Abstract
Sexual-stage glycoproteins of Eimeria are important components of the oocyst wall, a structure that ensures the efficient transmission of these and related parasites. In this study, the primary enzyme in the glycosylation pathway of Eimeria tenella, glucosamine:fructose-6-phosphate aminotransferase (EtGFAT), has been characterized as a macrogamete-specific protein. Although the transcription of EtGFAT was observed early in macrogamete development, protein expression was restricted to mature macrogametes, prior to their conversion into unsporulated oocysts. Genes coding for three other enzymes required for N-acetylgalactosamine (GalNAc) synthesis were also transcribed during E. tenella macrogamete development. Gene transcription of the enzyme responsible for the O-linked transfer of GalNAc to proteins, EtGalNAc-T, was upregulated primarily in unsporulated oocyst stages, and accordingly, a significant increase in GalNAc levels was observed in E. tenella gametocytes and oocysts. Gam56 and Gam82, two well-characterized glycoproteins of Eimeria macrogametes and the oocyst wall, contain high levels of GalNAc and represent probable targets of GalNAc O linkage. It appears that the glycosylation pathway, specifically relating to the formation of GalNAc O links, is dramatically upregulated in E. tenella sexual stages and may play a role in directing a number of macrogamete proteins to the developing oocyst wall.
Eimeria tenella is a protozoan parasite of chickens and a member of the Apicomplexa, a phylum that includes many important human pathogens, such as Plasmodium, Cryptosporidium, and Toxoplasma. Infection with one of seven species of Eimeria can cause coccidiosis, a highly contagious disease that is estimated to cost the broiler industry in excess of $1.5 billion per annum (36). The extremely infectious nature of these parasites can be attributed primarily to the durability of the excreted oocyst. Facilitating protection against detrimental forces, both physical and chemical (18), is the bilayered oocyst wall, a structure composed of a 40-nm outer layer and a 200-nm inner layer (4). The development of the oocyst and its protective wall is preceded by the formation and maturation of male and female gametes, known as microgametes and macrogametes, respectively.
Microscopic analyses have consistently demonstrated that wall-forming bodies I (WFBI) and II (WFBII) form the outer and inner layers of the oocyst wall, respectively (4, 13, 32). Furthermore, several proteins have been identified that are detected both in macrogamete WFBs and in the oocyst wall. The best characterized of these are EmGam56 and EmGam82, glycoproteins of Eimeria maxima, which are components of WFBII and, accordingly, the inner oocyst wall (13). EmGam56 and EmGam82 are processed into smaller peptides prior to incorporation into the oocyst wall, where proposed cross-linking occurs to ensure the formation of a stable extracellular matrix (5, 6). EtGam56, an E. tenella homologue of EmGam56, appears to undergo a similar process of truncation prior to oocyst wall formation (19, 25), implying that this mechanism is strongly conserved within the Eimeria genus.
The contents of the Eimeria oocyst wall were originally thought to consist of a high proportion of glycoproteins (37), a theory supported by biochemical analyses of EmGam56 and EmGam82 that revealed high concentrations of the amino sugar (glycan) galactosamine. It is unclear why these wall-forming proteins require glycosylation; however, the strong conservation of O-glycosylation motifs in both EmGam56 and EtGam56 (19) implies that it is an important process. N-Acetylgalactosamine (GalNAc) has also been implicated as the likely glycan moiety of glycoproteins present in the oocyst wall of the related apicomplexan, Cryptosporidium parvum, with a proposed role in host cell attachment (20, 35).
A key player in amino sugar biosynthesis and, potentially, glycosylation is glucosamine:fructose-6-phosphate aminotransferase (GFAT) (10), an enzyme found among all known organisms (12). Intriguingly, this enzyme has been found in Plasmodium berghei to be specific for macrogametes (17). We report here on the identification and characterization of a highly conserved E. tenella-specific GFAT (EtGFAT), proposing a role for the enzyme in macrogametocyte development and oocyst wall formation. We analyzed the gene transcription, protein expression, and localization within different developmental stages of E. tenella. In addition, the gametocyte-specific transcription of genes coding for other E. tenella glycosylation enzymes was also characterized to illustrate potential synchronicities. Finally, the relative levels of the two amino sugars, N-acetylglucosamine (GlcNAc) and GalNAc, were determined in the different E. tenella stages with additional emphasis on detecting unique glycosylated proteins.
MATERIALS AND METHODS
Bioinformatic analysis.
The P. berghei GFAT protein (PB000496.03.0) was identified as an exclusive protein of macrogametes through analysis of the supplementary data provided by Khan et al. (17) at http://www.cell.com/cgi/content/full/121/5/675/DC1/. The identification of putative glycosylation enzymes was carried out by using the BLASTp tool (1) available at the online predicted protein databases of E. tenella (http://www.genedb.org/genedb/etenella/), Toxoplasma gondii (http://toxodb.org), and C. parvum (http://cryptodb.org). The alignment of similar protein or nucleic acid sequences was performed by using CLUSTAL W (version 1.83) through the publicly available European Bioinformatics Institute website (http://www.ebi.ac.uk/Tools/clustalw/index.html) according to default settings. The calculation of molecular weights and prediction of antigenicity for protein sequences was performed by using Protean (Lasergene 7 Software Suite for Sequence Analysis; DNASTAR, Inc.). Catalytic domains of GFAT were identified by using the ScanProsite tool (release 20.22) (14), while the locations of signal peptides were predicted with the SignalP 3.0 Server (8).
Animals and parasites.
Australorp cockerels (Barter and Sons Pty., Ltd., Huntingwood, New South Wales, Australia) were purchased at 1 day of age and kept at 21°C with a 12-h light/dark cycle and with free access to food and water. Merozoites and unsporulated oocysts were purified from E. tenella-infected chicken ceca at 112 h postinfection (p.i.) and 168 h p.i, respectively, and oocysts sporulated where necessary, as described previously (34). Gametocytes were also purified from infected chicken ceca at 134 h p.i. by using the method described previously for E. maxima (30). Whole cecal samples were also taken at 134 and 144 h p.i. for immunolocalization studies. The samples were stored in 2% paraformaldehyde-0.1 M phosphate buffer (pH 7.4) until sectioning. Uninfected chicken cecal scrapings were also prepared as a negative control for reverse transcription-PCR (RT-PCR) and protein analyses.
RT-PCR.
Total RNA was purified from different E. tenella samples by using TRIzol reagent (Invitrogen). Unsporulated and sporulated oocysts were vortexed with an equal volume of glass beads (710 to 1,180 μm; Sigma) prior to RNA extraction until cell lysis could be observed for >90% of each sample. The analysis of major rRNA bands was performed with an Experion automated electrophoresis system (Bio-Rad) using the Experion RNA StdSens chip. Total RNA samples were treated with DNase I (RNase-free) (Roche), for 1 h at 37°C, and the reaction was stopped with the addition of 2.5 mM EDTA, followed by a 10-min incubation at 75°C. The synthesis of cDNA was carried out by using a SuperScript III first-strand synthesis system for RT-PCR using oligo(dT)20 primer. The E. tenella cDNA samples were standardized to comparable levels of EtActin transcription by amplifying the gene from different dilutions (1/5, 1/25, and 1/100) of each sample at 25 cycles. PCR was carried out with Advantage 2 polymerase (Clontech) using gene-specific primers at a final concentration of 0.2 μM. All PCR amplification was carried out using the following program: denaturation (95°C, 3 min) for 1 cycle and then denaturation (95°C, 30 s), annealing (65°C, 1 min), and extension (68°C, 3 min) for 25 to 35 cycles. The design of oligonucleotide primers was based on genomic sequence obtained from the E. tenella Genome Project (http://www.sanger.ac.uk/Projects/E_tenella/http://www.sciencedirect.com/science?_ob=RedirectURL&_method=externObjLink&_locator=url&_cdi=6626&_plusSign=%2B&_targetURL=http%253A%252F%252Fwww.sanger.ac.uk%252FProjects%252FE_tenella%252F) using the xx/xx/200x assembly. The names and sequences of forward and reverse primers used to amplify each E. tenella gene are detailed in Table 1. PCR products were analyzed by agarose gel electrophoresis using GelRed for nucleic acid visualization.
TABLE 1.
Primers used for the amplification of E. tenella gene transcripts
| Gene | Primer | Sequence (5′-3′) |
|---|---|---|
| EtGFAT | Et57F | GGGCGATGGGCAGCTTACTACAAC |
| Et58R | TCAAATGAACGCCCAAGGTCCAG | |
| EtAGM | Et155F | GCAGGTGGGAGTTGTACAGACAGC |
| Et156R | CTGCAACTGCGGGGAATTCG | |
| EtUAP | Et157F | GCTTTGAAGGCCCCAAAGG |
| Et158R | GGCAGTTGTCGACAGGGAGG | |
| EtUAE | Et167F | GCTACATCGGCAGCCACACG |
| Et168R | CACTGGCAGCAAATTGTTTGGC | |
| EtGalNAc-T | Et165F | GACGAGTACTTGCCTTACCTCCCC |
| Et166R | CACGCGGTTCTTCCACACG | |
| EtActin | E0043 | GGAATTCGTTGGCCGCCCAAAGAATCC |
| E0044 | GCTCTAGATTAGCTCGGCCCAGACTCATC | |
| EtGam56 | E0035 | CATATGGTAGAAGTGCCAATGGAC |
| E0036 | CACGTGTTAGTAGAAGCTGGAGTGGCT |
Sequencing.
Each PCR product was excised from an agarose gel and the DNA purified by using a QIAquick gel extraction kit (Qiagen). The purified DNA was then cloned into the sequencing vector, pCR4-TOPO, using a TOPO TA cloning kit (Invitrogen). The recombinant plasmids were sequenced by using the forward and reverse M13 primers (vector specific) at the Australian Genome Research Facility (Brisbane, Australia). Sequence data were analyzed by using the SeqMan program (Lasergene 7 Software Suite for Sequence Analysis; DNASTAR, Inc.) and aligned to the original gene sequences by using CLUSTAL W (described above).
Generation of antisera for EtGFAT detection.
A short peptide (CNPDKPRGLAKTVTVS-NH2) was designed to the last 15 amino acids of the EtGFAT protein sequence (C terminal). The peptide was synthesized by Auspep Pty., Ltd. (Melbourne, Australia) to a purity of 82%. The N-terminal cysteine residue was added to facilitate conjugation to keyhole limpet hemocyanin (KLH) carrier protein (also performed at Auspep). A single rabbit was immunized at 0, 3, 6, and 9 weeks with 500-μg doses of the KLH-conjugated EtGFAT peptide, administered by subcutaneous injection at several different sites, by the Institute of Medical Veterinary Sciences (Adelaide, Australia). Antigen used for the first inoculation was emulsified in complete Freund adjuvant, whereas antigen for subsequent inoculations was emulsified in incomplete Freund adjuvant. Prebleed sera was taken prior to the first inoculation (negative control), while the anti-EtGFAT sera were collected by exsanguination at 10.5 weeks.
SDS-PAGE and transfer.
Parasite purifications and negative control chicken ceca to be analyzed by Western blotting were prepared under reducing and alkylating conditions as follows: samples were resuspended in reducing buffer (6 M guanidinium HCl, 50 mM Tris-HCl, 10 mM EDTA, 10 mM dithiothreitol [pH 8.6]) and incubated for 2 h at 60°C, after which a 1/4 volume of alkylating buffer (reducing buffer with 200 mM iodoacetamide) was added, and samples incubated for an additional 30 min in the dark. The reaction was stopped by the addition of 250 mM dithiothreitol, and insoluble material removed by 15 min of centrifugation (10,000 × g). Biological samples to be analyzed by lectin blotting were prepared under reducing conditions by resuspending in 2× SDS reducing buffer (100 mM Tris-HCl [pH 6.8], 20% glycerol, 4% SDS, 200 mM dithiothreitol). The initial lysis of oocyst samples was achieved by vortexing with glass beads (as described earlier). All protein samples were supplemented with 0.02% bromophenol blue and heated for 5 min at 95°C prior to electrophoresis. Approximately 5 μg of each protein sample (quantified by using a NanoDrop ND-1000 spectrophotometer) was electrophoresed at 180 V for 1 h on a 4 to 12% polyacrylamide gel (NuPAGE Bis-Tris gel; Invitrogen) using morpholine ethanesulfonic acid (MES) buffer (Invitrogen). A SeeBlue Plus2 prestained standard (Invitrogen) was run for size estimation. One gel was stained with Flamingo fluorescent gel stain (Bio-Rad) to ensure that equal quantities of total protein had been loaded into each well. Proteins were transferred to polyvinylidene difluoride membrane for 1 h at 100 V according to protocols published previously (16).
Immunoblotting and lectin blotting.
Membranes used for immunoblotting were blocked, washed, and probed with primary and secondary antibodies as described previously (3). The rabbit anti-EtGFAT and mouse anti-EmGam56 (5) primary antibodies were used at dilutions of 1 in 100 and 1 in 200, respectively. The rabbit and mouse primary antibodies were detected with anti-rabbit and anti-mouse IgG alkaline phosphatase-conjugated secondary antibodies, respectively, at a dilution of 1 in 1,000. Membranes used for lectin blotting were blocked for 30 min at 37°C in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4 [pH 7.4]) supplemented with 5% gelatin. The membrane was washed twice for 4 min in PBS-0.05% Tween 20 and once for 4 min in PBS. Each membrane was probed with 10 μg of biotinylated lectin (Vector Laboratories)/ml in PBS-1% gelatin-0.05% Tween 20 (blocking buffer) for 1 h at room temperature with constant agitation. Wheat germ agglutinin (WGA) lectin was used to detect N-acetylglucosamine, and Jacalin (JAC) lectin was used to detect N-acetylgalactosamine. Each membrane was washed and probed (as described above) with ExtrAvidin alkaline phosphatase (Sigma) at 1 in 2,000 in blocking buffer. Membranes were washed and glycosylated proteins detected using Fast 5-bromo-4-chloro-3-indolylphosphate (BCIP)/nitroblue tetrazolium (NBT) substrate (Sigma) according to the manufacturer's instructions.
Immunofluorescence microscopy.
Paraffin sections were cut from fixed samples of E. tenella (134 and 144 h p.i) to a thickness of 4 μm. The tissue was rehydrated by soaking the slides in xylol three times for 10 min each time, 100% ethanol twice for 3 min each time, 95% ethanol for 3 min, 90% ethanol for 3 min, 70% ethanol for 3 min, and citrate buffer (10 mM [pH 6.0]) three times for 10 min each time. Antigen retrieval was carried out using a 900-W microwave oven as follows: samples were boiled three times for 5 min each time in citrate buffer on medium power and for 10 min in Tris-HCl buffer (100 mM, pH 10.0) on low power. Rabbit anti-EtGFAT (1 in 100) and mouse anti-EmGam56 (1 in 200) were diluted in PBS-3% bovine serum albumin-0.05% Tween 20 (blocking buffer), added to slides, and incubated overnight at 4°C in a humidified chamber. The slides were washed the following day with PBS three times for 10 min each time. Goat anti-rabbit IgG-fluorescein isothiocyanate (FITC) conjugate (Sigma) and goat anti-mouse IgG-Texas Red (Invitrogen) secondary antibodies were diluted 1 in 100 in blocking buffer and applied to tissue sections for 1 h at room temperature (in the dark). Slides were washed twice in PBS for 10 min, counterstained with 1 μg of DAPI (4′,6′-diamidino-2-phenylindole)/ml for 5 min, and washed twice more. The tissue sections were mounted in PBS-50% glycerol under a glass coverslip and visualized with a Research BX51 microscope (Olympus Optical Co., Ltd.). FITC-conjugated antibodies were visualized by using a 470- to 490-nm excitation filter, Texas Red-conjugated antibodies were visualized by using a 520- to 550-nm excitation filter, and DAPI-positive cells (i.e., E. tenella merozoites, microgametes, and host cell nuclei) were visualized by using a 330- to 385-nm excitation filter. The acquisition of images and the overlay of images visualized under different filters were performed using the DP Controller software (Olympus Optical).
Lectin microtiter assay.
Protein lysates, prepared for lectin blotting (as described above), were diluted to 5 ng/μl (for GalNAc detection) or 1 ng/μl (GlcNAc) in enzyme-linked immunosorbent assay (ELISA) buffer I (0.59% NaHCO3, 0.31% Na2CO3, 0.02% NaN3 [pH 9.6]) and added in quadruplicate (100 μl/well) to a 96-well, U-bottom, ELISA microplate (Griener Bio-One). After an overnight incubation at 4°C, wells were rinsed twice in blocking buffer (PBS, 0.05% Tween 20), once in PBS, and blocked for 1 h at room temperature with PBS-0.5% Tween 20. Wells were rinsed (as described above) and probed for 1 h at room temperature with WGA (diluted to 10 μg/ml in blocking buffer) or JAC (1 μg/ml) biotinylated lectin. Wells were rinsed (as described above) and probed for 1 h at room temperature with ExtrAvidin alkaline phosphatase conjugate (1 in 10,000 in blocking buffer). Plates were rinsed as described above, and 200 μl of p-nitrophenyl phosphate disodium substrate, prepared at 1 mg/ml in 1 M diethanolamine (pH 9.8), was added to each well. After 30 min, the absorbance values were read at 405 nm by using a PowerWave HT microplate scanning spectrophotometer (BioTek Instruments, Inc.).
Statistical analysis.
One-way and two-way analysis of variance (ANOVA) and Bonferroni's multiple comparison posttests were carried out on lectin microtiter assay results. All tests were conducted on quadruplicate samples using the GraphPad Prism version 5.00 program (GraphPad Software, Inc.).
Nucleotide sequence accession numbers.
The partial mRNA and corresponding amino acid sequences of EtGFAT, EtAGM, EtUAP, EtUAE, and EtGalNAc-T were deposited into GenBank under accession numbers GQ463153 to GQ463157, respectively.
RESULTS
Identification and conservation of EtGFAT.
The GFAT protein of P. berghei (PB000496.03.0 [PbGFAT]) has previously been detected in macrogametes through high-throughput proteomics (17). Here, homologues of PbGFAT were identified in E. tenella and the related Apicomplexa, Toxoplasma gondii and C. parvum, using BLASTp analysis of their respective predicted protein databases. A putative GFAT protein was identified in E. tenella showing a high level of homology to PbGFAT of 8.4 E−177 and is referred to as EtGFAT. The GFAT proteins in T. gondii (TgGFAT) and C. parvum (CpGFAT) also showed high levels of sequence homology to PbGFAT at 6.7 E−164 and 1.4 E−176, respectively.
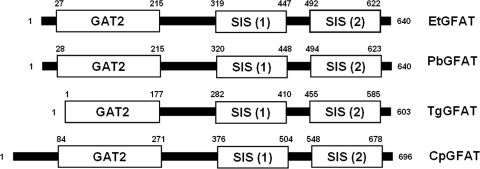
The EtGFAT predicted protein sequence is 640 amino acids long with a predicted N-terminal signal peptide truncating the predicted mature protein to 625 residues. PbGFAT, TgGFAT, or CpGFAT were not predicted to contain signal peptide sequences. The mature EtGFAT sequence was calculated to have a molecular mass of 67.7kDa. Pfam analysis of the EtGFAT protein sequence revealed the presence of a glutamine aminotransferase class II motif (GAT2; Pfam-A: PF00310) and two sugar isomerase motifs (SIS; Pfam-A: PF01380). These catalytic domains are strongly conserved in the four apicomplexan GFAT sequences (Fig. 1), and their relative positions are consistent with other GFAT enzymes (12), although the GAT domain of TgGFAT is incomplete.
FIG. 1.
Conservation of GFAT catalytic domains in the apicomplexan glucosamine:fructose-6-phosphate aminotransferases. Schematic representations of the predicted protein sequences for EtGFAT (E. tenella, SNAP00000005021), PbGFAT (P. berghei, PB000392.03.0), TgGFAT (T. gondii, TGGT1_116350), and CpGFAT (C. parvum, cgd1_3730) show the location of the class II glutamine aminotransferase, “GAT2,” and two sugar isomerase motifs, “SIS (1)” and “SIS (2),” marked as boxes with the first and last residue numbered accordingly.
Sexual-stage-specific expression of EtGFAT.
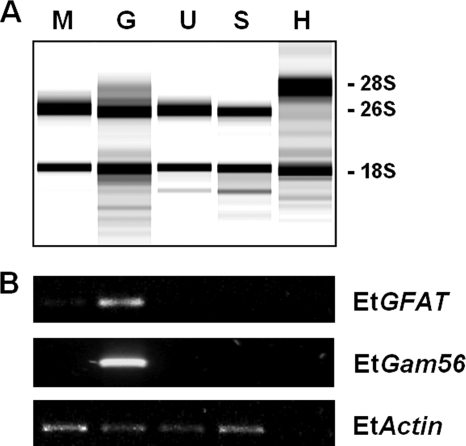
Further investigation was focused on testing the hypothesis that EtGFAT, like PbGFAT, is developmentally expressed in macrogametes. Total RNA was isolated from purified E. tenella merozoites, gametocytes, unsporulated oocysts, and sporulated oocysts and from uninfected host cecal scrapings (negative control). The size difference between the large rRNA subunit of Gallus gallus (28S) and E. tenella (26S) has been described previously (31) and was utilized here to assess the level of host contamination in the different parasite purifications. The presence of a faint host-specific 28S rRNA band in the gametocyte sample is consistent with microscopic examination of gametocyte purifications where host contamination was commonly observed (data not shown); however, purified merozoite and unsporulated and sporulated oocyst samples were free of contaminating host RNA (Fig. 2A).
FIG. 2.
Relative transcription of EtGFAT in different developmental stages of E. tenella. (A) Analysis of total RNA purified from E. tenella and host tissue using automated electrophoresis. The major rRNA bands, including the 28S (host-specific), 26S (parasite-specific), and 18S (parasite and host), rRNA bands, are marked accordingly. (B) RT-PCR analysis of EtGFAT and EtGam56 transcription. Prior to RT-PCR, each cDNA sample was standardized to comparable levels of EtActin transcription. Merozoites (M), gametocytes (G), unsporulated oocysts (U), sporulated oocysts (S), and uninfected host control (H) are as indicated.
The gene encoding EtGFAT (EtGFAT) was transcribed almost exclusively in E. tenella gametocytes, with only faint transcription detected in late merozoite stages (112 h p.i.) and none in unsporulated and sporulated oocyst stages (Fig. 2B). The EtGFAT gene amplified at the expected size of 339 bp, and sequencing of the purified DNA confirmed the identity of the amplicon (100% identity to SNAP00000005021). The transcription profile of EtGFAT was very similar to that of EtGam56, the gene encoding the macrogamete-specific protein EtGam56, although the transcription of EtGFAT is detected earlier than EtGam56. As expected, none of the E. tenella-specific genes were amplified from uninfected chicken cDNA.
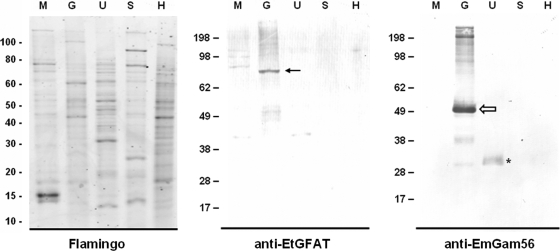
To determine whether the increased transcription of EtGFAT in E. tenella gametocytes corresponded to increased expression of the native EtGFAT protein, antiserum was developed against a synthetic peptide representing the last (i.e., C-terminal) 15 amino acids of EtGFAT. KLH was conjugated to the peptide antigen prior to immunization to promote immunostimulation and optimum antibody production. ELISA analysis of the anti-EtGFAT antisera revealed highly specific reactivity against the unconjugated EtGFAT synthetic peptide compared to the reactivity of prebleed sera from the same animal (data not shown). Western blot analysis revealed an 80-kDa protein exclusively in gametocytes (Fig. 3), a finding consistent with the RT-PCR results (Fig. 2B). This is interpreted as the native EtGFAT protein, albeit at a larger size than the 67.7 kDa calculated from the mature protein sequence. However, it should be noted that the antisera also reacted slightly with merozoite proteins at approximately 85, 100, and 120 kDa. As expected, the native EtGam56 (51 kDa) and truncated EtGam56 (32 kDa) were detected in E. tenella gametocytes and unsporulated oocysts, respectively (Fig. 3), using antisera previously developed against EmGam56, the E. maxima homologue of EtGam56 (5). No prominent bands were detected in any of the five protein samples when we used either prebleed antisera or anti-KLH antibody (data not shown).
FIG. 3.
Relative expression of EtGFAT in different developmental stages of E. tenella. Immunoblot analysis of whole-cell protein lysates from E. tenella was performed, and host tissues were probed with antisera developed against either EtGFAT or EmGam56. The native EtGFAT (closed black arrow), the native EtGam56 (open white arrow), and the truncated EtGam56 protein (asterisk) are marked accordingly. An identical SDS-PAGE gel was stained for total protein using Flamingo fluorescent gel stain to demonstrate comparable protein loading. Size markers (in kilodaltons) are displayed to the left of each immunoblot. Merozoites (M), gametocytes (G), unsporulated oocysts (U), sporulated oocysts (S), and uninfected host control (H) are as indicated.
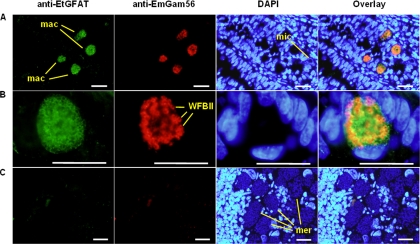
EtGFAT is localized to E. tenella macrogametes.
Sections prepared from E. tenella-infected chicken intestine were probed with anti-EtGFAT antisera in order to localize the expression of the native protein in situ. The sections were also probed with anti-EmGam56 to ensure the positive identification of E. tenella macrogametes. Like anti-EmGam56, anti-EtGFAT localized specifically to E. tenella macrogametes (DAPI negative) at 144 h p.i. and displayed negligible binding to surrounding host tissue and E. tenella microgametes, both of which were stained positively with DAPI (Fig. 4A). Closer examination of E. tenella macrogametes revealed that EtGam56 localized to WFBII (Fig. 4B), a finding consistent with anti-EmGam56 localization within E. maxima macrogametes (13). In contrast, EtGFAT localization was more diffuse, and it is not clear whether the protein is restricted to any specific organelle (Fig. 4B). Populations of merozoites (DAPI positive), also present in the 144-h p.i. section, were not detected with either the anti-EtGFAT or anti-EmGam56 antisera (Fig. 4C), which is consistent with the findings of Western blot analyses (Fig. 3). Although EtGam56 was also detected in macrogametes at the earlier postinfection time point of 134 h p.i., EtGFAT was detected only at 144 h p.i. (data not shown).
FIG. 4.
Immunolocalization of native EtGFAT in E. tenella. Paraffin sections of E. tenella-infected chicken intestine were taken at 144 h p.i. and stained with rabbit antisera developed against EtGFAT and mouse antisera developed against EmGam56. EtGFAT was detected with an FITC-conjugated anti-rabbit antibody (green) and EmGam56 with a Texas Red-labeled anti-mouse antibody (red). The sections were counterstained with DAPI (blue) for the detection of host, merozoite, and microgamete nuclei. An overlay of all three colors is shown for each image. (A) Region of tissue infected with E. tenella macrogametes (mac) and microgametes (mic); (B) closer examination of an E. tenella macrogamete; (C) region of infected tissue with E. tenella merozoites (mer). Bars, 10 μm.
Glycosylation enzymes downstream of EtGFAT also display gametocyte-specific transcription.
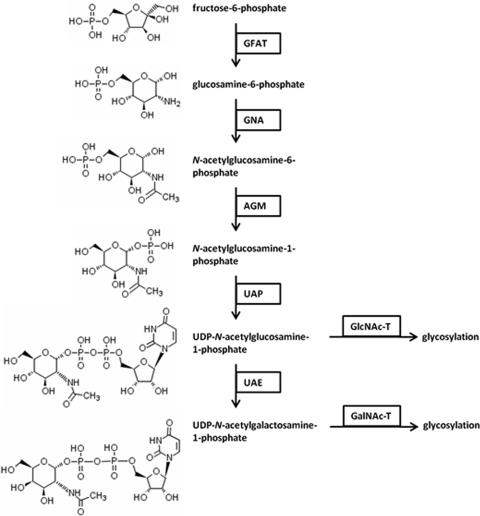
GFAT represents the first enzyme in the amino sugar biosynthesis and glycosylation pathway illustrated in Fig. 5. BLASTp analysis using previously characterized glycosylation enzymes from other organisms uncovered all but two of these enzymes (glucosamine-6-phosphate N-acetyltransferase [GNA] and GlcNAc-T) in the E. tenella predicted protein database (Table 2). No E. tenella homologue could be identified for either the Giardia intestinalis (21) or Mus musculus (9) GNA protein sequence. The primary E. tenella hit for the Caenorhabditis elegans GlcNAc-T (22) was not considered a true orthologue because it was already annotated as a putative stress-induced protein and showed only 24% identity. Although the primary E. tenella hit for the Dictyostelium discoideum GlcNAc-T (42) returned an E value of 2.2 E−34, a majority of the alignment was to a near continuous stretch of asparagine residues (approximately 60 Asp over a 70-amino-acid region), and when this region was removed from the BLASTp query sequence, no homologue was identified. The E. tenella phosphoacetylglucosamine mutase (EtAGM) and E. tenella UDP-N-acetylglucosamine 4-epimerase (EtUAE) protein sequences were identified by using reference sequences obtained from the annotated genome database of C. parvum, whereas the E. tenella UAP-N-acetylglucosamine pyrophosphorylase (EtUAP) and EtGalNAc-T were identified using reference proteins from G. intestinalis (21) and T. gondii (38), respectively.
FIG. 5.
Schematic representation of the amino sugar biosynthesis and glycosylation pathway. The name of each carbohydrate substrate and product is listed to the right of its chemical structure. The abbreviated name of the enzyme responsible for each step in the pathway is listed below the carbohydrate substrate in a black box. GFAT, glucosamine:fructose-6-phosphate aminotransferase; GNA, glucosamine-6-phosphate N-acetyltransferase; AGM, phosphoacetylglucosamine mutase; UAP, UDP-N-acetylglucosamine pyrophosphorylase; UAE, UDP-N-acetylglucosamine 4-epimerase; GlcNAc-T, N-acetylglucosaminyl transferase; GalNAc-T, N-acetylgalactosaminyl transferase.
TABLE 2.
Identification of putative amino sugar biosynthesis and glycosylation enzymes in E. tenella
| Enzymea | Reference sequence (organism)b | E. tenella homologue | BLAST E-valuec |
|---|---|---|---|
| GFAT | PB000496.03.0 (Plasmodium berghei) | SNAP00000005021 | 8.4 E−177 |
| GNA | AY185915 (Giardia intestinalis) | None | 0.53* |
| NP_062298 (Mus musculus) | None | 0.99* | |
| AGM | cgd4_3310 (Cryptosporidium parvum) | SNAP00000000160 | 7.5 E−28 |
| UAP | AY187035 (Giardia intestinalis) | SNAP00000006592 | 2.2 E−13 |
| UAE | cgd4_2600 (Cryptosporidium parvum) | EIMER_contig00031419.(Exonerate2BLASTCDS.Gene.15) | 4.5 E−91 |
| GlcNAc-T | U77412 (Caenorhabditis elegans) | NAd | 1.0 E−09* |
| AF375997 (Dictyostelium discoideum) | NA | 2.2 E−34* | |
| GalNAc-T | AY424890 (Toxoplasma gondii) | EIMER_contig_00030594 (Exonerate2BLASTCDS.Gene.387) | 3.0 E−101 |
Enzyme names have been abbreviated as listed in the legend to Fig. 5.
Reference enzyme protein sequences are from the organisms listed in parentheses.
*, BLAST hits not considered to represent true homologues (see the text).
NA, not applicable.
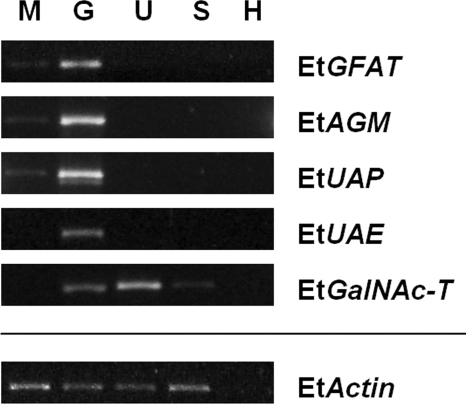
The developmental transcription of the genes encoding these newly uncovered E. tenella glycosylation enzymes was investigated and compared to that of EtGFAT. Interestingly, the gene transcription of all four enzymes (including EtGFAT) involved in the synthesis of the N-acetylgalactosamine (GalNAc) glycan displayed very similar profiles, with the most prominent transcription detected in the gametocyte stages (Fig. 6). Additional transcription was detected at much lower levels in merozoites for EtGFAT, EtAGM, and EtUAP but not for EtUAE, whereas no transcription of any of these four genes was observed in either unsporulated or sporulated oocysts. In contrast, the transcription of EtGalNAc-T, the gene encoding the enzyme responsible for the transfer of GalNAc to proteins, was strongest in unsporulated oocysts with lower levels of transcription also observed in gametocytes and sporulated oocysts. No transcription of EtGalNAc-T was observed in the merozoite stages. Again, no E. tenella genes were amplified from uninfected host cDNA.
FIG. 6.
Relative transcription of glycosylation genes in different developmental stages of E. tenella. RT-PCR was carried out on five E. tenella genes encoding enzymes involved in glycan synthesis (including EtGFAT, EtAGM, EtUAP, and EtUAE) and glycan transfer (EtGalNAc-T). Prior to RT-PCR, each cDNA sample was standardized to comparable levels of EtActin transcription. Merozoites (M), gametocytes (G), unsporulated oocysts (U), sporulated oocysts (S), and uninfected host control (H) are as indicated.
Sequencing of each of the PCR amplicons confirmed the identity of all E. tenella genes analyzed. The sequences obtained for EtAGM and EtUAP were 99 and 100% identical to the original predicted gene sequence (as listed in Table 2). Both EtUAE (99%) and EtGalNAc-T (100%) showed stronger homology to alternative gene predictions (EIMER_contig_00031419.Exonerate2BLASTCDS.Gene.2 and GLIMMERHMM190_V3.0.1_PHASES00000221606, respectively) than listed in Table 2. However, an even greater level of homology was observed between the sequenced EtUAE to cgd4_2600 (51% compared to 50% for the original sequence) and between the sequenced EtGalNAc-T and AY424890 (62% compared to 47%) using BLASTx analysis of the C. parvum and T. gondii predicted protein databases, respectively.
GlcNAc is transiently higher in gametocytes, while GalNAc remains higher in subsequent oocyst stages.
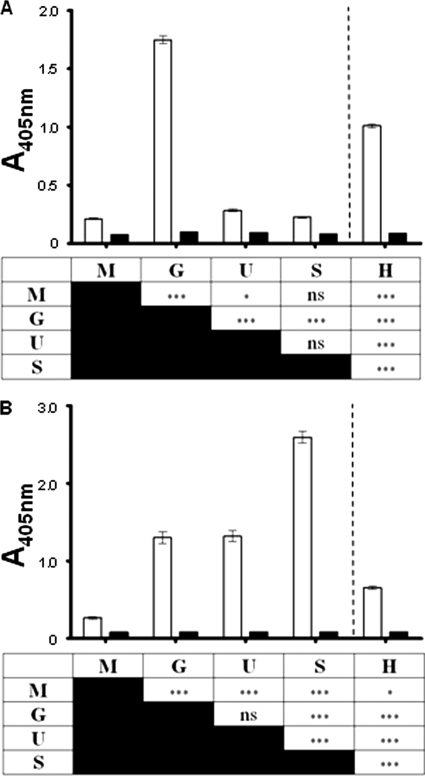
A microtiter assay was used to establish the relative levels of amino sugar (either as free glycans or glycoproteins) present in different developmental stages of E. tenella. GlcNAc and GalNAc were detected in each cell lysate using the lectins WGA and JAC, respectively. A significant increase (P < 0.001) in the level of GlcNAc was observed in E. tenella gametocytes (134 h p.i.) and was determined to be approximately eight times higher than the levels in merozoites and sporulated oocysts and approximately six times higher than the levels in unsporulated oocysts (Fig. 7A). The relatively moderate level of GlcNAc detected in uninfected host lysate was still significantly lower (P < 0.001) than that observed in E. tenella gametocytes, indicating that the unavoidable host contamination in this sample was not leading to an overestimation of GlcNAc levels. Similarly, a significant increase (P < 0.001) in GalNAc was observed in gametocytes compared to merozoites (Fig. 7B). However, the levels of this amino sugar were not significantly different (P > 0.05) in gametocytes and unsporulated oocysts, both of which were approximately five times higher than the levels in merozoites. Furthermore, a significant twofold increase (P < 0.001) was also observed in sporulated oocysts compared to unsporulated oocysts and gametocytes. Uninfected host GalNAc levels were significantly lower (P < 0.001) than that of the gametocyte sample, again indicating no overestimation of GalNAc in this parasite sample.
FIG. 7.
Relative amino sugar levels in different developmental stages of E. tenella. Cell lysates of each sample were assayed for the presence of the amino sugars, N-acetylglucosamine and N-acetylgalactosamine, using the lectins WGA (A) and JAC (B), respectively (white bars). All samples were also tested in the absence of lectin (black bars). A statistical comparison was made between each of the five protein samples (***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, P > 0.05 [not significant]). All samples were tested in quadruplicate. Merozoites (M), gametocytes (G), unsporulated oocysts (U), sporulated oocysts (S), and uninfected host control (H) are as indicated.
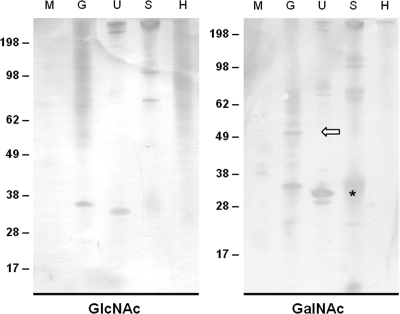
The protein lysates used for the lectin microtiter assay were also analyzed by lectin blotting. Again, WGA and JAC were used to detect proteins glycosylated with GlcNAc or GalNAc amino sugars, respectively. A comparison of both blots revealed a number of common protein bands, including a gametocyte-specific 35-kDa band, an unsporulated oocyst-specific 32-kDa band, and 120- and 75-kDa bands in sporulated oocysts (Fig. 8). However, a number of bands were detected only with JAC (i.e., GalNAc glycans present), including a gametocyte-specific band of ∼51 kDa. Furthermore, some common protein bands were more prominent when detected with JAC than with WGA, such as the unsporulated oocyst-specific 32-kDa band.
FIG. 8.
Detection of glycosylated proteins in different developmental stages of E. tenella. Immunoblots of whole-cell protein lysates from E. tenella and host tissue were probed with WGA or JAC for the detection of proteins glycosylated with N-acetylglucosamine or N-acetylgalactosamine, respectively. A unique gametocyte-specific band (open white arrow) and unsporulated oocyst-upregulated band (asterisk), both JAC positive, are marked accordingly. Size markers (in kilodaltons) are displayed to the left of each lectin blot. Merozoites (M), gametocytes (G), unsporulated oocysts (U), sporulated oocysts (S), and uninfected host control (H) are as indicated.
DISCUSSION
The development of micro- and macrogametocytes in the Eimeria parasite is an essential process that precedes the formation of the unsporulated oocyst, a life cycle stage adapted for efficient transmission between hosts. The identification of the macrogamete-specific GFAT in the fellow apicomplexan P. berghei (17) has provided the basis for an analysis of macrogamete-specific glycosylation in E. tenella, as outlined here.
A homologous group of GFAT enzymes was uncovered in E. tenella (EtGFAT), T. gondii (TgGFAT), and C. parvum (CpGFAT) using the P. berghei GFAT protein sequence as a BLAST query. The GFAT classification of these four sequences was also confirmed by the identification of the three characteristic GFAT catalytic domains (Fig. 1A). The absence of the complete GAT2 motif in TgGFAT was not addressed here but may be the result of incorrect coding sequence predictions from genomic sequence data.
PbGFAT was previously detected in purified samples of P. berghei macrogametes (17), and accordingly, EtGFAT gene transcription was restricted almost exclusively to gametocyte stages of E. tenella (Fig. 2), corresponding to the detection of native EtGFAT protein in E. tenella gametocytes by Western blotting (Fig. 3). The size discrepancy observed between the native EtGFAT and its calculated molecular weight was surprising but has been observed previously with GFAT proteins (27). The minimal transcription of EtGFAT in late-stage merozoites did not appear to correspond to protein expression in the same parasite stage. However, it is possible that the faint merozoite-specific protein bands (detected at a higher molecular weight than the gametocyte-specific EtGFAT) may represent different posttranslational modifications of EtGFAT.
Immunolocalization experiments successfully demonstrated exclusive expression of the native EtGFAT protein within E. tenella macrogametes but not microgametes, merozoites, or uninfected host (Fig. 4). EtGFAT was not, however, definitively localized to any known macrogamete organelles, such as wall-forming bodies (see anti-EmGam56, Fig. 4B); studies with human GFAT, by comparison, have demonstrated cytoplasmic localization of the native protein (27).
The observed synchronicity of EtGFAT, EtAGM, EtUAP, and EtUAE transcription in E. tenella gametocytes (Fig. 6) implies that the entire glycan synthesis pathway is “switched on” during gametocyte development. Like E. tenella, genes encoding the G. intestinalis enzymes responsible for GalNAc production are transcribed at higher levels during encystment (21), leading to the formation of a GalNAc-rich (63%) cyst wall (15, 29). The absence of a putative “EtGNA” was unexpected, but given the importance of this enzyme, it is unlikely that EtGNA is truly absent (perhaps indicating a higher level of sequence divergence with this enzyme).
Amino sugars from other excretable cyst-producing eukaryotes have been shown to be directly incorporated into the cyst wall as carbohydrate polymers. In addition to G. intestinalis, Entamoeba invadens utilizes newly produced GlcNAc as a substrate for chitin and chitosan, the two major components of the cyst wall (2, 11). However, the low levels of carbohydrate (1%) detected in purified oocyst wall samples of Eimeria (23) suggest that they are unlikely to be composed of similar amino sugar polymers. It would seem, therefore, the likely fate of amino sugars in Eimeria gametocytes is as substrates for protein glycosylation.
The absence of a putative GlcNAc-T in E. tenella (Table 2) suggests that GlcNAc is required as a substrate for GalNAc synthesis (via the activity of EtUAE) rather than direct protein glycosylation. This theory was supported by observations of a transient GalNAc increase in E. tenella gametocytes, with a return to lower levels in oocyst stages (Fig. 7). GlcNAc is also an essential substrate for the formation of glycosylphosphatidylinositol-anchored proteins (28, 41), which comprise many of the known surface molecules of E. tenella merozoites and sporozoites (40). Here, however, we have focused on the process of N- and O-linked glycosylation and its potential role with known O-linked glycoproteins of gametocytes, such as EtGam56.
The prolonged transcription of EtGalNAc-T (at its highest in unsporulated oocysts [Fig. 6]) was consistent with a prolonged GalNAc increase in E. tenella gametocytes and oocyst stages (Fig. 7), and this may have a direct link with the stockpiling and eventual cross-linking of the oocyst wall glycoproteins Gam56 and Gam82 (6, 7). It is possible that O glycosylation is required for trafficking of these proteins to the oocyst wall, a hypothesis supported by the conservation of an O glycosylation motif in both EmGam56 and EtGam56 and an additional oocyst wall protein of E. tenella, EtGam22 (19).
The synchronized production of oocyst wall-forming proteins and upregulation of glycosylation in E. tenella gametocytes is a trait unlikely to be shared with the P. berghei (the source of the GFAT sequence used for the original BLAST query), whose oocysts are not excreted and do not possess a rigid wall. However, the transcription of cyst wall proteins—CWP1, CPW2, and CWP3—and of glucosamine-6-phosphate isomerase (i.e., GFAT equivalent) in encysting Giardia lamblia is synchronized (21) and under the control of a single transcription factor, gMyb2, itself upregulated during encystation (39). The identification of a macrogamete-specific transcription factor and promoter (i.e., that might be controlling the transcription of glycosylation enzymes) could help identify other sexual stage proteins of E. tenella and have applications in developing transgenic yellow fluorescent protein-tagged macrogametes (33, 44).
Glycosylation is utilized by other eukaryotes for a multitude of functions, including protein folding, targeting, recognition, and adhesion (26). In fungi, GFAT has already been successfully targeted for selective inhibition (24, 43), and likewise, EtGFAT may represent an effective target for controlling E. tenella infections and the transmission of avian coccidiosis.
Acknowledgments
This project was made possible by the funding of a Ph.D. scholarship by the Institute for the Biotechnology of Infectious Diseases, University of Technology, Sydney, Australia. E. tenella whole-genome sequence data were produced by a BBSRC-funded collaboration between the Pathogen Sequencing Unit at the Wellcome Trust Sanger Institute and the Institute for Animal Health (United Kingdom).
The Houghton strain of E. tenella was originally provided by Martin Shirley (Institute for Animal Health, Compton Laboratory, Newbury, Berkshire, United Kingdom).
Footnotes
Published ahead of print on 6 November 2009.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arroyo-Begovich, A., A. Carabez-Trejo, and J. Ruiz-Herrera. 1980. Identification of the structural component in the cyst wall of Entamoeba invadens. J. Parasitol. 66:735-741. [PubMed] [Google Scholar]
- 3.Belli, S. I., M. Lee, P. Thebo, M. G. Wallach, B. Schwartsburd, and N. C. Smith. 2002. Biochemical characterisation of the 56 and 82-kDa immunodominant gametocyte antigens from Eimeria maxima. Int. J. Parasitol. 32:805-816. [DOI] [PubMed] [Google Scholar]
- 4.Belli, S. I., N. C. Smith, and D. Ferguson. 2006. The coccidian oocyst: a tough nut to crack! Trends Parasitol. 22:416-423. [DOI] [PubMed] [Google Scholar]
- 5.Belli, S. I., M. G. Wallach, C. Luxford, M. J. Davies, and N. C. Smith. 2003. Roles of tyrosine-rich precursor glycoproteins and dityrosine- and 3,4-dihydroxyphenylalanine-mediated protein cross-linking in development of the oocyst wall in the coccidian parasite Eimeria maxima. Eukaryot. Cell 2:456-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belli, S. I., M. G. Wallach, and N. C. Smith. 2003. Cloning and characterization of the 82-kDa tyrosine-rich sexual stage glycoprotein, GAM82, and its role in oocyst wall formation in the apicomplexan parasite, Eimeria maxima. Gene 307:201-212. [DOI] [PubMed] [Google Scholar]
- 7.Belli, S. I., D. Witcombe, M. G. Wallach, and N. C. Smith. 2002. Functional genomics of gam56: characterisation of the role of a 56 kilodalton sexual stage antigen in oocyst wall formation in Eimeria maxima. Int. J. Parasitol. 32:1727-1737. [DOI] [PubMed] [Google Scholar]
- 8.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 9.Boehmelt, G., I. Fialka, G. Brothers, M. D. McGinley, S. D. Patterson, R. Mo, C. C. Hui, S. Chung, L. A. Huber, T. W. Mak, and N. N. Iscove. 2000. Cloning and characterization of the murine glucosamine-6-phosphate acetyltransferase EMeg32: differential expression and intracellular membrane association. J. Biol. Chem. 275:12821-12832. [DOI] [PubMed] [Google Scholar]
- 10.Buchanan, J. M. 1973. The amidotransferases. Adv. Enzymol. Relat. Areas Mol. Biol. 39:91-183. [DOI] [PubMed] [Google Scholar]
- 11.Das, S., K. Van Dellen, D. Bulik, P. Magnelli, J. Cui, J. Head, P. W. Robbins, and J. Samuelson. 2006. The cyst wall of Entamoeba invadens contains chitosan (deacetylated chitin). Mol. Biochem. Parasitol. 148:86-92. [DOI] [PubMed] [Google Scholar]
- 12.Durand, P., B. Golinelli-Pimpaneau, S. Mouilleron, B. Badet, and M. A. Badet-Denisot. 2008. Highlights of glucosamine-6P synthase catalysis. Arch. Biochem. Biophys. 474:302-317. [DOI] [PubMed] [Google Scholar]
- 13.Ferguson, D. J., S. I. Belli, N. C. Smith, and M. G. Wallach. 2003. The development of the macrogamete and oocyst wall in Eimeria maxima: immuno-light and electron microscopy. Int. J. Parasitol. 33:1329-1340. [DOI] [PubMed] [Google Scholar]
- 14.Gattiker, A., E. Gasteiger, and A. Bairoch. 2002. ScanProsite: a reference implementation of a PROSITE scanning tool. Appl. Bioinformatics 1:107-108. [PubMed] [Google Scholar]
- 15.Gerwig, G. J., J. A. van Kuik, B. R. Leeflang, J. P. Kamerling, J. F. Vliegenthart, C. D. Karr, and E. L. Jarroll. 2002. The Giardia intestinalis filamentous cyst wall contains a novel β(1-3)-N-acetyl-d-galactosamine polymer: a structural and conformational study. Glycobiology 12:499-505. [DOI] [PubMed] [Google Scholar]
- 16.Harlow, E., and D. Lane. 1988. Immunoblotting and antibodies: a laboratory manual, p. 490-492. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 17.Khan, S. M., B. Franke-Fayard, G. R. Mair, E. Lasonder, C. J. Janse, M. Mann, and A. P. Waters. 2005. Proteome analysis of separated male and female gametocytes reveals novel sex-specific Plasmodium biology. Cell 121:675-687. [DOI] [PubMed] [Google Scholar]
- 18.Korich, D. G., J. R. Mead, M. S. Madore, N. A. Sinclair, and C. R. Sterling. 1990. Effects of ozone, chlorine dioxide, chlorine, and monochloramine on Cryptosporidium parvum oocyst viability. Appl. Environ. Microbiol. 56:1423-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krucken, J., R. J. Hosse, A. N. Mouafo, R. Entzeroth, S. Bierbaum, P. Marinovski, K. Hain, G. Greif, and F. Wunderlich. 2008. Excystation of Eimeria tenella sporozoites impaired by antibody recognizing gametocyte/oocyst antigens GAM22 and GAM56. Eukaryot. Cell 7:202-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Llovo, J., A. Lopez, J. Fabregas, and A. Munoz. 1993. Interaction of lectins with Cryptosporidium parvum. J. Infect. Dis. 167:1477-1480. [DOI] [PubMed] [Google Scholar]
- 21.Lopez, A. B., K. Sener, E. L. Jarroll, and H. van Keulen. 2003. Transcription regulation is demonstrated for five key enzymes in Giardia intestinalis cyst wall polysaccharide biosynthesis. Mol. Biochem. Parasitol. 128:51-57. [DOI] [PubMed] [Google Scholar]
- 22.Lubas, W. A., D. W. Frank, M. Krause, and J. A. Hanover. 1997. O-linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. J. Biol. Chem. 272:9316-9324. [DOI] [PubMed] [Google Scholar]
- 23.Mai, K., P. A. Sharman, R. A. Walker, M. Katrib, D. DeSouza, M. J. McConville, M. G. Wallach, S. I. Belli, D. J. Ferguson, and N. C. Smith. 2009. Oocyst wall formation and composition in coccidian parasites. Mem. Inst. Oswaldo Cruz 104:281-289. [DOI] [PubMed] [Google Scholar]
- 24.Milewski, S., H. Chmara, and E. Borowski. 1986. Antibiotic tetaine: a selective inhibitor of chitin and mannoprotein biosynthesis in Candida albicans. Arch. Microbiol. 145:234-240. [DOI] [PubMed] [Google Scholar]
- 25.Mouafo, A. N., A. Weck-Heimann, J. F. Dubremetz, and R. Entzeroth. 2002. Monoclonal antibodies specific for the two types of wall-forming bodies of Eimeria tenella macrogametes (Coccidia, Apicomplexa). Parasitol. Res. 88:217-224. [DOI] [PubMed] [Google Scholar]
- 26.Nairn, A. V., W. S. York, K. Harris, E. M. Hall, J. M. Pierce, and K. W. Moremen. 2008. Regulation of glycan structures in animal tissues: transcript profiling of glycan-related genes. J. Biol. Chem. 283:17298-17313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nerlich, A. G., U. Sauer, V. Kolm-Litty, E. Wagner, M. Koch, and E. D. Schleicher. 1998. Expression of glutamine:fructose-6-phosphate amidotransferase in human tissues: evidence for high variability and distinct regulation in diabetes. Diabetes 47:170-178. [DOI] [PubMed] [Google Scholar]
- 28.Orlean, P., and A. K. Menon. 2007. Thematic review series: lipid posttranslational modifications: GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J. Lipid Res. 48:993-1011. [DOI] [PubMed] [Google Scholar]
- 29.Ortega-Barria, E., H. D. Ward, J. E. Evans, and M. E. Pereira. 1990. N-Acetyl-d-glucosamine is present in cysts and trophozoites of Giardia lamblia and serves as receptor for wheat germ agglutinin. Mol. Biochem. Parasitol. 43:151-165. [DOI] [PubMed] [Google Scholar]
- 30.Pugatsch, T., D. Mencher, and M. Wallach. 1989. Eimeria maxima: isolation of gametocytes and their immunogenicity in mice, rabbits, and chickens. Exp. Parasitol. 68:127-134. [DOI] [PubMed] [Google Scholar]
- 31.Schaap, D., G. Arts, N. F. J. van Poppel, and A. N. Vermeulen. 2005. De novo ribosome biosynthesis is transcriptionally regulated in Eimeria tenella, dependent on its life cycle stage. Mol. Biochem. Parasitol. 139:239-248. [DOI] [PubMed] [Google Scholar]
- 32.Scholtyseck, E., H. Mehlhorn, and D. M. Hammond. 1971. Fine structure of macrogametes and oocysts of coccidia and related organisms. Z. Parasitenkd. 37:1-43. [DOI] [PubMed] [Google Scholar]
- 33.Shi, T. Y., X. Y. Liu, L. L. Hao, J. D. Li, A. N. Gh, M. H. Abdille, and X. Suo. 2008. Transfected Eimeria tenella could complete its endogenous development in vitro. J. Parasitol. 94:978-980. [DOI] [PubMed] [Google Scholar]
- 34.Shirley, M. W. 1995. Eimeria species and strains of chickens, p. 1-24. In J. Eckert, R. Braun, M. W. Shirley, and P. Coudert (ed.), Biotechnology: guidelines on techniques in coccidiosis research. European Commission, Luxembourg.
- 35.Stein, B., L. Stover, A. Gillem, K. Winters, J. H. Leet, and C. Chauret. 2006. The effect of lectins on Cryptosporidium parvum oocyst in vitro attachment to host cells. J. Parasitol. 92:1-9. [DOI] [PubMed] [Google Scholar]
- 36.Stephens, D. A. 1998. Coccidiosis, p. 591-593. In P. J. Delves and I. M. Roitt (ed.), Encyclopedia of immunology, 2nd ed. Academic Press, Inc., San Diego, CA.
- 37.Stotish, R. L., C. C. Wang, and M. Meyenhofer. 1978. Structure and composition of the oocyst wall of Eimeria tenella. J. Parasitol. 64:1074-1081. [PubMed] [Google Scholar]
- 38.Stwora-Wojczyk, M. M., J. C. Kissinger, S. L. Spitalnik, and B. S. Wojczyk. 2004. O glycosylation in Toxoplasma gondii: identification and analysis of a family of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases. Int. J. Parasitol. 34:309-322. [DOI] [PubMed] [Google Scholar]
- 39.Sun, C. H., D. Palm, A. G. McArthur, S. G. Svard, and F. D. Gillin. 2002. A novel Myb-related protein involved in transcriptional activation of encystation genes in Giardia lamblia. Mol. Microbiol. 46:971-984. [DOI] [PubMed] [Google Scholar]
- 40.Tabares, E., D. Ferguson, J. Clark, P. E. Soon, K. L. Wan, and F. Tomley. 2004. Eimeria tenella sporozoites and merozoites differentially express glycosylphosphatidylinositol-anchored variant surface proteins. Mol. Biochem. Parasitol. 135:123-132. [DOI] [PubMed] [Google Scholar]
- 41.Tiede, A., C. Nischan, J. Schubert, and R. E. Schmidt. 2000. Characterisation of the enzymatic complex for the first step in glycosylphosphatidylinositol biosynthesis. Int. J. Biochem. Cell Biol. 32:339-350. [DOI] [PubMed] [Google Scholar]
- 42.Van Der Wel, H., H. R. Morris, M. Panico, T. Paxton, A. Dell, L. Kaplan, and C. M. West. 2002. Molecular cloning and expression of a UDP-N-acetylglucosamine (GlcNAc):hydroxyproline polypeptide GlcNAc-transferase that modifies Skp1 in the cytoplasm of Dictyostelium. J. Biol. Chem. 277:46328-46337. [DOI] [PubMed] [Google Scholar]
- 43.Wojciechowski, M., S. Milewski, J. Mazerski, and E. Borowski. 2005. Glucosamine-6-phosphate synthase, a novel target for antifungal agents. Molecular modeling studies in drug design. Acta Biochim. Pol. 52:647-653. [PubMed] [Google Scholar]
- 44.Yan, W., X. Liu, T. Shi, L. Hao, F. M. Tomley, and X. Suo. 2009. Stable transfection of Eimeria tenella: constitutive expression of the YFP-YFP molecule throughout the life cycle. Int. J. Parasitol. 39:109-117. [DOI] [PubMed] [Google Scholar]