Abstract
Enterococci are among the most common human intestinal lactic acid bacteria, and they are known to produce bacteriocins. In this study, fecal enterococci were isolated from infants and screened for bacteriocin production. Bacteriocin-producing Enterococcus avium isolates were obtained, and a new pediocin-like bacteriocin was purified and characterized. This bacteriocin, termed avicin A, was found to be produced by isolates from two healthy infants. It was purified to homogeneity from culture supernatant by ion-exchange and reversed-phase chromatography, and part of its amino acid sequence was obtained. The sequence of a 7-kb DNA fragment of a bacteriocin locus was determined by PCR and DNA sequencing. The bacteriocin locus was organized into four operon-like structures consisting of (i) the structural genes encoding avicin A and its immunity protein, (ii) a divergicin-like bacteriocin (avicin B) gene, (iii) an ABC bacteriocin transporter gene and two regulatory genes (histamine protein kinase- and response regulator-encoding genes), and (iv) induction peptide pheromone- and transport accessory protein-encoding genes. It was shown that the production of avicin A was regulated by the peptide pheromone-inducible regulatory system. Avicin A shows very high levels of similarity to mundticin KS and enterocin CRL35. This bacteriocin showed strong antimicrobial activity against many species of Gram-positive bacteria, including the food-borne pathogen Listeria monocytogenes. The avicin A locus is the first bacteriocin locus identified in E. avium to be characterized at the molecular level.
Bacteriocins are ribosomally synthesized antimicrobial peptides and proteins. Production of these compounds is widespread in Gram-negative and Gram-positive bacteria (23). Bacteriocins produced by lactic acid bacteria (LAB) have recently been classified into two major categories: the lantibiotics or lanthionine-containing bacteriocins (class I) and the non-lanthionine-containing bacteriocins (class II) (5). According to this classification, the former class III bacteriocins (large heat-labile bacteriocins) were considered nonbacteriocins and hence designated bacteriolysins. The class II bacteriocins are further subdivided into four subclasses: subclass IIa (pediocin-like bacteriocins), subclass IIb (two-peptide bacteriocins), subclass IIc (cyclic bacteriocins), and subclass IId (nonpediocin linear peptide bacteriocins). Class II bacteriocins are most commonly found in enterococci. The subclass IIa bacteriocins are known for their strong antilisterial activity, and they are distinguished by their N-terminal conserved YYGNG motif and two covalently S-S-linked cysteines separated by four amino acid residues (11).
The production of subclass IIa bacteriocins usually requires four genes: a bacteriocin gene (which encodes the bacteriocin precursor), an immunity gene (which protects the producer from its bacteriocin), and the ABC transporter and transport accessory genes (31, 44). Bacteriocins are synthesized as biologically inactive prepeptides (precursors) containing an N-terminal leader peptide that is cleaved off during maturation and exported (23, 31) by a dedicated ABC transporter. The biosynthesis of subclass IIa bacteriocins is frequently regulated by a quorum-sensing regulatory mechanism that consists of a peptide pheromone (inducing peptide) that acts as a signal in a phosphorylation reaction with the receptor histidine protein kinase; this is followed by phosphorylation of the response regulator to activate gene expression in the various operons required for bacteriocin production (32).
Bacteriocins have a narrow spectrum of antimicrobial activity, killing only strains of the same species or closely related species (26, 27), but they can be very potent and are able to kill other bacteria at nanomolar concentrations. Most bacteriocin-producing LAB have been isolated from foods, and very few bacteriocin producers have been isolated from humans so far (14). It has been proposed that bacteriocins can be used not only in food preservation but also in medicine as selective antimicrobials to inhibit pathogens without affecting the normal flora (17). Bacteriocin production is considered a probiotic feature, and it has convincingly been shown that bacteriocin-producing LAB can effectively inhibit the growth of and kill listeriae in mice (4). Thus, identification and characterization of bacteriocin-producing LAB of human origin are needed in order to develop probiotic bacteria with diverse antimicrobial potential. In line with this goal, we aimed at isolating bacteriocin-producing LAB from healthy human infants. In two independent screenings for bacteriocin-producing LAB in fecal samples from healthy infants, a new bacteriocin from Enterococcus avium was purified and characterized at the molecular and genetic levels.
MATERIALS AND METHODS
Isolation and identification of bacteriocin-producing strains.
In order to screen for bacteriocin-producing LAB, we isolated LAB from feces of 13 healthy Norwegian infants in 2005 and 20 Ethiopian infants in 2007. Bacteriocin-producing strains were obtained from fecal samples from one Norwegian infant and one Ethiopian infant. The strains were isolated on reinforced clostridial medium or de Man-Rogosa-Sharpe medium (MRS) after fecal samples were diluted. The species was identified by using 16S rRNA gene sequencing and BLAST analysis.
Bacterial strains, plasmids, and culture conditions.
Strains and plasmids used in this study are shown in Table 1. Solid media and soft agar were prepared with 1.5% and 0.7% agar, respectively. All strains were maintained as frozen stocks at −80°C in 13% glycerol. Enterococcus species were grown in GM17 or MRS at 37°C. Lactobacillus, Pediococcus, Leuconostoc, and Carnobacterium species were grown in MRS at 25°C or 30°C. Lactococcus species were grown in GM17 or MRS at 30°C. Escherichia coli was grown in Luria-Bertani (LB) medium at 37°C. Listeria species were grown in GM17 at 37°C.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Source or referenceb |
|---|---|---|
| Strains | ||
| Carnobacterium divergens NCDO 2763T | Sensitive to avicin A | NCDO |
| Carnobacterium divergens NCDO 2306 | Sensitive to avicin A | NCDO |
| Carnobacterium piscicola NCDO 2762T | Sensitive to avicin A | NCDO |
| Carnobacterium piscicola NCDO 2764 | Sensitive to avicin A | NCDO |
| Carnobacterium piscicola UI 49 | Bacteriocin producer | 45 |
| Escherichia coli DH5α | φ80dlacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169 | Promega |
| Enterococcus avium 208 | Avicin A producer, Norwegian infant isolate | This study |
| Enterococcus avium GM62 | Sensitive to avicin A | This study |
| Enterococcus avium UA62 | Sensitive to avicin A | This study |
| Enterococcus avium UM83 | Sensitive to avicin A | This study |
| Enterococcus avium XA83 | Avicin A producer, Ethiopian infant isolate | This study |
| Enterococcus faecalis 185 | Infant isolate, sensitive to avicin A | This study |
| Enterococcus faecalis LMGT 2708RA | Pediocin-resistant mutant of E. faecalis EF BRIDGE (B) | LMGT |
| Enterococcus faecium P21 | LMGT | |
| Enterococcus maldoratus XM83 | Infant isolate, sensitive to avicin A | This study |
| Enterococcus faecalis EF BRIDGE (B) | Indicator sensitive to avicin A | LMGT |
| Lactobacillus fermentum KLD | LMGT | |
| Lactobacillus plantarum 965 | LMGT | |
| Lactobacillus rhamnosus 205 | Infant isolate, sensitive to avicin A | This study |
| Lactobacillus sakei NCDO 2714 | Indicator sensitive to avicin A | NCDO |
| Lactobacillus sakei 5 | Bacteriocin producer | V. Eijsink |
| Lactococcus lactis IL 1403 | Host for transformation, contains lcnC and lcnD analogues | 49 |
| Lactococcus lactis CM4 | ||
| Leuconostoc gelidum Ta 11a | LMGT | |
| Listeria innocua BL86/26 B | Sensitive to avicin A | LMGT |
| Pediococcus acidilactici | Pediocin producer | LMGT |
| Pediococcus pentosaceus NCDO 559 | NCDO | |
| Plasmids | ||
| pBluescript KS | 3.0-kb cloning vector, αlacZ, Apr | Stratagene |
| pMG36e | 3.7-kb lactococcal expression vector, Emr | 47 |
| pMG36eDV | pMG36e containing divergicin-like ORF, avcB | This study |
| pCR2.1-TOPO | 3.9-kb cloning vector, Apr | Invitrogen |
Apr, ampicillin resistant; Emr, erythromycin resistant.
NCDO, National Collection of Dairy Organisms (Reading, United Kingdom); LMGT, Laboratory of Microbial Gene Technology, Norwegian University of Life Sciences.
Bacteriocin antimicrobial activity assay.
Antimicrobial activity was detected by using a deferred antagonism assay (41). The isolated bacteria were spotted onto MRS agar plates and incubated for 16 h at 37°C. The colonies were overlaid with an overnight culture (diluted 400-fold) of the indicator strain (Table 1). After overnight incubation, the formation of growth inhibition zones around the colonies was used as an indication of antimicrobial activity. To investigate whether the antimicrobial activity observed was caused by a proteinaceous compound, the sensitivity to proteinase K was tested. Only proteinase K-sensitive activities were used in further studies.
Quantitative determination of the antimicrobial activity of the bacteriocin in cell-free culture supernatant (CFS) was performed by using a microtiter assay system (21). A twofold serial dilution (in MRS) of 100 μl CFS (with the pH adjusted to about 7) was prepared in a microtiter plate well containing 50 μl MRS to which 150 μl of a diluted (400-fold in MRS) overnight culture of the indicator strain Lactobacillus sakei NCDO 2714 was added. The plate was incubated for 16 h, after which growth inhibition was measured turbidometrically at 620 nm with a microtiter plate reader (Labsystems iEMS reader MF; Labsystems, Helsinki, Finland). One bacteriocin unit was defined as the amount of bacteriocin that inhibited the growth of the NCDO 2714 indicator strain by 50%. Likewise, the MICs of the purified bacteriocin for sensitive strains were determined by using a microtiter assay. Purified bacteriocin was used to determine the molar MICs for the individual indicator strains tested.
Bacteriocin induction assay.
Quorum-sensing-dependent regulation of bacteriocin production was tested by using a slight modification of a previously described method (7). In order to obtain non-bacteriocin-producing E. avium XA83 (bac− phenotype) from the bacteriocin-producing culture (bac+ phenotype), this culture was serially diluted in MRS and grown overnight, and a bac− culture was obtained with the highest dilutions. In order to induce bacteriocin production, 50 μl of CFS from a bacteriocin-producing overnight culture was added to 5 ml of a bac− strain-inoculated culture in MRS broth and incubated overnight. Also, the inducing peptide AvcP, which was synthesized and purified (>95% pure) by EZBiolab (United States), was used in induction experiments.
Purification of bacteriocin.
The supernatant from a 500-ml overnight culture (21 h at 37°C in MRS) of E. avium XA83 was collected. Ammonium sulfate (40 g per 100 ml) was added to the supernatant, which was then agitated for 30 min at 4°C. The bacteriocin was precipitated from the supernatant by centrifugation (14,000 rpm for 30 min at 4°C) and dissolved in 50 ml sterile distilled water, and the pH was adjusted to 3.5 with 1 M HCl.
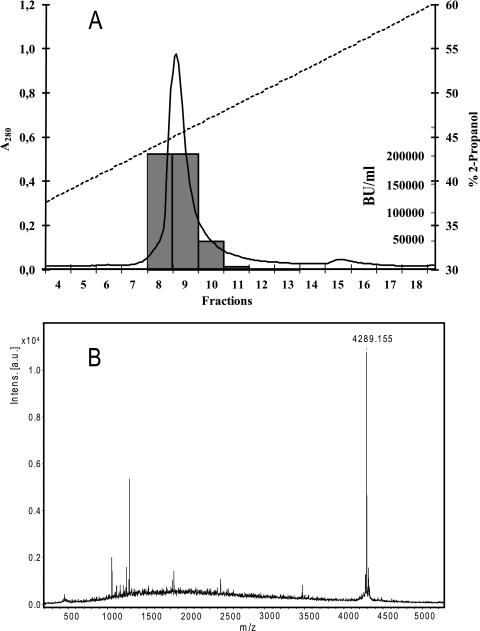
The preparation was then passed through a 5-ml SP Sepharose Fast Flow column (GE Healthcare Biosciences, Uppsala, Sweden) equilibrated with 10 mM acetic acid. The column was washed twice with 20 ml of 20 mM sodium phosphate (pH 6.8). The column was eluted with a stepwise gradient consisting of 10 ml of 0.1 NaCl (two runs), 10 ml of 0.3 NaCl (two runs), and 10 ml of 1 M NaCl (two runs) at a flow rate of 1 ml per min. All fractions except the second 10-ml 0.1 M NaCl fraction contained bacteriocin activity, and the most activity was found in the first 10-ml 0.3 M NaCl eluate. Subsequent purification was performed by using reversed-phase chromatography with an Äkta Purifier fast protein liquid chromatography system. The most active fraction (0.3 M eluate) from the ion-exchange chromatography step was applied to a reversed-phase column (Resource I; Pharmacia Biotechnology) equilibrated with 0.1% trifluoroacetic acid (TFA) in water. Elution was performed by using a 30-column volume (CV) linear gradient from 0 to 100% 2-propanol containing 0.1% TFA, and 2-ml fractions were collected. The two fractions with the most activity (fractions 9 and 10) were combined and diluted in sterile water (final volume, 20 ml), and then they were applied to a Source 5RPC ST 4.6/415 column (Pharmacia Biotechnology) and eluted with a 5-CV linear gradient as described above in 1-ml fractions (Fig. 1A). The fractions were assayed for antimicrobial activity, and the two most active fractions (fractions 8 and 9) coincided with the single peak of absorbance at 280 nm (Fig. 1A) and were stored at −20°C until further analysis.
FIG. 1.
(A) Results of second reversed-phase chromatography of avicin A. Elution was performed by using a 30-CV linear gradient of 0 to 100% 2-propanol containing 0.1% TFA. Solid line, absorbance at 280 nm; dashed line, isopropanol gradient; bars, bacteriocin units (BU) in eluted active fractions. A sample for mass spectroscopy analysis was obtained from fraction 9. (B) Mass spectrometry analysis of avicin A obtained from the second reversed-phase chromatography. Intens., intensity; a.u., arbitrary units.
Mass spectrometry and N-terminal amino acid sequencing.
The molecular weight of the purified bacteriocin was determined by mass spectrometry as previously described (6). Briefly, a bacteriocin sample (fraction 9 [Fig. 1A]) was mixed 1:1 with a saturated solution of α-cyano-4-hydroxycinnamic acid in 0.1% TFA-acetonitrile (2:1) and deposited on a ground steel matrix-assisted laser desorption ionization target. Mass spectra were recorded in reflection mode with an Ultraflex TOF/TOF (Daltonics), using a pulsed ion extraction setting of 40 ns and an acceleration voltage of 25 kV. The spectra displayed the accumulated signals of 200 laser shots with the laser power adjusted to just above the threshold level. A peptide mass similarity search was done at ExPASy with the TagIdent tool (http://au.expasy.org/tools/tagident). The N-terminal amino acid sequence of the peptide (bacteriocin) was determined by Edman degradation as previously described (21).
General molecular techniques.
Genomic DNA was isolated from LAB strains by the cetyltrimethylammonium bromide method (1), with some modifications, and was purified using a QIAquick PCR purification kit (Qiagen, Germany). Plasmid DNA was isolated from E. coli and Lactococcus lactis using a Qiagen plasmid miniprep kit (Qiagen, Germany). All enzymes used for DNA manipulation were purchased from New England Biolabs and were used according to the manufacturer's instructions. Primers were synthesized by Thermo Scientific (Germany). PCR products and DNA digests were purified with a QIAquick PCR product purification kit. PCR products were purified from an agarose gel using a Wizard SV gel and the PCR Clean-Up system (Promega, United States).
PCR and sequencing.
Genomic DNA of the bacteriocin producers XA83 and 208 were digested with DraI, RsaI, ClaI, EcoRV, HindIII, and SpeI, and purified fragments were ligated into plasmid pCR2.1-TOPO digested with DraI and RsaI and plasmid pBluescript KS digested with ClaI, EcoRV, HindIII and SpeI, respectively. Each ligation product in pCR2.1-TOPO was used as a PCR template to amplify part of the bacteriocin gene (113 bp) by using the degenerate primers AvFwr and AvRev (Table 2) deduced from the N-terminal peptide sequence. The PCR product was cloned into the vector pCR2.1-TOPO and sequenced with the T7 universal primer. T7 and new primers that were designed based on the sequence were used to amplify and sequence flanking regions of the bacteriocin gene using ligation products in pBluescript KS as PCR templates. This procedure was repeated until the bacteriocin locus was completely sequenced. Primers used in this study are listed in Table 2. Cycle sequencing was done by using a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, United States), and the PCR products were sequenced by using the ABI Prism 377 DNA sequencing system (Applied Biosystems, United States). Contigs were assembled by using the BioEdit software, version 7.0.0. The integrity of the contigs was confirmed by performing PCR with genomic DNA and sequencing the products. Both strands were sequenced to make sure that the whole sequence was error-free. Open reading frames (ORFs) were identified by ORF finder at NCBI (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). A similarity search was done by using blastn and/or blastx at NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
TABLE 2.
PCR primers
| Primer | Sequence (5′-3′) |
|---|---|
| Ur4 | AACTGCTTTAATTGTGGCTAA |
| F1 | GGAGTGTGTTGATATGACAAG |
| AvcF4 | GCTTTGCGAAATGATGGAGT |
| Df5 | TGCGACATTTAATATGAAGAG |
| R1 | TCATTCCATACCTGACGGTG |
| Dr8 | TCATGAATCCCACCACAATC |
| Df8 | ACAGGAGTTTTCGCACTTGG |
| Df10 | AGCATGACGAAAAGGATTGC |
| Df11 | GTATTAACAGCAGTCCCAGACG |
| Dr15 | CCCAATAATCCCAAATCCTG |
| Df12 | AGCAAACGAGGAACAGATGG |
| Dr11 | TTCCCAGAACCACTCATACC |
| Df13 | GAACTAGCGCAAATAAGGGAG |
| Df14 | CATGATAGGAAGTGAGTCTGTG |
| Df20 | GTTGCTTGGGTTAAAAGCTG |
| Dr12 | TTACAGTTAATCCCGTTTGCTC |
| Df16 | GATGTGATTAATATGGATGAGG |
| Df17 | CAGCTTTGGCTAATGTCGAG |
| Dr13 | CTCGACATTAGCCAAAGCTG |
| Df18 | AATTGTTTCGTTTTGCGACA |
| Dr14 | TAGAGAAATGCAACCCTTCAG |
| T7 | GTAATACGACTCACTATAGGG |
| M13rev | CAGGAAACAGCTATGAC |
| AvFwr | ACNTAYTAYGGNAAYGGNGT |
| AvRev | GCNCCNCCNGTNGCNARRTTNGC |
| TFF2 | TGACTGGGGAAAGCCATCAG |
| TFR2 | GGCTTTGCCCCAGTCACCTGA |
| aviB-F1a | TGGTGGTCTAGATAGTGCGACATTTAATATGAAGAGC |
| aviB-R1a | TGGTGGCTGCAGCTAACCAAAACACCCACCTG |
Primers used for cloning, not for primer walking. Underlining indicates XbaI and PstI restriction sites; bold type indicates sequences complementary to the template DNA.
Cloning of the divergicin-like ORF.
Primers aviB-F1 and aviB-R1 (Table 2) containing XbaI and PstI restriction sites were designed for the regions flanking the two ends of the divergicin-like ORF (avcB) to amplify a 309-bp product from genomic DNA. The PCR product was cloned in plasmid pMG36e using standard protocols (43). The recombinant plasmid was used to transform competent E. coli DH5α prepared by the CaCl2 method (43) and grown on LB plates containing 200 μg/ml erythromycin. The plasmid extracted from positive transformants was digested with XbaI and PstI to check for the presence of the insert. The presence and integrity of the insert were confirmed by PCR and sequencing. pMG36e with the correct avcB sequence was designated pMG36eDV. pMG36eDV was electroporated into competent L. lactis IL 1403 (19, 20), and the transformation mixture was grown on SR medium containing 1 μg/ml erythromycin. pMG36eDV was extracted from positive transformants after they were lysed as described below. Cells were pelleted by centrifugation, washed with 0.5 ml TES buffer (10 mM Tris-HCl [pH 8], 100 mM NaCl, 1 mM EDTA; pH 8), resuspended in 250 μl GTE buffer (50 mM glucose, 25 mM Tris-HCl [pH 8], 10 mM EDTA; pH 8) containing 5 mg/ml lysozyme and 100 μg/ml RNase A, and incubated at 37°C for 30 min. The presence of the insert in pMG36eDV was confirmed by PCR. In order to determine if the L. lactis IL 1403 avcB clone expressed antimicrobial activity, both a CFS and an ammonium sulfate precipitate of a CFS from an overnight culture were tested with strains sensitive to the E. avium bacteriocin (avicin A) and avicin A-resistant mutants of Enterococcus faecalis and E. avium.
Nucleotide sequence accession number.
The nucleotide sequence obtained in this study has been deposited in the NCBI database under accession number FJ851402.
RESULTS
Identification of bacteriocin-producing strains.
A total of 103 fecal LAB isolates were obtained from 20 Ethiopian infants, and E. avium isolates were obtained from 3 infants. The E. avium isolate from one infant produced bacteriocin-like activity. The 16S rRNA gene sequence of this strain showed 99% sequence identity to the 16S rRNA gene sequence of E. avium ATCC 14025 (type strain) (36). This isolate of E. avium (XA83) was selected for further study. Only 1 of 13 Norwegian infants was found to carry E. avium that produced a bacteriocin identical to the bacteriocin produced by the E. avium isolate from Ethiopia (15). Inactivation by protease K treatment suggested that the antimicrobial activity was a due to a proteinaceous substance.
Purification of avicin A.
A supernatant from a 0.5-liter overnight culture of E. avium XA83 grown in MRS was used for bacteriocin purification. This supernatant was precipitated with ammonium sulfate (40%) and subsequently purified by ion-exchange chromatography, followed by two-step reversed-phase chromatography (Table 3). The culture supernatant contained 1,280 bacteriocin units/ml as determined with the indicator strain L. sakei NCDO 2714. Ammonium sulfate precipitation resulted in 80% recovery and an approximately 26-fold increase in the specific activity. The level of recovery by ion-exchange and reversed-phase chromatography was low (due to loss of activity to other fractions). The results of the purification procedure are summarized in Table 3. The specific activity of the final purified elute from the second reversed-phase chromatography was about 29,000-fold higher than that of the culture supernatant (Fig. 1A). The monoisotopic molecular mass of the purified bacteriocin (termed avicin A) was determined by mass spectrometry to be 4,288.2 Da (M+1, 4,289.2) (Fig. 1B), which is close to the molecular mass of the subclass IIa bacteriocin mundticin, whose monoisotopic molecular mass is 4,285 Da (3). The following sequence was obtained by N-terminal peptide sequencing: TYYGNGVSCNKKGCSVDWGKAI.
TABLE 3.
Purification of avicin A
| Purification step | Vol (ml) | Recovery (%) | Protein concn (mg/ml)a | Antimicrobial activity (bacteriocin units/ml) | Sp act (bacteriocin units/mg) | Increase in sp act (fold) |
|---|---|---|---|---|---|---|
| Culture supernatant | 500 | 100 | 19.9 | 1.3 × 103 | 65 | 1.0 |
| Ammonium sulfate precipitate | 50 | 80 | 6.06 | 1.0 × 104 | 1,650 | 26 |
| Ion-exchange chromatography | 10 | 16 | 0.28 | 1.0 × 104 | 35,700 | 570 |
| First reversed-phase chromatography | 2 | 26 | 0.8 | 8.2 × 104 | 1.02 × 105 | 1,600 |
| Second reversed-phase chromatography | 1 | 32 | 0.11 | 2.05 × 105 | 1.86 × 106 | 29,000 |
The protein concentration was determined either by determining the optical density at 280 nm or by using the calculated absorbance value for purified avicin A (from the second reversed-phase chromatography fraction) (1 mg/ml = 3.257).
Inhibition spectrum of avicin A.
Members of several species of Gram-positive bacteria (Listeria, Enterococcus, Lactobacillus, Leuconostoc, Pediococcus, and Carnobacterium species) were susceptible to avicin A (Table 4), while the Lactococcus strains tested were not affected. Based on the MICs, Lactobacillus rhamnosus 205 and Listeria species seem to be the organisms most sensitive to avicin A, whereas pediococci appear to be less sensitive (Table 4). The subclass IIa bacteriocin-resistant strain E. faecalis LMGT 2708RA was also found to be resistant to avicin A (Table 4), as expected for a subclass IIa bacteriocin. The antimicrobial spectrum of the avicin A producer was also tested with the same indicators using the deferred overlay assay. The results are consistent with the results obtained using purified avicin A, which indicates that no other bacteriocins are produced by E. avium XA83.
TABLE 4.
Inhibition spectrum of avicin A
| Strain | Inhibitiona | MIC (nM)b |
|---|---|---|
| Carnobacterium divergens NCDO 2763T | + | 1.25 |
| Carnobacterium divergens NCDO 2306 | + | 2.51 |
| Carnobacterium piscicola NCDO 2762T | + | 0.31 |
| Carnobacterium piscicola NCDO 2764 | + | 0.31 |
| Enterococcus avium GM62 | + | 0.63 |
| Enterococcus avium UA62 | + | 5.02 |
| Enterococcus avium UM83 | + | 0.31 |
| Enterococcus faecalis EF BRIDGE (B) | + | 10.04 |
| Enterococcus faecalis LMGT 2708RA | − | |
| Enterococcus maldoratus XM83 | + | 5.02 |
| Lactobacillus plantarum LMG 2003 | − | |
| Lactobacillus rhamnosus 205 | + | <0.31 |
| Lactobacillus sakei NCDO 2714 | + | 1.25 |
| Lactobacillus sakei 5 | − | |
| Lactococcus lactis IL 1403 | − | |
| Lactococcus lactis CM4 | − | |
| Leuconostoc lactis NCDO 533 | + | 5.02 |
| Leuconostoc mesentereroides NCDO 529T | + | 2.51 |
| Leuconostoc gelidum Ta 11a | − | |
| Listeria innocua BL86/26 B | + | <0.31 |
| Listeria monocytogenes 223 serotype 1 | + | <0.31 |
| Listeria monocytogenes 400 serotype 4 | + | <0.31 |
| Pediococcus acidilactici NCDO 1851 | + | 40.16 |
| Pediococcus acidilactici PacI | − | |
| Pediococcus pentosaceus NCDO 814 | + | 20.08 |
+, inhibition; −, no inhibition. Antimicrobial tests with E. avium XA83 were performed by using the deferred assay. The MICs were not determined for insensitive indicators. The MICs varied within ±30%.
MICs of purified avicin A.
Identification of the bacteriocin gene locus.
Based on information for the N-terminal amino acid sequence, the gene sequence of avicin A was obtained by performing PCR with degenerate primers (AvFwr and AvRev) and sequencing the 113-bp PCR product. New primers were designed based on the nucleotide sequence for further sequencing by primer walking, and the avicin locus sequence was obtained. Specific fragments generated by restriction digestion and PCR were sequenced and assembled into an 8,660-bp contig. Southern blot analysis showed that the avicin A locus is located on chromosomal DNA (data not shown).
DNA sequence analysis.
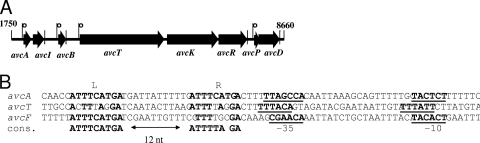
DNA sequence analysis of the 8,660-bp contig identified eight putative ORFs, seven of which are apparently bacteriocin related and localized in a 7-kb locus on the sequenced fragment. The unrelated ORF is probably involved in carbohydrate metabolism, as the deduced amino acid sequence encoded by it showed significant similarity (95% identity) to an annotated maltose-6′-phosphate glucosidase of Clostridium sp. 7_2_43FAA (accession number ZP_05129714). A similarity search in public databases revealed that the seven ORFs include the genes that encode a structural bacteriocin (avcA) and its immunity protein (avcI), a divergicin A-like bacteriocin (avcB), a dedicated ABC-type transporter (avcT), a peptide induced two-component regulatory system (histidine kinase [avcK], a response regulator [avcR], a peptide pheromone [avcP]), and a transport accessory protein (avcD) (Table 5). All ORFs were unidirectionally oriented (Fig. 2A) and, with the exception of avcR, preceded by a putative ribosome binding site. The bacteriocin gene cluster is organized into four predicted operon structures, all of which contain putative −10 and −35 sites in the upstream noncoding region of the operon (Fig. 2B).
TABLE 5.
Proteins similar to ORF products of the avicin A locus
| ORF | Length of product (amino acids) | Similar protein |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Designation | Length (amino acids) | Producer(s) | % Identity | % Similarity | E value(s) | Function | Reference(s) | ||
| avcA | 61 | MunA | 58 | Enterococcus mundtii strains | 87 | 89 | 9 × 10−20, 10−19 | Precursor of mundticin KS, enterocin CRL35 | 25, 42 |
| avcI | 97 | SakXIM | 97 | Lactobacillus sakei 5 | 65 | 80 | 2 × 10−27 | Sakacin X immunity protein | 48 |
| avcB | 67 | Divergicin A | 75 | Carnobacterium divergens LV13 | 45 | 60 | 7 × 10−10 | Divergicin A | 51 |
| avcT | 725 | StxT | 723 | Lactobacillus sakei 5 | 70 | 86 | 0 | ABC transporter | 48 |
| avcK | 442 | StxK | 437 | Lactobacillus sakei 5 | 58 | 80 | 4 × 10−144 | Histidine protein kinase | 48 |
| avcR | 244 | StxR | 252 | Lactobacillus sakei 5 | 77 | 90 | 6 × 10−113 | Response regulator | 48 |
| avcP | 45 | IP-TX | 45 | Lactobacillus sakei 5 | 62 | 77 | 10−8 | Inducer peptide | 48 |
| avcD | 171 | BrcD | 158 | Brochothrix campestris ATCC 43754 | 45 | 69 | 8 × 10−36 | Transport accessory protein | 28 |
FIG. 2.
(A) Genetic organization of avicin A locus (GenBank accession number FJ851402). avcA is the avicin A precursor gene (183 bp), avcI is the immunity gene (291 bp), avcB is the divergicin-like bacteriocin gene (201 bp), avcT is the ABC transporter gene (2,175 bp), avcK is the histidine kinase gene (1,326 bp), avcR is the response regulator gene (732 bp), avcP is the peptide-pheromone gene (135 bp), and avcD is the transport accessory protein gene (513 bp). p indicates promoter positions. The vertical bars immediately after arrows indicate inverted repeats. (B) Alignment of putative promoter sequences. Putative −10 and −35 sequences are underlined. Direct repeats (L and R) are enclosed in boxes, and the consensus sequence (cons.) is indicated below the boxes. The direct repeats were not identified in the promoter of avcB.
The first operon (avcAI) includes the structural gene for avicin A (avcA) and the immunity gene (avcI), which is located 40 nucleotides downstream of avcA. The promoter region upstream of avcA contains a pair of 9-nucleotide identical direct repeats that are separated by 12 nucleotides (Fig. 2B). Right after the stop codon of avcI, there is a pair of 9-nucleotide inverted repeats separated by 12 nucleotides.
The second operon was predicted to be a monosistronic transcript unit consisting of a single ORF (avcB), which is located 368 nucleotides downstream of avcI. The product shows significant similarity to divergicin A (51). No repeats were found in the promoter region of avcB. A pair of 10-nucleotide inverted repeats separated by 24 nucleotides is located 15 nucleotides downstream of the termination codon of avcB.
The third operon (avcTKR) contains genes encoding an ABC transporter (avcT), a histidine protein kinase (avcK), and a response regulator (avcR). As in the promoter of avcAI, a pair of 9-nucleotide direct repeats with one mismatch that were separated by 12 nucleotides were identified upstream of the −35 site of the avcTKR promoter (Fig. 3B). In addition, a pair of 17-nucleotide inverted repeats having four mismatches and separated by 12 nucleotides is located 42 nucleotides downstream of the termination codon of the ORF avcR.
FIG. 3.
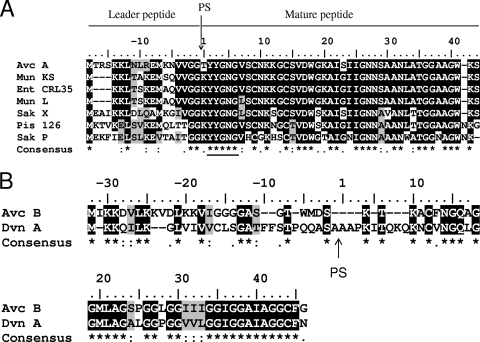
(A) Multiple alignment of avicin A precursor with precursors of closely related subclass IIa bacteriocins. Mun KS, precursor of mundticin KS; Mun L, precursor of mundticin L; Ent CRL35, precursor of enterocin CRL35; Sak X, precursor of sakacin X; Pis 126, precursor of piscolin 126; Sak P, precursor of sakacin P. The vertical arrow indicates a cleavage site where the leader peptide is removed from the mature peptide. The YYGNG motif is underlined. (B) Pairwise alignment of avicin B (Avc B) and divergicin A precursor (Dvn A). PS indicates the processing site for divergicin A.
The fourth operon comprises genes encoding a putative peptide pheromone (avcP) and a transport accessory protein (avcD). A pair of direct repeats similar to those identified in the promoter regions of the avcAI and avcTKR operons was also found in the promoter region of this operon (Fig. 2B). These repeats contain 9 nucleotides with four mismatches (three mismatches are consecutive), and one of the pairs is identical to the pairs that occur in avcAI. One nucleotide downstream of the termination codon of avcD, a perfect 16-nucleotide inverted repeats pair separated by 19 nucleotides was identified.
The inverted repeats at the 3′OH end of the mRNAs may function as rho-independent transcription terminators, although no obvious U tract follows the stem structure. The direct repeats identified in the promoter region may serve as binding sites for a response regulator (8, 39, 40).
Protein sequence analysis.
avcA encodes a 61-amino-acid prepeptide that includes an N-terminal double-glycine leader consisting of 18 amino acid residues and a mature peptide consisting of 43 residues. The pI is 9.18, and the theoretical monoisotopic molecular mass of avicin A (the two cysteines are assumed to form a disulfide bridge) was estimated to be 4,288.2 Da. The extinction coefficient at 280 nm (path length, 1 cm) was calculated to be 3.26 (1 U of optical density at 280 nm = 3.26 mg/ml).
The mature peptide (43 amino acids) contains the N-terminal conserved motif YYGNG (Fig. 3A), as well as the two conserved cysteine residues (at positions 9 and 14) typical of subclass IIa bacteriocins.
The deduced amino acid sequence of the prepeptide showed high levels of similarity to other IIa bacteriocins (Table 5). Mature avicin A is 97% identical to mundticin KS (25) and enterocin CRL35 (42) and differs only at amino acid residues 1 and 23. Also, mature avicin A is 95%, 85%, and 82% identical to mundticin L (12), sakacin X (48), and piscicolin 126 (24), respectively. The double-glycine leader peptide of avicin A exhibits high levels of similarity (64% identity) to the leader peptides of sakacin X (48), mundticin KS (25), mundticin L (12), and enterocin CRL35 (45).
avcI encodes a 97-amino-acid peptide that is significantly similar to the immunity proteins of sakacin X (48) and piscicolin 126 (Table 5) (18, 24). The immunity proteins of pediocin-like bacteriocins have recently been divided into subgroups A, B, and C (13). Avicin A immunity protein is a member of subgroup B, which includes the immunity proteins of sakacin X, piscicolin 126, sakacin P, and mundticin KS. Because of the similarity between AvcI and sakacin X immunity protein produced by L. sakei 5, we examined whether there is cross-protection between E. avium XA83 and L. sakei 5. The results showed that neither E. avium nor purified avicin A inhibited L. sakei 5 but L. sakei 5 inhibited E. avium XA83, demonstrating that the immunity protein of avicin A is not sufficient to provide protection against the plethora of bacteriocins produced by L. sakei 5.
avcB may code for a 67-amino-acid glycine-rich (30%) peptide that shares 45% identity (60% similarity) with divergicin A, a bacteriocin that inhibits Carnobacterium divergens (Table 5 and Fig. 3B) (51). Both avicin-producing strains contain the avcB gene, but neither strain had any apparent immunity-encoding ORF. In order to investigate whether avcB encodes a functionally expressed bacteriocin, we tested the inhibition of bac+ strain XA83 using pediocin-resistant mutants of E. avium and E. faecalis, but no activity was observed. Notably, no sec leader sequence was found in the N-terminal part of translated avcB; however, a potential double-glycine leader was identified (Fig. 3B.).
In order to investigate whether the avcB gene encodes a functional bacteriocin, it was cloned in pMG36e and expressed in L. lactis IL 1403, which contains the lactococcin A transport gene apparatus (49). The resulting clone did not show antibacterial activity against the bacteria tested. Neither CFS nor the ammonium sulfate precipitate fraction inhibited the growth of strains that are sensitive or resistant (E. avium and E. faecalis) to avicin A. However, we cannot exclude the possibility that the lack of antimicrobial activity could have been due to improper processing of the AvcB leader peptide in IL 1403 or due to lack of expression, although the host carried the proper processing and transport mechanisms for such bacteriocins. When a very small inoculum of E. avium XA83 is added to new broth (dilution factor, 10−6), avicin A is not produced because its quorum-sensing regulatory system is not turned on (see below). The avaB gene is apparently not regulated by quorum sensing since no binding site for the response regulator was found, but no antimicrobial activity was observed in this culture, which suggests that avcB was not expressed. Based on the cumulative evidence it is very unlikely that avicin B is a functional bacteriocin.
avcT encoded a 725-amino-acid protein that showed a high level of similarity to a putative ABC transporter of sakacin X and significant levels of similarity to other bacteriocin ABC transporters, suggesting that it is involved in the export of avicin A (Table 5) (48).
The avcK, avcR, and avcP genes, which constitute a three-component regulatory system, are located adjacent to one another but in two different operons (Fig. 2A), and they encode proteins composed of 442, 244, and 45 amino acids, respectively. These ORFs share significant similarity with genes encoding histidine protein kinase, a response regulator, and an inducing peptide of sakacin X, respectively (Table 5) (48).
avcD encodes a 171-amino-acid peptide that shares similarity with a transport accessory protein of brochocin C, a two-peptide bacteriocin (Table 5) (28). The N-terminal half of AvcD is 56% identical to the deduced amino acid sequence of ORF 5 in the TX locus of L. sakei 5 (48). This region of AvcD is only 38% identical to the corresponding region in the transport accessory protein of brochocin C.
Induction of avicin A production.
A bac− phenotype could be obtained by extensive dilution of a bacteriocin-producing culture; 10−6 dilution of an overnight bacteriocin-producing culture resulted in a non-bacteriocin-producing culture (bac−). Only addition of cell-free culture supernatant (CFS) from a bacteriocin-producing culture or a synthetic inducing peptide (AvcP) could then induce bacteriocin production in the bac− culture.
The induction activity was tested with a synthetic AvcP peptide pheromone. The minimum concentration of the synthetic peptide that induced bacteriocin production in a bac− culture was 1 ng/ml; 0.1 ng/ml could not induce bacteriocin production. This result confirms that avcP encodes the peptide pheromone that regulates avicin A production. Although the inducer peptides of avicin A and sakacin X share significant similarity, CFS from L. sakei 5 did not induce bacteriocin production in a strain with the avicin A-negative phenotype.
DISCUSSION
In this study, we report biochemical and genetic characterization of a novel subclass IIa bacteriocin produced by two different E. avium strains (XA83 and 208) isolated from two healthy human infants from two different countries, Ethiopia and Norway. The structural and immunity genes of the bacteriocins produced by these two strains are identical. PCR confirmed that the gene structures of the two strains are identical. For these reasons, their bacteriocins were given the same designation, avicin A. An interesting observation was that the levels of bacteriocin-producing isolates XA83 and 208 were higher (>10−6 CFU g−1 feces) than the levels of nonproducing isolates of E. avium (10−5 CFU g−1 feces). The bacteriocin producers also accounted for 25 to 45% of the total bacterial plate counts on the LAB selective growth media. Moreover, the frequent occurrence of E. avium in healthy infants (both Norwegian and Ethiopian) suggests that bacteriocin-producing E. avium may play an important role in the succession of a healthy gut microbiota in some neonates. It is not uncommon that different bacterial strains or species produce identical bacteriocins. For example, mundticin ATO6 and mundticin KS are two identical subclass IIa bacteriocins isolated from two different strains (ATO6 and NFRI 7393) of E. mundtii obtained from different sources (25, 42). To our knowledge, this the first report of a thorough elucidation of the activity spectrum, gene structure, and regulation of a bacteriocin system of E. avium.
The subclass IIa bacteriocin inhibition spectrum includes species of the genera Listeria, Lactobacillus, Leuconostoc, Pediococcus, Lactococcus, Carnobacterium, Enterococcus, Staphylococcus, Micrococcus, Streptococcus, Clostridium, Bacillus, and Brochotrix (10, 30). The inhibition spectrum of avicin A is similar and crosses species borders as this bacteriocin inhibits Listeria, Lactobacillus, Leuconostoc, Pediococcus, and Carnobacterium species in addition to Enterococcus.
Amino acid sequence analysis revealed that avicin A contains the N-terminal pediocin consensus motif (YYGNG) and two cysteine residues, indicating that avicin A belongs to bacteriocin subclass IIa. Recently, subclass IIa bacteriocins have been divided into four subgroups on the basis of similarities and differences in their C-terminal regions (33). Avicin A belongs to subgroup 1, which encompasses mundticin KS, piscicolin 126, sakacin X, sakacin P, enterocin A, and pediocin PA-1. Mundticin KS, which is produced by E. mundtii NFRI 7393 isolated from grass silage (25), is the bacteriocin most closely related to avicin A both in the leader sequence and in the mature peptide. However, unlike the avicin A gene, which is located on a chromosome, the mundticin KS gene is located on a plasmid. In addition, the structural gene of mundticin A (munA) and the immunity gene (munC) are located in different operons, whereas in the case of avicin A the structural gene and the immunity gene are in the same operon, like the genes of most subclass IIa bacteriocins characterized to date (23). Moreover, it has been shown by heterologous expression of mundticin KS in E. faecium and Lactobacillus curvatus that an accessory protein is not needed for transport of mundticin KS, but the data did not confirm the absence of an accessory gene in the cloning host. Other studies have shown by using deletion analysis that an accessory protein is required for export of leucocin A and pediocin PA-1 (46, 50). Since the gene encoding the accessory protein is present in the avicin A locus, this protein might be crucial for bacteriocin externalization.
The biosynthesis of many class II bacteriocins is regulated by a quorum-sensing mechanism through a three-component regulatory system which consists of an inducing peptide, a histidine protein kinase, and a response regulator (31). The presence of genes encoding these three components in the avicin A locus indicates that biosynthesis of avicin A is regulated by a quorum-sensing system. However, some differences have been observed between the genetic organization of the avicin locus and the genetic organization of the loci of other class II bacteriocins. The most surprising difference is the difference in the organization of the three-component regulatory system, where the histidine protein kinase and response regulator genes are in an operon together with the ABC transporter gene, while the peptide pheromone gene is located with the gene encoding the accessory transporter protein AvcD in another operon (Fig. 2A). This organization is different from the organization of most of the regulated class II bacteriocins, where the three genes responsible for regulation are located in the same operon (2, 16, 18, 22, 34, 38). Our results conclusively show that avicin P functions as a pheromone to induce avicin A production in a nonproducing culture, which is consistent with a quorum-sensing regulatory mechanism (32).
Our experiments indicate that avcB is not functional and is probably a relic of a previous functional bacteriocin. This hypothesis is also supported by the absence of a dedicated immunity protein, and the fact that avcB clones in L. lactis IL 1403 did not show antimicrobial activity may support this notion.
Almost all strains that produce subclass IIa bacteriocins characterized thus far were isolated from foods or related products (9). In contrast, avicin A is produced by a bacterium isolated from human feces, suggesting that it is capable of establishing and proliferating in the gut. Human origin and survival in the gut are two of the several selection criteria that must be met by probiotics. In addition, our strains produce a bacteriocin that inhibits pathogenic bacteria, and bacteriocin production is an important probiotic feature (4). Diseases associated with E. avium (such as brain abscesses, endocarditis, and bacteremia) are rarely reported (29, 35, 37). In contrast, our strains might be proven to be beneficial as intestinal LAB and possibly also as probiotics; however, further studies have to be performed to determine this.
Acknowledgments
This project was funded by the Norwegian Research Council.
Footnotes
Published ahead of print on 20 November 2009.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1999. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, NY.
- 2.Axelsson, L., and A. Holck. 1995. The genes involved in production of and immunity to sakacin A, a bacteriocin from Lactobacillus sake Lb706. J. Bacteriol. 177:2125-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennik, M. H., B. Vanloo, R. Brasseur, L. G. Gorris, and E. J. Smid. 1998. A novel bacteriocin with a YGNGV motif from vegetable-associated Enterococcus mundtii: full characterization and interaction with target organisms. Biochim. Biophys. Acta 1373:47-58. [DOI] [PubMed] [Google Scholar]
- 4.Corr, S. C., Y. Li, C. U. Riedel, P. W. O'Toole, C. Hill, and C. G. Gahan. 2007. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl. Acad. Sci. U. S. A. 104:7617-7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotter, P. D., C. Hill, and R. P. Ross. 2005. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3:777-788. [DOI] [PubMed] [Google Scholar]
- 6.Diep, D. B., L. Godager, D. Brede, and I. F. Nes. 2006. Data mining and characterization of a novel pediocin-like bacteriocin system from the genome of Pediococcus pentosaceus ATCC 25745. Microbiology 152:1649-1659. [DOI] [PubMed] [Google Scholar]
- 7.Diep, D. B., L. S. Havarstein, and I. F. Nes. 1995. A bacteriocin-like peptide induces bacteriocin synthesis in Lactobacillus plantarum C11. Mol. Microbiol. 18:631-639. [DOI] [PubMed] [Google Scholar]
- 8.Diep, D. B., L. S. Havarstein, and I. F. Nes. 1996. Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J. Bacteriol. 178:4472-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drider, D., G. Fimland, Y. Hechard, L. M. McMullen, and H. Prevost. 2006. The continuing story of class IIa bacteriocins. Microbiol. Mol. Biol. Rev. 70:564-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eijsink, V. G., M. Skeie, P. H. Middelhoven, M. B. Brurberg, and I. F. Nes. 1998. Comparative studies of class IIa bacteriocins of lactic acid bacteria. Appl. Environ. Microbiol. 64:3275-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ennahar, S., T. Sashihara, K. Sonomoto, and A. Ishizaki. 2000. Class IIa bacteriocins: biosynthesis, structure and activity. FEMS Microbiol. Rev. 24:85-106. [DOI] [PubMed] [Google Scholar]
- 12.Feng, G., G. K. Guron, J. J. Churey, and R. W. Worobo. 2009. Characterization of mundticin L, a class IIa anti-Listeria bacteriocin from Enterococcus mundtii CUGF08. Appl. Environ. Microbiol. 75:5708-5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fimland, G., L. Johnsen, B. Dalhus, and J. Nissen-Meyer. 2005. Pediocin-like antimicrobial peptides (class IIa bacteriocins) and their immunity proteins: biosynthesis, structure, and mode of action. J. Pept. Sci. 11:688-696. [DOI] [PubMed] [Google Scholar]
- 14.Flynn, S., D. van Sinderen, G. M. Thornton, H. Holo, I. F. Nes, and J. K. Collins. 2002. Characterization of the genetic locus responsible for the production of ABP-118, a novel bacteriocin produced by the probiotic bacterium Lactobacillus salivarius subsp. salivarius UCC118. Microbiology 148:973-984. [DOI] [PubMed] [Google Scholar]
- 15.Forberg, T. 2005. Lactic acid bacteria of different origin, production of antimicrobial substances and distribution of bacteriocin genes. Norwegian University of Life Sciences, Aas, Norway.
- 16.Franz, C. M., M. J. van Belkum, R. W. Worobo, J. C. Vederas, and M. E. Stiles. 2000. Characterization of the genetic locus responsible for production and immunity of carnobacteriocin A: the immunity gene confers cross-protection to enterocin B. Microbiology 146:621-631. [DOI] [PubMed] [Google Scholar]
- 17.Gillor, O., A. Etzion, and M. A. Riley. 2008. The dual role of bacteriocins as anti- and probiotics. Appl. Microbiol. Biotechnol. 81:591-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gursky, L. J., N. I. Martin, D. J. Derksen, M. J. van Belkum, K. Kaur, J. C. Vederas, M. E. Stiles, and L. M. McMullen. 2006. Production of piscicolin 126 by Carnobacterium maltaromaticum UAL26 is controlled by temperature and induction peptide concentration. Arch. Microbiol. 186:317-325. [DOI] [PubMed] [Google Scholar]
- 19.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holo, H., and I. F. Nes. 1995. Transformation of Lactococcus by electroporation, p. 195-199. In J. A. Nickolof (ed.), Electroporation protocols for microorganisms. Humana Press Inc., Totowa, NJ. [DOI] [PubMed]
- 21.Holo, H., O. Nilssen, and I. F. Nes. 1991. Lactococcin A, a new bacteriocin from Lactococcus lactis subsp. cremoris: isolation and characterization of the protein and its gene. J. Bacteriol. 173:3879-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huhne, K., L. Axelsson, A. Holck, and L. Krockel. 1996. Analysis of the sakacin P gene cluster from Lactobacillus sake Lb674 and its expression in sakacin-negative Lb. sake strains. Microbiology 142:1437-1448. [DOI] [PubMed] [Google Scholar]
- 23.Jack, R. W., J. R. Tagg, and B. Ray. 1995. Bacteriocins of gram-positive bacteria. Microbiol. Rev. 59:171-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jack, R. W., J. Wan, J. Gordon, K. Harmark, B. E. Davidson, A. J. Hillier, R. E. Wettenhall, M. W. Hickey, and M. J. Coventry. 1996. Characterization of the chemical and antimicrobial properties of piscicolin 126, a bacteriocin produced by Carnobacterium piscicola JG126. Appl. Environ. Microbiol. 62:2897-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawamoto, S., J. Shima, R. Sato, T. Eguchi, S. Ohmomo, J. Shibato, N. Horikoshi, K. Takeshita, and T. Sameshima. 2002. Biochemical and genetic characterization of mundticin KS, an antilisterial peptide produced by Enterococcus mundtii NFRI 7393. Appl. Environ. Microbiol. 68:3830-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klaenhammer, T. R. 1988. Bacteriocins of lactic acid bacteria. Biochimie 70:337-349. [DOI] [PubMed] [Google Scholar]
- 27.Klaenhammer, T. R. 1993. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12:39-85. [DOI] [PubMed] [Google Scholar]
- 28.McCormick, J. K., A. Poon, M. Sailer, Y. Gao, K. L. Roy, L. M. McMullen, J. C. Vederas, M. E. Stiles, and M. J. Van Belkum. 1998. Genetic characterization and heterologous expression of brochocin-C, an antibotulinal, two-peptide bacteriocin produced by Brochothrix campestris ATCC 43754. Appl. Environ. Microbiol. 64:4757-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohanty, S., B. Dhawan, A. Kapil, B. K. Das, P. Pandey, and A. Gupta. 2005. Brain abscess due to Enterococcus avium. Am. J. Med. Sci. 329:161-162. [DOI] [PubMed] [Google Scholar]
- 30.Nes, I. F., D. A. Brede, and H. Holo. 2006. The nonlantibiotic heat-stable bacteriocins of gram-positive bacteria, p. 107-114. In A. Kastin (ed.), Handbook of biologically active peptides. Elsevier, Amsterdam, The Netherlands.
- 31.Nes, I. F., D. B. Diep, L. S. Havarstein, M. B. Brurberg, V. Eijsink, and H. Holo. 1996. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Van Leeuwenhoek 70:113-128. [DOI] [PubMed] [Google Scholar]
- 32.Nes, I. F., and V. G. H. Eijsink. 1999. Regulation of group II peptide bacteriocin synthesis by quorum-sensing mechanisms, p. 175-192. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. American Society for Microbiology, Washington, DC.
- 33.Nissen-Meyer, J., P. Rogne, C. Oppegard, H. S. Haugen, and P. E. Kristiansen. 2009. Structure-function relationships of the non-lanthionine-containing peptide (class II) bacteriocins produced by gram-positive bacteria. Curr. Pharm. Biotechnol. 10:19-37. [DOI] [PubMed] [Google Scholar]
- 34.O'Keeffe, T., C. Hill, and R. P. Ross. 1999. Characterization and heterologous expression of the genes encoding enterocin A production, immunity, and regulation in Enterococcus faecium DPC1146. Appl. Environ. Microbiol. 65:1506-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel, R., M. R. Keating, F. R. Cockerill III, and J. M. Steckelberg. 1993. Bacteremia due to Enterococcus avium. Clin. Infect. Dis. 17:1006-1011. [DOI] [PubMed] [Google Scholar]
- 36.Patel, R., K. E. Piper, M. S. Rouse, J. M. Steckelberg, J. R. Uhl, P. Kohner, M. K. Hopkins, F. R. Cockerill III, and B. C. Kline. 1998. Determination of 16S rRNA sequences of enterococci and application to species identification of nonmotile Enterococcus gallinarum isolates. J. Clin. Microbiol. 36:3399-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perez-Castrillon, J. L., M. Martin-Luquero, J. C. Martin-Escudero, P. Pascual, A. Casero, and V. Herreros. 1997. Endocarditis caused by Enterococcus avium. Scand. J. Infect. Dis. 29:530. [DOI] [PubMed] [Google Scholar]
- 38.Quadri, L. E., M. Kleerebezem, O. P. Kuipers, W. M. de Vos, K. L. Roy, J. C. Vederas, and M. E. Stiles. 1997. Characterization of a locus from Carnobacterium piscicola LV17B involved in bacteriocin production and immunity: evidence for global inducer-mediated transcriptional regulation. J. Bacteriol. 179:6163-6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Risoen, P. A., L. S. Havarstein, D. B. Diep, and I. F. Nes. 1998. Identification of the DNA-binding sites for two response regulators involved in control of bacteriocin synthesis in Lactobacillus plantarum C11. Mol. Gen. Genet. 259:224-232. [DOI] [PubMed] [Google Scholar]
- 40.Risoen, P. A., O. Johnsborg, D. B. Diep, L. Hamoen, G. Venema, and I. F. Nes. 2001. Regulation of bacteriocin production in Lactobacillus plantarum depends on a conserved promoter arrangement with consensus binding sequence. Mol. Genet. Genomics 265:198-206. [DOI] [PubMed] [Google Scholar]
- 41.Ryan, M. P., M. C. Rea, C. Hill, and R. P. Ross. 1996. An application in cheddar cheese manufacture for a strain of Lactococcus lactis producing a novel broad-spectrum bacteriocin, lacticin 3147. Appl. Environ. Microbiol. 62:612-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saavedra, L., C. Minahk, A. P. de Ruiz Holgado, and F. Sesma. 2004. Enhancement of the enterocin CRL35 activity by a synthetic peptide derived from the NH2-terminal sequence. Antimicrob. Agents. Chemother. 48:2778-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 44.Skaugen, M., L. M. Cintas, and I. F. Nes. 2003. Genetics of bacteriocin production in lactic acid bacteria, p. 225-260. In B. J. B. Wood and P. J. Warner (ed.), Genetics of lactic acid bacteria. Kluwer Academic/Plenum Publishers, New York, NY.
- 45.Stoffels, G., I. F. Nes, and A. Guthmundsdottir. 1992. Isolation and properties of a bacteriocin-producing Carnobacterium piscicola isolated from fish. J. Appl. Bacteriol. 73:309-316. [DOI] [PubMed] [Google Scholar]
- 46.van Belkum, M. J., and M. E. Stiles. 1995. Molecular characterization of genes involved in the production of the bacteriocin leucocin A from Leuconostoc gelidum. Appl. Environ. Microbiol. 61:3573-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van de Guchte, M., J. M. van der Vossen, J. Kok, and G. Venema. 1989. Construction of a lactococcal expression vector: expression of hen egg white lysozyme in Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 55:224-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vaughan, A., V. G. Eijsink, and D. Van Sinderen. 2003. Functional characterization of a composite bacteriocin locus from malt isolate Lactobacillus sakei 5. Appl. Environ. Microbiol. 69:7194-7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Venema, K., M. H. Dost, P. A. Beun, A. J. Haandrikman, G. Venema, and J. Kok. 1996. The genes for secretion and maturation of lactococcins are located on the chromosome of Lactococcus lactis IL1403. Appl. Environ. Microbiol. 62:1689-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venema, K., J. Kok, J. D. Marugg, M. Y. Toonen, A. M. Ledeboer, G. Venema, and M. L. Chikindas. 1995. Functional analysis of the pediocin operon of Pediococcus acidilactici PAC1.0: PedB is the immunity protein and PedD is the precursor processing enzyme. Mol. Microbiol. 17:515-522. [DOI] [PubMed] [Google Scholar]
- 51.Worobo, R. W., M. J. Van Belkum, M. Sailer, K. L. Roy, J. C. Vederas, and M. E. Stiles. 1995. A signal peptide secretion-dependent bacteriocin from Carnobacterium divergens. J. Bacteriol. 177:3143-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]