Abstract
In Saccharomyces cerevisiae, the pheromone-induced ubiquitylation and degradation of the filamentation pathway-specific activator, Tec1, suppresses cross talk between the mating and filamentous growth mitogen-activated protein kinase (MAPK) pathways. The mating pathway MAPK, Fus3, phosphorylates Tec1, resulting in its recognition by the SCF (for Skp1, Cullin, F-box containing) E3 ubiquitin ligase complex, leading to its proteolysis. Previously, it was found that Tec1 destruction requires phosphorylation on threonine 273 (T273). T273 is embedded in the sequence LLpTP, which is identical to the canonical binding site for Cdc4, a conserved F-box substrate adaptor for the SCF complex. However, recent work on both Cdc4 and the human Cdc4 ortholog Fbw7 has shown that a second substrate phosphorylation can be required for optimal Cdc4 binding in vitro. We report here that high-affinity binding of recombinant Cdc4 to Tec1 phosphopeptides requires phosphorylation of not only T273 but also a second site, T276. Significantly, both phospho-sites on Tec1 and a conserved basic pocket on Cdc4 are critical for Tec1 proteolysis in response to pheromone treatment of cells, establishing a role for two-phosphate recognition by yeast Cdc4 in substrate targeting in vivo.
Cellular differentiation and survival depend on soliciting the correct intracellular response to environmental cues. Paradoxically, the signaling pathways in eukaryotic cells that perform these crucial functions often share components. How insulation of these interconnected signaling pathways is achieved to ensure a specific and accurate internal response to extracellular signals remains a central question in cell biology. In the budding yeast Saccharomyces cerevisiae, two conserved mitogen-activated protein kinase (MAPK) cascades share numerous components but regulate two distinct developmental programs, namely, mating and filamentous growth (FG). Each pathway contains a pathway-specific MAPK, Fus3 for the mating pathway and Kss1 for the FG pathway (6). However, since there are groups of shared components upstream and downstream of these pathway-specific kinases, additional mechanisms must exist to maintain signaling fidelity.
In a significant advance for the field, one such mechanism has been reported that maintains signaling specificity between the mating and FG pathway. Specifically, we and others found that, upon activation of the mating pathway, the mating pathway-specific MAPK, Fus3, directs the destruction of the FG pathway-specific transcription factor, Tec1 (1, 2). During pheromone signaling, the FG pathway MAPK, Kss1, is activated, but the destruction of Tec1 prevents it from promoting transcription of FG pathway targets. Tec1 is phosphorylated by active Fus3, marking it as a substrate for the SCF ubiquitin ligase complex that in turn ubiquitylates Tec1 and targets it for proteolysis. This mechanism is operative under basal signaling conditions in the absence of pheromone (where receptor-independent signal transduction through the MAPK cascade occurs), as well as in cells exposed to mating pheromone. Although this previous work provided an important insight into a mechanism that maintains signaling fidelity, several details remain unclear. In particular, there is conflicting evidence for which proteins are involved in Tec1 recognition and degradation, and the exact residues on Tec1 that form the phosphodegron are incompletely defined. First, the identity of the F-box protein that interacts with phospho-Tec1 has not been definitively resolved. F-box proteins are substrate-specific adaptor subunits of the SCF complex that directly bind to the phosphorylated moiety of substrates (3). There are data that support a role in Tec1 destruction for two F-box proteins: Cdc4, a WD-40 repeat containing protein (2), and Dia2, a leucine-rich repeat-containing protein (1). Cdc4 was shown to be required for Tec1 ubiquitylation in vivo, and recombinant Cdc4 was shown to bind to phosphorylated recombinant Tec1 in vitro (2). However, the dependence of Cdc4 binding on T273 was not reported. Dia2 is a nonessential F-box protein identified in a screen for repressors of invasive growth (9). Tec1 degradation was blocked in cells lacking Dia2, suggesting that it plays a role in mediating Tec1 proteolysis (1). However, no biochemical evidence was shown that Dia2 directly binds or ubiquitylates Tec1, leaving open the possibility that it may act indirectly.
Second, several lines of evidence suggest that other phosphorylations, in addition to T273, may be involved in Tec1 degradation. Mass spectrometry-based phosphopeptide mapping studies have shown that, in addition to T273, active Fus3 phosphorylates multiple sites on Tec1 in vitro, even though the loss of phosphorylation at T273 was sufficient to block Tec1 degradation in response to pheromone (1, 2). In vivo labeling studies using [γ-32P]ATP to monitor the phosphorylation of Tec1 in the presence of pheromone further suggest that mutation of T273 was not sufficient to abolish all phosphorylation on Tec1 (M. Z. Bao and H. D. Madhani, unpublished data). Whether phosphorylation on residues other than T273 is required for Tec1 destruction has not previously been investigated. Recent structural studies of the human Cdc4 ortholog, Fbw7, have shown that it contains a second phosphate-binding pocket that can recognize a second substrate phosphorylation (4). Moreover, yeast Cdc4 itself was shown in the same study to bind diphosphorylated peptides from its substrate Sic1 in vitro with a Kd of up to 4 μM, although the relevance of this binding mode to Sic1 destruction in vivo was not reported (4).
Here, we describe an analysis of the residues in Tec1 required for its destruction and provide further evidence that Cdc4 is the F-box protein that directly interacts with the Tec1 phosphodegron. We identify a second phosphorylated residue, T276, as being required for Tec1 destruction and signaling specificity in vivo. Using peptide binding studies, we show that Cdc4 directly recognizes this region of Tec1 and requires phosphorylation of both T273 and T276 for tight binding (Kd = 80 nM). We also show that residues in Cdc4 orthologous to those in the second phosphate-binding pocket defined structurally in Fbw7 are required for efficient pheromone-induced Tec1 destruction and signaling specificity in vivo. Together, these data support the assignment of Cdc4 as the receptor for the Tec1 phosphodegron.
MATERIALS AND METHODS
Yeast strains, plasmids, and epitope tags.
The strains used in the present study are of the Sigma 1278b background and are listed in Table S1 in the supplemental material. The plasmids used in the present study are listed in Table S2 in the supplemental material. Cdc4 strains harboring mutant Cdc4 alleles in a cdc4Δ background were generated through sporulation and tetrad dissection of cdc4Δ/CDC4 diploid cells transformed with corresponding plasmids.
β-Galactosidase assay.
Liquid β-galactosidase assays were performed as previously described (1). Error bars represent standard deviation from three replicates.
Pheromone time course experiments.
Evaluation of Tec1 protein level in the presence of pheromone in Sigma 1278b strains was performed as previously described (1) with the exception of the immunoblot visualization technique. Indicated immunoblots were visualized digitally by LiCor Biosciences Odyssey Infrared Imaging System. Antibodies to the myc-epitope tag and tubulin were incubated simultaneously with the immunoblot. IRDye infrared secondary antibodies to the myc-tagged proteins (IRDye 680-goat anti-mouse antibody, LiCor 926-32220) and tubulin (IRDye 800-goat anti-rat antibody) were used.
Cdc4-Skp1 protein purification.
Cdc4-Skp1 was isolated from BL21-Codon Plus (DE)-RIPL cells (catalog no. 230280; Stratagene). Cells transformed with pMT3169 (BHM 1193) were induced at mid-log phase with 0.2 M IPTG (isopropyl-β-d-thiogalactopyranoside) for 10 to 12 h at 18°C prior to harvest. Per 1 liter of cells, lysis was performed in 50 ml of buffer A (50 mM Tris [pH 7.6], 500 mM NaCl, 10% glycerol, 0.1% NP-40, 5 mM β-mercaptoethanol) containing one tablet of protease inhibitor cocktail (Complete TM EDTA-free protease inhibitor, catalog no. 11873580001; Roche Applied Science) using 0.2 g of lysozyme. After incubation at 4°C for 35 min, the cell lysate was brought to 0.1 mM CaCl2 and 6 mM MgCl2 and treated with DNase I for an additional 35 min. Lysate was then clarified at 15,000 rpm for 15 min. Buffer A-washed Ni-NTA agarose was added to the supernatant for binding in batch at 4°C for 1 h. Agarose-lysate slurry was then applied to a fritted column. The column was washed three times with one column volume of buffer A prior to elution by the application of one column volume of buffer B (50 mM Tris [pH 7.6], 500 mM NaCl, 10% glycerol, 0.1% NP-40, 250 mM imidazole, 5 mM β-mercaptoethanol). To the eluate, an equal volume buffer B without NaCl and imidazole was added. The eluate was brought to 1 mM EDTA and 1 mM dithiothreitol before the addition of washed glutathione S-transferase (GST) agarose. Binding was performed in batch at 4°C for 1 h. Slurry was then applied to fritted column and washed twice with one column volume of phosphate-buffered saline-EDTA-dithiothreitol (DTT) buffer prior to elution with one column volume of Buffer C (150 mM NaCl, 50 mM Tris-HCl [pH 8.0], 20 mM glutathione, 1 mM DTT). Elution is then concentrated on a YM-30 Centriprep centrifugal filter unit (Millipore catalog no. 4307) to 3 ml. This volume was then applied to a Slide-A-Lyzer dialysis cassette (Pierce Biotechnology) for dialysis overnight at 4°C in 600 ml of storage buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 25% glycerol, 0.1% NP-40, 5 mM β-mercaptoethanol). Buffer was changed after 12 h, and the cassette dialyzed for a further 4 h prior to the removal of liquid for protein quantification by the Bradford assay and storage at −20°C.
Fluorescence anisotropy.
Peptides used in the study were labeled at the C terminus with 5-iodoacetamido fluorescein (Sigma catalog no. I9271-25MG), purified by high-pressure liquid chromatography (HPLC), and confirmed by matrix-assisted laser desorption ionization (MALDI) mass spectrometry. Binding studies were performed in 384-well Corning opaque plate in triplicate. Each reaction was 40 μl of total volume with various protein concentrations (0.078 to 5 μM) and 5 nM labeled peptide in polarization buffer (50 mM Tris, 100 mM NaCl, 5 mM β-mercaptoethanol, 5% glycerol, 0.1 mg of bovine serum albumin/ml). Polarization data were collected by an Analyst HT microplate reader (FPS mode; five reads/well, 10 ms between readings; z-height, 5.775 [middle]; emission, 485 nm; excitation, 530 nm). The Kd for each peptide was calculated by using KaleidoGraph software for best-fit curve to the polarization data according to the following formula: y = m1 + m2·{([peptide] + M0 + m3) − sqrt(([peptide] + M0 + m3)·([peptide] + M0 + m3) − (4·[peptide]·M0))}, where M0 = x, m1 = scaling factor (y intercept) = 0.1, m2 = scaling factor = 0.02, and m3 = Kd.
RESULTS AND DISCUSSION
Two phosphorylation sites on Tec1 are essential for its destruction in response to signaling through the mating pathway.
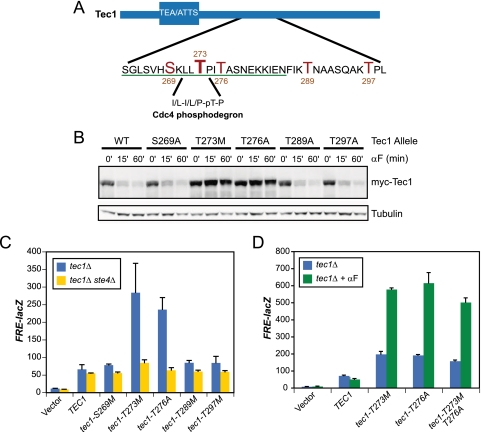
Previous mass spectrometry-based phosphopeptide mapping of a recombinant MBP-Tec1 fusion protein phosphorylated in vitro by Fus3 immunoprecipitated from pheromone-treated cells revealed multiple sites of phosphorylation in addition to T273 (Fig. 1A) (1, 2). To test whether phosphorylated residues in the vicinity of T273 play a role in Tec1 regulation in vivo, we generated nonphosphorylatable mutants of each site (S269A, T276A, T289A, and T297A) and assayed Tec1 degradation in response to activation of the mating pathway. Each mutant allele of myc epitope-tagged Tec1 was expressed from its native promoter on a centromeric plasmid in cells lacking the endogenous copy of the TEC1 gene. Cells were treated with synthetic mating pheromone (α-factor), and samples were collected over the course of an hour. Although mutation of S269, T289, and T297 had no effect on Tec1 degradation, the T276A mutation blocked Tec1 degradation and appeared indistinguishable from the strain mutated at T273 (Fig. 1B).
FIG. 1.
Two phosphorylation sites on Tec1 are required both for mediating Tec1 degradation in response to pheromone and for maintaining signaling specificity. (A) Schematic of the region on Tec1 containing the residues identified as phosphorylated by mass spectrometry. Phosphorylated residues are indicated in red. The residues of Tec1 in the mutagenesis studies are underlined in green. The sequence of the previously identified Cdc4 phosphodegron (CPD) and its location are indicated. (B) Time course experiments of Tec1 phospho-site alleles. Immunoblot of extracts from tec1Δ cells expressing the indicated myc-epitope tagged allele from a centromeric plasmid. Cells were treated for the indicated amount of time with 5 μM pheromone before harvest. Immunoblot was probed with anti-myc and anti-tubulin (loading control) antibodies and was visualized by the LiCor Odyssey infrared imaging system. (C) FRE(TEC1)::lacZ expression in tec1Δ and tec1Δ ste4Δ cells transformed with the indicated plasmids. (D) FRE(TEC1)::lacZ expression in tec1Δ cells transformed with the indicated plasmids. Exponentially growing cultures were split, and 5 μM pheromone was added to one. Both were then grown at 30°C for two additional hours prior to harvesting of cells.
To determine whether mutation of T276 produced the expected loss of signaling specificity, we examined the expression of a filamentation pathway-specific reporter gene (FRE-lacZ) in cells harboring the mutant allele. A loss of signaling specificity between the mating and FG pathways would result in mating pathway-dependent activation of the filamentation reporter, both under basal signaling conditions in which the pheromone response pathway is partially active in the absence of pheromone and under conditions of pheromone-mediated stimulation. Indeed, cells expressing tec1-T276A also demonstrated increased expression of FRE-lacZ, which was eliminated by mutational inactivation of the gene coding for the pheromone receptor-coupled G-protein beta subunit, Ste4 (Fig. 1C). FRE-lacZ expression increased further upon addition of pheromone to the cells expressing tec1-T276A, again yielding an effect that was quantitatively indistinguishable from that produced by mutation of T273 (Fig. 1D). In contrast, mutation of residues that are not required for Tec1 degradation does not affect the basal (Fig. 1C) or pheromone-stimulated (Fig. 1D) expression of FRE-lacZ. We also constructed an allele of TEC1 in which the threonine residues at both 273 and 276 were mutated. Cells expressing the resulting tec1-T273M T276A double mutant behaved similarly to the single mutants (Fig. 1D).
In addition to more thoroughly investigate the required sequence elements of the Tec1 phosphodegron, we mutated most of the residues flanking T273 and T276, from S263 to N285. Lysine residues were mutated to arginines, alanines to valines, while all other residues were changed to alanines. tec1Δ cells expressing these myc epitope-tagged Tec1 mutants were assayed for (i) Tec1 degradation during induction of the mating pathway and (ii) cross talk between the mating and FG pathways (as evidence by an increase in basal FRE-lacZ activity). A summary of these data is presented in Table 1. Interestingly, T273 and T276, which are both phosphorylated by Fus3 in vitro, were essential for both pheromone-induced destruction of Tec1 and cross talk suppression. tec1-P274S and tec1-A277V cells displayed some stabilization of Tec1 and also produced various levels of cross talk. Notably, neither of the two leucine residues preceding T273 (L271 and L272) were required. These leucines form part of the Cdc4 substrate recognition motif identified by Nash et al. (7), who reported that mutation of the leucines abrogates Cdc4 binding. This suggested that if Cdc4 does recognize Tec1 in vivo, it does so via a mechanism that does not require this leucine dipeptide.
TABLE 1.
Summary of analysis of Tec1 mutantsa
| Tec1 allele | Protein stability in pheromone-treated cellsb | Mean β-galactosidase activity ± SD (FRE-lacZ) |
|---|---|---|
| Wild type | - | 1 |
| S263A | - | 1.19 ± 0.25 |
| G264A | - | 0.96 ± 0.2 |
| L265A | - | 0.84 ± 0.02 |
| S266A | - | 1.01 ± 0.14 |
| V267A | - | 0.97 ± 0.03 |
| H268A | - | 1.02 ± 0.09 |
| S269A | - | 1.18 ± 0.40 |
| K270R | - | 0.97 ± 0.26 |
| L271A | -/+ | 1.01 ± 0.12 |
| L272A | -/+ | 1.10 ± 0.04 |
| T273 M | ++ | 3.27 ± 0.24 |
| P274S | ++ | 2.76 ± 0.41 |
| T276A | ++ | 3.55 ± 0.52 |
| A277V | + | 1.74 ± 0.16 |
| S278A | + | 1.25 ± 0.11 |
| N279A | - | 0.82 ± 0.18 |
| E280A | + | 1.31 ± 0.17 |
| K281R | - | 0.95 ± 0.12 |
| I283A | + | 1.05 ± 0.13 |
| E284A | - | 0.85 ± 0.08 |
| N285A | - | 0.94 ± 0.05 |
| T289A | - | 1.27 ± 0.10 |
| T297A | - | 1.27 ± 0.28 |
Shown is a summary of immunoblotting time course experiments of cells treated with mating pheromone (second column), as well as an examination of basal FRE-lacZ expression (third column). The wild type was set to a value of 1.
-, Wild-type destruction of myc-Tec1 in pheromone-treated cells over a 1-h time course; ++, no destruction; +, partial destruction; +/-, slight destruction.
Role of Cdc4 in Sigma 1278b strain background and in cells expressing Tec1 from the native promoter.
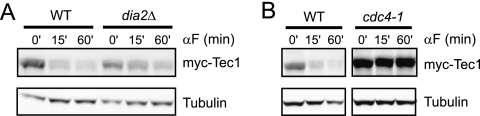
As described in the introduction, Cdc4 was implicated in Tec1 destruction in cells overexpressing Tec1 from the strong pGAL1 promoter (2). These experiments were also performed in the filamentation-defective strain background W303. Therefore, we engineered the cdc4-1 mutation in the filamentation-competent Sigma 1278b strain background and examined the destruction of Tec1 in strains expressing Tec1 from its native promoter on a centromeric plasmid. We observed that cdc4-1 cells displayed high basal levels of Tec1 and that Tec1 was not degraded when the cells were treated with pheromone at the nonpermissive temperature (Fig. 2A). cdc4-1 cells exhibit cell cycle arrest due to the well-characterized interaction between Cdc4 and Sic1, a Cdk inhibitor whose Cdc4-mediated degradation is essential for the onset of S phase (10). To determine whether stabilization of Tec1 was indirectly due to this cell cycle arrest, we examined Tec1 degradation in cells arrested in late G1 by overexpression of a nondegradable mutant of Sic1, sic1Δaa215-284 (5). Under these conditions Tec1 was still degraded (see Fig. S1 in the supplemental material). Taken together, these data suggest that the defect in Tec1 degradation in cdc4-1 cells reported previously (2) is not peculiar to Tec1 overexpression, the W303 strain background or cell cycle-arrested cells. We also reexamined the role of the alternative F-box protein, Dia2, in Tec1 destruction and found that cells lacking Dia2 display a weaker defect in degradation than cdc4-1 cells (Fig. 2B).
FIG. 2.
Direct comparison of Dia2 and Cdc4 indicate that Cdc4 is the F-box that recognizes phosphorylated Tec1. (A) Shown is an immunoblot of a pheromone time course performed at 37°C with wild-type and cdc4-1 cells transformed with myc-Tec1 expressed from its native promoter on a CEN plasmid. Cells were shifted to the nonpermissive temperature for 1 h prior to treatment with pheromone. (B) Shown is an immunoblot of a pheromone time course with wild-type and dia2Δ strains.
Two phospho-sites on Tec1, T273 and T276, are required for Cdc4 binding.
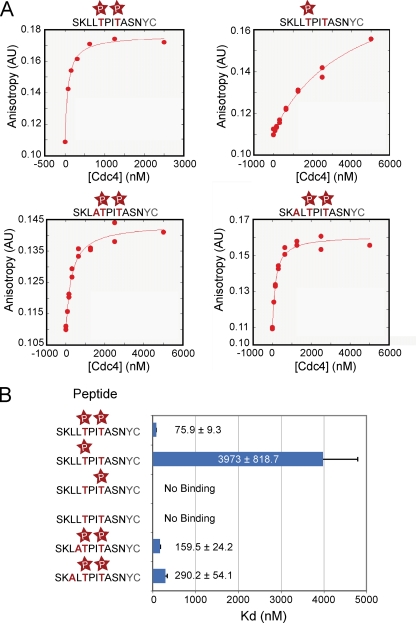
Given the strong in vivo phenotype of cdc4-1 mutant cells in Tec1 destruction, we tested whether Cdc4 directly recognizes the Tec1 phosphodegron we defined in vivo and whether this degron must be singly or doubly phosphorylated for such recognition to occur. As described in the introduction, it has been reported that Cdc4 can recognize dually phosphorylated peptides from the Sic1 protein with micromolar affinity (4). Given the requirement for two phosphorylated residues in Tec1, we hypothesized that these could be involved in an analogous binding reaction with Cdc4. We used fluorescence anisotropy to measure the binding of purified recombinant Cdc4, in complex with the SCF component Skp1, to C-terminally fluorescein-labeled peptides corresponding to an 11-amino-acid region of Tec1 containing the LLpTP motif (Fig. 3). These peptides were synthesized in various combinations with T273 or T276 phosphorylated (Fig. 3B). We found that whereas the wild-type peptide phosphorylated on both T273 and T276 bound to Cdc4 with a high affinity (75.9 ± 9.3 nM), removal of either phosphorylation significantly affected binding. Loss of the phosphorylation at T276 resulted in an ∼52-fold increase in Kd to nearly 4 μM. Notably, loss of the phosphorylation at T273 decreased binding to levels undetectable by the assay, indicating that the two phosphates make different contributions to binding affinity. As expected, peptides lacking both phosphorylations displayed no Cdc4 binding. Consistent with our in vivo data, loss of the leucines upstream of T273 showed little effect on Cdc4 binding: leucine to alanine changes of either L271 or L272 produced only modest increases of two- to threefold in the Kd. The close correspondence between the in vivo and in vitro data provides strong evidence for a direct role for Cdc4 in targeting Tec1 for destruction.
FIG. 3.
The phosphorylations at T273 and T276 make large contributions to the binding affinity to Cdc4. (A) KaleidoGraph best-fit curves of peptide binding data to Cdc4-Skp1. All data for each peptide are plotted. (B) Summary of peptide binding affinity to Cdc4-Skp1.
A second pocket on Cdc4 mediates binding to the dually phosphorylated Tec1 degron.
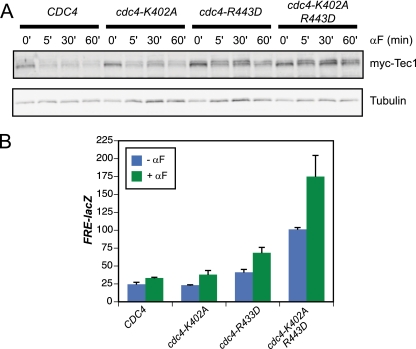
Next, we wanted to identify the residues on Cdc4 that are required for binding to dually phosphorylated Tec1. From the previously published crystal structure of S. cerevisiae Cdc4 bound to a peptide containing the LLpTP motif, it is known that a phosphothreonine fits into an electropositive binding pocket in Cdc4 (8). Consistent with subsequent structural work on the human Cdc4 ortholog Fbw7 (4), examination of a surface charge representation of Cdc4 revealed a second electropositive patch adjacent to the core binding pocket. This patch corresponds to the region of Fbw7 reported to recognize a second substrate phosphate. Residues in this patch on Cdc4 were previously characterized and found not to be essential for Sic1 degradation during cell cycle progression (8). However, to determine whether this region of Cdc4 might be important for the degradation of Tec1, we examined the effect of mutating two residues, K402 and R443, within the electropositive patch.
Cells harboring a cdc4-R443D allele or the cdc4-K402A R443D double allele show a defect in pheromone-induced degradation of Tec1 relative to cells harboring wild-type Cdc4 (Fig. 4A). Mutation of K402 alone did not show a strong stabilization phenotype, but combining the K402A mutation with R443D did show a somewhat stronger phenotype than R443D alone, indicating that K402A is involved in recognition of the Tec1 phosphodegron (Fig. 4A). Consistent with the observed effects on Tec1 protein levels, mutation of R443 or both K402 and R443 also resulted in hyperactivation of the FRE-lacZ reporter gene during both basal signaling and pheromone-induced signaling (Fig. 4B).
FIG. 4.
Residues outside of the Cdc4 canonical substrate binding pocket are required for Tec1 degradation and inhibition of cross talk. (A) Immunoblot of a pheromone time course with a cdc4Δ strain transformed with the indicated plasmids; (B) FRE(TEC1)::lacZ expression of a cdc4Δ strain transformed with the indicated plasmids.
Conclusion.
We have presented here an analysis of the molecular details of Tec1 destruction during pheromone signaling. We have identified an additional residue on Tec1, T276, which is phosphorylated by Fus3 and required to signal for Tec1 proteolysis in response to signaling. Significantly, cells expressing the tec1-T276A allele exhibit cross talk from the mating pheromone response pathway to the filamentous growth pathway. We further show through direct binding experiments that this second phosphorylation site is recognized with phospho-T273 by the Cdc4 F-box protein. Moreover, we demonstrate that residues of a basic pocket on the surface of Cdc4 are important for Tec1 destruction and signaling specificity. Together, these data provide compelling evidence for the in vivo recognition of a SCF substrate via recognition of a two-phosphate degron by Cdc4. These data support the view put forth by Hao et al. (4) that proposes that the Cdc4 family of proteins generally recognizes substrates through the recognition of two phosphates.
Supplementary Material
Acknowledgments
We thank Hans-Ulrich Mösch for the TEC1 constructs, Toshi Tsukiyama for the 3X-FLAG-kanMX cassette, Brian Green for the initial pRS306-pGAL-Sic1-aa215-284 construct, and Stephen Orlicky and Mike Tyers for the Cdc4 constructs. We also thank Matt Daugherty and Sigurd Braun for help and advice regarding the HPLC and MALDI mass spectrometry work, as well as Margot Quinlan for help regarding KaleidoGraph use. Finally, we thank Anupama Seshan, David Morgan, Sandy Johnson, and Jonathan Weissman for advice.
This study was supported by an NIH grant to H.D.M. (GM63670). M.Z.B. was supported by a National Science Foundation Predoctoral Fellowship. T.R.S. was supported by an American Heart Association predoctoral fellowship.
Footnotes
Published ahead of print on 6 November 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Bao, M. Z., M. A. Schwartz, G. T. Cantin, J. R. Yates III, and H. D. Madhani. 2004. Pheromone-dependent destruction of the Tec1 transcription factor is required for MAP kinase signaling specificity in yeast. Cell 119:991-1000. [DOI] [PubMed] [Google Scholar]
- 2.Chou, S., L. Huang, and H. Liu. 2004. Fus3-regulated Tec1 degradation through SCF(Cdc4) determines MAPK signaling specificity during mating in yeast. Cell 119:981-990. [DOI] [PubMed] [Google Scholar]
- 3.Deshaies, R. J., and C. A. Joazeiro. 2009. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78:399-434. [DOI] [PubMed] [Google Scholar]
- 4.Hao, B., S. Oehlmann, M. E. Sowa, J. W. Harper, and N. P. Pavletich. 2007. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell 26:131-143. [DOI] [PubMed] [Google Scholar]
- 5.Hodge, A., and M. Mendenhall. 1999. The cyclin-dependent kinase inhibitory domain of the yeast Sic1 protein is contained within the C-terminal 70 amino acids. Mol. Gen. Genet. 262:55-64. [DOI] [PubMed] [Google Scholar]
- 6.Madhani, H. D., C. A. Styles, and G. R. Fink. 1997. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 91:673-684. [DOI] [PubMed] [Google Scholar]
- 7.Nash, P., X. Tang, S. Orlicky, Q. Chen, F. B. Gertler, M. D. Mendenhall, F. Sicheri, T. Pawson, and M. Tyers. 2001. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414:514-521. [DOI] [PubMed] [Google Scholar]
- 8.Orlicky, S., X. Tang, A. Willems, M. Tyers, and F. Sicheri. 2003. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112:243-256. [DOI] [PubMed] [Google Scholar]
- 9.Pan, X., and J. Heitman. 1999. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4874-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwob, E., T. Bohm, M. D. Mendenhall, and K. Nasmyth. 1994. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in Saccharomyces cerevisiae. Cell 79:233-244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.