by a wide range of bacterial pathogens (24), and this structural modification appears to play an important role in host-pathogen interactions. In many cases, the O-acetyl groups constitute prominent immunogenic epitopes critical for the host immune responses against the microorganism and for the development of protective vaccines (24, 35). The role of cepacian acetylation remains unknown, although one can expect a role similar to that observed for other acetylated polysaccharides.
The last steps in extracellular polysaccharide biosynthesis are the export of the repeat units to the periplasmic side of the inner membrane, their polymerization, and export of the nascent polymer. All evidence indicates that cepacian biosynthesis proceeds via the Wzy-dependent pathway. In this model, the lipid carrier-linked heptasaccharide repeat units are exported across the inner membrane by the putative flippase BceQ, being polymerized at the periplasmic face by the putative polymerase BceI (Fig. 6). BceQ and BceI are integral membrane proteins, and their involvement in cepacian biosynthesis is demonstrated by the EPS-deficient phenotype of the respective insertion mutants (31). In many bacteria, polymerization activity is influenced by an additional protein, referred to as polysaccharide copolymerase (PCP) (reviewed in reference 12). BceF is a PCP protein, having an N-terminal domain that spans twice the inner membrane with a large periplasmic loop and a C-terminal cytoplasmic domain with tyrosine kinase activity (15). The precise role(s) of this type of protein in polysaccharide biosynthesis is still unknown, but these proteins seem to play a critical role in the translocation of the polysaccharide chains from the periplasm to the cell surface through interaction with an outer membrane protein (12), which is likely to be BceE (Fig. 6).
Although several studies report the ability of Bcc clinical and environmental isolates to synthesize EPS (2, 11, 37, 52), not much is known on EPS biosynthesis by non-Bcc strains. This study demonstrates that, with the exception of B. mallei strains, all the Burkholderia strains with their genomes sequenced do have the bce gene cluster (together or fragmented) and most likely produce cepacian. This is the case of the rhizosphere non-Bcc species B. xenovorans, B. phymatum, and B. phytofirmans, which produced large amounts of cepacian. Most of the Bcc clinical isolates are EPS producers, with the exception of isolates of the B. cenocepacia species (52).
Polysaccharides secreted by bacteria play different roles in their biology and are frequently essential virulence determinants in pathogens of humans, livestock, and plants. Extracellular polysaccharides are also important in symbiotic interactions such as biological nitrogen fixation symbiosis between bacteria and plants, in adhesion to soil particles or roots, and as a barrier to harmful compounds, among other functions. In natural environments, bacterial polysaccharides may have different roles, depending on the ecological niche. One of the functions ascribed to EPSs is a role in the initial plant colonization and enhancement of survival of bacteria such as those of the genera Agrobacterium, Erwinia, and Pseudomonas, among others (13). The EPS produced by the alfalfa-symbiotic bacterium Sinorhizobium meliloti also functions as a signaling molecule, triggering a developmental response or suppressing defense responses by the plant (17). The EPS from the plant growth-promoting species Burkholderia gladioli was shown to elicit induced systemic resistance on cucumber (33). Bcc clinical and environmental strains were assessed for EPS production and onion tissue maceration ability, but no correlation could be established between EPS production and the ability to cause maceration of onion tissue (2). Other functions attributed to EPSs are of a protective nature, namely, barriers against desiccation, predation, antibiotics, or binding of toxic metal ions. In this study, we demonstrate that cepacian seems to play a protective role against desiccation. Moreover, only the soil isolates B. xenovorans LB400 and B. multivorans ATCC 17616, but not the clinical isolate B. cepacia IST408, were desiccation resistant. Due to the limited number of tested strains, no broader conclusions can be drawn regarding the fitness of environmental versus clinical strains in relation to desiccation. According to Vriezen et al. (51), the desiccation process comprises 3 phases. Phase I is the drying of the cells where metabolic processes slow down due to the lack of water. Phase II is the storage phase, where the decline of cell viability occurs. Phase III is the rehydration of the cells. It is possible that, due to cepacian hygroscopic properties, it retains more water, retarding the loss of cell viability during phase II, explaining the observed results. It is generally stated that the presence of EPS confers desiccation resistance to bacterial cells. Yet, only a few studies have been done correlating the presence of EPS with drying tolerance. One such study was performed by Tamaru et al. (45), showing that EPS-producing Nostoc commune colonies were highly stress tolerant, while EPS-depleted cells lost most of this ability. In addition, Tamaru and colleagues showed that the amount of EPS could be correlated with the degrees of both desiccation and freezing tolerance (45). In a recent work, Knowles and Castenholz (21) lend additional support to the role of EPS in stress tolerance by examining the effect of released EPS on neighboring cells within rock-inhabiting microbial communities. These authors showed that the addition of EPS significantly enhanced the desiccation tolerance of Chlorella sp. CCMEE 6038 and of Chroococcidiopsis sp. CCMEE 5056 (21). Another condition here imposed to Burkholderia strains was metal ion stress. Although appropriate concentrations of metals are required for bacterial growth, excessive concentrations can be lethal. In this study, the presence of high concentrations of two metal ions showed that the EPS plays an important role in survival against iron stress but provides only a slight protection from zinc stress. Overall, the results obtained in our study clearly show the importance of cepacian in desiccation and ion metal tolerance induced by Burkholderia, confirming a crucial function of the EPS in stress resistance.
The role of the EPS of Burkholderia as a virulence determinant has been demonstrated with mice (10, 43). In those studies, strains producing EPS and non-EPS derivative producers were compared, and the latter ones turned out to be less virulent. Nevertheless, there are no available data indicating whether EPS is produced in vivo by the Bcc. In addition, a limited survey of the clinical outcome in patients colonized with EPS- and non-EPS-producing Burkholderia strains concluded that no correlation could be established between EPS production ability and the persistence or virulence of bacteria (11). In a more recent study, Zlosnik et al. (52) carried out a larger survey of clinical isolates from 100 CF patients, showing that strains of B. cenocepacia, one of the most virulent Bcc species, are frequently nonmucoid. Additionally, they observed that isolates from chronically infected CF patients convert predominantly from mucoid to nonmucoid, opposite to the P. aeruginosa nonmucoid-to-mucoid conversion associated with the acute-to-chronic transition of infection (22). Whether the Bcc mucoid-to-nonmucoid conversion leads to a poorer clinical outcome is not yet known. Nevertheless, these authors suggested that nonmucoid isolates may be associated with increased disease severity and that the mucoid phenotype may be associated with persistence. These observations indicate that cepacian is unlikely to be a good marker for assessing pathogenicity in Burkholderia which depends on both bacterial and host determinants.
This study demonstrated the power of using comparative genomics in combination with genetics and microbial physiology to link EPS production by Burkholderia strains to the genes required for its biosynthesis. Furthermore, the widespread distribution of cepacian biosynthesis ability among clinical and environmental Burkholderia strains points out a multitude of possible roles of the EPS in host colonization, biofilm formation, or protection against hazardous compounds, making the elucidation of cepacian biosynthesis regulation crucial for understanding the biology of these microorganisms.
Acknowledgments
We gratefully acknowledge David P. Speert for B. multivorans strains D2095 and D2214.
This work was supported by FEDER and Fundação para a Ciência e a Tecnologia, Portugal (contract PTDC/BIA-MIC/66977/2006); Ph.D. grants to A.S.F. and I.N.S.; and a postdoctoral grant to S.A.S. C.G.R. acknowledges a Ph.D. grant from Fundação Calouste Gulbenkian, Portugal.
Footnotes
Published ahead of print on 30 November 2009.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartholdson, S. J., A. R. Brown, B. R. Mewburn, D. J. Clarke, S. C. Fry, D. J. Campopiano, and J. R. Govan. 2008. Plant host and sugar alcohol induced exopolysaccharide biosynthesis in the Burkholderia cepacia complex. Microbiology 154:2513-2521. [DOI] [PubMed] [Google Scholar]
- 3.Buendia, A. M., B. Enenkel, R. Koplin, K. Niehaus, W. Arnold, and A. Puhler. 1991. The Rhizobium meliloti exoZl exoB fragment of megaplasmid 2: ExoB functions as a UDP-glucose 4-epimerase and ExoZ shows homology to NodX of Rhizobium leguminosarum biovar viciae strain TOM. Mol. Microbiol. 5:1519-1530. [DOI] [PubMed] [Google Scholar]
- 4.Bullock, W. C., J. M. Fernandz, and J. M. Short. 1987. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. Biotechniques 5:376-379. [Google Scholar]
- 5.Bylund, J., L. A. Burgess, P. Cescutti, R. K. Ernst, and D. P. Speert. 2006. Exopolysaccharides from Burkholderia cenocepacia inhibit neutrophil chemotaxis and scavenge reactive oxygen species. J. Biol. Chem. 281:2526-2532. [DOI] [PubMed] [Google Scholar]
- 6.Cescutti, P., M. Bosco, F. Picotti, G. Impallomeni, J. H. Leitão, J. A. Richau, and I. Sá-Correia. 2000. Structural study of the exopolysaccharide produced by a clinical isolate of Burkholderia cepacia. Biochem. Biophys. Res. Commun. 273:1088-1094. [DOI] [PubMed] [Google Scholar]
- 7.Chiarini, L., P. Cescutti, L. Drigo, G. Impallomeni, Y. Herasimenka, A. Bevivino, C. Dalmastri, S. Tabacchioni, G. Manno, F. Zanetti, and R. Rizzo. 2004. Exopolysaccharides produced by Burkholderia cenocepacia recA lineages IIIA and IIIB. J. Cyst. Fibros. 3:165-172. [DOI] [PubMed] [Google Scholar]
- 8.Coenye, T., J. J. LiPuma, D. Henry, B. Hoste, K. Vandemeulebroecke, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia cepacia genomovar VI, a new member of the Burkholderia cepacia complex isolated from cystic fibrosis patients. Int. J. Syst. Evol. Microbiol. 51:271-279. [DOI] [PubMed] [Google Scholar]
- 9.Coenye, T., E. Mahenthiralingam, D. Henry, J. J. LiPuma, S. Laevens, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia ambifaria sp. nov., a novel member of the Burkholderia cepacia complex including biocontrol and cystic fibrosis-related isolates. Int. J. Syst. Evol. Microbiol. 51:1481-1490. [DOI] [PubMed] [Google Scholar]
- 10.Conway, B. A., K. K. Chu, J. Bylund, E. Altman, and D. P. Speert. 2004. Production of exopolysaccharide by Burkholderia cenocepacia results in altered cell-surface interactions and altered bacterial clearance in mice. J. Infect. Dis. 190:957-966. [DOI] [PubMed] [Google Scholar]
- 11.Cunha, M. V., S. A. Sousa, J. H. Leitão, L. M. Moreira, P. A. Videira, and I. Sá-Correia. 2004. Studies on the involvement of the exopolysaccharide produced by cystic fibrosis-associated isolates of the Burkholderia cepacia complex in biofilm formation and in persistence of respiratory infections. J. Clin. Microbiol. 42:3052-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuthbertson, L., I. L. Mainprize, J. H. Naismith, and C. Whitfield. 2009. Pivotal roles of the outer membrane polysaccharide export and polysaccharide copolymerase protein families in export of extracellular polysaccharides in gram-negative bacteria. Microbiol. Mol. Biol. Rev. 73:155-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denny, T. P. 1995. Involvement of bacterial polysaccharides in plant pathogenesis. Annu. Rev. Phytopathol. 33:173-197. [DOI] [PubMed] [Google Scholar]
- 14.Dudman, W. F. 1977. The role of surface polysaccharides in natural environments, p. 357-414. In I. W. Sutherland (ed.), Surface carbohydrates of the prokaryotic cell. Academic Press, New York, NY.
- 15.Ferreira, A. S., J. H. Leitão, S. A. Sousa, A. M. Cosme, I. Sá-Correia, and L. M. Moreira. 2007. Functional analysis of Burkholderia cepacia genes bceD and bceF, encoding a phosphotyrosine phosphatase and a tyrosine autokinase, respectively: role in exopolysaccharide biosynthesis and biofilm formation. Appl. Environ. Microbiol. 73:524-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Firmin, J. L., K. E. Wilson, R. W. Carlson, A. E. Davies, and J. A. Downie. 1993. Resistance to nodulation of cv. Afghanistan peas is overcome by nodX, which mediates an O-acetylation of the Rhizobium leguminosarum lipo-oligosaccharide nodulation factor. Mol. Microbiol. 10:351-360. [DOI] [PubMed] [Google Scholar]
- 17.Gibson, K. E., H. Kobayashi, and G. C. Walker. 2008. Molecular determinants of a symbiotic chronic infection. Annu. Rev. Genet. 42:413-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goris, J., P. De Vos, J. Caballero-Mellado, J. Park, E. Falsen, J. F. Quensen III, J. M. Tiedje, and P. Vandamme. 2004. Classification of the biphenyl- and polychlorinated biphenyl-degrading strain LB400T and relatives as Burkholderia xenovorans sp. nov. Int. J. Syst. Evol. Microbiol. 54:1677-1681. [DOI] [PubMed] [Google Scholar]
- 19.Govan, J. R., P. H. Brown, J. Maddison, C. J. Doherty, J. W. Nelson, M. Dodd, A. P. Greening, and A. K. Webb. 1993. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet 342:15-19. [DOI] [PubMed] [Google Scholar]
- 20.Herasimenka, Y., P. Cescutti, G. Impallomeni, S. Campana, G. Taccetti, N. Ravenni, F. Zanetti, and R. Rizzo. 2007. Exopolysaccharides produced by clinical strains belonging to the Burkholderia cepacia complex. J. Cyst. Fibros. 6:145-152. [DOI] [PubMed] [Google Scholar]
- 21.Knowles, E. J., and R. W. Castenholz. 2008. Effect of exogenous extracellular polysaccharides on the desiccation and freezing tolerance of rock-inhabiting phototrophic microorganisms. FEMS Microbiol. Ecol. 66:261-270. [DOI] [PubMed] [Google Scholar]
- 22.Koch, C., and N. Hoiby. 1993. Pathogenesis of cystic fibrosis. Lancet 341:1065-1069. [DOI] [PubMed] [Google Scholar]
- 23.Komlos, J., A. B. Cunningham, A. K. Camper, and R. R. Sharp. 2005. Interaction of Klebsiella oxytoca and Burkholderia cepacia in dual-species batch cultures and biofilms as a function of growth rate and substrate concentration. Microb. Ecol. 49:114-125. [DOI] [PubMed] [Google Scholar]
- 24.Kooistra, O., E. Luneberg, B. Lindner, Y. A. Knirel, M. Frosch, and U. Zahringer. 2001. Complex O-acetylation in Legionella pneumophila serogroup 1 lipopolysaccharide. Evidence for two genes involved in 8-O-acetylation of legionaminic acid. Biochemistry 40:7630-7640. [DOI] [PubMed] [Google Scholar]
- 25.Lefebre, M. D., and M. A. Valvano. 2002. Construction and evaluation of plasmid vectors optimized for constitutive and regulated gene expression in Burkholderia cepacia complex isolates. Appl. Environ. Microbiol. 68:5956-5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loutet, S. A., S. J. Bartholdson, J. R. Govan, D. J. Campopiano, and M. A. Valvano. 2009. Contributions of two UDP-glucose dehydrogenases to viability and polymyxin B resistance of Burkholderia cenocepacia. Microbiology 155:2029-2039. [DOI] [PubMed] [Google Scholar]
- 27.Mahenthiralingam, E., T. A. Urban, and J. B. Goldberg. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3:144-156. [DOI] [PubMed] [Google Scholar]
- 28.Markowitz, V. M., E. Szeto, K. Palaniappan, Y. Grechkin, K. Chu, I. M. Chen, I. Dubchak, I. Anderson, A. Lykidis, K. Mavromatis, N. N. Ivanova, and N. C. Kyrpides. 2008. The integrated microbial genomes (IMG) system in 2007: data content and analysis tool extensions. Nucleic Acids Res. 36:D528-D533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marolda, C. L., J. Vicarioli, and M. A. Valvano. 2004. Wzx proteins involved in biosynthesis of O antigen function in association with the first sugar of the O-specific lipopolysaccharide subunit. Microbiology 150:4095-4105. [DOI] [PubMed] [Google Scholar]
- 30.McComb, E. A., and R. M. McCready. 1957. Determination of acetyl in pectin and acetylated carbohydrate polymers. Anal. Chem. 29:819-821. [Google Scholar]
- 31.Moreira, L. M., P. A. Videira, S. A. Sousa, J. H. Leitão, M. V. Cunha, and I. Sá-Correia. 2003. Identification and physical organization of the gene cluster involved in the biosynthesis of Burkholderia cepacia complex exopolysaccharide. Biochem. Biophys. Res. Commun. 312:323-333. [DOI] [PubMed] [Google Scholar]
- 32.Nelson, M. J., S. O. Montgomery, W. R. Mahaffey, and P. H. Pritchard. 1987. Biodegradation of trichloroethylene and involvement of an aromatic biodegradative pathway. Appl. Environ. Microbiol. 53:949-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park, K., J. W. Kloepper, and C. M. Ryu. 2008. Rhizobacterial exopolysaccharides elicit induced resistance on cucumber. J. Microbiol. Biotechnol. 18:1095-1100. [PubMed] [Google Scholar]
- 34.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pier, G. B., F. Coleman, M. Grout, M. Franklin, and D. E. Ohman. 2001. Role of alginate O acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect. Immun. 69:1895-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Potts, M. 1994. Desiccation tolerance of prokaryotes. Microbiol. Rev. 58:755-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richau, J. A., J. H. Leitão, M. Correia, L. Lito, M. J. Salgado, C. Barreto, P. Cescutti, and I. Sá-Correia. 2000. Molecular typing and exopolysaccharide biosynthesis of Burkholderia cepacia isolates from a Portuguese cystic fibrosis center. J. Clin. Microbiol. 38:1651-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richau, J. A., J. H. Leitão, and I. Sá-Correia. 2000. Enzymes leading to the nucleotide sugar precursors for exopolysaccharide synthesis in Burkholderia cepacia. Biochem. Biophys. Res. Commun. 276:71-76. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. CSHL Press, New York, NY.
- 40.Sessitsch, A., T. Coenye, A. V. Sturz, P. Vandamme, E. A. Barka, J. F. Salles, J. D. Van Elsas, D. Faure, B. Reiter, B. R. Glick, G. Wang-Pruski, and J. Nowak. 2005. Burkholderia phytofirmans sp. nov., a novel plant-associated bacterium with plant-beneficial properties. Int. J. Syst. Evol. Microbiol. 55:1187-1192. [DOI] [PubMed] [Google Scholar]
- 41.Slauch, J. M., A. A. Lee, M. J. Mahan, and J. J. Mekalanos. 1996. Molecular characterization of the oafA locus responsible for acetylation of Salmonella typhimurium O-antigen: oafA is a member of a family of integral membrane trans-acylases. J. Bacteriol. 178:5904-5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sousa, S. A., L. M. Moreira, J. Wopperer, L. Eberl, I. Sá-Correia, and J. H. Leitão. 2007. The Burkholderia cepacia bceA gene encodes a protein with phosphomannose isomerase and GDP-D-mannose pyrophosphorylase activities. Biochem. Biophys. Res. Commun. 353:200-206. [DOI] [PubMed] [Google Scholar]
- 43.Sousa, S. A., M. Ulrich, A. Bragonzi, M. Burke, D. Worlitzsch, J. H. Leitão, C. Meisner, L. Eberl, I. Sá-Correia, and G. Doring. 2007. Virulence of Burkholderia cepacia complex strains in gp91phox-/- mice. Cell. Microbiol. 9:2817-2825. [DOI] [PubMed] [Google Scholar]
- 44.Stanier, R. Y., N. J. Palleroni, and M. Doudoroff. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43:159-271. [DOI] [PubMed] [Google Scholar]
- 45.Tamaru, Y., Y. Takani, T. Yoshida, and T. Sakamoto. 2005. Crucial role of extracellular polysaccharides in desiccation and freezing tolerance in the terrestrial cyanobacterium Nostoc commune. Appl. Environ. Microbiol. 71:7327-7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vandamme, P., J. Goris, W. M. Chen, P. de Vos, and A. Willems. 2002. Burkholderia tuberum sp. nov. and Burkholderia phymatum sp. nov., nodulate the roots of tropical legumes. Syst. Appl. Microbiol. 25:507-512. [DOI] [PubMed] [Google Scholar]
- 48.Vandamme, P., B. Holmes, M. Vancanneyt, T. Coenye, B. Hoste, R. Coopman, H. Revets, S. Lauwers, M. Gillis, K. Kersters, and J. R. Govan. 1997. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 47:1188-1200. [DOI] [PubMed] [Google Scholar]
- 49.Vanlaere, E., A. Baldwin, D. Gevers, D. Henry, E. De Brandt, J. J. LiPuma, E. Mahenthiralingam, D. P. Speert, C. Dowson, and P. Vandamme. 2009. Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int. J. Syst. Evol. Microbiol. 59:102-111. [DOI] [PubMed] [Google Scholar]
- 50.Videira, P. A., A. P. Garcia, and I. Sá-Correia. 2005. Functional and topological analysis of the Burkholderia cenocepacia priming glucosyltransferase BceB, involved in the biosynthesis of the cepacian exopolysaccharide. J. Bacteriol. 187:5013-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vriezen, J. A., F. J. de Bruijn, and K. Nusslein. 2006. Desiccation responses and survival of Sinorhizobium meliloti USDA 1021 in relation to growth phase, temperature, chloride and sulfate availability. Lett. Appl. Microbiol. 42:172-178. [DOI] [PubMed] [Google Scholar]
- 52.Zlosnik, J. E., T. J. Hird, M. C. Fraenkel, L. M. Moreira, D. A. Henry, and D. P. Speert. 2008. Differential mucoid exopolysaccharide production by members of the Burkholderia cepacia complex. J. Clin. Microbiol. 46:1470-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]