Abstract
Eukaryotic cells store oils in the chemical form of triacylglycerols in distinct organelles, often called lipid droplets. These dynamic storage compartments have been intensely studied in the context of human health and also in plants as a source of vegetable oils for human consumption and for chemical or biofuel feedstocks. Many microalgae accumulate oils, particularly under conditions limiting to growth, and thus have gained renewed attention as a potentially sustainable feedstock for biofuel production. However, little is currently known at the cellular or molecular levels with regard to oil accumulation in microalgae, and the structural proteins and enzymes involved in the biogenesis, maintenance, and degradation of algal oil storage compartments are not well studied. Focusing on the model green alga Chlamydomonas reinhardtii, the accumulation of triacylglycerols and the formation of lipid droplets during nitrogen deprivation were investigated. Mass spectrometry identified 259 proteins in a lipid droplet-enriched fraction, among them a major protein, tentatively designated major lipid droplet protein (MLDP). This protein is specific to the green algal lineage of photosynthetic organisms. Repression of MLDP gene expression using an RNA interference approach led to increased lipid droplet size, but no change in triacylglycerol content or metabolism was observed.
Triacylglycerols (TAGs) are stored in lipid droplets which are subcellular structures in specialized cells ubiquitous to eukaryotes but have more recently also been identified in some prokaryotes (26). In plants and animals, lipid droplets are surrounded by cytosol and are believed to bud off the endoplasmic reticulum (ER) (15, 26). While traditionally considered merely as storage compartments, recent studies suggest that lipid droplets in animals play important additional roles in lipid homeostasis and protein storage (8). In oilseed plants, TAG accumulated in seeds is used as a reservoir of energy and membrane lipid building blocks to support rapid growth after germination (15). Many green algae are capable of accumulating large amounts of TAG in lipid droplets, particularly as a result of abiotic stresses, such as nutrient deprivation or high-light exposure. Although TAG metabolism in algae has not yet been extensively studied at the biochemical or molecular level, it is proposed that TAG turnover contributes primarily to the assembly of membrane lipids to facilitate rapid cell division after the cessation of nutrient limitation (14, 38).
The general structure of lipid droplets is conserved in different species with a globular neutral lipid core enclosed by a membrane lipid monolayer (26). In addition, specific proteins are associated with lipid droplets and play important roles in lipid droplet structure and function. A number of recent proteomic studies of lipid droplets from different animals and tissues (8, 40), Saccharomyces cerevisiae (4), and plants (16, 17) have revealed that the lipid droplet-associated proteins of these organisms are quite distinct. For example, the abundant lipid droplet proteins in animals—the so-called “PAT” family of proteins comprised of perilipin, adipose differentiation-related protein (ADRP), and TIP47 (21)—have no apparent orthologs in the desiccating seed plant Arabidopsis thaliana; conversely, the oleosins which coat the oil bodies of Arabidopsis and many other seed plants are not found in animals (26). Reverse genetic studies of these proteins have helped to elucidate the role of A. thaliana oleosins in regulating lipid droplet size and preventing droplet fusion (35, 36) or that of mouse adipocyte perilipin in regulating lipolytic activity at the lipid droplet surface (37). Moreover, recent genomewide RNAi screens in Drosophila cells implicated 1.5 to 3.0% of all genes as directly or indirectly involved in lipid droplet formation and/or regulation and resulted in the identification of a new role for the Arf1-COPI vesicular transport machinery in regulating droplet morphology and lipid utilization (7, 12). In contrast, few molecular details are known about algal lipid droplet biogenesis although many TAG-rich algal species have been described (14).
Our efforts to identify proteins related to the PAT protein family or oleosins in the Chlamydomonas reinhardtii genome (24) or genomes of other green algal and diatom species, including Thalassiosira pseudonana, Volvox carteri, and Chlorella sp. NC64A, revealed no putative algal orthologs. In order to identify both potentially novel and conserved proteins which function in algal lipid droplet biogenesis, we studied the accumulation of TAG in lipid droplets of nitrogen-limited C. reinhardtii cells and identified candidate lipid droplet-associated proteins by mass spectrometry.
MATERIALS AND METHODS
Strains and growth conditions.
Chlamydomonas reinhardtii dw15 (cw15 nit1 mt+) and CC-125 (nit1 nit2 mt+) strains were grown in Tris-acetate-phosphate (TAP) medium with NH4Cl at 10 mM in liquid cultures or on agar-solidified plates under continuous light (70 to 80 μmol m−2 s−1) as previously described (30). Strain CC-125 was used in generating the data shown in Fig. 1B and Fig. 2, and the dw15 strain or transgenic derivatives thereof were used in the remainder of this study. To induce N limitation, mid-log-phase cells grown in 10 mM NH4Cl TAP were centrifuged for 5 min at 5,000 × g, washed once in TAP lacking NH4Cl or any other N source (TAP-N), and resuspended in TAP-N. Cells were counted with a hematocytometer.
FIG. 1.
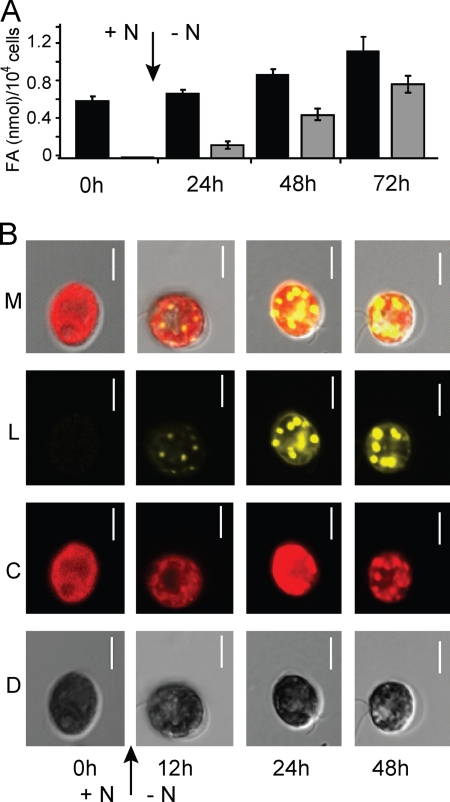
Accumulation of TAGs in lipid droplets during N limitation. (A) Quantification of total cellular fatty acids (black bars) and fatty acids in TAG (gray bars) of dw15 cells grown in medium with 10 mM NH4+ (+N), or cells switched (arrow) to medium with no NH4+ (−N) and grown for 24, 48, or 72 h. (B) Confocal microscopy images of Nile red-stained CC-125 cells grown in medium with 10 mM NH4+ or cells switched (arrow) to medium with 0 mM NH4+ and grown for 12, 24, or 48 h. Differential interference contrast (D), chlorophyll autofluorescence (C), Nile red fluorescence indicating lipid droplets (L), and all three images merged (M) are shown. The scale bars indicate 5 μm.
FIG. 2.
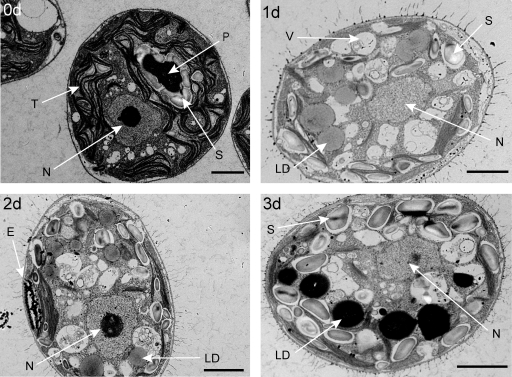
Ultrastructure of cells during N limitation. Electron micrographs of a representative CC-125 cell grown in medium containing 10 mM NH4+ (0d) and representative cells switched to 0 mM NH4+ and grown for 1 (1d), 2 (2d), or 3 (3d) days, respectively, are shown. Scale bars represent 2 μm. E, eyespot; LD, lipid droplets; N, nucleus; P, pyranoid; S, starch granules; T, thylakoid membranes; V, vacuoles.
Isolation of lipid droplets from N-limited C. reinhardtii.
Cells grown in 500 ml TAP-N medium for 24 h were centrifuged at 3,000 × g for 5 min and resuspended in 10 ml of isolation buffer (50 mM HEPES [pH 7.5], 5 mM MgCl2, 5 mM KCl, 0.5 M sucrose, 1 mM phenylmethylsulfonylfluoride, 1 mM 2,2′-dipyridyl, with 1× protease inhibitor cocktail [Roche, Branchburg, NJ]). Cells were disrupted with the aid of a Potter homogenizer, using 20 strokes as well as sonication at the lowest energy setting (3-W output; Misonex Sonicator 3000; Newton, CT) for 5 s. The homogenate was overlaid with buffer 2 (same as the isolation buffer described above but lacking 0.5 M sucrose) and centrifuged at 100,000 × g for 30 min. Lipid droplets floating on the overlay buffer were removed with a bent Pasteur pipette and diluted 10-fold with washing buffer (same as the isolation buffer but with KCl concentration increased to 150 mM). The suspension was overlaid with buffer 2 and centrifuged as described above. This washing process was repeated three times, and lipid droplets were suspended in buffer 2 and stored at −80°C. Preliminary attempts to isolate lipid droplets with this method from cells deprived of nitrogen for 2 or 3 days resulted in lipid droplets with significantly increased chlorophyll contamination compared to the lipid droplets from cells deprived of N for 24 h (data not shown); as such, lipid droplets from cells deprived of N for 24 h were used throughout in this study. Thylakoid membranes were isolated according to Chua and Bennoun (9), and enriched eyespots were prepared as previously described (33).
Mass spectrometric identification of lipid droplet proteins.
Proteins were precipitated as previously described (33) and solubilized in sodium dodecyl sulfate (SDS) sample buffer by sonication. Proteins were separated on 4 to 20% SDS-PAGE gels (Bio-Rad, Hercules, CA) and stained with EZBlue gel staining reagent (Sigma-Aldrich, St. Louis, MO). The lane containing lipid droplet proteins was sliced into 15 bands, which were processed for in-gel trypsin digestion as previously described (34) and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) by the Michigan State University Research and Technology Facility. The extracted peptides were resuspended in a solution of 2% acetonitrile-0.1% trifluoroacetic acid to 20 μl. From this, 10 μl were automatically injected by a Waters nanoAcquity sample manager (Waters, Milford, MA) and loaded for 5 min onto a Waters Symmetry C18 peptide trap (5 μm, 180 μm by 20 mm) at 4 μl/min in 2% acetonitrile-0.1% formic acid. The bound peptides were then eluted onto a Waters BEH C18 nanoAcquity column (1.7 μm, 100 μm by 100 mm) and eluted over 35 min with a gradient of 2% B to 35% B for 21 min, 90% B from 21 to 24 min and 5% B after 24.1 min at a flow rate of 0.6 μl/min. Separations were done using a Waters nanoAcquity UPLC (buffer A, 99.9% water-0.1% formic acid; buffer B, 99.9% acetonitrile-0.1% formic acid), and the separated peptides were sprayed into a Thermo Fisher Scientific (Waltham, MA) LTQ mass spectrometer by using a Michrom Advance nanospray source. Survey scans were taken in the ion trap, and the top five ions from each survey scan were automatically selected for high-resolution zoom scans followed by low-energy collision-induced dissociation (CID). The resulting MS/MS spectra were converted to peak lists by the use of BioWorks browser version 3.3.1 (Thermo Fisher Scientific) using the default parameters and searched against the Chlamydomonas reinhardtii database, version 3.0, downloaded from the DOE Joint Genome Institute (http://www.jgi.doe.gov), using the Mascot searching algorithm, version 2.1 (Matrix Science). The Mascot output was then analyzed using Scaffold (Proteome Software) to probabilistically validate protein identifications by using the ProteinProphet computer algorithm (27). Assignments validated above the Scaffold 95% confidence filter were considered true. Mascot parameters for database searching allowed up to two missed tryptic sites, variable modification of oxidation of methionine and carbamidomethyl cysteine, a peptide tolerance of ±100 ppm, an MS/MS tolerance of 0.8 Da, and a peptide charge state limited to +2/+3.
Fatty acid and lipid analysis.
Various cellular and subcellular fractions were extracted into methanol-chloroform-formic acid (2:1:0.1 [vol/vol/vol]) followed by phase separation by the addition of 1 M KCl and 0.2 M H3PO4. The organic phase was spotted onto ammonium sulfate-impregnated Si250 silica plates, and TAG was separated with petroleum ether-diethyl ether-acetic acid (80:20:1 [vol/vol/vol]). After visualization by brief iodine staining, TAG bands were quantitatively scraped from the plates and processed for fatty acid methyl esterification (FAME) and quantified by gas chromatography as previously described (31). For total cellular fatty acids, the total cellular extract from a known number of cells was processed directly for FAME as described above.
Construction of vectors and transformation of C. reinhardtii.
All PCRs were performed using the Phusion high-fidelity DNA polymerase (Finnzymes, Woburn, MA) according to the manufacturer's instructions; all ligation reactions were performed with T4 DNA ligase (Invitrogen, Carlsbad, CA); and the sequences of the modified regions of all final expression vectors were confirmed by DNA sequencing at the Michigan State University Research Technology Support Facility. In order to construct the pNITPRO1 vector, the Chlamydomonas RBCS2 3′ untranslated region (UTR) was amplified from genomic DNA with primers P1 (5′-GTTGGTACCTGATAAGGATCCCCGCTC-3′) and P2 (5′-GTTGTAAAACGACGGCCAGTGCC-3′) and ligated into KpnI-digested pBluescript II KS+ (Stratagene, Agilent Technologies, Santa Clara, CA) after digestion with the same enzyme; a clone containing the desired orientation of the insert was identified by PCR amplification using P2 (5′-GTTGTAAAACGACGGCCAGTGCC-3′) and P3 (5′-TAATACGACTCACTATAGGG-3′; the T7 sequencing primer), and the resulting plasmid was designated pEM1. The Chlamydomonas NIT1 promoter region was amplified from the vector pMN24 (11) by using primers P4 (5′-CGGTTCTAGACGTGTATGGCTTTGGCTAC-3′) and P5 (5′-GGTAACTAGTACTGGCAGGATTCGGCTG-3′). This amplicon was cut with PciI and SpeI and ligated into pRBCS2 after its digestion with the same enzymes; the resulting plasmid was designated pEM2. PCR primers P6 (5′-TTTGCTCACATGTACAGGTG-3′) and P7 (5′-CCCCCATATGACCCGCTTCAAATACGC-3′) were used to produce a NIT1 promoter-MCS-RBCS2 3′ UTR of pERM2 amplicon, which was cut with PciI and ligated into a PciI/SwaI-digested pSP124S—a plasmid containing the ble selectable marker gene for Chlamydomonas transformation.
To generate an MLDP RNAi construct, primers P8 (5′-GCGCACTAGTCTGGAAAGCCTCTGAAGCAC-3′) and P9 (5′-CCTTCCATGGGGTCGTAGTACTTGTCGGCG-3′) were used to amplify a sense amplicon from genomic DNA, while P10 (5′-CCCCCTGCAGCTGGAAAGCCTCTGAAGCAC-3′) and P11 (5′-GGAACCATGGTCGGTGCTCCGGAGAATCTT-3′) were used to amplify the antisense amplicon from cDNA. The sense and antisense amplicons were double digested with SpeI/NcoI and PstI/NcoI, respectively, and ligated into SpeI/PstI-digested pBluescript II KS+. The formation of the triple ligation product was confirmed by DNA sequencing, and the resulting hairpin insert was excised by SpeI/PstI digestion and ligated into pNITPRO1 digested with the same enzymes. The resulting plasmid was designated pMLDP-RNAi. The C. reinhardtii dw15 strain was transformed as previously described (22), except that vectors were linearized with PacI, and transformants were selected and maintained on TAP agar plates supplemented with 1% yeast extract and 10 μg ml−1 zeocin.
Quantitative real-time reverse transcription-PCR (RT-PCR).
Total RNA was purified using the RNeasy plant minikit (Qiagen, Germantown, MA) including the on-column DNase digestion. Five hundred nanograms of RNA was reverse transcribed using the RETROscript kit (Ambion, Austin, TX) in a 10-μl reaction mixture. Real-time PCR was performed on an ABI Prism 7000 (Applied Biosystems, Foster City, CA) using the SYBR green PCR master mix with an equivalent input of 50 ng RNA and 0.3 μM of each primer (MLDP-F [5′-GGATGCCTGGACCAAGTTC-3′] and MLDP-R [5′-GAGTGCACCAGGAGGTCGT-3′]; RACK1-F [5′-GACCACCAACCCCATCATC-3′] and RACK1-R [5′-AGACGGTCACGGTGTTGAC-3′]). In all experiments, expression of MLDP was normalized to RACK1 (CBLP) expression, a gene commonly used for normalization in C. reinhardtii (2). Analysis of RACK1 expression by the 2−ΔCT method (20) resulted in the following fold changes (averages and standard deviations are indicated) in expression when using the average threshold cycle (CT) value of cells grown in 10 mM N: 1.01 ± 0.17 for cells grown in medium containing 10 mM NH4Cl, and 1.14 ± 0.11, 1.79 ± 0.72, and 1.62 ± 0.28 for cells switched to 0 mM N for 1, 2, and 3 days, respectively. The standard curve method for quantification of mRNA abundance was used as previously described (19). Melting curves were performed after PCR to confirm that the amplification product was unique.
Fluorescent, confocal, and electron microscopy analyses.
For confocal microscopy analysis, cells were resuspended in phosphate-buffered saline (PBS). Cells were stained with Nile red at a final concentration of 2.5 μg ml−1 (from a stock of 50 μg ml−1 in methanol), followed by a 10-min incubation in the dark. Stained cells were then mixed 1:1 (vol/vol) with 2% low-temperature melting point agarose kept at 38°C and applied to the microscope slide. After the solidification of the agarose, images were captured using a Zeiss 510 Meta ConforCor3 LSM microscope (Zeiss, Maple Grove, MN). For neutral-lipid-specific detection of Nile red fluorescence, the 488-nm argon laser was used in combination with a 560- to 615-nm filter; for combined chlorophyll autofluorescence and Nile red fluorescence of polar lipids, a 647- to 753-nm filter was used. Samples were viewed with a 63× oil immersion lens objective. Postacquisition image handling was done with Zeiss AIM software. For fluorescence microscopy analysis, a Zeiss Axio Imager M1 microscope was used. Nile red fluorescence in neutral lipids was detected using a UV light source with a 500 ± 24 nm/542 ± 27 nm excitation/emission filter set. For the measurement of apparent lipid droplet diameter, micrographs were analyzed using Image J software (1). For electron microscopy analysis, centrifuged cell pellets were processed as previously described (13), and transmission electron micrographs were captured using a JEOL100 CXII instrument (Japan Electron Optics Laboratories, Tokyo, Japan).
Bioinformatics.
Sequences were routinely searched using BLAST (3). Sequences were aligned using the Tcoffee multiple sequence alignment program (29). Hydropathy plots were generated employing the Kyte-Doolittle algorithm (18), using the ProtScale program at http://www.expasy.ch/tools/protscale.html. The value G in each graph is the grand average of hydropathy value (GRAVY) for each protein and was calculated using the GRAVY calculator program at http://gravy.laborfrust.de/.
RESULTS
TAG accumulation following shift to N-limited medium.
While C. reinhardtii does not accumulate TAG under nutrient-replete growth conditions, TAG has been reported to accumulate in this genus after N or phosphorus limitation (39). In order to optimize conditions for the isolation of lipid droplets from C. reinhardtii cells, the time course of N deprivation and its effect on TAG accumulation and lipid droplet formation was analyzed. The amount of fatty acid esterified in TAG steadily increased over time to approximately 12%, 45%, and then 65% of total cellular fatty acids 24, 48, and 72 h, respectively, after resuspending the cells in TAP medium lacking a source of N, as shown in Fig. 1A. Total fatty acid profiles of N-limited cells showed moderate changes in comparison to N-replete cells, with a slight increase in 16:0 (carbon number:number of double bonds) and a concomitant decrease in 18:3Δ9,12,15 (superscript indicates the position of the double bond counting from the carboxyl end); TAG fatty acid profiles of N-replete and N-limited cells were more distinct, with a decrease in 16:0 and an increase in 18:1/16:4 in N-limited cells (Table 1). Total cellular fatty acids increased on a per-cell basis 48 and 72 h after imposing N-limiting conditions (Fig. 1A), indicating that de novo fatty acid synthesis contributes at least in part to TAG accumulation.
TABLE 1.
Total fatty acid content of whole cells compared to fatty acids bound in TAGs isolated from +N cells and −N cellsb
| Fatty acid | Fatty acid content ofa: |
|||
|---|---|---|---|---|
| Whole cells |
TAG |
|||
| +N | −N | +N | −N | |
| 16:0 | 22.1 ± 0.14 | 26.2 ± 0.70 | 51.3 ± 1.30 | 37.6 ± 3.99 |
| 16:1 | 3.4 ± 0.02 | 2.9 ± 0.01 | 5.4 ± 2.37 | 3.7 ± 0.54 |
| 16:2 | 1.2 ± 0.00 | 1.4 ± 0.06 | 1.6 ± 0.96 | 1.4 ± 0.17 |
| 16:3 | 4.5 ± 0.15 | 3.4 ± 0.25 | 13.9 ± 6.88 | 3.2 ± 0.27 |
| 18:0 | 2.4 ± 0.02 | 2.5 ± 0.06 | 0.8 ± 0.84 | 1.4 ± 0.17 |
| 18:1, 16:4 | 21.9 ± 0.13 | 21.2 ± 0.14 | 7.9 ± 2.46 | 22.1 ± 8.45 |
| 18:2 | 8.0 ± 0.02 | 9.9 ± 0.06 | 6.1 ± 3.33 | 13.6 ± 1.51 |
| 18:3Δ5,9,12 | 6.7 ± 0.01 | 6.5 ± 0.12 | 4.6 ± 1.62 | 5.7 ± 0.63 |
| 18:3Δ9,12,15 | 27.9 ± 0.10 | 24.7 ± 0.13 | 7.3 ± 3.69 | 9.9 ± 1.20 |
| 18:4 | 1.9 ± 0.02 | 1.5 ± 0.02 | 1.1 ± 0.16 | 1.3 ± 0.16 |
Averages of three replicates and the standard deviation are shown.
+N indicates cells grown in medium containing 10 mM NH4+; −N indicates cells switched to 0 mM NH4+ and grown for 1 day.
A time course of lipid droplet formation in N-deprived C. reinhardtii cells is shown in Fig. 1B. Using the lipid-specific fluorescent dye, Nile red, lipid droplet formation was observed by confocal microscopy, adjusting excitation and emission filters to predominately detect fluorescence from the neutral lipid fraction (10). The first lipid droplets were observed after 12 h of N deprivation in distinct locations within the cell, and prolonged exposure of the cells to 24 or 48 h of N deprivation caused an increase in size and number of lipid droplets. Similarly, transmission electron micrographs revealed several ultra-structural changes that occur over a 72-h time course of N limitation, including a reduction of stacked thylakoid membranes, accumulation of starch granules, and the appearance of lipid droplets (Fig. 2).
Properties of lipid droplets.
Lipid droplets were prepared from cells that were deprived of N for 24 h and compared to isolated thylakoid membranes and a fraction highly enriched in eyespots—a chloroplast-localized, carotenoid- and lipid-rich organelle (33). Results of spectral analyses of the various subcellular fractions indicated the presence of carotenoids (Fig. 3A). These results were similar to what was previously observed for an enriched eyespot fraction (33), although when normalized on a per-fatty-acid basis, carotenoids were much less abundant in lipid droplets. The near absence of chlorophylls in isolated lipid droplets represented by the absorbance maximum at approximately 660 nm visible in the thylakoid fraction (Fig. 3A) suggested low contamination of the lipid droplets with thylakoid membranes. The ratios of TAG-esterified fatty acids to total fatty acids of lipid droplets and eyespots were approximately 88% and 25%, respectively (Fig. 3B), corresponding to the higher relative membrane abundance in the eyespots. The TAG fatty acid profile of isolated lipid droplets was very similar to the profile of TAGs isolated from cells deprived of N for 1 day, whereas the TAG fatty acid profile of eyespots was quite distinct, being much more enriched with palmitic acid on a mole percentage basis (Table 2). Taken together, these results indicated that the lipid droplets were distinct from the lipid globules found in the eyespot fraction, with minimal contamination of thylakoid membranes, the predominant membrane fraction in C. reinhardtii.
FIG. 3.
Characteristics of isolated lipid droplets. (A) Absorbance spectra of ethanol extracts (96%) normalized on an equal fatty acid basis from thylakoids of cells grown in medium containing 10 mM NH4+ (+N) or cells switched to 0 mM NH4+ and grown for 1 day (−N 1d) from enriched eyespot fraction and from lipid droplets were recorded. (B) Ratios of fatty acids derived from TAG (TAG FA) to total fatty acids (Total FA) for extracts from whole cells grown in medium with 10 mM NH4+ (+N) or whole cells switched to and grown in medium with no NH4+ (−N) for 24, 48, or 72 h, the enriched eyespot fraction (Eye), and isolated lipid droplets (LD) as indicated. (C) Proteins (30 μg) separated by gradient (4 to 20%) SDS-PAGE from whole cells (Tot. Prot., total protein), thylakoids, eyespots, and lipid droplets as indicated. The positions of molecular mass markers in kDa are indicated on the left, and sections excised from the gel separating lipid droplet-associated protein are indicated on the right.
TABLE 2.
Fatty acid content of whole organelles (eyespot and lipid droplet) compared to fatty acids bound in TAGs isolated from indicated organelles
| Fatty acid | Fatty acid content ofa: |
|||
|---|---|---|---|---|
| Eyespot |
Lipid droplet |
|||
| Whole organelle | TAG | Whole organelle | TAG | |
| 16:0 | 35.5 ± 0.67 | 67.0 ± 7.55 | 32.2 ± 1.24 | 41.8 ± 4.33 |
| 16:1 | 3.0 ± 0.01 | 4.5 ± 1.08 | 3.4 ± 0.37 | 3.1 ± 0.58 |
| 16:2 | 3.4 ± 0.02 | 3.0 ± 0.30 | 1.2 ± 0.15 | 0.9 ± 0.06 |
| 16:3 | 15.7 ± 0.18 | 7.8 ± 0.45 | 6.4 ± 3.03 | 5.1 ± 1.70 |
| 18:0 | 3.0 ± 0.01 | 1.9 ± 0.50 | 2.1 ± 0.87 | 1.5 ± 0.77 |
| 18:1, 16:4 | 18.4 ± 0.39 | 7.9 ± 1.99 | 27.2 ± 1.74 | 27.3 ± 2.86 |
| 18:2 | 10.9 ± 0.71 | 4.7 ± 1.77 | 10.7 ± 0.67 | 10.0 ± 1.67 |
| 18:3Δ5,9,12 | 5.6 ± 0.09 | 1.4 ± 0.57 | 7.7 ± 1.59 | 5.9 ± 2.62 |
| 18:3Δ9,12,15 | 3.3 ± 0.47 | 0.8 ± 0.41 | 6.8 ± 4.69 | 2.9 ± 0.34 |
| 18:4 | 1.1 ± 0.02 | 1.1 ± 0.47 | 2.3 ± 0.92 | 1.4 ± 0.38 |
Organelles were isolated from cells that were switched to medium containing 0 mM NH4+ and grown for 1 day. Two replicates were averaged for eyespot samples and three for lipid droplet samples. The averages and the standard deviation are shown.
Analysis of proteins from whole cells, thylakoids, the eyespots, and lipid droplets by SDS-PAGE followed by total protein staining (Fig. 3C) revealed a pattern for lipid droplets that was distinct from the other protein fractions, with one major protein of approximately 27 kDa, as well as several less-abundant proteins, which were specifically enriched in lipid droplets compared to thylakoids and the eyespots. The lipid droplet proteins were excised from the gel (as indicated in Fig. 3C) and subjected to MS for protein identification.
Proteins identified in the lipid droplet fraction.
As summarized in Table S1 in the supplemental material, 259 proteins with two or more unique peptides were identified and fell into a broad range of protein functional groups (see Fig. S1A in the supplemental material). Sixteen proteins predicted to be involved in lipid metabolism, including three presumed acyl coenzyme A (acyl-CoA) synthetases and a predicted lipoxygenase—proteins previously identified as associated with lipid droplets by studies of other organisms (6, 8)—were among those proteins. BTA1, the enzyme responsible for the synthesis of a major extraplastidic membrane lipid, diacylglyceryl-N-trimethylhomoserine (30), was also present. Several predicted components of vesicular trafficking pathways were identified, including subunits of the coat protein complex I and its putative regulator, ARF1a, as well as other small Rab-type GTPases. Orthologs of these proteins have also been identified in lipid droplet proteomics studies done on different animal species (6, 8), and recent Drosophila cell RNAi inactivation studies targeting Arf79a (ARF1a ortholog) and the coat protein complex I-encoding genes revealed a previously unknown role for these proteins in lipid droplet formation and utilization (7, 12). The presence of presumed orthologs in C. reinhardtii lipid droplets suggests the possibility of a similar role in algal lipid droplet biogenesis.
Other functional groups which were represented by many proteins included 54 predicted ribosomal and translation-related proteins, 26 predicted mitochondrial proteins, and 24 predicted proteins of the photosynthetic apparatus. Ribosomal and mitochondrial proteins have been identified in lipid droplet proteomic studies of different animal species (8, 40) and plant oil bodies (in plants from mitochondria only) (17), and could be common contaminating proteins in lipid droplet preparations or, alternatively, could indicate the presence of ribosomal/mitochondrial-lipid droplet physical interactions, as has been previously postulated (8). The identification of 24 photosynthetic apparatus proteins was surprising, given the near absence of chlorophyll in purified lipid droplets, and it is likely that these hydrophobic apoproteins are contaminating artifacts that become associated during the isolation procedure; however, the most abundant stromal enzyme, ribulose-1,5-bisphosphate carboxylase/oxygenase, was not detected. A more stringent analysis of 93 proteins (identified by more than 12 spectral counts) resulted in an ∼38% decrease in the number of lipid metabolism proteins retained in the “identified” group (from 16 to 10), whereas relative decreases in the more predominant contaminating groups were generally higher (∼78% for ribosome/DNA/translation-related proteins, ∼69% for mitochondrial proteins, and ∼50% for photosynthetic apparatus proteins [see Fig. S1A and S1B in the supplemental material]). In addition, 45 proteins of unknown function were identified. Based on spectral counts for all unique peptides for a given protein (see Table S2 in the supplemental material), one of these was, by a factor of 10, the most abundant protein in gel slice 9 (Fig. 3C), which also contained the most abundant protein in the lipid droplet fraction based on staining. Its calculated molecular mass is 28 kDa, which approximately corresponded to its position in the gel (Fig. 3C). Therefore, this protein was presumed to be the most abundant protein in the lipid droplet fraction and was tentatively designated the major lipid droplet protein (MLDP; protein identification no. 192823 in the C. reinhardtii v.3.0 database [http://genome.jgi-psf.org/Chlre3/Chlre3.home.html]; NCBI accession number XP_001697668). Notably, the second-most abundant protein based on spectral counts was BTA1, the enzyme catalyzing the biosynthesis of betaine lipid. Both proteins were also identified in studies of C. reinhardtii lipid droplets conducted in parallel but were dismissed as contamination (41).
MLDP suppression affects lipid droplet size.
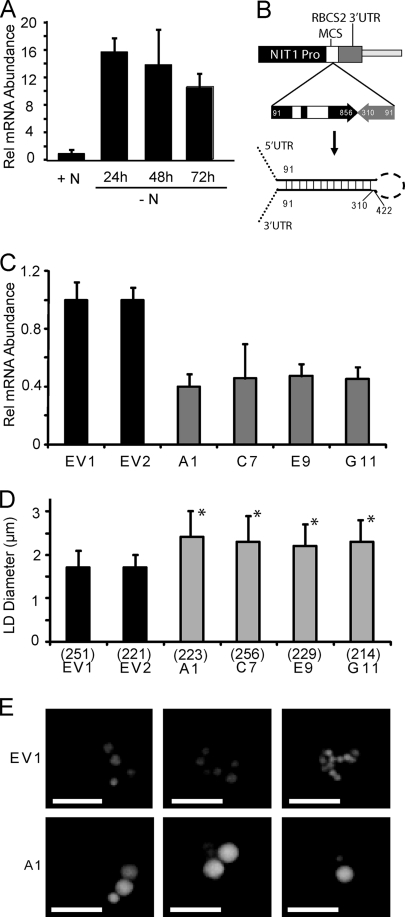
Steady-state mRNA abundance of MLDP was determined by quantitative real-time RT-PCR in cells grown in 10 mM N and cells deprived of N for 24, 48, and 72 h as shown in Fig. 4A. The widely used C. reinhardtii control gene RACK1 (CBLP) was used as an internal control. Analysis of apparent fold change in RACK1 expression by the 2−ΔCT method (see Materials and Methods) indicated that changes in its expression based on total RNA input to the reverse transcriptase reaction were minor if there were any (∼1.5-fold increases at 48 and 72 h with no N). Previous studies have shown that during N deprivation in C. reinhardtii, the amount of chloroplast and cytoplasmic ribosomes and total RNA content decrease (24). While this can potentially add an additional level of difficulty in identifying a suitable control gene in these experiments, the degree to which this contributes to the apparent minor fold changes in expression for RACK1 is unknown. With the above in mind, RACK1 was deemed a suitable control gene for these experiments, with the caveat that it may slightly underestimate the fold changes for genes induced by 48 or 72 h of N deprivation, and vice versa for downregulated transcripts. When normalized to RACK1, MLDP mRNA showed a 16-fold increase in abundance after 24 h of N deprivation and continued to be abundant even after 72 h of N deprivation. Moreover, MLDP mRNA abundance qualitatively corresponded during a time course of N deprivation with the accumulation of TAG (Fig. 1A). The large increase in MLDP transcript abundance in N-deprived cells was further corroborated by Northern blotting (Fig. 5). To begin the functional analysis of MLDP in C. reinhardtii, MLDP RNAi lines were generated. For this purpose, a plasmid carrying an expression cassette which is inducible by N deprivation, pNITPRO1, was constructed based on the nitrate reductase (NIT1) promoter. The pNITPRO1 vector was used to drive the expression of an MLDP genomic sense cDNA antisense RNAi hairpin (Fig. 4B). Zeocin-resistant transformants were screened for MLDP mRNA reduction at 48 or 72 h of N deprivation. Of 64 lines screened, nine lines were identified with a range of MLDP transcript reduction between 20% and 60% of empty vector controls. The four strongest MLDP-RNAi lines (A1, C7, E9, and G11) showed a consistent 55 to 60% reduction in MLDP transcript measured after 72 h of N deprivation from three independently grown cultures (Fig. 4C).
FIG. 4.
Induction and RNAi suppression of MLDP gene expression. (A) Relative abundance of MLDP mRNA normalized to RACK1 (CBLP) in wild-type dw15 cells grown in medium with 10 mM NH4+ (+N), or dw15 cells switched to and grown in medium with no NH4+ (−N) for 24, 48, or 72 h measured by quantitative RT-PCR. Data are expressed as fold changes compared to the +N growth condition, which is given a value of 1. The standard deviation of three biological repeats, each done in triplicate, is shown. (B) Diagram showing the MLDP-RNAi construct designed with the NIT1 inducible promoter (Pro) of vector pNITPRO1 driving the expression of an MLDP genomic inverted repeat (numbers indicate base pairs from predicted 5′ UTR), and the predicted transcribed RNA hairpin product. (C) QRT-PCR analysis of MLDP mRNA levels in two empty vector control lines (EV1 and EV2), and four MLDP-RNAi lines isolated from four independent transformation events (A1, C7, E9, G11). Data are represented as percent change compared to EV1 and normalized to RACK1 as in panel A. Three replicates were averaged and standard deviation is shown. (D) Analysis of apparent lipid droplet diameter in empty vector controls and MLDP-RNAi lines described in (C). The average and standard deviation are shown for n (indicated in parentheses below each bar) lipid droplets counted, and cell lines are marked with an asterisk, for which P < 0.01 compared to EV1 in a two-tailed Student t test. (E) Fluorescent micrographs of representative cells of lines EV1 and A1 stained for lipid droplets with Nile red.
FIG. 5.
Northern blot analysis of MLDP. Five micrograms of total RNA isolated from cells grown in 10 mM NH4Cl (10 mM N) and cells deprived of N for 2 days (−N 2 d) was processed for Northern blotting as previously described (23). MLDP mRNA was specifically detected using a 32P-radiolabeled probe amplified from cDNA with forward and reverse primers 5′-ATGGTGGTGATCTTGTTTGACAGAG-3′ and 5′-CATTCCGCTGTACCTCCAGG-3′. Total RNA was stained with methylene blue (23) and shown as a loading control (rRNA). The results for two biological replicates of this experiment are shown (panels 1 and 2).
The MLDP-RNAi lines were analyzed for possible lipid droplet morphological phenotypes by fluorescence microscopy analysis of Nile red-stained cells deprived of N for 3 days. An apparent increase in the average size of lipid droplets in the MLDP-RNAi lines was observed (Fig. 4D and E). The apparent individual diameters of a population of >200 random and in-focus lipid droplets per RNAi or control line were measured after 3 days of N deprivation (Fig. 4D). An approximately 40% increase in the average lipid droplet diameter was determined in the MLDP-RNAi lines.
MLDP has orthologs only in green algae.
The newly discovered MLDP protein has few orthologs, none of which have been functionally characterized. When searched against the National Center for Biological Information database of nonredundant proteins by the use of BLAST (3), no sequence with significant similarity is identified. However, several genome projects of green algal species are currently under way, and by searching against green algal genomes at the respective websites, potential orthologs with high sequence identity were identified in Chlorella sp. NC64A, Chlorella vulgaris, and Volvox carteri (Fig. 6). No discernible ortholog was present in available diatom or red algal genomes or the genomes of Ostreococcus species, which belong to a green algal lineage distinct from C. reinhardtii. Apparently, MLDP is a rare protein in nature as we currently know it at the molecular level and presumably arose during the evolution of green algae. As such, it also has no discernible domains to which functions have been assigned.
FIG. 6.
Multiple sequence alignment of MLDP with other green algal orthologs and hydropathy plots of MLDP and other known lipid droplet binding proteins. (A) MLDP and putative orthologs identified in the Chlorella sp. NC64A (http://genome.jgi-psf.org/ChlNC64A_1/ChlNC64A_1.home.html), Chlorella vulgaris C-169 (http://genome.jgi-psf.org/Chlvu1/Chlvu1.home.html), and Volvox carteri (http://genome.jgi-psf.org/Volca1/Volca1.home.html) genomes. Conserved residues are indicated with asterisks, and the coloring indicates the CORE index, where red and blue indicate the highest and lowest probability of the alignment being correct at a given residue, respectively, with colors in between the spectrum of red and blue representing a range of intermediate probabilities. Hydropathy plots for the proteins (followed by NCBI accession numbers in parentheses) MLDP (XP_001697668) (B), mouse ADRP (NP_031434) (C), mouse perilipin (NP_783571) (D), and Arabidopsis OLE1 (NP_194244) (E).
Grand average of hydropathy (GRAVY) indices and Kyte-Doolittle (18) hydropathy plots were generated for MLDP, as well as for mouse ADRP and perilipin, and Arabidopsis oleosin 1 (OLE1) (Fig. 6B to E). All four proteins were distinct with respect to extent and region of hydrophobicity, with the most hydrophobic region for MLDP occurring approximately between amino acid residues 160 and 215. The proteins ranked according to their GRAVY index (highest to lowest) are MLDP (0.11), OLE1 (−0.17), ADRP (−0.28), and perilipin (−0.40). Thus, MLDP is a very hydrophobic protein although it lacks a specific hydrophobic core characteristic for oleosin.
DISCUSSION
The potential of microalgae to provide sustainable feedstock for biofuel production has led to renewed interest in the biology of this diverse group of organisms (14). While C. reinhardtii is unlikely to become the microalga of choice for the production of biofuels, it is a well-established genetic and genomic model for the study of unicellular green algae (25). However, very little is known about lipid metabolism or the cell biology of lipid droplets and its regulation in this organism. The time course and conditions for the induction of oil accumulation described here (Fig. 1 and 2) and the detailed analysis of lipid droplets provide the basis for a molecular analysis of the mechanisms of TAG accumulation in C. reinhardtii and possibly unicellular green algae in general. As a logical first step, we focused on the TAG-storing compartment itself, the lipid droplet. Its fatty acid and pigment composition is clearly distinct from that of other organelles (Fig. 3). This distinct composition and the very low chlorophyll content in view of the predominance of thylakoid membranes in the cell suggested that the obtained oil droplet preparation was only slightly contaminated with thylakoid membranes, the largest membrane system in the cell. However, we cannot rule out some level of contamination with ER-derived microsomes especially since lipid droplets are generally thought to be derived from the ER. An increase in the total amount of fatty acids in N-deprived cells indicated that the biosynthesis of TAGs is not merely due to conversion of membrane lipids but the result of de novo synthesis. The specific fatty acid composition of lipid droplet lipids tended toward shorter and more saturated fatty acids, suggesting that newly synthesized fatty acids were quickly incorporated into TAGs before fatty acid modification by desaturases or elongases could occur; alternatively, reduced activities of these fatty acid metabolic enzymes could account for this observation.
Lipid droplets in different organisms are increasingly recognized as dynamic organelles that provide important nodes of cellular metabolic networks and participate in intracellular lipid trafficking (e.g., see reference 12). In algae, the induction of lipid droplet formation is intricately regulated by nutrient deprivation and other abiotic stresses that lead to a cessation of cell division or an overreduction of the photosynthetic electron transport chain (14). In this study, we sought to identify the proteins associated with lipid droplets to determine factors involved in the biogenesis or turnover of this organelle in C. reinhardtii. Aside from those of unknown function, of particular interest are lipid metabolic enzymes and proteins involved in vesicular transport. Intriguingly, the betaine lipid (diacylglyceryl-N,N,N-trimethylhomoserine) biosynthetic enzyme, BTA1 (30), had the second highest spectral count of proteins in the lipid droplet fraction. This protein was also identified as one of few in the lipid droplet fraction in a parallel study but was considered a contamination (41). This parallel study concluded that no protein was specifically associated with the respective lipid droplet fraction prepared and treated differently than in this study.
Here, a number of predicted acyl-CoA synthetases and acyltransferases were present as well that could be involved in the biosynthesis of TAGs or the transfer of acyl groups between membrane lipid and neutral lipid pools. In animal cells, enzymes involved in phosphatidylcholine biosynthesis have been found to be critical for lipid droplet formation and turnover. Lipid droplets in animal cells are surrounded by a monolayer of phospholipids, and the availability of those phospholipids affects lipid droplet size. When the rate-limiting enzyme of phosphatidylcholine biosynthesis was inactivated in Drosophila cells, lipid droplets increased in size presumably because the larger droplets have a lower surface area-to-volume ratio and require less phospholipids to form the phospholipid monolayer (12). As C. reinhardtii lacks phosphatidylcholine, it seems plausible that its role in the formation of lipid droplets is taken by betaine lipid, which has similar properties and is thought to functionally replace phosphatidylcholine in this green alga (32). A rich complement of proteins predicted to be involved in vesicular trafficking, including COP1 homologs, Rab GTPase, or ARF-related GTPase, were also identified in the proteomics data set. Repression of the respective orthologs in Drosophila cells was shown to affect lipid droplet formation and dynamics and TAG utilization (12). Our understanding of the interaction of lipid droplets with the vesicular transport machinery is only rudimentary at this time. The presence of the respective proteins in C. reinhardtii follows a common theme also observed for animal cells and provides an opportunity to study the role of the vesicular transport machinery in lipid droplet formation in a unicellular model organism. However, the specific association of these proteins with lipid droplets in C. reinhardtii remains to be confirmed.
The identification of proteins in the ribosome-/translation-/DNA-related, mitochondrial, and photosynthetic apparatus functional groups suggests a basic level of contamination in our lipid droplet protein fraction. While more-stringent filtering of the proteomics data set to include proteins with more than 12 spectral counts resulted in a larger decrease in the relative proportion of these apparent contaminating functional groups, compared to lipid metabolism proteins (see Fig. S1B and Table S2 in the supplemental material), the significance of such analyses should be regarded as circumstantial, given the potential for protein size bias in the method. Comparing the results of the analysis of the lipid droplet proteins presented here with proteomic studies of the eyespot (33), flagella (28), and mitochondria (5) (see Table S1 in the supplemental material) reveals that some degree of contamination with proteins from specific functional groups is common (e.g., ribosome-/translation-/DNA-related, mitochondrial, and photosynthetic apparatus proteins found in the eyespot study, or ribosome contamination in the mitochondrial and flagellar studies). As such, apparent contamination of subcellular preparations in C. reinhardtii seems to be a common challenge.
To begin the functional analysis of lipid droplet-associated proteins in C. reinhardtii, we focused on the most abundant protein in the lipid droplet fraction based on Coomassie brilliant blue staining (Fig. 3C) and spectral counts (see Table S2 in the supplemental material). The identity of this protein was deduced by MS analysis of all proteins in the respective gel slice, and MLDP was identified based on 10-fold overabundance of spectral counts for its peptides. Repeated attempts to localize an MLDP-green fluorescent protein (GFP) fusion protein to the lipid droplets failed because none of the tested constructs were expressed in C. reinhardtii. Unlike plants, C. reinhardtii does not seem to have an ortholog of oleosins, distinct structural proteins surrounding the lipid bodies in plant oil seeds (15, 36). Interestingly, oleosins are also absent from oil-rich mesocarp tissues of plants like oil palm or olives, and their presence may be related to special needs of seeds. Indeed, reduced oleosin content leads to delayed germination and reduced freezing tolerance (35, 36). At the cellular level, a reduction in oleosin caused an increase in oil body size. Likewise, when MLDP was inactivated in C. reinhardtii, an increase in lipid droplet size was observed (Fig. 4). Despite an extensive screening of transgenic lines, expression of the MLDP gene could be reduced by only approximately 60% in the best RNAi lines leading to a moderate but statistically significant increase in oil droplet size in these lines. Because MLDP lacks any features that readily classify it as a protein with known catalytic activity, it may function as a structural protein affecting lipid droplet dynamics and size in C. reinhardtii. An increase in lipid droplet size has been shown in different systems to delay turnover of TAGs. We measured TAG levels (at 3 days of N deprivation) and TAG turnover in these lines following the resupply of N in the medium but could not detect a statistically significant difference in the levels of TAG or rate of TAG metabolization in these lines compared to the empty vector control lines. It seemed likely that the observed moderate change in lipid droplet size would lead to only a small change in the rate of TAG metabolization, too small for it to be detected within the statistical limitations of our analytical method. This does not rule out the possibility that MLDP is important in preventing lipid droplet fusion to provide a larger surface area-per-volume ratio to increase the accessibility of enzymes metabolizing TAGs.
The newly discovered MLDP is a rare protein in nature unique to green algae. The expression of its gene is strictly limited to conditions favoring TAG biosynthesis, providing indirect evidence for a role of MLDP in lipid droplet formation or maintenance. These properties potentially make MLDP a marker for lipid droplets and TAG accumulation.
Supplementary Material
Acknowledgments
This work was supported by a grant from the U.S. Air Force Office of Scientific Research Grant FA9550-08-0165 and the Michigan Agriculture Experiment Station.
Footnotes
Published ahead of print on 13 November 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 11:36-42. [Google Scholar]
- 2.Allen, M. D., J. Kropat, S. Tottey, J. A. Del Campo, and S. S. Merchant. 2007. Manganese deficiency in Chlamydomonas results in loss of photosystem II and MnSOD function, sensitivity to peroxides, and secondary phosphorus and iron deficiency. Plant Physiol. 143:263-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Athenstaedt, K., D. Zweytick, A. Jandrositz, S. D. Kohlwein, and G. Daum. 1999. Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J. Bacteriol. 181:6441-6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atteia, A., A. Adrait, S. Brugiere, M. Tardif, L. R. van, O. Deusch, T. Dagan, L. Kuhn, B. Gontero, W. Martin, J. Garin, J. Joyard, and N. Rolland. 2009. A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the alpha-proteobacterial mitochondrial ancestor. Mol. Biol. Evol. 26:1533-1548. [DOI] [PubMed] [Google Scholar]
- 6.Bartz, R., J. K. Zehmer, M. Zhu, Y. Chen, G. Serrero, Y. Zhao, and P. Liu. 2007. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J. Proteome Res. 6:3256-3265. [DOI] [PubMed] [Google Scholar]
- 7.Beller, M., C. Sztalryd, N. Southall, M. Bell, H. Jackle, D. S. Auld, and B. Oliver. 2008. COPI complex is a regulator of lipid homeostasis. PLoS Biol. 6:e292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cermelli, S., Y. Guo, S. P. Gross, and M. A. Welte. 2006. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr. Biol. 16:1783-1795. [DOI] [PubMed] [Google Scholar]
- 9.Chua, N. H., and P. Bennoun. 1975. Thylakoid membrane polypeptides of Chlamydomonas reinhardtii: wild-type and mutant strains deficient in photosystem II reaction center. Proc. Natl. Acad. Sci. U. S. A. 72:2175-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elsey, D., D. Jameson, B. Raleigh, and M. J. Cooney. 2007. Fluorescent measurement of microalgal neutral lipids. J. Microbiol. Methods 68:639-642. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez, E., R. Schnell, L. P. Ranum, S. C. Hussey, C. D. Silflow, and P. A. Lefebvre. 1989. Isolation and characterization of the nitrate reductase structural gene of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U. S. A. 86:6449-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo, Y., T. C. Walther, M. Rao, N. Stuurman, G. Goshima, K. Terayama, J. S. Wong, R. D. Vale, P. Walter, and R. V. Farese. 2008. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature 453:657-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris, E. H. 1989. The Chlamydomonas sourcebook: a comprehensive guide to biology and laboratory use. Academic Press, San Diego, CA. [DOI] [PubMed]
- 14.Hu, Q., M. Sommerfeld, E. Jarvis, M. Ghirardi, M. Posewitz, M. Seibert, and A. Darzins. 2008. Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 54:621-639. [DOI] [PubMed] [Google Scholar]
- 15.Huang, A. H. C. 1992. Oil bodies and oleosins in seeds. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43:177-200. [Google Scholar]
- 16.Jolivet, P., E. Roux, S. d'Andrea, M. Davanture, L. Negroni, M. Zivy, and T. Chardot. 2004. Protein composition of oil bodies in Arabidopsis thaliana ecotype WS. Plant Physiol. Biochem. 42:501-509. [DOI] [PubMed] [Google Scholar]
- 17.Katavic, V., G. K. Agrawal, M. Hajduch, S. L. Harris, and J. J. Thelen. 2006. Protein and lipid composition analysis of oil bodies from two Brassica napus cultivars. Proteomics 6:4586-4598. [DOI] [PubMed] [Google Scholar]
- 18.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 19.Larionov, A., A. Krause, and W. Miller. 2005. A standard curve based method for relative real time PCR data processing. BMC Bioinformatics 6:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)). Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 21.Londos, C., C. Sztalryd, J. T. Tansey, and A. R. Kimmel. 2005. Role of PAT proteins in lipid metabolism. Biochimie 87:45-49. [DOI] [PubMed] [Google Scholar]
- 22.Lumbreras, V., D. R. Stevens, and S. Purton. 2008. Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J. 14:441-447. [Google Scholar]
- 23.Mamedov, T. G., E. R. Moellering, and R. Chollet. 2005. Identification and expression analysis of two inorganic C- and N-responsive genes encoding novel and distinct molecular forms of eukaryotic phosphoenolpyruvate carboxylase in the green microalga Chlamydomonas reinhardtii. Plant J. 42:832-843. [DOI] [PubMed] [Google Scholar]
- 24.Martin, N. C., K. Chiang, and U. W. Goodenough. 1976. Turnover of chloroplast and cytoplasmic ribomsomes during gametogenesis in Chlamydomonas reinhardi. Dev. Biol. 51:190-201. [DOI] [PubMed] [Google Scholar]
- 25.Merchant, S. S., S. E. Prochnik, O. Vallon, E. H. Harris, S. J. Karpowicz, G. B. Witman, A. Terry, A. Salamov, L. K. Fritz-Laylin, L. Marechal- Drouard, W. F. Marshall, L. H. Qu, D. R. Nelson, A. A. Sanderfoot, M. H. Spalding, V. V. Kapitonov, Q. Ren, P. Ferris, E. Lindquist, H. Shapiro, S. M. Lucas, J. Grimwood, J. Schmutz, P. Cardol, H. Cerutti, G. Chanfreau, C. L. Chen, V. Cognat, M. T. Croft, R. Dent, S. Dutcher, E. Fernandez, H. Fukuzawa, D. Gonzalez-Ballester, D. Gonzalez-Halphen, A. Hallmann, M. Hanikenne, M. Hippler, W. Inwood, K. Jabbari, M. Kalanon, R. Kuras, P. A. Lefebvre, S. D. Lemaire, A. V. Lobanov, M. Lohr, A. Manuell, I. Meier, L. Mets, M. Mittag, T. Mittelmeier, J. V. Moroney, J. Moseley, C. Napoli, A. M. Nedelcu, K. Niyogi, S. V. Novoselov, I. T. Paulsen, G. Pazour, S. Purton, J. P. Ral, D. M. Riano-Pachon, W. Riekhof, L. Rymarquis, M. Schroda, D. Stern, J. Umen, R. Willows, N. Wilson, S. L. Zimmer, J. Allmer, J. Balk, K. Bisova, C. J. Chen, M. Elias, K. Gendler, C. Hauser, M. R. Lamb, H. Ledford, J. C. Long, J. Minagawa, M. D. Page, J. Pan, W. Pootakham, S. Roje, A. Rose, E. Stahlberg, A. M. Terauchi, P. Yang, S. Ball, C. Bowler, C. L. Dieckmann, V. N. Gladyshev, P. Green, R. Jorgensen, S. Mayfield, B. Mueller-Roeber, S. Rajamani, R. T. Sayre, P. Brokstein, I. Dubchak, D. Goodstein, L. Hornick, Y. W. Huang, J. Jhaveri, Y. Luo, D. Martinez, W. C. Ngau, B. Otillar, A. Poliakov, A. Porter, L. Szajkowski, G. Werner, K. Zhou, I. V. Grigoriev, D. S. Rokhsar, and A. R. Grossman. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318:245-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy, D. J. 2001. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog. Lipid Res. 40:325-438. [DOI] [PubMed] [Google Scholar]
- 27.Nesvizhskii, A. I., A. Keller, E. Kolker, and R. Aebersold. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75:4646-4658. [DOI] [PubMed] [Google Scholar]
- 28.Pazour, G. J., N. Agrin, J. Leszyk, and G. B. Witman. 2005. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170:103-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poirot, O., E. O'Toole, and C. Notredame. 2003. Tcoffee@igs: a web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 31:3503-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riekhof, W. R., B. B. Sears, and C. Benning. 2005. Annotation of genes involved in glycerolipid biosynthesis in Chlamydomonas reinhardtii: discovery of the betaine lipid synthase BTA1Cr. Eukaryot. Cell 4:242-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossak, M., A. Schäfer, N. Xu, D. A. Gage, and C. Benning. 1997. Accumulation of sulfoquinovosyl-1-O-dihydroxyacetone in a sulfolipid-deficient mutant of Rhodobacter sphaeroides inactivated in sqdC. Arch. Biochem. Biophys. 340:219-230. [DOI] [PubMed] [Google Scholar]
- 32.Sato, N., and N. Murata. 1991. Transition of lipid phase in aqueous dispersions of diacylglyceryltrimethylhomoserine. Biochim. Biophys. Acta 1082:108-111. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt, M., G. Gessner, M. Luff, I. Heiland, V. Wagner, M. Kaminski, S. Geimer, N. Eitzinger, T. Reissenweber, O. Voytsekh, M. Fiedler, M. Mittag, and G. Kreimer. 2006. Proteomic analysis of the eyespot of Chlamydomonas reinhardtii provides novel insights into its components and tactic movements. Plant Cell 18:1908-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 35.Shimada, T. L., T. Shimada, H. Takahashi, Y. Fukao, and I. Hara-Nishimura. 2008. A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J. 55:798-809. [DOI] [PubMed] [Google Scholar]
- 36.Siloto, R. M., K. Findlay, A. Lopez-Villalobos, E. C. Yeung, C. L. Nykiforuk, and M. M. Moloney. 2006. The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18:1961-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tansey, J. T., C. Sztalryd, J. Gruia-Gray, D. L. Roush, J. V. Zee, O. Gavrilova, M. L. Reitman, C. X. Deng, C. Li, A. R. Kimmel, and C. Londos. 2001. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc. Natl. Acad. Sci. U. S. A. 98:6494-6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson, G. A. 1996. Lipids and membrane function in green algae. Biochim. Biophys. Acta 1302:17-45. [DOI] [PubMed] [Google Scholar]
- 39.Van Donk, E., M. Lurling, D. O. Hessen, and G. M. Lokhorst. 1997. Altered cell wall morphology in nutrient-deficient phytoplankton and its impact on grazers. Limnol. Oceanogr. 42:357-364. [Google Scholar]
- 40.Walther, T. C., and R. V. Farese, Jr. 2009. The life of lipid droplets. Biochim. Biophys. Acta 1791:459-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, Z. T., N. Ullrich, S. Joo, S. Waffenschmidt, and U. Goodenough. 30 October 2009. Algal lipid bodies: stress induction, purification and biochemical characterization in wild-type and starch-less Chlamydomonas reinhardtii. Eukaryot. Cell doi: 10.1128/EC.00272-09. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.