Abstract
c-Jun N-terminal kinases (JNKs) are potently activated by a number of cellular stimuli. Small GTPases, in particular Rac, are responsible for initiating the activation of the JNK pathways. So far, the signals leading from extracellular stimuli to the activation of Rac have remained elusive. Recent studies have demonstrated that the Src homology 2 (SH2)- and Src homology 3 (SH3)-containing adaptor protein Crk is capable of activating JNK when ectopically expressed. We found here that transient expression of Crk induces JNK activation, and this activation was dependent on both the SH2- and SH3-domains of Crk. Expression of p130Cas (Cas), a major binding protein for the Crk SH2-domain, also induced JNK activation, which was blocked by the SH2-mutant of Crk. JNK activation by Cas and Crk was effectively blocked by a dominant-negative form of Rac, suggesting for a linear pathway from the Cas-Crk-complex to the Rac-JNK activation. Many of the stimuli that activate the Rac-JNK pathway enhance engagement of the Crk SH2-domain. JNK activation by these stimuli, such as epidermal growth factor, integrin ligand binding and v-Src, was efficiently blocked by dominant-negative mutants of Crk. A dominant-negative form of Cas in turn blocked the integrin-, but not epidermal growth factor - nor v-Src-mediated JNK activation. Together, these results demonstrate an important role for Crk in connecting multiple cellular stimuli to the Rac-JNK pathway, and a role for the Cas-Crk complex in integrin-mediated JNK activation.
Interaction of cells with a variety of agonists results in a stimulation of mitogen-activated protein kinases (MAPKs), which control the expression of genes that are important for many cell functions including proliferation and differentiation. c-Jun N-terminal kinases (JNKs) are members of the MAPK family that are activated in response to cellular stresses and other stimuli, such as growth factors, changes in osmolarity and ultraviolet irradiation. Substrates for activated JNK include the transcription factors c-Jun, ATF2, and Elk-1; JNK phosphorylates the transactivating domain within each of these molecules thereby increasing their transcriptional activity. As with all the MAPK cascades, the JNK pathways align themselves into a well conserved, three component, sequential kinase cascade (1–3). Evidence supports a pivotal role for small GTP-binding proteins in initiating the activation of the MAPK pathways. In particular, Rac and Cdc42, which are members of the Rho-family of small GTPases, have been shown to lie upstream of the JNK cascades (4, 5). So far, the topology of the signaling path leading to the activation of these GTPases has remained elusive.
Crk is a member of an adaptor protein family consisting mostly of the Src homology 2 and 3 (SH2 and SH3) domains, and was originally identified as an avian retrovirus encoding the oncogene product v-Crk (6). Two cellular homologs, c-CrkI and c-CrkII, are produced from the same gene by alternative splicing (7). In v-Crk-transformed cells, tyrosine phosphorylation levels of intracellular proteins are significantly increased (8), which correlates with the transforming activity of v-Crk (9). Among the highly phosphorylated proteins is Cas, which has been shown to interact with the SH2-domain of v-Crk (10). Interestingly, Cas is known to become tyrosine phosphorylated in nontransformed cells in response to a number of cellular stimuli, such as growth factor stimulation and integrin-mediated cell adhesion (for a review, see ref. 11). Under these conditions, tyrosine-phosphorylated Cas appears to be the major binding protein for the SH2-domain of c-Crk, and it readily coimmunoprecipitates with c-Crk (12, 13). The SH3-domain of c-Crk can bind to multiple target molecules, including two guanine nucleotide exchange proteins, C3G (14) and Sos (15), and DOCK 180, a 180-kDa protein of unknown function (16). These proteins are efficiently recruited by c-Crk to the Cas-Crk signaling complex upon cellular stimulation (12). Hence, c-Crk has been suggested to be involved in growth factor- and integrin-mediated signaling pathways, but a physiological role for c-Crk has remained largely unknown. Recently, Tanaka et al. (17) reported that JNK is activated in v-Crk-transformed cells, and can be activated by a transient overexpression of v-Crk and c-Crk. These studies prompted us to examine the physiological role of c-Crk and we demonstrate here that c-Crk has a previously unidentified crucial role in connecting multiple cellular stimuli to the activation of JNK via the small GTP-binding protein Rac.
MATERIALS AND METHODS
Plasmids.
The Crk-constructs used in this study have been described in refs. 7 and 18. pCAGGS-expression vectors for DOCK180, C3G and Sos have been described in refs. 14–16. pCAGGS-(Myc)Rap1N17 was constructed in the laboratory of M.M. Wild-type Cas-expression construct has been described (10). The expression vector for CasΔSD lacking the substrate domain (amino acids 213–514) was constructed as in ref. 19. pSRα3-(HA)JNK1, pCMV5-(M2)JNK1, pSRα3-(HA)ERK2, pSRα3-RasN17, pSRα3-Rac1N17, pEXV-Rac1V12, pCMV5-(HA)MEKKΔ, and pSG-v-Src have been described (5, 20, 21). pEBB-constructs for wild-type Grb2 and the SH2-mutant of Grb2 (R86K) are described in ref. 22. pEGFP-N1-vector coding for the green fluorescent protein is from CLONTECH.
Cell Lines and Transfections.
COS and HeLa cells were maintained in DMEM supplemented with 10% fetal calf serum. Transient transfections were performed using either the lipofectamine or the calcium phosphate method. Lipofectamine was used following the manufacturer’s instructions (GIBCO/BRL). For the calcium phosphate method, 2.4 μg of purified DNA were used for a 35-mm cell dish containing 3 × 105 cells per well. 24 hr after transfections, the cells were rinsed with PBS and refed with serum-free media for 18–24 hr before lysis.
Antibodies, Immunoblot Analysis, Immunoprecipitations.
The following antibodies were from commercial sources: Anti-hemagglutinin (HA, clone Y-11; Santa Cruz Biotechnology), anti-Flag (M2; Kodak, IBI), anti-Myc and anti-Ras (Calbiochem), and anti-c-CrkII, anti-Cas, and anti-Grb2 (Transduction Laboratories, Lexington, KY). Antisera against DOCK180 and C3G are described (16). Immunoblotting and immunoprecipitations were carried out as described (12).
JNK and Erk Kinase Assays and c-Jun Luciferase Assay.
JNK and Erk activities were measured by immunocomplex kinase assays as in ref. 20. In parallel with the kinase assays, equal aliquots of the unlabeled immunoprecipitates were analyzed with the antibodies indicated in the figure legends to determine the expression levels of the transfected proteins. For the luciferase assay, HeLa cells were cotransfected with 1 μg of the pFR-Luc reporter plasmid, 50 ng of pFA-c-Jun (1–223) (PathDetect Reporting System, Stratagene) and with the plasmids indicated in the text. After transfection, the cells were kept in serum-free medium for 24 hr, lysed, and assayed for luciferase expression by using the Luciferase Assay System (Promega) according to manufacturer’s protocol. Unless otherwise indicated, the figures are representatives of three independently performed experiments.
Immunocytochemistry.
Cos-7 cells were transfected with the indicated plasmids together with pEGFP-1 as a marker for transfected cells. After 24 hr incubation in serum-free medium, cells were fixed with paraformaldehyde and permeabilized with Triton X-100. Staining of filamentous actin was done with rhodamine-conjugated phalloidin (Sigma), and cells were analyzed with a Nikon Axiovert system at 100× magnification. Cells expressing green fluorescent protein were studied under excitation-emission filter for rhodamine to analyze actin fiber organization.
Epidermal Growth Factor (EGF) Stimulation and Cell Attachment.
To stimulate cells with EGF, COS and HeLa cells were kept in serum-free medium for 24 hr after stimulation with 100 ng/ml of EGF (Calbiochem). Before serum-starvation, the cells had been transfected with the plasmids indicated. Luciferase assay was carried out 7 hr after the addition of EGF and JNK immunocomplex assay was done as above. To analyze Erk activation in response to EGF, COS-7 cells were transiently transfected with the plasmids indicated in the figure legend, followed by stimulation with 100 ng/ml of EGF for 10 min. Erk activity was measured by immunocomplex kinase assay as above. When cells were stimulated with integrin-mediated cell adhesion, the cells were first transfected with the indicated plasmids as described above. Forty-eight hours after transfection, the cells were detached by trypsinization and either immediately lysed or reattached on 20 μg/ml fibronectin-coated plates for 15 min at 37°C. Immunocomplex kinase assays were performed as above.
RESULTS
c-Crk Activates the JNK Pathway in an SH2- and SH3-Dependent Manner.
To examine the capability of c-CrkII to activate JNK, COS-7 cells were cotransfected with the c-CrkII expression vector and a plasmid encoding a HA epitope-tagged JNK1. JNK1 kinase activity was assayed in immunoprecipitates with glutathione S-transferase-c-Jun (1–79) as the substrate. Consistent with an earlier report (17), c-Crk was found to efficiently activate JNK (Fig. 1A). The activation of JNK by Crk should in turn result in stimulation of c-Jun transcriptional activity. To study this, activation of GAL4-c-Jun (1–223) fusion protein containing the c-Jun activation domain was analyzed by using luciferase as a reporter gene. As shown in Fig. 1B, expression of c-Crk efficiently induced GAL4-c-Jun activation, which is strictly dependent on the upstream JNK activation. As a control, overexpression of another adaptor protein, Grb2, failed to activate the JNK pathway (Fig. 1B). Cotransfection of c-Crk with HA-tagged Erk, which is another member of the MAPK family, failed to result in Erk activation, whereas EGF readily activated Erk in the same cells (see Fig. 3D). This is in agreement with the earlier finding that Erk activity is not increased in fibroblasts stably expressing v-Crk (17), and suggests that Crk is a specific activator of the JNK pathway.
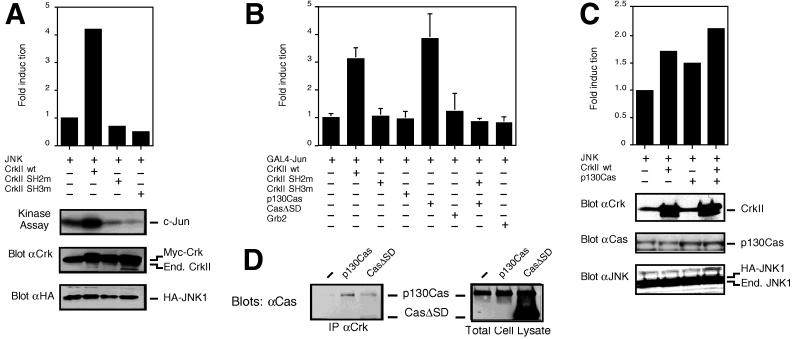
Figure 1.
Effects of c-CrkII and Cas on JNK activation. (A) COS cells were cotransfected with the pSRα3-(HA)JNK1-vector (JNK) together with a pCAGGS-expression vector containing the cDNA either for the wild-type c-CrkII (CrkII wt), the SH2-mutant of c-CrkII (the R38V-mutation) (CrkII SH2m), the SH3-mutant of c-CrkII (the W169L-mutation) (CrkII SH3m), or with the empty pCAGGS-expression vector, and JNK immunocomplex kinase assays were performed. Expression levels of the wild-type and mutant forms of c-CrkII and of the HA-tagged JNK were analyzed by immunoblotting with anti-c-CrkII and anti-HA antibodies, respectively. Note that the pCAGGS-constructs contain a myc-epitope in the N terminus of the produced protein. (B) HeLa cells were cotransfected with pFR-Luc and pFA-c-Jun (1–223) (GAL4-Jun), together with expression vectors for either the wild-type or the mutant forms of c-CrkII, with pSSRα-expression vectors coding for the wild-type form of Cas (p130Cas) or the form of Cas in which the c-Crk-binding site (also known as the substrate domain) had been deleted (CasΔSD), or with the wild-type Grb2 (Grb2). Transcriptional activity of GAL4-c-Jun (1–223) was analyzed using luciferase as a reporter gene. (C) COS cells were cotransfected with pSRα3-(HA)JNK1, together with pACT-c-CrkII (90 ng per well) or pSSRα-p130Cas (500 ng per well). The parental pSRα3-plasmid was added to the transfection mixture to keep the total amount of DNA constant. JNK activity was determined by immunocomplex kinase assays. The amounts of Cas- and Crk-plasmids used result in a submaximal activation of JNK as determined by titration experiments, explaining the lower JNK activation in this experiment compared with that observed in A and B. Expression levels of c-CrkII, Cas, and the HA-tagged JNK were analyzed by immunoblotting with anti-c-CrkII, anti-Cas, and anti-HA antibodies, respectively. (D) The substrate domain of Cas is required for Cas binding to c-Crk. COS cells were cotransfected with an expression vector coding for the wild-type c-CrkII, together with either the empty pSSRα-vector or pSSRα-vector coding for the wild-type form of Cas (p130Cas) or the substrate domain-deleted form of Cas (CasΔSD). Lysates containing same amounts of proteins were immunoprecipitated with anti-c-CrkII antibodies, and coprecipitation of Cas was studied by immunoblotting with anti-Cas antibodies (Left). Expression levels of the wild-type and mutant forms of Cas were analyzed by immunoblotting with the same Cas-antibody, which recognizes equally well both the full length and the truncated form of the protein (Right).
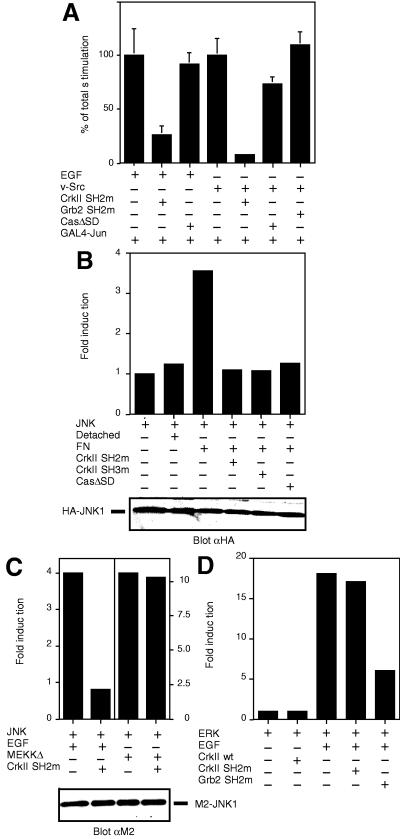
Figure 3.
Dominant negative forms of Crk block EGF-, integrin- and v-Src-induced JNK activation. (A) HeLa cells were transfected with pFR-Luc, pFA-c-Jun (1–223) (GAL4-Jun) and the indicated plasmids, and luciferase assay was carried out. Grb2 SH2m denotes for the dominant negative SH2-mutant (R86K) of Grb2. v-Src-induced JNK activation was assessed in cells transfected with the expression plasmid pSG-v-Src (v-Src). Where indicated, EGF stimulation was carried out as in Materials and Methods. (B) COS-7 cells were cotransfected with pSRα3-(HA)JNK1 (JNK) and the indicated plasmids. The cells were either directly lysed on the dish and subjected to immunoprecipitations, or the cells were trypsinized and either lysed immediately (Detached) or reattached on dishes coated with fibronectin (FN). JNK activity was studied by an immunocomplex kinase assay. (C) COS-7 cells were transfected with pCMV5-(M2)JNK1 (JNK) and the plasmids indicated, and JNK immunocomplex kinase assays were performed. Cells were treated with or without EGF as in (A). As a control, the effect of Crk-mutant expression on MEKK-induced JNK activation was studied by the expression of pCMV5-(HA)MEKKΔ (MEKKΔ). (D) COS-7 cells were transfected with pSRα3-(HA)ERK2 (ERK) and the indicated plasmids and treated or not with EGF as described in Materials and Methods. Erk activity was measured by immunocomplex kinase assay using myelin basic protein as a substrate.
c-CrkII consists of one SH2 and two SH3-domains: whereas the SH2-domain binds to tyrosine-phosphorylated proteins and the N-terminal SH3-domain recognizes proline-rich motifs in cytoplasmic proteins, the C-terminal c-Crk SH3-domain has no identified binding proteins or function (23). To investigate the significance of the SH2 and SH3-domains in c-Crk-induced JNK activation, mutant forms of c-Crk were cotransfected with HA-JNK1 in COS-7 cells. Neither c-Crk mutant with nonfunctional SH2, c-CrkII-R38V, nor that with nonfunctional SH3, c-CrkII-W169L, enhanced JNK activation (Fig. 1A). Likewise, mutant forms of c-Crk failed to stimulate the transcriptional activity of c-Jun (Fig. 1B). We confirmed that similar amounts of wild-type and mutant c-Crk proteins were expressed in the transfected cells. These results demonstrate that both the SH2- and SH3-domains of c-Crk are required for the enhancement of JNK activity by c-Crk.
Crk Connects Cas to the JNK Pathway.
One of the predominant binding proteins for the SH2-domain of c-Crk is tyrosine-phosphorylated Cas (see above). To further investigate the role of the Crk SH2-domain engagement in JNK activation, transient transfection experiments with an expression plasmid for Cas alone or together with a c-Crk expression plasmid were carried out, and JNK kinase activity and c-Jun transcriptional activation were analyzed. Expression of Cas alone at a level that was 2-fold higher than the endogenous protein resulted in a corresponding increase in the Cas-Crk complex formation (Fig. 1D), and also stimulated JNK activation in the transfected cells (Fig. 1B). Coexpression of Cas together with c-Crk further enhanced JNK activation compared with when either one of the molecules was expressed alone (Fig. 1C). Several lines of evidence suggested that Cas and Crk are in the same pathway with respect to JNK activation. A mutant form of Cas (CasΔSD), in which a region known as the substrate domain is deleted, is not capable of interacting with Crk (Fig. 1D, ref. 10). When expressed in COS-7 cells, CasΔSD did not induce JNK activation (Fig. 1B), and neither did CasΔSD potentiate the c-Crk-induced JNK activation. In addition, the SH2-mutant form of c-Crk (c-CrkII-R38V), which is not capable of interacting with Cas, functioned as a dominant-negative with respect to Cas-induced JNK activation (Fig. 1B). Importantly, these results demonstrate that Crk can function as an intermediate in connecting tyrosine-phosphorylated binding partners, such as Cas, to the JNK pathway, and further support the significance of the engagement of the Crk SH2-domain in JNK activation.
Rac Is a Downstream Mediator for Cas- and Crk-Induced JNK Activation.
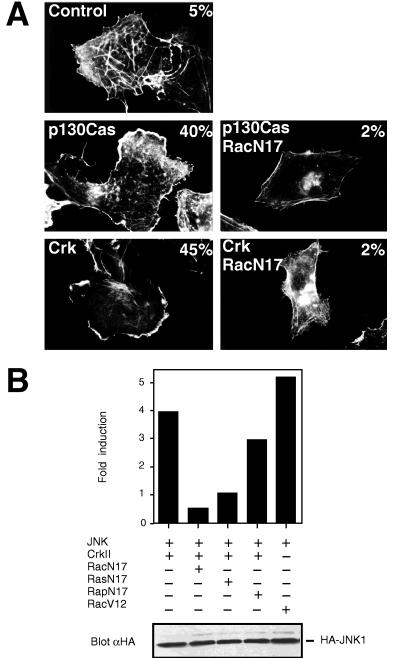
During the course of our studies, we observed an extensive membrane ruffling in cells that were transfected with an expression plasmid for c-Crk or Cas (Fig. 2A). The small GTP-binding protein Rac, when activated, has been shown to induce membrane ruffle formation (24), suggesting that Crk and Cas efficiently activate Rac. Indeed, Crk and Cas-induced membrane ruffling was blocked by a dominant negative form of Rac (Rac1N17) (Fig. 2A), but not by dominant negative forms of other GTPases, such as RasN17 (not shown). This observation together with the fact that many of the Crk SH3-binding proteins are activators of small GTPases suggested that Crk might function as a molecular switch between tyrosine-phosphorylated proteins and the small GTP-binding proteins, in particular Rac, which control the activation of the JNK pathways. Dominant inhibitory forms of various GTPases were therefore cotransfected with c-Crk and HA-JNK1, and JNK kinase activity was determined. As shown in Fig. 2B, dominant negative form of Rac (Rac1N17) completely abolished Crk-induced JNK activation. In contrast, dominant negative Rap1 (Rap1N17) had a negligible effect on Crk-induced JNK activation. Dominant-negative Rac also blocked Cas-induced JNK activation (not shown). As reported previously (4, 5), activated form of Rac (Rac1V12) readily activated JNK (Fig. 2B). Together, these findings demonstrate that Crk can efficiently activate Rac, which in turn is an obligatory intermediate step connecting Crk to JNK activation. Although we found no evidence that c-Crk would affect the Ras-Erk pathway (Fig. 3D), expression of a dominant negative Ras (RasN17) had a significant inhibitory effect on Crk-induced JNK activation (Fig. 2B). Crk therefore requires a functional Ras molecule to fully activate JNK via Rac.
Figure 2.
(A) Expression of c-Crk and Cas induces membrane ruffling in a Rac-dependent manner. Cos-7 cells were cotransfected with the pEGFP-N1-vector coding for a green fluorescent protein and with an empty control plasmid, pSSRα-p130Cas or pCAGGS-c-CrkII without (Left) or with pSRα3-Rac1N17 (Right) as indicated in the figures. Transfected cells were identified for the expression of the green fluorescent protein and rhodamine-conjugated phalloidin was used to visualize filamentous actin, as described in Materials and Methods. The number in the upper right-hand corner of each panel denotes for the percentage of the transfected cells displaying membrane ruffles. (B) c-Crk activates JNK via the GTPase Rac. COS cells were transfected with pSRα3-(HA)JNK1 (JNK) and pCAGGS-CrkII (CrkII), together with either pSRα3-Rac1N17 (RacN17), pCAGGS-Rap1N17 (RapN17), pSRα3-RasN17 (RasN17) or an empty expression plasmid. JNK activation in response to activated Rac1 (RacV12) expression is also shown. JNK activity was assayed in an immunocomplex kinase assay.
Crk Connects Multiple Cellular Stimuli to the Rac-JNK Pathway.
Our finding that Crk can connect upstream tyrosine-phosphorylated proteins, such as Cas, to the downstream Rac-JNK pathway prompted us to investigate whether Crk might have a previously unidentified role in mediating JNK activation in response to cellular stimuli. In support of this notion, many of the stimuli that induce JNK activation also increase the engagement of the Crk SH2-domain. Most notably, oncogenic v-Src and ligand binding of integrins enhance Crk binding to tyrosine-phosphorylated cytoplasmic proteins, such as Cas (10, 12, 25–27), whereas ligand-activated EGF receptor can directly bind and engage the SH2-domain of Crk (28, 29). In separate studies, all these stimuli have been shown to efficiently activate JNK (4, 5, 30, 31). Significantly, it has also been demonstrated that JNK activation induced by v-Src and EGF stimulation can be blocked with dominant-negative forms of Rac and Ras to the same extent as we observed above for Crk-induced JNK activation (4, 5). To determine whether c-Crk is a necessary intermediate in the signaling cascade leading from the EGF receptor, v-Src and integrins to JNK activation, we made use of the Crk SH2- and SH3-mutants described above, which have been shown to function as specific dominant negative inhibitors for Crk signaling (7, 15, 18, 22). As shown in Fig. 3, transient expression of v-Src, stimulation of cells with EGF and attachment of cells to an integrin ligand fibronectin all stimulated JNK activation. When either the SH2- or the SH3-mutant of Crk was expressed in the cells, both the transcriptional activation of c-Jun (Fig. 3A) and JNK catalytic activation (Fig. 3C) in response to EGF stimulation was significantly blocked. The Crk-mutants also efficiently blocked v-Src- and integrin-induced activation of the JNK pathway (Fig. 3 A and B). Several control experiments were performed to verify the specificity of the dominant-negative Crk-constructs. Expression of MEKKΔ, which is an activated form of the upstream kinase in the JNK cascade and functions downstream of Rac (1–3), elicited a marked increase of JNK activity that was not blocked by dominant negative forms of Crk (Fig. 3C). Similarly, no effect was observed on EGF-induced Erk activation when the dominant-negative mutants of Crk were expressed (Fig. 3D). In contrast, the corresponding dominant-negative mutants of Grb2 readily blocked EGF-induced Erk activation (Fig. 3D, see also ref. 22), but they had no effect on v-Src-induced JNK activation (Fig. 3A) and only a moderate effect on EGF- and integrin-mediated JNK activation (not shown). Together, these results establish a specific role for Crk in connecting a variety of different cellular stimuli to JNK activation.
As mentioned above, some of the cellular stimuli that activate the JNK pathway induce Cas-Crk interaction. We therefore studied whether tyrosine-phosphorylated Cas is an upstream component in connecting integrins, EGF receptor and v-Src via Crk to the JNK pathway. To this end, the mutant form of Cas (CasΔSD) that is not capable of interacting with Crk was used as a dominant-negative construct. As shown in Fig. 3B, expression of CasΔSD readily blocked integrin-induced JNK activation. In contrast, expression of CasΔSD had a negligible effect on v-Src or EGF-induced JNK activation (Fig. 3A) or on EGF-induced Erk activation (data not shown). Together, these results indicate that Cas is responsible for connecting integrins to the Crk-JNK pathway, whereas additional Crk SH2-binding proteins may be used downstream of v-Src and EGF receptor stimulation.
Characterization of the Role of Crk SH3-Binding Proteins in JNK Activation.
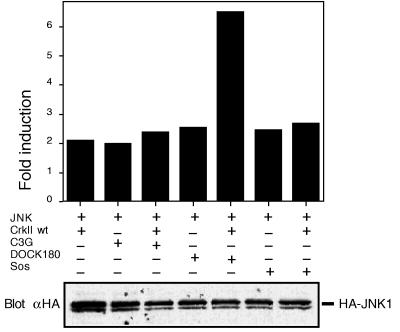
To get an insight into how Crk might connect the upstream stimuli to the Rac-JNK pathway, we studied which of the Crk SH3-binding proteins could induce activation of the JNK pathway. Expression vectors coding for Sos, C3G, and DOCK180 were expressed in COS cells, and their capability to activate the JNK pathway was studied by JNK kinase assay and by monitoring c-Jun transcriptional activation. As shown in Fig. 4, expression of Sos and C3G readily induced activation of the JNK pathway; similar observations for Sos and C3G have recently been made by other investigators (32). Interestingly, we found that expression of DOCK180 also resulted in a robust activation of the JNK pathway (Fig. 4), suggesting a potential role for any one of the three proteins in connecting Crk to the JNK pathway. Additional studies indicated that coexpression of Crk with DOCK180, but not with C3G or Sos, resulted in a synergistic activation of the JNK pathway as determined by JNK immunocomplex kinase assay (Fig. 4). In a more long-term c-Jun transcriptional activity assay, coexpression of Crk with C3G also demonstrated synergistic activation of the pathway, whereas coexpression of Sos again did not have any effect on Crk-induced JNK pathway activation (not shown). Lack of dominant negative forms specifically acting on one of the Crk SH3-binding proteins but not on the others has prevented us from directly determining their role in Crk signaling. We found, however, that dominant-negative Rac is an efficient inhibitor of Sos- and DOCK180-induced JNK activation, but it has no effect on C3G-induced JNK activation (data not shown, see also ref. 32). Taken together with our finding that Rac is an obligatory intermediate in Crk-induced JNK activation, our results suggest that DOCK180 or Sos, rather than C3G, may connect Crk to the Rac-JNK pathway.
Figure 4.
Crk SH3-domain binding proteins C3G, DOCK180 and Sos activate JNK. COS cells were transfected with pSRα3-(HA)JNK1 (JNK) and pCAGGS-expression vector coding for either DOCK180, Sos or C3G, with or without simultaneous wild-type c-Crk II expression. JNK kinase activity was measured as above by immunocomplex kinase assay, and the results are expressed as fold-induction compared with the JNK kinase activity observed in cells transfected with the pSRα3-(HA)JNK1-vector and an empty pCAGGS-vector.
DISCUSSION
The adaptor protein Crk was the first identified SH2/SH3-containing molecule, but its role in intracellular signaling pathways has remained obscure. Recent findings by Tanaka et al. (17) suggested the putative connection between the Crk molecules and the JNK pathway, and our findings reported here confirm these observations. Importantly, our studies indicate that the connection from Crk to the JNK pathway is not limited to an exogenously expressed Crk, but the endogenous c-Crk molecule has a physiological role in connecting a number of cellular stimuli to the JNK pathway via the small GTP-binding protein Rac. These studies also suggest that the modular, conserved nature of the MAPK pathways can be extended beyond the sequential, three kinase cascade: in analogy to the Ras-Erk-pathway, in which Grb2 connects to the small GTPase Ras via an exchange factor Sos, Crk may have a similar role upstream of the Rac-JNK pathway.
Among the stimuli that were found to activate the Rac-JNK pathway in a Crk-dependent manner were EGF receptor activation, oncogenic v-Src, and integrin-mediated cell adhesion. Our results suggest that the role of the SH2-domain of Crk is to connect these cellular stimuli to the Rac-JNK pathway by binding to tyrosine-phosphorylated proteins. Our studies further indicate that tyrosine-phosphorylated Cas is a necessary component in the pathway leading from integrins to JNK activation via Crk, whereas a different mechanism appears to be employed in the case of EGF receptor activation and v-Src expression. Our preliminary data indicate that Crk binding to several tyrosine-phosphorylated docking proteins may connect these stimuli to the Rac-JNK pathway. In the Grb2/Sos/Ras/MAPK pathway, translocation of Sos by Grb2 to the plasma membrane, where its substrate Ras is located, is an essential event. Similarly, the role of Cas and other potential Crk SH2-binding molecules may be to localize Crk in the proximity of the plasma membrane, which could be necessary for the activation of the downstream JNK pathway. It is possible that the role of Crk as an activator of the JNK pathway is not limited to v-Src, cell adhesion, and EGF stimulation. The tyrosine kinase Pyk2 was recently demonstrated to be activated by tumor necrosis factor α, ultraviolet irradiation, and osmolarity changes, and to function as an upstream mediator of the JNK signaling pathway by these stimuli (33). As activated Pyk2 has been shown to induce tyrosine phosphorylation of Crk SH2-target proteins, such as Cas (34–36), it remains to be seen whether Crk functions downstream of Pyk2 to couple these stress signals with the JNK pathway, as well.
Our findings reported here identify a signaling cascade downstream from Crk through its SH3-binding proteins, possibly DOCK180 or Sos, to the Rac-mediated JNK activation. It remains to be determined how DOCK180 might enhance Rac activation; interestingly, preliminary results indicate that DOCK180 may have exchange activity on small GTP-binding proteins as its expression increases the GTP/GDP ratio in the cells (M.M., unpublished data). Recent results by Nimnual et al. (37) in turn suggest that Sos may directly function as an exchange factor for Rac. Studies by Nimnual et al. also demonstrated that Sos couples the Ras and Rac signaling pathways, as prior activation of Ras by Sos was required for Sos-induced Rac activation; this type of coupling might explain the widely observed requirement for both Ras and Rac in JNK activation. Studies by Tanaka and coworkers have highlighted the role of the Crk SH3-binding protein C3G as an activator of the JNK pathway (17, 32). Interestingly, Tanaka et al. found that neither dominant negative Ras nor dominant negative Rac is able to block C3G-induced JNK activation. Instead, C3G appears to activate JNK through a pathway involving the mixed lineage kinase family of proteins (32). It remains to be seen if under some conditions c-Crk mediates JNK activation through the Rac-independent C3G-mixed lineage kinase pathway, and, on the other hand, what are the physiological cellular stimuli that connect to the C3G-mixed lineage kinase signaling cascade.
Recent findings by Klemke et al. (38) have demonstrated that the Cas-Crk signaling complex functions as a “molecular switch” for cell migration. Overexpression of the two molecules resulted in enhanced cell migration, and interestingly, this was blocked by a dominant negative form of Rac. These results suggest that Rac activation downstream of the Cas-Crk complex is mediating not only JNK activation, but also cell migration. The potential functional connection between JNK activation and cell migration remains to be studied. Significantly, Klemke et al. found that dominant negative forms of Cas and Crk were able to block both integrin-mediated haptotactic cell migration, as well as cytokine-induced migration. These findings further emphasize the role of the Crk signaling complex as a point of convergence in the actions of a variety of factors known to influence cell locomotion, growth, and differentiation. Taken together with the increasing evidence showing a role for JNK in a number of physiological and pathological cellular processes (1–3), the Crk signaling complex may prove to be a useful target for the design of therapeutic strategies.
Acknowledgments
We thank M. Russello for expert technical assistance and B. Mayer for providing the Grb2-constructs. This work was supported by National Institutes of Health grant CA71560 (to K.V.). F.D. and M.G.-G. were supported by fellowships from the American-Italian Cancer Foundation and Human Frontier Science Program, respectively. K.V. is a Pew Scholar in biomedical sciences.
ABBREVIATIONS
- EGF
epidermal growth factor
- HA
hemagglutinin
- JNK
c-Jun N-terminal kinase
- MAPK
mitogen-activated protein kinase
- SH2 and SH3
Src homology 2 and 3
References
- 1.Davis R J. Trends Biochem Sci. 1994;19:470–473. doi: 10.1016/0968-0004(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 2.Cano E, Mahadevan L C. Trends Biochem Sci. 1995;20:117–122. doi: 10.1016/s0968-0004(00)88978-1. [DOI] [PubMed] [Google Scholar]
- 3.Minden A, Karin M. Biochim Biophys Acta. 1997;1333:F85–104. doi: 10.1016/s0304-419x(97)00018-8. [DOI] [PubMed] [Google Scholar]
- 4.Coso O A, Chiariello M, Yu J C, Teramoto H, Crespo P, Xu N, Miki T, Gutkind J S. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 5.Minden A, Lin A, Claret F X, Abo A, Karin M. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 6.Mayer B J, Hamaguchi M, Hanafusa H. Nature (London) 1988;332:272–275. doi: 10.1038/332272a0. [DOI] [PubMed] [Google Scholar]
- 7.Matsuda M, Tanaka S, Nagata S, Kojima A, Kurata T, Shibuya M. Mol Cell Biol. 1992;12:3482–3489. doi: 10.1128/mcb.12.8.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsuda M, Mayer B J, Fukui Y, Hanafusa H. Science. 1990;248:1537–1539. doi: 10.1126/science.1694307. [DOI] [PubMed] [Google Scholar]
- 9.Mayer B J, Hanafusa H. J Virol. 1990;64:3581–3589. doi: 10.1128/jvi.64.8.3581-3589.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakai R, Iwamatsu A, Hirano N, Ogawa S, Tanaka T, Mano H, Yazaki Y, Hirai H. EMBO J. 1994;13:3748–3756. doi: 10.1002/j.1460-2075.1994.tb06684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burridge K, Chrzanowska-Wodnicka M. Annu Rev Cell Dev Biol. 1996;12:463–518. doi: 10.1146/annurev.cellbio.12.1.463. [DOI] [PubMed] [Google Scholar]
- 12.Vuori K, Hirai H, Aizawa S, Ruoslahti E. Mol Cell Biol. 1996;16:2606–2613. doi: 10.1128/mcb.16.6.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ribon V, Saltiel A R. J Biol Chem. 1996;271:7375–7380. doi: 10.1074/jbc.271.13.7375. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka S, Morishita T, Hashimoto Y, Hattori S, Nakamura S, Shibuya M, Matuoka K, Takenawa T, Kurata T, Nagashima K, et al. Proc Natl Acad Sci USA. 1994;91:3443–3447. doi: 10.1073/pnas.91.8.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuda M, Hashimoto Y, Muroya K, Hasegawa H, Kurata T, Tanaka S, Nakamura S, Hattori S. Mol Cell Biol. 1994;14:5495–5500. doi: 10.1128/mcb.14.8.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasegawa H, Kiyokawa E, Tanaka S, Nagashima K, Gotoh N, Shibuya M, Kurata T, Matsuda M. Mol Cell Biol. 1996;16:1770–1776. doi: 10.1128/mcb.16.4.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanaka S, Ouchi T, Hanafusa H. Proc Natl Acad Sci USA. 1997;94:2356–2361. doi: 10.1073/pnas.94.6.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka S, Hattori S, Kurata T, Nagashima K, Fukui Y, Nakamura S, Matsuda M. Mol Cell Biol. 1993;13:4409–4415. doi: 10.1128/mcb.13.7.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamoto T, Sakai R, Ozawa K, Yazaki Y, Hirai H. J Biol Chem. 1996;271:8959–8965. doi: 10.1074/jbc.271.15.8959. [DOI] [PubMed] [Google Scholar]
- 20.Cavigelli M, Dolfi F, Claret F X, Karin M. EMBO J. 1995;14:5957–5964. doi: 10.1002/j.1460-2075.1995.tb00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minden A, Lin A, McMahon M, Lange-Carter C, Derijard B, Davis R J, Johnson G L, Karin M. Science. 1994;266:1719–1723. doi: 10.1126/science.7992057. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka M, Gupta R, Mayer B J. Mol Cell Biol. 1995;15:6829–6837. doi: 10.1128/mcb.15.12.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feller S M, Ren R, Hanafusa H, Baltimore D. Trends Biochem Sci. 1994;19:453–458. doi: 10.1016/0968-0004(94)90129-5. [DOI] [PubMed] [Google Scholar]
- 24.Ridley A J, Paterson H F, Johnston C L, Diekmann D, Hall A. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 25.Matsuda M, Mayer B J, Hanafusa H. Mol Cell Biol. 1991;11:1607–1613. doi: 10.1128/mcb.11.3.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koch C A, Moran M F, Anderson D, Liu X Q, Mbamalu G, Pawson T. Mol Cell Biol. 1992;12:1366–1374. doi: 10.1128/mcb.12.3.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Birge R B, Fajardo J E, Reichman C, Shoelson S E, Songyang Z, Cantley L C, Hanafusa H. Mol Cell Biol. 1993;13:4648–4656. doi: 10.1128/mcb.13.8.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Birge R B, Fajardo J E, Mayer B J, Hanafusa H. J Biol Chem. 1992;267:10588–10595. [PubMed] [Google Scholar]
- 29.Fajardo J E, Birge R B, Hanafusa H. Mol Cell Biol. 1993;13:7295–7302. doi: 10.1128/mcb.13.12.7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyamoto S, Teramoto H, Coso O A, Gutkind J S, Burbelo P D, Akiyama S K, Yamada K M. J Cell Biol. 1995;131:791–805. doi: 10.1083/jcb.131.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mainiero F, Murgia C, Wary K K, Curatola A M, Pepe A, Blumemberg M, Westwick J K, Der C J, Giancotti F G. EMBO J. 1997;16:2365–2375. doi: 10.1093/emboj/16.9.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka S, Hanafusa H. J Biol Chem. 1998;273:1281–1284. doi: 10.1074/jbc.273.3.1281. [DOI] [PubMed] [Google Scholar]
- 33.Tokiwa G, Dikic I, Lev S, Schlessinger J. Science. 1996;273:792–794. doi: 10.1126/science.273.5276.792. [DOI] [PubMed] [Google Scholar]
- 34.Astier A, Avraham H, Manie S N, Groopman J, Canty T, Avraham S, Freedman A S. J Biol Chem. 1997;272:228–232. doi: 10.1074/jbc.272.1.228. [DOI] [PubMed] [Google Scholar]
- 35.Astier A, Manie S N, Avraham H, Hirai H, Law S F, Zhang Y, Golemis E A, Fu Y, Druker B J, Haghayeghi N, et al. J Biol Chem. 1997;272:19719–19724. doi: 10.1074/jbc.272.32.19719. [DOI] [PubMed] [Google Scholar]
- 36.Schaller M D, Sasaki T. J Biol Chem. 1997;272:25319–25325. doi: 10.1074/jbc.272.40.25319. [DOI] [PubMed] [Google Scholar]
- 37.Nimnual A S, Yatsula B A, Bar-Sagi D. Science. 1998;279:560–563. doi: 10.1126/science.279.5350.560. [DOI] [PubMed] [Google Scholar]
- 38.Klemke R L, Leng J, Molander R, Brooks P C, Vuori K, Cheresh D A. J Cell Biol. 1998;140:961–972. doi: 10.1083/jcb.140.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]