Abstract
Background
The objectives were to measure the levels of gingival crevicular fluid (GCF) biomarkers and subgingival bacterial species in periodontally healthy and periodontitis subjects in order to explore relations among these biomarkers, the subgingival microbiota, and clinical parameters of periodontal disease.
Material and methods
Clinical periodontal parameters were measured at 6 sites per tooth in 20 periodontitis and 20 periodontally healthy subjects. GCF and subgingival plaque samples were obtained from the mesiobuccal aspect of every tooth. GCF levels of interleukin-1β (IL-1β), matrix metalloproteinase-8 (MMP-8) and IL-8 were measured using checkerboard immunoblotting and the levels of 40 bacterial taxa quantified using checkerboard DNA-DNA hybridization. A subset of “clinically healthy” (CH) sites from each group was analyzed separately. Significance of differences between groups was determined using the unpaired t-test or the Mann-Whitney test. Correlations among immunological, microbiological and clinical data were determined using the Spearman rank correlation coefficient.
Results
There were positive correlations among mean clinical parameters and mean levels of the 3 biomarkers and proportions of Orange and Red complex species (p<0.05). CH sites from periodontitis subjects had higher levels of IL-1β and IL-8 and higher proportions of Orange and Red complex species (p<0.05) than CH sites from periodontally healthy subjects. Red complex species were positively associated with the expression of all biomarkers (p<0.05), while Purple and Yellow complex species had negative correlations with IL-1β and IL-8 (p<0.05).
Conclusions
CH sites from periodontitis subjects present higher levels of GCF biomarkers and periodontal pathogens than CH sites from periodontally healthy subjects. Different microbial complexes demonstrated distinct associations with specific GCF biomarkers.
Keywords: Periodontal diseases, Microbiology, Gingival crevicular fluid, Cytokines, Matrix metalloproteinases
Introduction
It is widely recognized that there is an interplay between the colonizing species of subgingival biofilms and the adjacent periodontal tissues.1,2 This interplay is often marked by an increase in gingival inflammation and the release of biologically active substances into the gingival crevicular fluid (GCF). Therefore, GCF provides a non-invasive, easily collected fluid to measure in vivo the inflammatory mediators released during disease processes that affect the periodontium.3 The relationships among the subgingival microbiota and host mediators can be examined by analyzing samples of the subgingival biofilm and the GCF collected from the same periodontal site(s). However, few studies have attempted to correlate the levels of GCF biomarkers with microbiological data obtained from the same site and most such studies were limited in the number of bacterial species investigated.4–7
Based on studies examining associations among GCF biomarkers and subgingival microbial species, it has been reported, for instance, that Tannerella forsythia might induce the release of GCF MMP-8 and MMP-9.4 Higher levels of periodontal pathogens have also been associated with total amounts in GCF of vascular endothelial growth factor (VEGF), monocyte chemotactic protein-1 (MCP-1) and IL-8.8 Strong correlations between GCF levels of C-terminal telopeptide of type I collagen (ICTP) and mean levels of several periodontal pathogens including T. forsythia, Porphyromonas gingivalis, Prevotella intermedia, Prevotella nigrescens, and Treponema denticola have also been found.7 We have recently described a checkerboard immunoblotting (CBIB) technique for the high-throughput quantification of different GCF biomarkers in a large number of samples simultaneously.9 The objectives of the present study were to measure the levels of 3 inflammatory mediators in GCF samples from periodontally healthy and chronic periodontitis subjects and to characterize the subgingival microbiota associated with the sampled sites in order to explore relationships among these biomarkers, the subgingival biofilm composition, and clinical parameters of periodontal disease. The hypothesis to be tested was that periodontal disease subjects would differ in levels of GCF biomarkers and levels of subgingival species from periodontally healthy subjects, even at sites with similar clinical characteristics in the two clinical groups. In addition, we examined the relation of the subgingival microbial composition with the expression of GCF biomarkers in samples from the same sites.
Material and methods
Subject population and clinical examination
The study population comprised 20 chronic periodontitis subjects and 20 periodontally healthy subjects, recruited from the clinic of the Department of Periodontology, Dental School, Aristotle University of Thessaloniki, Greece. The Ethics Committee of the Dental School at Thessaloniki University approved the protocol, including the clinical examination and the taking of gingival crevicular fluid and plaque samples. All subjects signed informed consent prior to their enrollment into the study. To be included in the study, the periodontally healthy subjects had to be ≥ 20 years of age, have at least 24 natural teeth and no probing pocket depth (PD) > 3mm and no attachment level > 2 mm. The subjects with chronic periodontitis were at least 20 years old, had a minimum of 20 teeth and a minimum of 8 sites with PD > 4 mm and attachment level > 2 mm. Subjects were excluded if they had any systemic condition that would influence the course of periodontal disease or treatment, or medical conditions that would require antibiotic prophylaxis for routine dental procedures. Individuals who had taken antibiotics in the previous 3 months or were either pregnant or nursing were also excluded.
A calibrated clinician measured periodontal parameters at 6 sites per tooth (mesiobuccal, buccal, distobuccal, mesiolingual, lingual and distolingual), of all teeth present, with the exclusion of third molars. The clinical measurements included: visible plaque accumulation (0 or 1), gingival redness (0 or 1), bleeding on probing (BOP; 0 or 1), suppuration (0 or 1), recession (mm) and PD (mm). Recession and PD were measured using a North Carolina probe‡ with mm intervals. The values were rounded to the nearest upper mm. The clinical and demographic parameters of the study subjects are presented in Table 1.
Table 1.
Mean (± SD) demographic, immunological and clinical parameters for both the clinical groups
| Health (n = 20) | Periodontitis (n = 20) | |
|---|---|---|
| age (years)‡§ | 38 ± 13 | 55 ± 12 |
| % males‖ | 40 | 35 |
| % smokers‖ | 50 | 40 |
| cigarettes per day¶ | 7.5 ± 8.3 | 12.1 ± 21.1 |
| pocket depth (mm)‡§ | 1.82 ± 0.24 | 3.66 ± 0.86 |
| recession (mm)‡§ | 0.11 ± 0.11 | 1.38 ± 0.71 |
| % sites with: | ||
| plaque‡¶ | 22 ± 18 | 75 ± 23 |
| bleeding on probing‡§ | 10 ± 8 | 66 ± 16 |
| gingival redness‡§ | 3 ± 5 | 49 ± 32 |
| suppuration*¶ | 0 ± 0 | 3 ± 4 |
| GCF volume (ul)‡§ | 0.16 ± 0.07 | 0.49 ± 0.21 |
| IL-1β (pg/site)‡§ | 7.4 ± 4.8 | 29.9 ± 14.6 |
| IL-8 (pg/site)‡§ | 45.6 ± 35.0 | 98.8 ± 42.4 |
| MMP-8 (ng/site)†§ | 14.1 ± 15.1 | 34.7 ± 30.0 |
p<0.05,
p<0.01 and
p<0.001
unpaired t-test
Fisher's exact test
Mann-Whitney test
Gingival crevicular fluid sampling
Gingival crevicular fluid samples were obtained from the mesiobuccal site of every tooth present (excluding third molars) from each subject, for a total of 931 samples. Following isolation of the site with cotton rolls to prevent contamination with saliva, supragingival plaque was removed, the tooth air dried and a 30-second GCF sample was collected on filter strips§. Periopaper strips were gently inserted into the orifice of the periodontal pocket, 1–2 mm subgingivally. GCF volume was determined using a GCF meter‖, calibrated following the protocol described by Chapple et al. 1999.10 The samples were immediately placed in Eppendorf tubes containing 150 µl of elution buffer [PBST (0.05% Tween 20 in phosphate-buffered saline)], transported to the laboratory and stored at −80°C until assayed. Samples visibly contaminated with blood were discarded. GCF samples were collected prior to clinical measurements.
Checkerboard Immunoblotting (CBIB)
Details of the use of the CBIB technique for the quantification of biomarkers in GCF have recently been published.9 Briefly: PVDF membranes¶ were assembled in a blotting apparatus#. A mixture of capture monoclonal antibodies for IL-1β, MMP-8 and IL-8**, was loaded into each slot of the blotting apparatus and bound to the surface of the membrane. The membrane was then blocked with 0.5% Hammersten casein†† in TBS-T buffer. Standards and samples were loaded into the blotting apparatus and incubated for 2 h at room temperature. After incubation, samples and standards were aspirated from the slots, the blotting apparatus was disassembled, the PVDF membrane released from the apparatus and washed. Nonspecific binding sites on the remaining surface of the membrane were then blocked with 120 ml of 1% Hammersten casein in TBS-T buffer by incubation overnight.
Since GCF samples contain biotin derived from bacteria present in the gingival crevice, endogenous biotin was blocked with a Streptavidin/Biotin blocking kit‡‡. The membrane was then loaded in another blotting apparatus§§, perpendicularly to the original orientation. Biotinylated antibodies to IL-1β, MMP-8 and IL-8**, diluted in blocking buffer, were pipetted into different channels of the blotting apparatus. After 1 h of incubation, the membrane was washed using a vacuum system‖‖ and incubated with streptavidin conjugated with HRP** diluted in blocking buffer. Detection of signals was performed using a chemiluminescent substrate kit¶¶. A sensitive autoradiography film## was used to capture light emission from the membrane and developed in an automated developing machine***. Films were scanned using a flatbed scanner††† and the scanned image analyzed using software for array analysis‡‡‡. Arbitrary units (Volume), proportional to the intensity of the signals were assigned to standards and unknowns. The amounts of the unknown samples (antigens in GCF samples) were estimated from the standard curve using a logit-log model and expressed as pg/site (IL-1β and IL-8) or ng/site (MMP-8).
Microbiological assessment
Subgingival plaque samples were taken from the mesiobuccal aspect of all teeth, excluding third molars immediately after GCF sampling. The subgingival plaque samples were collected using sterile Gracey curettes‡ after removal of supragingival plaque (if present). The samples were placed in separate microcentrifuge tubes containing 0.15 ml TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.6), and 0.1 ml of 0.5 M NaOH was added. All samples were processed at The Forsyth Institute. Samples were individually analyzed for their content of 40 bacterial species using the checkerboard DNA-DNA hybridization technique.11 In brief, the samples were lysed and the DNA placed in lanes on a nylon membrane using a blotting device#. After fixation of the DNA to the membrane, the membrane was placed in a different blotting apparatus §§, with the lanes of DNA at 90° to the lanes of the device. Digoxigenin-labeled whole genomic DNA probes to 40 subgingival species were hybridized in individual lanes of the blotting apparatus. After hybridization, the membranes were washed at high stringency and the DNA probes detected using antibody to digoxigenin, conjugated with alkaline phosphatase. Signals were detected using a chemifluorescent substrate§§§ and read using a computer-linked instrument that read the intensity of the fluorescence signals resulting from the probe-target hybridization‖‖‖. Two lanes in each run contained standards at the concentration of 105 and 106 cells of each species. The sensitivity of the assay was adjusted to permit the detection of 104 cells of a given species by adjusting the concentration of each DNA probe. Signals were converted to absolute counts by comparison with standards on the same membrane. Failure to detect a signal was recorded as zero. A total of 931 subgingival samples were evaluated.
Data analysis
Clinical data were available from 6 sites per tooth for visible plaque, gingival redness, bleeding on probing, suppuration, recession and pocket depth. GCF volume was measured at each mesiobuccal site. The levels of each inflammatory mediator were measured for up to 28 GCF samples per subject and expressed as pg/site or ng/site. Microbiological data available for each subject were the counts of each of the 40 test species for up to 28 subgingival plaque samples per subject. The data for each species were expressed as counts × 105 and as percentage of total DNA probe count at each site. Percentage of DNA probe count data for specific subgingival species were grouped into the microbial complexes described by Socransky et al. (1998).12 For overall analyses, the data for each clinical, microbiological and immunological parameter were averaged within a subject and then averaged across subjects in each clinical group separately. The distribution of the values for each variable in each group was tested using the D’Agostino-Pearson normality test and, depending on the result, significance of differences between healthy and chronic periodontitis subjects for each clinical, microbiological and immunological parameter was determined using either the unpaired t-test or the Mann-Whitney test. Significance of statistical difference between groups for percentage of males and percentage of smokers was tested using Fisher’s exact test. For certain analyses, the data were subset according to pocket depth categories into < 4 mm, 4–6 mm and > 6 mm, averaged within a subject within each category and then averaged across subjects. Significance of differences among the 3 pocket depth categories within the periodontitis group was determined using the repeated measures one-way ANOVA test. Significance of differences between the samples from shallow sites in healthy subjects and the samples from shallow sites in periodontitis subjects was tested using the unpaired t-test. A subset of sites considered “clinically healthy” (PD equal to 2 or 3 mm, no loss of attachment, no redness and no BOP), were averaged within a subject and then averaged across subjects in the healthy and periodontitis groups separately. Significance of differences between the mean levels of the inflammatory mediators, the mean GCF volume, and the mean % of DNA probe count of the microbial complexes for “clinically healthy” sites from the two groups was also tested using either the t-test or the Mann-Whitney test, depending on the result of the D’Agostino-Pearson normality test.
Correlations among the mean values for clinical parameters, mean levels of each biomarker and mean % of DNA probe count of the microbial complexes were investigated using the Spearman rank correlation coefficient. Values for clinical, microbiological and immunological parameters were averaged within each subject in both groups prior to computing the Spearman rank correlation coefficients. Associations among mean levels of the biomarkers and mean % of DNA probe count of the microbial complexes were also examined using the Spearman rank correlation coefficient.
Results
The mean demographic, clinical and immunological parameters for the two groups are displayed in Table 1. The periodontitis subjects exhibited significantly higher mean PD, recession, GCF volume and percentage of sites with BOP, plaque, redness and suppuration. The percentage of smoking subjects was not different between groups. The numbers of cigarettes smoked per day was higher in chronic periodontitis subjects but the difference between clinical groups was not statistically significant. The periodontitis group was also significantly older than the periodontally healthy group. Thus, the clinical parameters were analyzed using ANCOVA adjusting for age. Significant differences remained for the adjusted values (data not shown). The periodontitis subjects also had significantly higher mean levels of all 3 biomarkers when compared to healthy subjects.
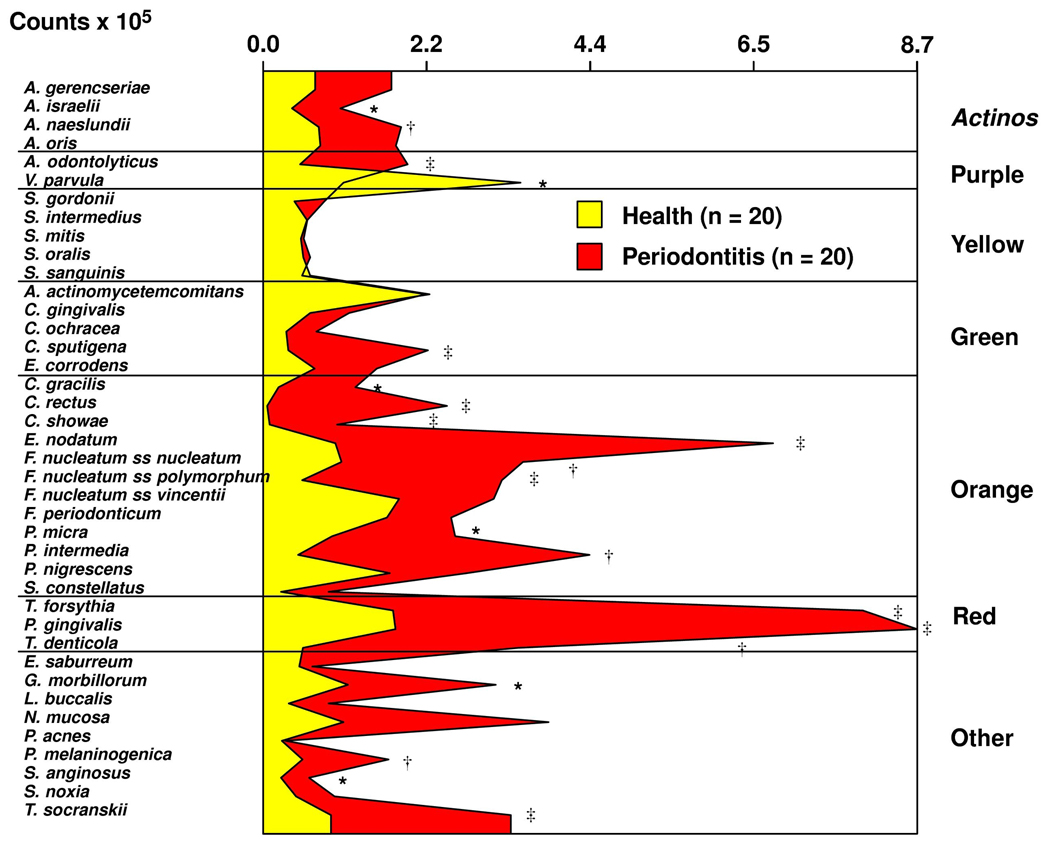
Figure 1 presents the mean counts × 105 of the 40 species for the two groups. Periodontitis subjects had statistically significantly higher levels of 18/40 species, including the periodontal pathogens: Eubacterium nodatum, T. forsythia, P. gingivalis and Treponema denticola. There was a strong positive correlation between mean clinical parameters (PD, recession, % BOP, % plaque % redness and GCF volume) and the mean levels of IL-1β and IL-8 (Table 2). MMP-8 showed weaker correlations, primarily with clinical signs of inflammation (% BOP, % redness and GCF volume; Table 2). There were also significant positive correlations among all mean clinical parameters and the mean percentage of DNA probe count for species of the Orange and Red complexes. The Spearman correlation coefficients were particularly high for the associations with the Red complex. High proportions of early colonizers, members of the Purple, Yellow and Green complexes, were statistically significantly negatively correlated with clinical measurements of periodontal disease.
Fig. 1.
Mean counts (× 105) of the 40 test species from up to 28 subgingival biofilm samples from each periodontally healthy and chronic periodontitis subject. Mean counts of each species were computed by averaging the data for each subject, and then averaging across subjects in each group separately. Significance of differences for each species between clinical groups was sought using the Mann-Whitney test; *p<0.05, †p<0.01, ‡p<0.001 and adjusted for 40 comparisons. 13 The species were ordered and grouped according to the complexes described by Socransky et al. (1998). 12
Table 2.
Relationships among mean clinical parameters, mean levels of GCF biomarkers, and mean % of total DNA probe count of each subgingival microbial complex
| Spearman rank correlation coeficients | ||||||||
|---|---|---|---|---|---|---|---|---|
| Clinical parameters: | GCF Biomarkers | Microbial Complexes | ||||||
| IL-1 | IL-8 | MMP-8 | Purple | Yellow | Green | Orange | Red | |
| mean pocket depth (mm) | 0.75 ‡ | 0.54 ‡ | NS | −0.60 | −0.58 ‡ | −0.46 † | 0.40 † | 0.73 ‡ |
| mean recession (mm) | 0.74 ‡ | 0.52 ‡ | 0.41 † | −0.57 | −0.57 ‡ | −0.47 † | 0.53 ‡ | 0.63 ‡ |
| % BOP | 0.75 ‡ | 0.65 ‡ | 0.39 * | −0.65 ‡ | −0.64 ‡ | −0.48 † | 0.48 † | 0.72 ‡ |
| % plaque | 0.81 ‡ | 0.63 ‡ | NS | −0.51 | −0.54 ‡ | −0.43 † | 0.54 ‡ | 0.67 ‡ |
| % gingival redness | 0.74 ‡ | 0.70 ‡ | 0.50 ‡ | −0.59 ‡ | −0.62 ‡ | −0.35 * | 0.52 ‡ | 0.67 ‡ |
| GCF volume (ul) | 0.86 ‡ | 0.66 ‡ | 0.39 * | −0.51 ‡ | −0.74 ‡ | −0.51 ‡ | 0.49 † | 0.80 ‡ |
p<0.05,
p<0.01 and
p<0.001
NS - non significant.
Differences among levels of IL-1β, IL-8 and MMP-8 in 3 PD categories (<4, 4–6, > 6 mm) were examined in the periodontitis group. Using repeated measures ANOVA, none of the differences among site categories was statistically significant (data not shown). The mean values for the 3 mediators at shallow sites (PD <4 mm) in the periodontitis group were statistically significantly higher compared with the mean values for shallow sites in the periodontally healthy group using the unpaired t-test: IL-1β (25.8 ± 16.5 vs. 7.4 ± 4.8 pg/site; p<0.001), IL-8 (92.6 ± 56.0 vs. 45.7 ± 35.0 pg/site; p<0.01) and MMP-8 (33.7 ± 32.5 vs. 14.1 ± 15.1 ng/site; p<0.05), mean ± SD for chronic periodontitis vs. periodontally healthy subjects, respectively. However, when the mean values for different clinical parameters at these shallow sites in both groups were compared using the unpaired t-test, sites in the periodontitis subjects still had statistically significantly deeper pockets, more recession, and higher prevalence of redness and BOP than periodontally healthy subjects (p < 0.001 [data not shown]).
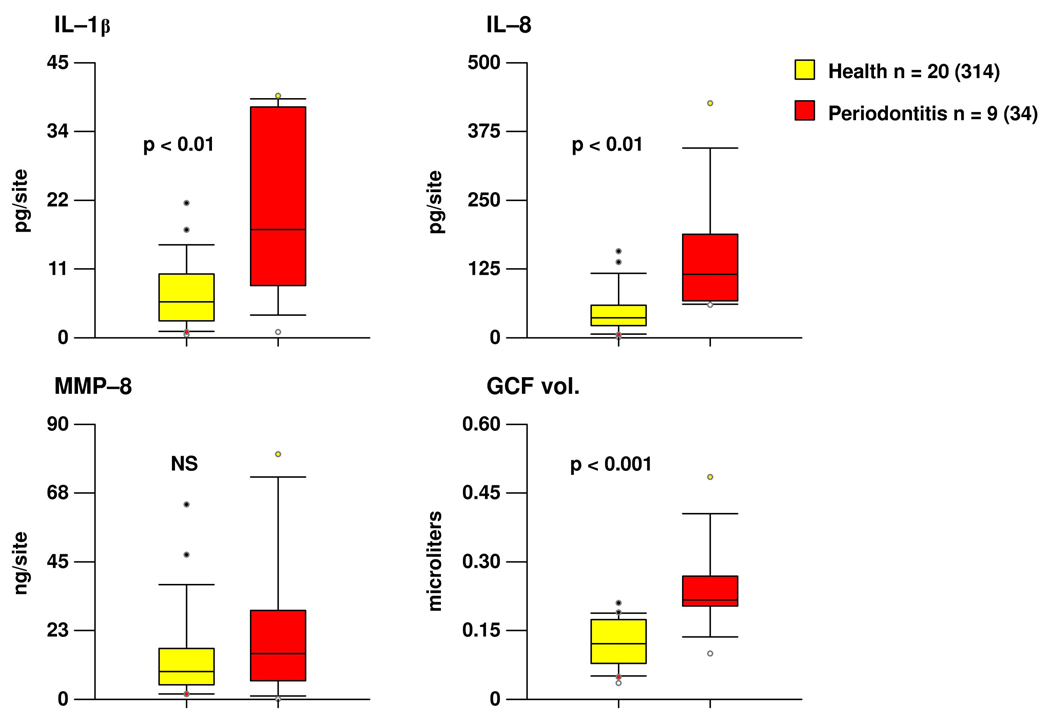
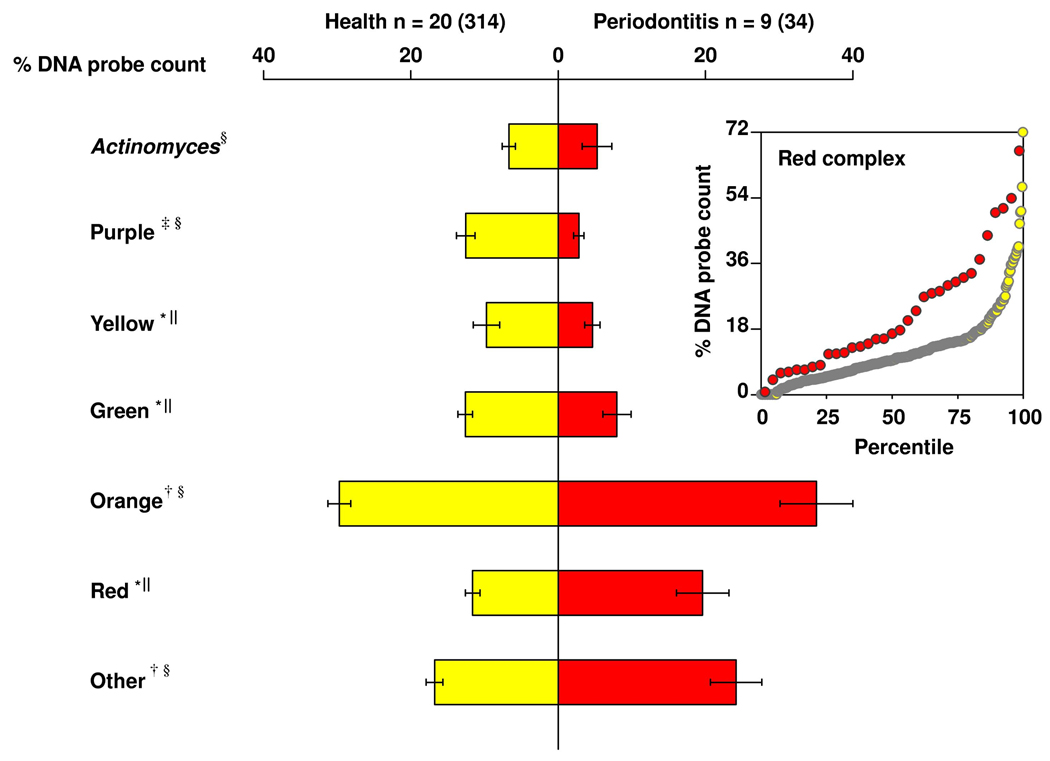
Since the shallow sites in periodontitis subjects were clinically worse than the shallow sites in periodontally healthy subjects, a comparable set of “clinically healthy” sites were selected from each group. These were selected based on PD equal to 2 or 3 and absence of loss of attachment, BOP and gingival redness. Despite the clinical similarity of sites in the 2 subject groups, the periodontitis subjects had statistically significantly higher levels of IL-1β and IL-8 and also a larger volume of GCF at these sites (Fig. 2). This “clinically healthy” site category also differed between groups in its subgingival microbial composition. Periodontally healthy subjects had statistically significantly higher mean proportions of Purple, Yellow and Green complex species, while periodontitis individuals had statistically significantly higher mean proportions of Orange and Red complex species in their subgingival biofilms (Fig. 3). The percentile plot insert of Fig. 3 illustrates that the mean % of DNA probe count for Red complex species was higher in the periodonitis subjects compared to periodontally healthy subjects throughout the entire distribution of values.
Fig. 2.
Tukey box plots of the levels of IL-1β, IL-8, MMP-8 and GCF volume at “clinically healthy” sites (i.e. sites with pocket depth equal to 2 or 3 mm, no loss of attachment, no BOP, and no redness) in periodontally healthy and periodontitis subjects. The box indicates the lower and upper quartiles, the whiskers the 10th and 90th percentiles and the horizontal line in each box is the median. The circles indicate “outlier” values. The levels of each inflammatory mediator and the volume of GCF for this site category were averaged within a subject and then median determined across subjects in the 2 clinical groups separately. Significance of difference between the two groups was determined using the Mann-Whitney test. The numbers in parentheses represent the number of sites analyzed in each group.
Fig. 3.
Bilateral bar chart of the mean percentage of DNA probe count for the 7 subgingival microbial complexes described by Socransky et al. (1998)12 in periodontally healthy and periodontitis subjects. Mean % DNA probe count of each complex was computed by averaging the data for each subject, and then averaging across subjects in each group separately. The bars indicate the means and the whiskers the SEM. Significance of differences between clinical groups (*p<0.05, †p<0.01, ‡p<0.001) was sought using the unpaired t-test§ or the Mann-Whitney test‖ depending on the result of the D’Agostino-Pearson normality test. Insert: percentile plot of mean % DNA probe count of the Red complex species for periodontally healthy (white circles) and periodontitis (red circles) subjects. The y-axis represents the mean % DNA probe count and the x-axis represents the percentiles. Each circle depicts the mean value for an individual subject and the horizontal lines indicate the median values.
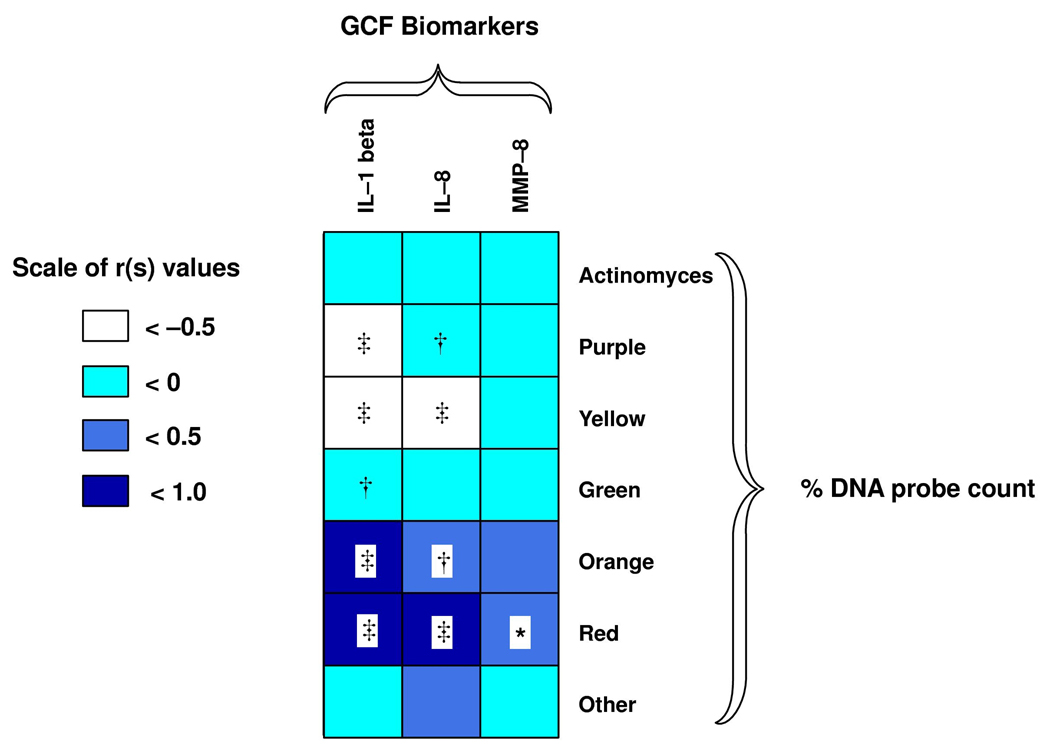
Associations among the mean proportions of the different subgingival microbial complexes and the mean GCF levels of the 3 biomarkers for the entire study population revealed that Red complex species were statistically significantly positively associated with the expression of all biomarkers (rs: 0.72, p<0.001; rs: 0.53, p<0.001; rs: 0.34, p<0.05 for IL-1 β, IL-8 and MMP-8, respectively). Orange complex species also demonstrated statistically significant positive associations with IL-1β (rs: 0.56, p<0.001) and IL-8 (rs: 0.45, p<0.01). Conversely, high proportions of the health associated species of the Purple and Yellow complexes presented statistically significant negative correlations with IL-1β (rs: −0.56, p<0.001; rs: −0.62, p<0.001 for Purple and Yellow complexes, respectively) and IL-8 (rs: −0.43, p<0.01; rs: −0.61, p<0.001 for Purple and Yellow complexes, respectively). Green complex species had a statistically significant negative association with IL-1β (rs: −0.46, p<0.01) (Fig. 4).
Fig. 4.
Grid-plot of the Spearman rank correlation coefficients among the mean proportions of the subgingival microbial complexes and the mean levels of GCF biomarkers in periodontally healthy and periodontitis subjects (*p<0.05, †p<0.01, ‡p<0.001).
Discussion
The goals of this investigation were to compare the mean levels of IL-1β, MMP-8 and IL-8 in GCF samples from periodontally healthy and chronic periodontitis subjects and examine the correlations among these biomarkers, clinical parameters of periodontal disease and the microbial composition of subgingival biofilms. The results demonstrated that the periodontitis subjects had significantly higher levels of the 3 inflammatory mediators than the periodontally healthy subjects. There were statistically significant positive associations among clinical parameters of periodontal disease and all 3 biomarkers. These findings reinforce previous reports of associations between the levels of IL-1β and clinical signs of periodontal disease.14–16 Of the 3 mediators tested, IL-1β displayed the strongest relationship with clinical signs of periodontitis. IL-8 has also been previously detected in GCF at higher levels in periodontitis subjects compared to gingivitis and healthy subjects.17–19
The elevated levels of MMP-8 in GCF samples from periodontitis subjects in the present study support its association with clinical manifestations of periodontal disease previously reported.20 However, MMP-8 had the weakest correlation with clinical signs of periodontitis and it was primarily associated with signs of inflammation rather than parameters of tissue destruction. These findings corroborate previous studies reporting the correlation between GCF levels of MMP-8 with gingival and bleeding indices.21,22 It should be noted that metalloproteinases are secreted in a latent form, which requires partial proteolysis for its activation in the extracellular compartment. The CBIB does not distinguish between the active and latent forms of MMP-8. It is possible that measurements of the active form of MMP-8 would result in more significant associations with signs of tissue destruction.23
When different pocket depth categories were examined, the differences between groups were still present when only shallow sites from the two groups were compared. However, the so-called “shallow” sites differed considerably between the healthy and periodontitis subjects in other mean clinical parameters, such as recession, percentage of sites that bled on probing or exhibited gingival redness and even in mean pocket depth (data not shown). Therefore, we compared sites that would be considered “clinically healthy” i.e. sites with 2 or 3 mm of pocket depth, no loss of attachment no BOP, and no gingival redness. There were statistically significantly higher levels of IL-1β and IL-8 in the periodontitis subjects compared to periodontally healthy subjects at such sites. The GCF volume was also elevated at these “clinically healthy” sites from periodontitis subjects.
The higher levels of IL-1β and IL-8 in “clinically healthy” sites suggest that inflammatory mechanisms take place within the gingival tissues before they can be clinically detected. In agreement with these findings, Zhang et al. (2002)24 demonstrated in a gingivitis model that levels of IL-1β in GCF increased after only 3 days of plaque accumulation, before clinical signs of inflammation appeared. Our results are also in accord with data from Engebretson et al. (2002)15, who reported that subjects with severe periodontal disease had higher levels of GCF IL-1β in shallow sites compared to shallow sites in subjects with mild/moderate disease. They concluded that GCF IL-1β expression was in part a host trait, possibly explained by genetic mechanisms. Data from Figueredo et al. (1999)25 were consistent with this interpretation, indicating an increased level of IL-1β in periodontitis subjects regardless of the disease severity of the site. However, in chronic periodontitis, IL-1β is produced in response to bacterial stimuli and in order to distinguish between IL-1β elevations as a result of a host trait from IL-1β generated in response to the microbial burden, characterization of the bacterial challenge is necessary.
When the microbiota of the “clinically healthy” sites was compared between groups, it became clear that they also differed considerably. The proportions of beneficial species of the Purple, Yellow and Green complexes were statistically significantly higher in the periodontally healthy subjects, while the percentage of Orange and Red complex species was statistically significantly higher in the periodontitis group. These findings suggest that the higher expression of GCF inflammatory mediators in “clinically healthy” sites from periodontitis subjects could be explained by an elevated burden of periodontal pathogens in the adjacent subgingival biofilm.
The results of our microbiological analysis were consistent with previous reports of higher levels of Orange and Red complex species in chronic periodontitis subjects compared to periodontally healthy individuals. In addition, we confirmed the well established positive associations between Red and Orange complex species and clinical parameters of periodontitis. Interestingly, associations among GCF biomarkers and the proportion of different bacterial complexes in subgingival plaque samples revealed that the composition of the subgingival microbiota clearly was related to the expression of GCF cytokines. Early colonizers such as members of the Purple, Yellow and Green complexes presented statistically significant negative associations with the levels of the 3 biomarkers. Conversely, the percentage of Red and Orange complex species presented statistically significant positive correlations with the levels of GCF biomarkers.
Since we only examined cross-sectional associational data, it is premature to make inferences on cause-effect relationships; however, our data are in accord with the concept of “reciprocal interaction” described by Socransky & Haffajee (2005) 26. According to their hypothesis, addition of early colonizing species during microbial succession would lead to the development of gingival inflammation. In turn, the increased inflammation would alter the local environment resulting in further multiplication of colonizing species and fostering the growth of more pathogenic ones, leading to additional inflammation and, potentially, tissue destruction.
Smoking is a well establish environmental risk factor for periodontal diseases known to influence GCF flow and composition 17 and to affect the subgingival microbiota 27. In our study population, approximately the same proportion of subjects smoked in both groups. Although the mean number of cigarettes smoked per day was higher in the periodontitis group, the differences were not statistically significant. Therefore, smoking probably had a minor impact on the clinical, microbiological and immunological differences between groups. When smokers were compared with non-smokers in each group separately, in general, there were no statistically significant differences for the clinical, microbiological and immunological parameters (data not shown). This surprising finding suggests that, in our population, smoking was not a major modifier of periodontal disease manifestation, a finding at odds with the current literature. A potential explanation for these findings resides in the fact that the chronic periodontitis subjects came from a population with very little access to dental care and high levels of local irritants and gingival inflammation. The presence of local irritants and inflammation might have outweighed the impact of smoking on disease outcome. A similar mechanism was suggested by Neely et al. (2001) 28 to explain the fact that history of smoking was not significantly associated with attachment loss over time in Sri Lankan tea laborers receiving no oral health care.
In summary, our data strengthened the association described in the literature between GCF levels of these 3 inflammatory mediators and clinical signs of periodontitis. We also demonstrated that clinically healthy sites in periodontitis subjects had significantly higher levels of IL-1β and IL-8, an increased volume of GCF and a higher proportion of Orange and Red complex species than clinically healthy sites in healthy subjects. These results suggest that clinically healthy sites in periodontitis subjects might be colonized by higher levels of periodontal pathogens and be exposed to a sub-clinical level of inflammation, potentially leading to a higher risk for periodontal disease initiation and progression. In addition, our findings suggest that the expression of GCF biomarkers is influenced by the microbial composition of the adjacent subgingival biofilm.
Acknowledgments
This work was supported in part by research grants DE-016700, T32 DE-07327 and DE-14242 from the National Institute of Dental and Craniofacial Research.
Support: Research grants DE-016700, DE-012861 and DE-014242 from the National Institute of Dental and Craniofacial Research
Footnotes
Key findings: Clinically healthy sites from periodontitis subjects present higher levels of GCF biomarkers and periodontal pathogens than clinically healthy sites from periodontally healthy subjects.
The authors have no conflicts of interest to report related to this study.
Hu-Friedy®, Chicago, IL.
Periopaper®, Interstate Drug Exchange, Amityville, NY.
Periotron 8000®, Oraflow Inc., Plainview, NY.
Hybond-P, Amersham Biosciences, Arlington Heights, IL.
MiniSlot®, Immunetics, Cambridge, MA.
R&D systems, Minneapolis, MN.
VWR International Ltd. Poole, England.
Vector Laboratories, Burlingame, CA.
Miniblotter 45®, Immunetics, Cambridge, MA.
Manifold, Immunetics, Cambridge, MA.
LumiGLO ELITE®, KPL, Gaithersburg, MD.
Hyperfil ECL, Amersham, Arlington Heights, IL.
Kodak M35A X-OMAT Processor, Kodak, Rochester, NY.
EPSON 1640 Excel, Epson, Long Beach, CA.
TotalLab, Nonlinear Dynamics, Durham, NC.
AttoPhos, Amersham Life Sciences, Arlington Heights, Illinois, USA.
Storm Fluorimager, Molecular Dynamics, Sunnyvale, CA, USA.
References
- 1.Gursoy UK, Kononen E, Uitto VJ. Stimulation of epithelial cell matrix metalloproteinase (MMP-2, -9, -13) and interleukin-8 secretion by fusobacteria. Oral Microbiol Immunol. 2008;23:432–434. doi: 10.1111/j.1399-302X.2008.00453.x. [DOI] [PubMed] [Google Scholar]
- 2.Yumoto H, Nakae H, Yamada M, et al. Soluble products from Eikenella corrodens stimulate oral epithelial cells to induce inflammatory mediators. Oral Microbiol Immunol. 2001;16:296–305. doi: 10.1034/j.1399-302x.2001.016005296.x. [DOI] [PubMed] [Google Scholar]
- 3.Champagne CM, Buchanan W, Reddy MS, Preisser JS, Beck JD, Offenbacher S. Potential for gingival crevice fluid measures as predictors of risk for periodontal diseases. Periodontol 2000. 2003;31:167–180. doi: 10.1034/j.1600-0757.2003.03110.x. [DOI] [PubMed] [Google Scholar]
- 4.Soder B, Airila MS, Soder PO, Kari K, Meurman J. Levels of matrix metalloproteinases-8 and -9 with simultaneous presence of periodontal pathogens in gingival crevicular fluid as well as matrix metalloproteinase-9 and cholesterol in blood. J Periodontal Res. 2006;41:411–417. doi: 10.1111/j.1600-0765.2006.00888.x. [DOI] [PubMed] [Google Scholar]
- 5.Jin L, Soder B, Corbet EF. Interleukin-8 and granulocyte elastase in gingival crevicular fluid in relation to periodontopathogens in untreated adult periodontitis. J Periodontol. 2000;71:929–939. doi: 10.1902/jop.2000.71.6.929. [DOI] [PubMed] [Google Scholar]
- 6.Jin LJ, Soder PO, Leung WK, et al. Granulocyte elastase activity and PGE2 levels in gingival crevicular fluid in relation to the presence of subgingival periodontopathogens in subjects with untreated adult periodontitis. J Clin Periodontol. 1999;26:531–540. doi: 10.1034/j.1600-051x.1999.260807.x. [DOI] [PubMed] [Google Scholar]
- 7.Palys MD, Haffajee AD, Socransky SS, Giannobile WV. Relationship between C-telopeptide pyridinoline cross-links (ICTP) and putative periodontal pathogens in periodontitis. J Clin Periodontol. 1998;25:865–871. doi: 10.1111/j.1600-051x.1998.tb02383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee E, Yang YH, Ho YP, Ho KY, Tsai CC. Potential role of vascular endothelial growth factor, interleukin-8 and monocyte chemoattractant protein-1 in periodontal diseases 1. Kaohsiung J Med Sci. 2003;19:406–415. doi: 10.1016/S1607-551X(09)70484-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teles RP, Sakellari D, Konstantinidis A, Socransky SS, Haffajee AD. Application of the checkerboard immunoblotting technique to the quantification of host biomarkers in gingival crevicular fluid. J Periodontol. 2009;80:447–456. doi: 10.1902/jop.2009.080440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapple IL, Landini G, Griffiths GS, Patel NC, Ward RS. Calibration of the Periotron 8000 and 6000 by polynomial regression. J Periodontal Res. 1999;34:79–86. doi: 10.1111/j.1600-0765.1999.tb02226.x. [DOI] [PubMed] [Google Scholar]
- 11.Socransky SS, Smith C, Martin L, Paster BJ, Dewhirst FE, Levin AE. "Checkerboard" DNA-DNA hybridization. Biotechniques. 1994;17:788–792. [PubMed] [Google Scholar]
- 12.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 13.Socransky SS, Haffajee AD, Smith C, Dibart S. Relation of counts of microbial species to clinical status at the sampled site. J Clin Periodontol. 1991;18:766–775. doi: 10.1111/j.1600-051x.1991.tb00070.x. [DOI] [PubMed] [Google Scholar]
- 14.Engebretson SP, Lamster IB, Herrera-Abreu M, et al. The influence of interleukin gene polymorphism on expression of interleukin-1beta and tumor necrosis factor-alpha in periodontal tissue and gingival crevicular fluid. J Periodontol. 1999;70:567–573. doi: 10.1902/jop.1999.70.6.567. [DOI] [PubMed] [Google Scholar]
- 15.Engebretson SP, Grbic JT, Singer R, Lamster IB. GCF IL-1beta profiles in periodontal disease. J Clin Periodontol. 2002;29:48–53. doi: 10.1034/j.1600-051x.2002.290108.x. [DOI] [PubMed] [Google Scholar]
- 16.Hou LT, Liu CM, Rossomando EF. Crevicular interleukin-1 beta in moderate and severe periodontitis patients and the effect of phase I periodontal treatment. J Clin Periodontol. 1995;22:162–167. doi: 10.1111/j.1600-051x.1995.tb00128.x. [DOI] [PubMed] [Google Scholar]
- 17.Giannopoulou C, Cappuyns I, Mombelli A. Effect of smoking on gingival crevicular fluid cytokine profile during experimental gingivitis. J Clin Periodontol. 2003;30:996–1002. doi: 10.1034/j.1600-051x.2003.00416.x. [DOI] [PubMed] [Google Scholar]
- 18.Mathur A, Michalowicz B, Castillo M, Aeppli D. Interleukin-1 alpha, interleukin-8 and interferon-alpha levels in gingival crevicular fluid. J Periodontal Res. 1996;31:489–495. doi: 10.1111/j.1600-0765.1996.tb01414.x. [DOI] [PubMed] [Google Scholar]
- 19.Tsai CC, Ho YP, Chen CC. Levels of interleukin-1 beta and interleukin-8 in gingival crevicular fluids in adult periodontitis. J Periodontol. 1995;66:852–859. doi: 10.1902/jop.1995.66.10.852. [DOI] [PubMed] [Google Scholar]
- 20.Ingman T, Tervahartiala T, Ding Y, et al. Matrix metalloproteinases and their inhibitors in gingival crevicular fluid and saliva of periodontitis patients. J Clin Periodontol. 1996;23:1127–1132. doi: 10.1111/j.1600-051x.1996.tb01814.x. [DOI] [PubMed] [Google Scholar]
- 21.Chen HY, Cox SW, Eley BM, Mantyla P, Ronka H, Sorsa T. Matrix metalloproteinase-8 levels and elastase activities in gingival crevicular fluid from chronic adult periodontitis patients. J Clin Periodontol. 2000;27:366–369. doi: 10.1034/j.1600-051x.2000.027005366.x. [DOI] [PubMed] [Google Scholar]
- 22.Kiili M, Cox SW, Chen HY, et al. Collagenase-2 (MMP-8) and collagenase-3 (MMP-13) in adult periodontitis: molecular forms and levels in gingival crevicular fluid and immunolocalisation in gingival tissue. J Clin Periodontol. 2002;29:224–232. doi: 10.1034/j.1600-051x.2002.290308.x. [DOI] [PubMed] [Google Scholar]
- 23.Mancini S, Romanelli R, Laschinger CA, Overall CM, Sodek J, McCulloch CA. Assessment of a novel screening test for neutrophil collagenase activity in the diagnosis of periodontal diseases. J Periodontol. 1999;70:1292–1302. doi: 10.1902/jop.1999.70.11.1292. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Kashket S, Lingstrom P. Evidence for the early onset of gingival inflammation following short-term plaque accumulation. J Clin Periodontol. 2002;29:1082–1085. doi: 10.1034/j.1600-051x.2002.291206.x. [DOI] [PubMed] [Google Scholar]
- 25.Figueredo CM, Ribeiro MS, Fischer RG, Gustafsson A. Increased interleukin-1beta concentration in gingival crevicular fluid as a characteristic of periodontitis. J Periodontol. 1999;70:1457–1463. doi: 10.1902/jop.1999.70.12.1457. [DOI] [PubMed] [Google Scholar]
- 26.Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol 2000. 2005;38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- 27.Haffajee AD, Socransky SS. Relationship of cigarette smoking to the subgingival microbiota. J Clin Periodontol. 2001;28:377–388. doi: 10.1034/j.1600-051x.2001.028005377.x. [DOI] [PubMed] [Google Scholar]
- 28.Neely AL, Holford TR, Loe H, Anerud A, Boysen H. The natural history of periodontal disease in man. Risk factors for progression of attachment loss in individuals receiving no oral health care. J Periodontol. 2001;72:1006–1015. doi: 10.1902/jop.2001.72.8.1006. [DOI] [PubMed] [Google Scholar]