Abstract
A new method for on-plate protein digestion and MALDI MS analysis is proposed involving an automated one-step sample separation using nanoflow-HPLC followed by nanoliter fraction collection and on-plate digestion with trypsin. This procedure uses a commercial automatic nanoliter fraction collection system for on-line spotting of the eluent onto a MALDI target. After protein digestion, the reaction is stopped by the addition of acidified matrix using the same automated system. Collected spots are subsequently analyzed using a MALDI-TOF/TOF mass spectrometer for protein sequencing and identification.
Keywords: acquired enamel pellicle, LC-MS/MS, on-plate digestion, proteomic, MALDI-TOF/TOF
Gel-free methods have been developed based on liquid separations coupled on-line with mass spectrometry (LC–MS) for proteomic analyses of complex biological systems (1);(2)). The sequence information obtained from peptide fragment fingerprints and database searching is often sufficient for identification (1–3). However, one of the limitations of this approach is the difficulty in identification of protein isoforms or highly homologous proteins. One approach to circumvent this problem proposed by Zheng et al. (4), is based on protein separation using a capillary column followed by fraction collection on MALDI targets pre-coated with trypsin for on-plate digestion, followed by MALDI-TOF/TOF analysis.
We have developed a new “hands-off” method for on-line protein digestion and MALDI analysis based on a robotic system coupled to a nano-HPLC. This system offers improved reproducibility and ease of handling for routine proteomics studies using standard gel-free mass spectrometry and tandem mass spectrometry screening procedures. In order to assess this approach, we performed on-plate plate digestion with standard proteins such as equine myoglobin (Sigma M0630), bovine insulin (Sigma, St. Louis, MO, I5500) and bovine serum albumin (Sigma L3908). Protein solutions were prepared at an initial concentration of 1mg/mL and then diluted, so that 0.5 μL of the protein solution corresponded to 20 pmol, 100 pmol, 200 pmol, 600 pmol and 12 nmol of protein. Aliquots, 0.5 μL were spotted onto a MALDI target, and the spot was then incubated with 0.2 μL of a trypsin solution (0.2 μg/μL in 25mM ammonium bicarbonate). The target was maintained in a humidifier chamber at 37oC. After incubation (Table 1), 0.5 μL of a MALDI matrix solution (α-CHCA, 5 mg/mL in 50% acetonitrile/0.3% TFA) was added to each spot. The same standard amounts were digested with trypsin (1:20) in-solution. A 0.5 μL aliquot of the digest was spotted onto a 384-well MALDI plate with 0.5 μL of α-CHCA (5mg/mL in 50% acetonitrile/0.3% TFA). The digestion of aliquots for both on-target and in-solution procedures were carried out for 10, 30, 60, 180 and 360 minutes.
Table 1.
Comparison of in-solution and on-plate digestion for bovine serum albumin, equine myoglobin and bovine insulin standards at 600 ng, 300 ng, 100 ng, 50 ng and 10 ng levels incubated for 5min, 30min, 1h, 3h and 6h. Pept refers to the number of tryptic peptides used for protein identification; Δ refers to the standard deviation in respect to the number of tryptic peptides for the different assays; Score refers to the MASCOT score and % Cov refers to the percentage of coverage from the MASCOT search.
| On-solution digestion | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 600ng | 300ng | 100ng | 50ng | 10ng | |||||||||||||||||
| Pept | Δ | Score | % Cov | Pept | Δ | Score | % Cov | Pept | Δ | Score | % Cov | Pept | Δ | Score | % Cov | Pept | Δ | Score | % Cov | ||
| BSA | 5 min | 7 | 2 | 82 | 13 | 6 | 2 | 75 | 8 | 8 | 2 | 84 | 8 | 6 | 2 | 76 | 8 | 6 | 2 | 4 | 4 |
| 30min | 17 | 3 | 184 | 27 | 12 | 3 | 162 | 14 | 11 | 2 | 121 | 14 | 8 | 3 | 104 | 7 | 7 | 2 | 3 | 3 | |
| 1 h | 17 | 3 | 196 | 23 | 12 | 3 | 193 | 14 | 17 | 3 | 232 | 18 | 11 | 2 | 149 | 11 | 10 | 2 | 14 | 14 | |
| 3 h | 19 | 3 | 400 | 36 | 16 | 3 | 346 | 18 | 15 | 3 | 193 | 22 | 11 | 3 | 146 | 14 | 9 | 3 | 11 | 11 | |
| 6 h | 18 | 4 | 335 | 43 | 18 | 3 | 213 | 27 | 14 | 4 | 224 | 23 | 12 | 3 | 112 | 13 | 10 | 2 | 8 | 8 | |
| Myoglobin | 5 min | 9 | 2 | 150 | 43 | 9 | 2 | 164 | 41 | 6 | 2 | 153 | 41 | 5 | 2 | 63 | 20 | 3 | 2 | 11 | 11 |
| 30 min | 12 | 2 | 194 | 81 | 12 | 2 | 113 | 80 | 6 | 2 | 162 | 43 | 7 | 2 | 121 | 80 | 6 | 2 | 17 | 17 | |
| 1 h | 12 | 3 | 278 | 81 | 10 | 2 | 194 | 68 | 7 | 2 | 198 | 69 | 5 | 3 | 155 | 69 | 6 | 2 | 20 | 20 | |
| 3 h | 15 | 3 | 226 | 81 | 14 | 3 | 311 | 80 | 14 | 3 | 345 | 80 | 4 | 2 | 114 | 24 | 5 | 3 | 20 | 20 | |
| 6 h | 15 | 2 | 500 | 81 | 15 | 3 | 568 | 80 | 13 | 3 | 313 | 80 | 8 | 3 | 167 | 59 | 5 | 2 | 24 | 24 | |
| Insulin | 5 min | 4 | 1 | 114 | 100 | 3 | 1 | 94 | 56 | 2 | 1 | 88 | 56 | 2 | 1 | 66 | 56 | 2 | 1 | 56 | 56 |
| 30 min | 3 | 1 | 113 | 100 | 4 | 1 | 69 | 100 | 3 | 2 | 81 | 56 | 2 | 1 | 63 | 56 | 2 | 1 | 56 | 56 | |
| 1 h | 4 | 2 | 89 | 100 | 2 | 1 | 91 | 56 | 2 | 1 | 75 | 56 | 2 | 1 | 71 | 56 | 2 | 1 | 56 | 56 | |
| 3 h | 4 | 1 | 132 | 100 | 3 | 1 | 104 | 100 | 2 | 1 | 75 | 56 | 3 | 1 | 74 | 56 | 2 | 1 | 56 | 56 | |
| 6 h | 3 | 1 | 119 | 100 | 3 | 1 | 109 | 56 | 2 | 1 | 71 | 56 | 2 | 1 | 84 | 56 | 2 | 1 | 56 | 56 | |
| On-Plate digestion | |||||||||||||||||||||
| 600ng | 300ng | 100ng | 50ng | 10ng | |||||||||||||||||
| Pept | Δ | Score | % Cov | Pept | Δ | Score | % Cov | Pept | Δ | Score | % Cov | Pept | Δ | Score | % Cov | Pept | Δ | Score | % Cov | ||
| BSA | 5 min | 8 | 2 | 94 | 8 | 7 | 2 | 84 | 8 | 7 | 2 | 74 | 6 | 6 | 2 | 76 | 7 | 6 | 2 | 71 | 7 |
| 30 min | 17 | 3 | 112 | 21 | 16 | 2 | 130 | 17 | 12 | 3 | 121 | 28 | 10 | 3 | 89 | 13 | 8 | 3 | 104 | 8 | |
| 1 h | 18 | 4 | 265 | 24 | 18 | 3 | 224 | 29 | 17 | 3 | 221 | 29 | 14 | 2 | 93 | 18 | 11 | 4 | 109 | 11 | |
| 3 h | 19 | 4 | 256 | 41 | 19 | 3 | 298 | 41 | 17 | 4 | 362 | 33 | 16 | 3 | 130 | 23 | 12 | 3 | 111 | 8 | |
| 6 h | 19 | 3 | 411 | 41 | 19 | 3 | 389 | 41 | 16 | 3 | 246 | 34 | 14 | 2 | 111 | 18 | 9 | 2 | 107 | 14 | |
| Myoglobin | 5 min | 9 | 2 | 143 | 45 | 10 | 2 | 174 | 33 | 6 | 2 | 254 | 36 | 5 | 2 | 70 | 24 | 4 | 2 | 62 | 12 |
| 30 min | 11 | 3 | 211 | 45 | 13 | 3 | 169 | 43 | 5 | 2 | 193 | 36 | 7 | 2 | 119 | 33 | 6 | 1 | 124 | 16 | |
| 1 h | 12 | 2 | 198 | 81 | 10 | 2 | 168 | 81 | 7 | 2 | 162 | 43 | 5 | 2 | 153 | 33 | 6 | 1 | 149 | 18 | |
| 3 h | 15 | 5 | 329 | 81 | 15 | 4 | 346 | 81 | 13 | 3 | 384 | 80 | 7 | 2 | 191 | 33 | 5 | 3 | 88 | 24 | |
| 6 h | 15 | 4 | 483 | 81 | 15 | 3 | 449 | 81 | 14 | 2 | 511 | 80 | 8 | 3 | 253 | 33 | 5 | 1 | 134 | 24 | |
| Insulin | 5 min | 4 | 2 | 99 | 100 | 2 | 1 | 114 | 56 | 2 | 1 | 69 | 56 | 2 | 1 | 88 | 56 | 2 | 1 | 71 | 56 |
| 30 min | 3 | 1 | 105 | 100 | 3 | 1 | 162 | 56 | 3 | 1 | 94 | 56 | 2 | 1 | 71 | 56 | 2 | 1 | 68 | 56 | |
| 1 h | 4 | 2 | 154 | 100 | 3 | 1 | 121 | 56 | 3 | 1 | 142 | 56 | 3 | 1 | 82 | 56 | 2 | 1 | 74 | 56 | |
| 3 h | 4 | 2 | 123 | 100 | 4 | 1 | 130 | 100 | 3 | 1 | 121 | 56 | 3 | 1 | 91 | 56 | 2 | 1 | 89 | 56 | |
| 6 h | 3 | 1 | 142 | 100 | 3 | 1 | 129 | 56 | 3 | 1 | 132 | 56 | 3 | 2 | 84 | 56 | 2 | 1 | 82 | 56 | |
MALDI-TOF/TOF MS analyses were performed on a 4800 MALDI TOF/TOF Analyzer (Applied Biosystems, Foster City, CA) and the data were processed and analysed by the Global Protein Server Workstation (Applied Biosystems, Foster City, CA, USA), which uses internal Mascot software (Matrix Science Ltd, U.K) to search peptide mass fingerprints and MS/MS data. Searches were performed against the SwissProt protein database (2006.09.28) for Homo Sapiens. The database search parameters were: mass tolerance of 40 ppm for precursor ions and 0.3 Da for fragment ions; trypsin digestion with two missed cleavages; variable modifications such as N - terminal Gln, pyroGlutamic acid, methionine oxidation, N-terminal acetylation and phosphorylation of serine, threonine and tyrosine residues. A positive identification was accepted at the 95% confidence level.
As expected, good protein coverage was achieved through the approach of in-solution digestion of the protein standards at all tested concentrations. Similar results were obtained using our protein on-plate digestion approach based on the number of detected peaks and percentage of sequence coverage (Table 1). However, differences in peak sensitivity and relative abundance were observed for BSA and myoglobin spectra obtained via in-solution digestion compared to on-plate digestion. This can be attributed to differences in the spot crystallization conditions. Nevertheless, it is possible to observe a significant number of tryptic peptides after 30 min of digestion using either approach. In both cases, protein identification was made at a 100% confidence level. Data from Table 1 also show that longer periods of digestion do not lead to a great increase the number of peptides identified for either set of conditions.
The online HPLC-MALDI digestion procedure was tested by injection of 1:1 mixtures of bovine insulin and myoglobin, using 5 pmol or 25 pmol of each protein. Protein separations were performed using an Ultimate 3000 (Dionex/LC Packings, Sunnyvale, CA) equipped with a C18 trapping column (Zorbax 300SB-C18, 5 μm particle size, 5 × 0.3 mm, Agilent Technologies) and a 150 mm × 75 μm Zorbax 300SB capillary analytical C18 column with 3.5 μm particle size (Agilent Technologies). After loading the sample onto the trapping column, the trapping column was washed for 5 min with 95% buffer A (water, 0.1% TFA), 5% Buffer B (acetonitrile, 0.05% TFA) at a flow rate of 300 nl/min. Flow was then reversed over the trapping column, and the sample was eluted onto the analytical capillary column at a flow rate of 300 nl/min. A linear gradient of 5% Buffer B to 50% Buffer B was run over a period of 35 min, followed by a 5 min gradient from 50% to 85% Buffer B and 5 min hold at 85% Buffer B. The column was then re-equilibrated in 5% Buffer B prior to further analyses. The separation was monitored at 214 nm using a UV detector (Dionex/LC Packings, Sunnyvale, CA) equipped with a 3 nL flow cell. The peptides eluting off the C18 column, starting 5 min after the beginning of the separation process, were directly deposited using a MALDI-plate collector (Probot, Dionex/LC Packings) onto LC-MALDI plates at 20 second intervals for each spot (100 nL/fraction). A trypsin solution (0.1 ug/mL prepared in 50 mM NH4HCO3/ACN 10%) was simultaneously added to the eluent stream at a flow rate of 800 nL/min (0.1 ug/mL prepared in 50 mM NH4HCO3/ACN 10%). This resulted in the deposition of a final volume of 370 nL per spot. For each separation run, a total of 156 fractions were collected. Following sample collection, the plate was kept at 37°C in a humidity chamber (greater than 80% relative humidity) for digestion. After digesting for 10 min, the plate was replaced into the Probot for deposition of a solution of α-CHCA matrix (2 mg/mL in 70% acetonitrile/0.3% TFA and Glu-Fib (15 fmol) for use as an internal standard) at a continuous flow rate of 270 nL/min. The spots were then analyzed by MALDI/TOF/TOF MS. The MALDI results were searched against the SwissProt Database and a similar number of peptides were matched from both mixtures, with 3 peptides for insulin at 5 pmol and at 25 pmol and 6 and 7 peptides for myoglobin at 5 pmol and 25 pmol, respectively. This corresponds to 56% and 33% sequence coverage for insulin and myoglobin, respectively. Excellent spectral reproducibility was obtained with the on-plate digestion (peak relative abundance < 15%). Thus, our approach allowed the use of conventional nanoflow conditions (300 nL/min) combined with chromatographic collection of well-separated spots using a simple commercial system.
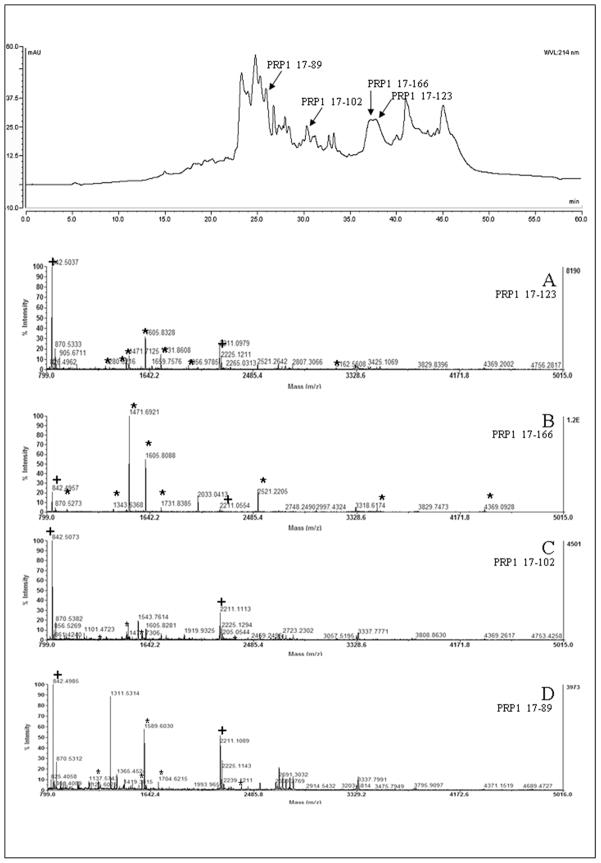
In order to test our methodology with a biological sample, the on-line digestion method was used in the characterization of the in vivo acquired enamel pellicle peptidome, as previously described (5). Acquired enamel pellicle is the tooth surface protein film formed by the selective adsorption of salivary proteins and peptides (5–7). Pellicle formation is considered a dynamic process influenced by several factors inherent to the individual subject, such as circadian cycle, oral microflora, proteolytic activity, physical and chemical properties of the tooth, as well as, the location of the teeth in the mouth (6). Several proteins have been identified as major components of enamel pellicle, such as proline-rich proteins (PRPs), statherin, amylase, albumin, S-IgA, cystatins, lysozyme, carbonic anhydrase, lactoferrin and histatins (8–11). The small amount of sample obtained from each individual, associated with its biological importance, makes the characterization of acquired enamel pellicle proteins an analytical challenge. A TFA extract of acquired enamel pellicle was analyzed by MALDI/TOF MS without digestion using the automated nanoflow fraction collection onto the MALDI target. The MALDI/TOF MS of the fractions collected over the chromatographic retention time window between 25 and 42 minutes were obtained in the linear mode (Figure 1). These spectra showed the presence of several components over a molecular weight range of 5000 to 17000 Da. Based on our previous work and (5, 9, 10) and that of Oppenheim (9);, the majority of ions observed in the spectra were assigned as arising from several known intact saliva proteins and known salivary peptides. In addition, several of the ions arose from unknown proteins. Using our proposed methodology of on-plate digestion more than 100 different tryptic peptides were characterised (12). This led to the identification of 12 distinct salivary proteins. As expected, a comparison of the results from the on-plate digestion with those from global digestion showed that a similar number of proteins were identified in each (a total of 11 positive identification using global digestion and 12 using on-plate digestion). However, with this new approach, we were able to characterize 40 more peptides and 2 additional proteins (MMP-20, O60882 and cystatin S, P01036). Another distinct advantage of the proposed methodology is the possibility of correlating the chromatographic data obtained from the injection of the untreated samples with the data obtained from the corresponding on-plate digested samples. In fact, it is possible to obtain the mass spectra of the intact protein forms (and their respective MW) and of their respective tryptic digest, allowing the positive identification of known and unknown species. This is particularly advantageous when the majority of different protein species, such as salivary acidic proline-rich proteins, give identical protein identification (data/results) as when a total digest is analyzed (PRP1/2-contains PRP3/4, P02810). As an example, the non-digested pellicle extract chromatogram (Figure 1) shows four different peaks (retention times of 38, 36, 30 and 25.4 minutes) that correspond to proteins identified as acidic proline-rich proteins. The protein masses identified in the corresponding mass spectra are 11158.5 Da (PRP1 (17–123)/PRP3, diphosphorylated intact form), 15513.2 Da (PRP1, diphosphorylated intact form), 9034.0 Da, and 7727.1 Da.
Figure 1.
Upper-Representative UV-chromatogram of a TFA protein extract separation with detection at 214 nm. Lower-. MALDI-MS spectra obtained from fractions eluting at 38 min (A), 36 min (B); 30 min (C) and 25.4 min (D). + indicates the m/z corresponding to the peptides of trypsin autolysis. * indicates the m/z used for protein identification using MASCOT.
The sequences identified as being consistent with the masses obtained from the MS analyses of the on-plate tryptic peptides were: m/z 1589,612 (35–47); m/z 1731,868 (93–109); m/z 4369,064 (123–166); m/z 1471.711 (123–136); m/z 2521.228 (123–147); m/z 1068.569 (137–147); m/z 1343.635 (123–135) m/z 1605.839 (108–122) and m/z 3492.682 (123–157) for PRP1 (Figure 1B) and m/z 1280.608 (47–59); m/z 1471.697 (13–136); m/z 2335.033 (47–70); m/z 1956.977 (91–109); m/z 1449.737 (108–121); m/z 3162.599 (91–121); m/z 1731.868 (93–109) and m/z 1605.839 (108–122) for PRP3 (Figure 1A). MS/MS data also enabled the identification of peptide fragments such as GRPQGPPQQGG (m/z 1471.710; 123–126) and GPPQQGGHPRPP (m/z 1284.210; 123–126) (Figure 1A and 1B). Taking into consideration the known sequence modifications (2 phosphate groups (Ser 17, Ser 34) and 1 pyroglutamic acid (Q17)) as described in literature, it is possible to assign the ion of m/z 15513.2 to the major sequence of PRP1 (17–166) and the ion of m/z 11158.5 to PRP3. Using the same approach, the two initially unidentified species of m/z 9034.0 and of m/z 7727.0 were identified as products of PRP1 proteolytic cleavage, corresponding to the amino acid sequences 17–102 and 17–89, respectively. These assignments are supported by the tryptic digest MALDI-MS data which showed ions of m/z 1461.553 (35–36), m/z 1280.608 (47–59), m/z 1589.612 (35–47), m/z 2335.033 (47–70), m/z 1704.621 (34–47), 1576.583 (34–46). The corresponding MS/MS spectra corroborated the identifications (data not shown). In summary, our approach offers several distinct advantages for protein analysis: The greater number of protein identifications obtained compared to the traditional procedure; The ability to obtain a correlation between ions detected from a non-digested sample chromatogram with ions detected from the corresponding trypsin digest, and; The ability to identify highly homologous individual proteins.
Supplementary Material
Acknowledgments
The authors express their appreciation for the financial support provided by the “Fundação para a Ciência e Tecnologia” (FCT- Grant n° SFRH/BPD/34752/2007 SFRH/BD/19769/2004 and POCTI/QUI/5890/2004) and, in part, from the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences
References
- 1.Ferguson PL, Smith RD. Annu Rev Biophys Biomol Struct. 2003;32:399–424. doi: 10.1146/annurev.biophys.32.110601.141854. [DOI] [PubMed] [Google Scholar]
- 2.Chen EI, Hewel J, Felding-Habermann B, Yates JR., 3rd Mol Cell Proteomics. 2006;5:53–6. doi: 10.1074/mcp.T500013-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Perlman DH, Huang H, Dauly C, Costello CE, McComb ME. Anal Chem. 2007;79:2058–66. doi: 10.1021/ac061919p. [DOI] [PubMed] [Google Scholar]
- 4.Zheng S, Yoo C, Delmotte N, Miller FR, Huber CG, Lubman DM. Anal Chem. 2006;78:5198–204. doi: 10.1021/ac052284h. [DOI] [PubMed] [Google Scholar]
- 5.Vitorino R, Calheiros-Lobo MJ, Williams J, Ferrer-Correia AJ, Tomer KB, Duarte JA, Domingues PM, Amado FM. Biomed Chromatogr. 2007;21:1107–17. doi: 10.1002/bmc.830. [DOI] [PubMed] [Google Scholar]
- 6.Lendenmann U, Grogan J, Oppenheim FG. Adv Dent Res. 2000;14:22–8. doi: 10.1177/08959374000140010301. [DOI] [PubMed] [Google Scholar]
- 7.Siqueira WL, Helmerhorst EJ, Zhang W, Salih E, Oppenheim FG. Ann N Y Acad Sci. 2007;1098:504–9. doi: 10.1196/annals.1384.023. [DOI] [PubMed] [Google Scholar]
- 8.Carlen A, Borjesson AC, Nikdel K, Olsson J. Caries Res. 1998;32:447–55. doi: 10.1159/000016486. [DOI] [PubMed] [Google Scholar]
- 9.Siqueira WL, Zhang W, Helmerhorst EJ, Gygi SP, Oppenheim FG. J Proteome Res. 2007;6:2152–60. doi: 10.1021/pr060580k. [DOI] [PubMed] [Google Scholar]
- 10.Vitorino R, Lobo MJ, Duarte J, Ferrer-Correia AJ, Tomer KB, Dubin JR, Domingues PM, Amado FM. Biochem Biophys Res Commun. 2004;320:342–6. doi: 10.1016/j.bbrc.2004.05.169. [DOI] [PubMed] [Google Scholar]
- 11.Yao Y, Berg EA, Costello CE, Troxler RF, Oppenheim FG. J Biol Chem. 2003;278:5300–8. doi: 10.1074/jbc.M206333200. [DOI] [PubMed] [Google Scholar]
- 12.Vitorino R, Calheiros-Lobo MJ, Duarte JA, Domingues PM, Amado FM. J Sep Sci. 2008;31:523–37. doi: 10.1002/jssc.200700486. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.