Abstract
Disinfection is an essential measure for interrupting human norovirus (HuNoV) transmission, but it is difficult to evaluate the efficacy of disinfectants due to the absence of a practicable cell culture system for these viruses. The purpose of this study was to screen sodium hypochlorite and ethanol for efficacy against Norwalk virus (NV) and expand the studies to evaluate the efficacy of antibacterial liquid soap and alcohol-based hand sanitizer for the inactivation of NV on human finger pads. Samples were tested by real-time reverse transcription-quantitative PCR (RT-qPCR) both with and without a prior RNase treatment. In suspension assay, sodium hypochlorite concentrations of ≥160 ppm effectively eliminated RT-qPCR detection signal, while ethanol, regardless of concentration, was relatively ineffective, giving at most a 0.5 log10 reduction in genomic copies of NV cDNA. Using the American Society for Testing and Materials (ASTM) standard finger pad method and a modification thereof (with rubbing), we observed the greatest reduction in genomic copies of NV cDNA with the antibacterial liquid soap treatment (0.67 to 1.20 log10 reduction) and water rinse only (0.58 to 1.58 log10 reduction). The alcohol-based hand sanitizer was relatively ineffective, reducing the genomic copies of NV cDNA by only 0.14 to 0.34 log10 compared to baseline. Although the concentrations of genomic copies of NV cDNA were consistently lower on finger pad eluates pretreated with RNase compared to those without prior RNase treatment, these differences were not statistically significant. Despite the promise of alcohol-based sanitizers for the control of pathogen transmission, they may be relatively ineffective against the HuNoV, reinforcing the need to develop and evaluate new products against this important group of viruses.
Human noroviruses (HuNoV) are the leading cause of acute nonbacterial gastroenteritis (11, 21). The U.S. Centers for Disease Control and Prevention estimate that approximately 23 million people suffer from HuNoV gastroenteritis each year. Indeed, 81% of outbreaks of nonbacterial gastroenteritis are caused by this agent (7, 8). These viruses may contaminate food, water, hands, and inanimate surfaces and are readily transmitted by contact with contaminated objects, by consumption of fecally contaminated food or water, or between people. In particular, hands are thought to be a principal vehicle for HuNoV transmission. Although many different hand hygiene agents are available, these generally do not have specific virucidal claims against the HuNoV. Two widely used types of hand hygiene products are antibacterial liquid soaps and alcohol-based hand sanitizers.
Little is known about the effectiveness of hand hygiene agents in reducing HuNoV on contaminated hands. One reason for this is that the HuNoV cannot be routinely cultured in vitro, a factor that complicates evaluation of the efficacy of disinfection strategies. As an alternative, investigators have used cultivable surrogates such as feline calicivirus (FCV) and murine norovirus (MNV) (10, 15, 22), but questions continue regarding their relevance. Quantitative real-time nucleic acid amplification technologies allow us to quantify RNA and can be used to estimate HuNoV titer, although the relationship to virus infectivity is not clear (5, 18, 26, 30). In this study, we used real-time reverse transcription-quantitative PCR (RT-qPCR) to test the effectiveness of sodium hypochlorite and ethanol against Norwalk virus (NV) in a suspension assay. This was followed by an evaluation of the efficacy of an antibacterial liquid soap, alcohol-based hand sanitizer, and water rinsing for the removal and/or inactivation of NV on the finger pads of human volunteers. To examine the likelihood that detection of residual HuNoV RNA was associated with intact and, hence, infectious virus, we also tested finger pad eluates by RT-qPCR both with and without prior RNase treatment.
MATERIALS AND METHODS
Norwalk virus inoculum.
NV was obtained from the stool of an experimentally infected human volunteer from a previous study (20). The stool was diluted 20% (wt/vol) in RNase-free water prior to inoculation on the finger pads.
Study subjects.
The study protocol was reviewed and approved by the Institutional Review Board of Emory University. A total of 10 adult volunteers were enrolled in this study after informed consent was obtained. Prior to each experiment, both hands of each volunteer were carefully inspected to ensure that they were free of any cuts, abrasions, or rashes.
Suspension assays.
Suspension tests for virucidal activity were performed in accordance with a modification of the method reported by Macinga et al. (22). Different concentrations of ethanol (3%, 17%, 31%, 47%, 62%, and 95%) were prepared using deionized water and were analyzed by gas chromatography. Similarly, various sodium hypochlorite concentrations (3 ppm, 22 ppm, 51 ppm, 160 ppm, and 1,600 ppm) were prepared using potassium phosphate buffer (pH 7.4), and total chlorine concentrations were determined by a digital titrator (Hach, Loveland, CO). Ten microliters of the 20% NV stool suspension was added to 990 μl of each test disinfectant solution or water (as a baseline control), quickly vortexed, and held for 30 s. Immediately following the exposure period, a 100-μl aliquot was removed from the tube and added to a tube containing 900 μl of 10% fetal bovine serum to neutralize the reaction. Then, a 100-μl aliquot was further diluted in 900 μl Hanks balanced salt solution (HBSS). Finally, 1 ml of the diluted virus-disinfectant mixture was concentrated to 50 μl by precipitation with 12% polyethylene glycol (Sigma, St. Louis, MO), followed by processing for RT-qPCR.
Hand sanitizer and liquid soap.
A hand sanitizer, containing 62% ethyl alcohol as the active ingredient, was purchased from a local retail source. The antibacterial liquid soap was obtained from Fisher Scientific International (Hampton, NH). The latter contained 0.5% triclosan as the active ingredient, with inactive ingredients including sodium lauryl ether sulfate, cocamidopropyl betaine, solulan, propylene glycol, sodium chloride, dyes, and fragrances. To screen for PCR inhibition, 10 μl of the test product was mixed with 980 μl of HBSS and 10 μl of 20% NV stool suspension, 10- and 100-fold serially diluted, and then tested by RT-qPCR.
Inoculation of NV onto human finger pads.
The volunteers first washed their hands with a nonmedicated soap for at least 10 s, rinsed with tap water, and dried them with a single-use paper towel. Approximately 4 ml of 70% ethanol was placed in the palm of one washed hand, and the volunteers were instructed to rub it over the entire surface of both hands until the alcohol and water had evaporated completely. The volunteers pressed each finger pad over the mouth of an empty 2.0-ml plastic vial (Sarstedt, Newton, NC) to demarcate the area (about 1 cm2) to receive the NV inoculum. Ten microliters of the 20% NV suspension was slowly placed at the center of each demarcated area using a pipettor.
Recovery of NV from contaminated finger pads.
Two controls consisted of (i) a single finger pad where the NV inoculum was recovered immediately after inoculation (input control) and (ii) a baseline (dried) control where the inoculum was recovered from another finger pad immediately after drying for 20 min. For collection of the input control, the inoculum on the thumb pad was eluted using a 10-μl aliquot of a 1-ml volume of HBSS by pipetting up and down three to four times. This eluate was returned to the collection tube, and the process was repeated three times. To recover the baseline control, the finger pad with the dried inoculum was placed over the mouth of a 2.0-ml plastic vial containing 1 ml of the HBSS, inverted so that the eluent was in contact with the contaminated area for 10 s, and then inverted rapidly in succession for a total of 20 inversions with the finger pad still in place. This soak-and-inversion process was repeated once. The vial was then turned upright, and any eluate remaining in contact with the finger pad was scraped against the inside rim of the vial to recover as much of the fluid as possible. After sample collection, the volunteers were instructed to decontaminate all their finger pads by pressing them onto a paper towel soaked with 10% bleach and then washing their hands with antibacterial liquid soap.
Treatment of NV on human finger pads.
Two experiments were designed to evaluate the effectiveness of the candidate hand hygiene agents against NV on finger pads. The first experimental protocol followed the method of the American Society for Testing and Materials (ASTM; E 1838-02) for determining the efficacy of the antibacterial liquid soap and the ethanol-based hand sanitizer against NV using the finger pads of adult volunteers (28). All finger pads on both hands were inoculated with approximately 5.6 × 106 NV genomic copies per finger pad. The two thumb pads were used for the input control (time zero), and the index fingers corresponded to the dried inoculum control. After inoculation and drying, the middle and ring finger pads of both hands were exposed to 1.0 ml of hand sanitizer or liquid soap in an open vial, respectively, for a contact time of 20 s. After exposure, the finger pads were gently scraped on the inside rim of the vial to remove residual sanitizer. In the case of the hand sanitizer treatment, the remaining virus was eluted using the soak-and-inversion method with HBSS as described above. For the liquid soap treatment, the finger pad was rinsed by placing it over the mouth of a vial containing 1.0 ml of tap water for a contact time of 10 s, after which the virus was eluted by soaking and inversion with HBSS. The two little finger pads were used to determine the extent of NV eliminated after rinsing with tap water alone. After inoculation and drying, the finger pad was placed over a vial containing 1.0 ml of tap water for 10 s, which was repeated for a total of 10 full inversions. This was followed by soaking and inversion with HBSS for virus elution. All finger pad eluates were frozen at −80°C until the assay. Five volunteers were asked to participate in two trials (on separate days), and the right and left hands of each volunteer served as duplicate treatments per trial, giving a total of 20 replicate samples for each exposure.
To determine if rubbing, which occurs during hand sanitizing and washing with soap, impacts virus removal, we repeated the experiments with a minor modification to the ASTM method. This was done for the hand sanitizer and liquid soap only. Briefly, approximately 5.6 × 106 genomic copies of NV were placed on the center of the finger pads of both hands except for the little finger. The two thumb pads constituted the input control (time zero), and the index fingers corresponded to the dried inoculum control. The two middle finger pads and two ring fingers were used to test the hand sanitizer and water rinse, respectively, after the virus inoculum had visibly dried. After placing a drop of hand sanitizer on the center of one middle finger pad, the volunteers were asked to rub the sanitizer-inoculated finger pad with another uninoculated finger pad for 10 s. This was followed by elution in HBSS using the soak-and-inversion technique. For the liquid soap treatment, the same protocol was used. After the virus inoculum had dried, the product was deposited on the finger pads, and the volunteers rubbed the liquid soap-inoculated finger pad with an uninoculated finger pad for 10 s, followed by contact with 1.0 ml tap water for 10 s (without inversion) and then final elution of remaining virus by soaking and inversion with HBSS. The two ring fingers were used to test water rinse using the methods described above and without rubbing. All finger pad eluates were frozen at −80°C until the assay.
TaqMan real-time RT-PCR.
Eluates were tested by RT-qPCR both with and without a prior RNase treatment using the method of Topping et al. (32). Briefly, a 100-μl volume of finger pad eluate was mixed with 1 μl RNase One (Promega, WI) and 11 μl 10× reaction buffer followed by incubation at 37°C for 15 min. Immediately after, 1 μl of finger pad eluate was diluted with 8.8 μl of diethyl pyrocarbonate (DEPC)-treated water and 0.2 μl of RNase inhibitor (Promega, WI), heated to 100°C for 5 min, and chilled on ice for 2 min to release the viral RNA (29). Primers targeting the RNA polymerase region of the Norwalk virus (sense primer, NVKS1, 5′-ACAGCATGGGACTCAACACA-3′; antisense primer, NVKS2, 5′-GGGAAGTACATGGGAATCCA-3′; and probe, NVKS3, 5′-TCACCAGAATTGGCCGAGGTTGT-3′) (4, 31) were used for amplification. The probe was dually labeled with the 5′ reporter dye FAM (6-carboxyfluorescein) and 3′ quencher dye TAMRA (6-carboxytetramethylrhodamine). RT-qPCRs were done in duplicate using the Qiagen one-step RT-PCR kit (Qiagen, Valencia, CA) and a Stratagene Mx3000P (Stratagene, La Jolla, CA) real-time PCR instrument. The 25-μl RT-qPCR mixture consisted of 10 μl of heat-released RNA or diluted Norwalk virus RNA standard, 0.4 mM deoxynucleoside triphosphate (dNTP) mixture, 0.4 μM both sense and antisense primer, 0.4 μM fluorescently labeled TaqMan probe, 1× Qiagen one-step RT-PCR buffer, 10 U RNase inhibitor, and 1 μl Qiagen RT-PCR enzyme mixture of HotStart Taq DNA polymerase and reverse transcriptase. Reverse transcription was done at 50°C for 32 min followed by 15 min of HotStart Taq DNA polymerase activation. A total of 45 amplification cycles consisting of 94°C for 15 s, 55°C for 15 s, and 70°C for 45 s were completed. For quantification, a full-length Norwalk virus cDNA that was cloned into a pSPORT1 vector (generous gift from K. Green, NIH) was used as a standard. The plasmid was serially diluted (corresponding to 20 to ∼2 × 104 genomic copies) and amplified. The log10-transformed genomic copy number was plotted against the threshold cycle (CT) value and analyzed by linear regression to make a standard curve.
Statistical analysis.
The NV log10 reduction for each hand hygiene agent was calculated by subtracting the log10-transformed NV genomic copy number for each agent from the log10-transformed baseline control (after 20 min of drying). A paired t test using Statistical Analysis Software program 9.2 (SAS, Cary, NC) was performed to examine the difference between NV reduction on the left hand and that on the right hand for each subject and between the two replicate trials. To examine the differences between the baseline control, hand sanitizer, liquid soap, and water rinse, we performed an analysis of variance (ANOVA) test using the SAS PROC GLM procedure. For the multiple comparisons, Tukey's test was employed to examine the differences between the baseline control, hand sanitizer, liquid soap, and water rinse for either standard or modified ASTM experiments.
RESULTS
Reproducibility of NV TaqMan real-time RT-PCR.
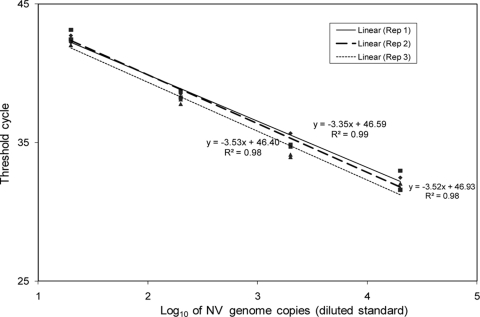
To determine the reproducibility of the NV RT-qPCR assay, 10-fold serial dilutions of previously quantified NV plasmid DNA were tested in duplicate in three independent experiments. The assay was log linear in the range of 1.3 to 4.3 log10 NV genomic copies of DNA, and the standard curve was reproducible, with no significant differences observed between replicate runs (Fig. 1). When the assay was applied to 20% NV stool suspensions with or without added disinfecting agents, no RT-qPCR inhibition was observed (data not shown).
FIG. 1.
The reproducibility of the NV RT-qPCR assay was determined using a 10-fold serially diluted NV plasmid DNA standard (2 × 104 to ∼20 genomic copies per reaction). The threshold cycle versus log10 transformed NV genomic copy number is shown for three replicate experiments analyzed using linear regression.
Assessment of disinfection efficacy using real-time RT-PCR.
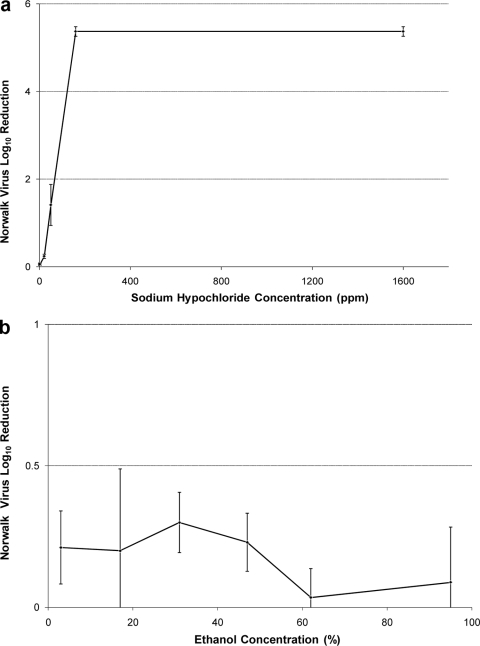
The results of the suspension assays indicate striking differences in the efficacy of sodium hypochlorite and ethanol against NV as evaluated by RT-qPCR (Fig. 2). A clear concentration effect was observed for sodium hypochlorite, with what appears to be log-linear inactivation between 22 ppm and 160 ppm after an exposure of 30 s. A 5-log10 reduction in genomic copies of NV cDNA was observed at sodium hypochlorite concentrations of 160 and 1,600 ppm. No clear concentration effect was observed for ethanol, and the maximum drop in titer did not exceed 0.5 log10 genomic copies of NV cDNA.
FIG. 2.
In vitro inactivation of Norwalk virus at various concentrations of sodium hypochlorite and ethanol. The suspension assay was used to measure virucidal activities of 3 ppm, 22 ppm, 51 ppm, 160 ppm, and 1,600 ppm of sodium hypochlorite (a) and 3%, 17%, 31%, 47%, 62%, and 95% ethanol (b) after a 30-s exposure time. Results indicate the mean NV log10 reduction with standard deviation from two replicate trials.
Efficacy of various hand hygiene agents against NV using standard and modified ASTM methods.
Although the concentrations of genomic copies of NV cDNA were consistently lower on finger pad eluates pretreated with RNase compared to those without prior RNase treatment, these differences were not statistically significant. Because the samples processed by RNase pretreatment may be considered more relevant with respect to virus infectivity, these data are further described below.
For the standard ASTM method, statistical analyses comparing replicate volunteer hands (left versus right) and experimental replicates (day 1 versus day 2) using the paired t test showed no significant differences (data not shown) (P > 0.05), allowing us to pool all replicates for further analysis. A slight reduction in detectable NV cDNA was observed after inoculum drying compared to the time zero input control, with an average 0.16 ± 0.06 log10 reduction (Table 1). Compared to the baseline control, the greatest mean reduction in genomic copies of NV cDNA was observed for the antibacterial liquid soap treatment (0.67 ± 0.47 log10 reduction), followed by the mean reduction associated with water rinsing (0.58 ± 0.37 log10 reduction). The alcohol-based hand sanitizer reduced the genomic copies of NV cDNA by an average of only 0.27 ± 0.12 log10 compared to the baseline control. There was a statistically significant difference in log10 reduction in comparing the three treatments (F test, P < 0.01). More specifically, paired statistical comparisons demonstrated significant differences (P < 0.05) in mean log10 reduction of genomic copies of NV cDNA in comparing the liquid soap or water rinse to the baseline control but no statistically significant difference in comparing the hand sanitizer to the baseline control (P = 0.053).
TABLE 1.
In vivo efficacies of hand wash agents against NV evaluated by standard and modified ASTM methodsa
| Exposure | Standard ASTM method |
Modified ASTM method |
||||
|---|---|---|---|---|---|---|
| n | Avg log reduction (SD) |
n | Avg log reduction (SD) |
|||
| No RNase | RNase | No RNase | RNase | |||
| Dry control | 20 | 0.20 (0.28)A | 0.16 (0.06)X | 20 | 0.22 (0.25)A | 0.20 (0.24)X |
| Hand sanitizer | 20 | 0.14 (0.31)A | 0.27 (0.12)X | 10 | 0.22 (0.22)A | 0.34 (0.22)X |
| Liquid soapb | 20 | 0.94 (0.46)B | 0.67 (0.47)Y | 10 | 1.20 (0.64)B | 1.10 (0.49)Y |
| Water rinsec | 20 | 0.75 (0.63)B | 0.58 (0.37)Y | 20 | 1.58 (0.48)B | 1.38 (0.49)Y |
Different superscript capital letter designations in a column indicate statistically significant differences (P < 0.05) between mean log10 reductions by disinfection/removal treatments for the finger pad eluates with no RNase treatment (A and B) or with RNase treatment (X and Y) prior to RT-qPCR.
There was a marginal statistical difference (P = 0.048) between the results from the standard and modified ASTM methods for samples that received the RNase treatment.
In comparing the results of the standard and modified ASTM methods for each disinfection/removal treatment, statistically significant differences (P < 0.05) were observed for treatments either with no RNase treatment or with RNase treatment prior to RT-qPCR.
The results obtained using the ASTM method with the rubbing modification were similar to those obtained using the standard ASTM method, albeit with slightly higher log10 reductions of NV genomic cDNA copy number. The greatest mean log10 reductions were observed for the water rinse only (1.38 ± 0.49 log10 reduction), followed by the antibacterial liquid soap (1.10 ± 0.49 log10 reduction); again, the ethanol-based hand sanitizer was the least effective (0.34 ± 0.22 log10 reduction) (Table 1). As was the case for the ASTM standard method, the log10 reduction in NV cDNA genomic copies for the ethanol-based hand sanitizer was not statistically significant compared to the baseline control (P = 0.21). In comparing the data using the ASTM standard method versus the method with the rubbing modification for any one treatment, statistically significant differences were observed for the liquid soap and the water rinse (P < 0.001).
DISCUSSION
In this study, we examined the efficacy of several disinfectants against Norwalk virus with virus reduction evaluated by RT-qPCR both with and without sample treatment with RNase. In initial screening studies using the suspension assay, we confirmed a concentration-dependent reduction in virus genome copy number after a 30-s exposure to sodium hypochlorite, with virus elimination at concentrations of >160 ppm. In contrast, NV appeared to be quite resistant to ethanol. Previous studies indicate that sodium hypochlorite affects the integrity of both capsid protein and viral RNA, while ethanol affects the viral capsid protein but not the nucleic acid (12, 24, 25). The fact that virus suspensions treated with higher concentrations of sodium hypochlorite were nondetectable by RT-qPCR is consistent with the proposed mode of action of this disinfectant. However, because the viral RNA does not appear to be degraded by ethanol, the true impact of this disinfectant on the infectivity of the virus cannot be definitively confirmed.
The relationship between detection of residual viral RNA and virus infectivity remains an important consideration in these studies. Some investigators have sought to address this issue by pretreating samples with RNase H with or without proteinase K, hypothesizing that naked (noninfectious) RNA will be degraded by RNase prior to the application of RT-qPCR. Hence, detectable amplicons will be more indicative of infectious virus. In our study, finger pad eluates were screened by RT-qPCR both with and without RNase H treatment prior to nucleic acid amplification; for samples pretreated with RNase H, the detection of viral RNA should be more indicative of the presence of infectious virus. Although overall residual virus titers after exposure to antibacterial soap or hand sanitizer were lower for samples that received the RNase pretreatment, the differences between RNase-treated and untreated samples were not statistically significant. We conclude that it is likely that at least some of the residual RNA after treatment was associated with intact virions (resistant to RNase treatment) and hence more indicative of infectious virus particles that were not affected by hand wash product treatment.
The efficacy of sanitizers against target microbes is impacted by a variety of factors, including disinfectant type and concentration, test method (in vitro and in vivo), target organism, and matrix. This complicates comparisons between studies. Studies of other nonenveloped enteric viruses and HuNoV surrogates probably provide the most relevant insights. For example, Sattar et al. (27) assessed the activity of a 60% ethanol-based hand gel against human adenoviruses, rhinoviruses, and rotaviruses on finger pads, using an infectivity assay, and reported that the product reduced the titers of all three viruses by 3 to >4 log10 compared to a hard-water rinse. However, others have observed that 70% ethanol was able to reduce the infectivity titer of hepatitis A virus and poliovirus type 1 inoculated onto human skin by only about 1 log10 (23). Kampf et al. (15) reported that a higher concentration of ethanol (>70%) was associated with better virucidal efficacy against FCV, although these results have been disputed by others (13). Two other studies examined the in vivo efficacy of 62% alcohol-based hand sanitizer against FCV and observed a <0.5 log10 reduction (17, 19). Other studies confirm the relatively poor efficacy of 60 to 70% ethanol for the inactivation of HuNoV surrogates (6, 22). Taken together, ethanol appears to have relatively poor efficacy against the nonenveloped enteric viruses.
Comparative studies have shown that some disinfectants intended for direct skin contact may be more effective than ethanol. While some investigators have shown that ethanol has comparatively better efficacy than isopropanol (15, 17), Gehrke et al. (13) found that 1-propanol was more effective than ethanol for FCV inactivation. Recently, two studies (16, 22) reported separately that ethanol-based formulas in combination with other ingredients not only significantly improved the efficacy of hand sanitizers but also exhibited a broad spectrum of virucidal activity. Specifically, Macinga et al. (22) reported on a hand sanitizer containing 70% ethanol with a synergistic blend of polyquaternium-37 and citric acid. In an in vitro study, each ingredient alone had limited efficacy against bacteriophage MS2, but the combination of the three agents produced >3 log10 virus reductions when applied to a variety of human enteric viruses and surrogates. Similarly, the formulation of Kramer et al. (16) consisting of 55% ethanol in combination with 10% propan-1-ol, 5.9% propan-1,2-diol, 5.7% butan-1,3-diol, and 0.7% phosphoric acid showed synergistic, broad-spectrum virucidal activity in both in vitro and in vivo studies. Agents such as these are promising alternatives to simple ethanol sanitizers and merit further evaluation and commercial development.
Consistent with other reports that have tested hand hygiene products against a variety of microorganisms (1, 2, 9, 19, 23), we found that a simple water rinse or use of an antibacterial liquid soap was more effective than alcohol-based agents for reducing NV contamination on human hands. We also observed that rubbing fingers provided better reduction of residual virus titer, suggesting that physical removal plays a role in the efficacy of these hand hygiene methods—especially for a water rinse where there is no disinfection effect.
Clearly, ethanol-based hand sanitizers are an effective deterrent to the transmission of enveloped viruses such as influenza virus, hepatitis B virus, and herpes simplex viruses 1 and 2 (14). However, they appear to be less effective for controlling the transmission of human enteric viruses, particularly the epidemiologically important HuNoV group. The relatively poor efficacy of ethanol-based hand sanitizers against NV, and perhaps other HuNoV, has important implications for infection control in health care settings and food service establishments where these products are commonly used and where HuNoV outbreaks are most frequent (11). Even though water washing, with or without the addition of an antibacterial soap, can remove some NV from fingers, it should not be relied upon to eliminate this virus that can be shed in titers of up to 1012 genome copies per gram of stool (3). Clearly, there remains a need to develop commercial hand hygiene agents with specific, high-level activity (≥4 log10 reduction) against human gastrointestinal viruses. The search for effective products continues.
Acknowledgments
This study was supported in part by a grant to L.-A. Jaykus and C. Moe from the International Life Sciences Institute—North America (ILSI-NA).
We thank K. J. Schwab at the Johns Hopkins Bloomberg School of Public Health for assistance with the TaqMan real-time PCR and K. Green from NIH/NIAID for her generous gift of the full-length Norwalk virus clone. We are grateful to Juan Leon and Helen Tang for their critical review of the manuscript.
Footnotes
Published ahead of print on 20 November 2009.
REFERENCES
- 1.Ansari, S. A., S. A. Sattar, V. S. Springthorpe, G. A. Wells, and W. Tostowaryk. 1988. Rotavirus survival on human hands and transfer of infectious virus to animate and nonporous inanimate surfaces. J. Clin. Microbiol. 26:1513-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansari, S. A., S. A. Sattar, V. S. Springthorpe, G. A. Wells, and W. Tostowaryk. 1989. In vivo protocol for testing efficacy of hand-washing agents against viruses and bacteria: experiments with rotavirus and Escherichia coli. Appl. Environ. Microbiol. 55:3113-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atmar, R. L., A. R. Opekun, M. A. Gilger, M. K. Estes, S. E. Crawford, F. H. Neill, and D. Y. Graham. 2008. Norwalk virus shedding after experimental human infection. Emerg. Infect. Dis. 14:1553-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bae, J., and K. J. Schwab. 2008. Evaluation of murine norovirus, feline calicivirus, poliovirus, and MS2 as surrogates for human norovirus in a model of viral persistence in surface water and groundwater. Appl. Environ. Microbiol. 74:477-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baert, L., C. E. Wobus, E. Van Coillie, L. B. Thackray, J. Debevere, and M. Uyttendaele. 2008. Detection of murine norovirus 1 by using plaque assay, transfection assay, and real-time reverse transcription-PCR before and after heat exposure. Appl. Environ. Microbiol. 74:543-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belliot, G., A. Lavaux, D. Souihel, D. Agnello, and P. Pothier. 2008. Use of murine norovirus as a surrogate to evaluate resistance of human norovirus to disinfectants. Appl. Environ. Microbiol. 74:3315-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanton, L. H., S. M. Adams, R. S. Beard, G. Wei, S. N. Bulens, M. A. Widdowson, R. I. Glass, and S. S. Monroe. 2006. Molecular and epidemiologic trends of caliciviruses associated with outbreaks of acute gastroenteritis in the United States, 2000-2004. J. Infect. Dis. 193:413-421. [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 2000. Outbreaks of Norwalk-like viral gastroenteritis—Alaska and Wisconsin, 1999. MMWR Morb. Mortal. Wkly. Rep. 49:207-211. [PubMed] [Google Scholar]
- 9.Davis, M. A., H. Sheng, J. Newman, D. D. Hancock, and C. J. Hovde. 2006. Comparison of a waterless hand-hygiene preparation and soap-and-water hand washing to reduce coliforms on hands in animal exhibit settings. Epidemiol. Infect. 134:1024-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doultree, J. C., J. D. Druce, C. J. Birch, D. S. Bowden, and J. A. Marshall. 1999. Inactivation of feline calicivirus, a Norwalk virus surrogate. J. Hosp. Infect. 41:51-57. [DOI] [PubMed] [Google Scholar]
- 11.Fankhauser, R. L., S. S. Monroe, J. S. Noel, C. D. Humphrey, J. S. Bresee, U. D. Parashar, T. Ando, and R. I. Glass. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 186:1-7. [DOI] [PubMed] [Google Scholar]
- 12.Fukuzaki, S. 2006. Mechanisms of actions of sodium hypochlorite in cleaning and disinfection processes. Biocontrol Sci. 11:147-157. [DOI] [PubMed] [Google Scholar]
- 13.Gehrke, C., J. Steinmann, and P. Goroncy-Bermes. 2004. Inactivation of feline calicivirus, a surrogate of norovirus (formerly Norwalk-like viruses), by different types of alcohol in vitro and in vivo. J. Hosp. Infect. 56:49-55. [DOI] [PubMed] [Google Scholar]
- 14.Kampf, G., and A. Kramer. 2004. Epidemiologic background of hand hygiene and evaluation of the most important agents for scrubs and rubs. Clin. Microbiol. Rev. 17:863-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kampf, G., D. Grotheer, and J. Steinmann. 2005. Efficacy of three ethanol-based hand rubs against feline calicivirus, a surrogate virus for norovirus. J. Hosp. Infect. 60:144-149. [DOI] [PubMed] [Google Scholar]
- 16.Kramer, A., A. S. Galabov, S. A. Sattar, L. Dohner, A. Pivert, C. Payan, M. H. Wolff, A. Yilmaz, and J. Steinmann. 2006. Virucidal activity of a new hand disinfectant with reduced ethanol content: comparison with other alcohol-based formulations. J. Hosp. Infect. 62:98-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lages, S. L., M. A. Ramakrishnan, and S. M. Goyal. 2008. In-vivo efficacy of hand sanitisers against feline calicivirus: a surrogate for norovirus. J. Hosp. Infect. 68:159-163. [DOI] [PubMed] [Google Scholar]
- 18.Lee, J., K. Zoh, and G. Ko. 2008. Inactivation and UV disinfection of murine norovirus with TiO2 under various environmental conditions. Appl. Environ. Microbiol. 74:2111-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin, C. M., F. M. Wu, H. K. Kim, M. P. Doyle, B. S. Michael, and L. K. Williams. 2003. A comparison of hand washing techniques to remove Escherichia coli and caliciviruses under natural or artificial fingernails. J. Food Prot. 66:2296-2301. [DOI] [PubMed] [Google Scholar]
- 20.Lindesmith, L., C. Moe, S. Marionneau, N. Ruvoen, X. Jiang, L. Lindblad, P. Stewart, J. LePendu, and R. Baric. 2003. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 9:548-553. [DOI] [PubMed] [Google Scholar]
- 21.Lopman, B. A., D. W. Brown, and M. Koopmans. 2002. Human caliciviruses in Europe. J. Clin. Virol. 24:137-160. [DOI] [PubMed] [Google Scholar]
- 22.Macinga, D. R., S. A. Sattar, L. A. Jaykus, and J. W. Arbogast. 2008. Improved inactivation of non-enveloped enteric viruses and their surrogates by a novel alcohol-based hand sanitizer. Appl. Environ. Microbiol. 74:5047-5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mbithi, J. N., V. S. Springthorpe, and S. A. Sattar. 1993. Comparative in vivo efficiencies of hand-washing agents against hepatitis A virus (HM-175) and poliovirus type 1 (Sabin). Appl. Environ. Microbiol. 59:3463-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDonnell, G., and A. D. Russell. 1999. Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 12:147-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nuanualsuwan, S., and D. O. Cliver. 2003. Capsid functions of inactivated human picornaviruses and feline calicivirus. Appl. Environ. Microbiol. 69:350-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ojeh, C. K., T. M. Cusack, and R. H. Yolken. 1995. Evaluation of the effects of disinfectants on rotavirus RNA and infectivity by the polymerase chain reaction and cell-culture methods. Mol. Cell. Probes 9:341-346. [DOI] [PubMed] [Google Scholar]
- 27.Sattar, S. A., M. Abebe, A. J. Bueti, H. Jampani, J. Newman, and S. Hua. 2000. Activity of an alcohol-based hand gel against human adeno-, rhino-, and rotaviruses using the fingerpad method. Infect. Control Hosp. Epidemiol. 21:516-519. [DOI] [PubMed] [Google Scholar]
- 28.Sattar, S. A., and S. A. Ansari. 2002. The fingerpad protocol to assess hygienic hand antiseptics against viruses. J. Virol. Methods 103:171-181. [DOI] [PubMed] [Google Scholar]
- 29.Schwab, K. J., M. K. Estes, F. H. Neill, and R. L. Atmar. 1997. Use of heat release and an internal RNA standard control in reverse transcription-PCR detection of Norwalk virus from stool samples. J. Clin. Microbiol. 35:511-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slomka, M. J., and H. Appleton. 1998. Feline calicivirus as a model system for heat inactivation studies of small round structured viruses in shellfish. Epidemiol. Infect. 121:401-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teunis, P. F., C. L. Moe, P. Liu, S. E. Miller, L. Lindesmith, R. S. Baric, J. Le Pendu, and R. L. Calderon. 2008. Norwalk virus: how infectious is it? J. Med. Virol. 80:1468-1476. [DOI] [PubMed] [Google Scholar]
- 32.Topping, J. R., H. Schnerr, J. Haines, M. Scott, M. J. Carter, M. M. Willcocks, K. Bellamy, D. W. Brown, J. J. Gray, C. I. Gallimore, and A. I. Knight. 2009. Temperature inactivation of feline calicivirus vaccine strain FCV F-9 in comparison with human noroviruses using an RNA exposure assay and reverse transcribed quantitative real-time polymerase chain reaction—a novel method for predicting virus infectivity. J. Virol. Methods 156:89-95. [DOI] [PubMed] [Google Scholar]