Abstract
African trypanosomes regulate transcription differently from other eukaryotes. Most of the trypanosome genome is constitutively transcribed by RNA polymerase II (Pol II) as large polycistronic transcription units while the genes encoding the major surface proteins are transcribed by RNA polymerase I (Pol I). In bloodstream form Trypanosoma brucei, the gene encoding the variant surface glycoprotein (VSG) coat is expressed in a monoallelic fashion from one of about 15 VSG bloodstream form expression sites (BESs). Little is known about the chromatin structure of the trypanosome genome, and the chromatin state of active versus silent VSG BESs remains controversial. Here, we determined histone H3 occupancy within the genome of T. brucei, focusing on active versus silent VSG BESs in the bloodstream form. We found that histone H3 was most enriched in the nontranscribed 50-bp and 177-bp repeats and relatively depleted in Pol I, II, and III transcription units, with particular depletion over promoter regions. Using two isogenic T. brucei lines containing marker genes in different VSG BESs, we determined that histone H3 is 11- to 40-fold depleted from active VSG BESs compared with silent VSG BESs. Quantitative PCR analysis of fractionated micrococcal nuclease-digested chromatin revealed that the active VSG BES is depleted of nucleosomes. Therefore, in contrast to earlier views, nucleosome positioning appears to be involved in the monoalleleic control of VSG BESs in T. brucei. This may provide a level of epigenetic regulation enabling bloodstream form trypanosomes to efficiently pass on the transcriptional state of active and silent BESs to daughter cells.
In eukaryotes nuclear DNA is packaged into linear arrays of nucleosomes, which provides one of the main determinants of accessibility to DNA binding proteins (37, 60, 68, 82). Nucleosomes consist of ∼146 bp of DNA wrapped around a histone octamer composed of two copies each of histones H2A, H2B, H3, and H4 (44). The ability to change how DNA is packaged within nucleosomes allows variation in the accessibility of different DNA binding sites and permits fine modulation of promoter activity (33). Recently, there have been major advances in our understanding of how chromatin structure impacts the regulation of RNA polymerase II (Pol II) transcription units. New developments (72) have enabled the determination of the global organization of nucleosomes in organisms including budding yeast, Drosophila, and humans (3, 35, 36, 48, 71, 83). These studies have shown evidence for nucleosome depletion at transcriptionally active regulatory regions, with the level of nucleosome occupancy inversely proportional to the rate of transcription initiation at the promoter (3, 36, 83). However, nucleosome remodeling at promoters does not always appear to be simply a consequence of transcriptional activity but is also thought to mechanistically regulate transcription through modulating the access of trans-acting factors (74).
Our understanding of the role that chromatin structure plays in the regulation of RNA Pol I transcription has relatively lagged behind that of Pol II (reviewed in references 6, 21, and 49). This is in part due to the repetitive nature of the nearly identical ribosomal DNA (rDNA) transcription units, which in Saccharomyces cerevisiae (and presumably most eukaryotes) are the only transcription units transcribed by Pol I (54). Approximately half of the rDNA repeats are transcriptionally active at any time (11-13). Silencing of the inactive rDNA units is mediated by the nucleolar remodeling complex (NoRC) (69), which silences the inactive rRNA genes by changing nucleosome positioning (38, 77). Despite the clear role that chromatin remodeling plays in transcriptional regulation of the rRNA, the precise nature of the chromatin present in active versus silent rRNA transcription units remains unclear (49). Studies using psoralen cross-linking (11) or more recent analyses combining this with chromatin endogenous cleavage (ChEC) (50) have suggested that transcriptionally active rDNA units are essentially devoid of nucleosomes. However, other recent studies have argued that active rDNA has a nucleosomal structure (30).
In African trypanosomes the majority of the trypanosome genome is constitutively transcribed as extensive Pol II transcription units. Very few regulatory regions have been defined, and only a few promoters have been well characterized. Trypanosomes appear to largely lack gene regulation at the level of transcription and, instead, rely on posttranscriptional control acting at the level of RNA processing and stability (10). Due to this very different way of regulating expression of their genes, it is unclear whether nucleosome positioning plays a role in trypanosomes similar to that of other eukaryotes.
Trypanosomes are further unconventional in that they use Pol I to transcribe not only the multicopy rDNA but also the genes encoding their major surface proteins: variant surface glycoprotein (VSG) in the bloodstream form or procyclin in the procyclic (insect mid-gut stage) form (22, 32, 64). This unique ability to use an unorthodox RNA polymerase to transcribe some of their protein-coding genes is presumably made possible by trans-splicing, which adds a capped 39-nucleotide spliced leader (SL) RNA to the 5′ end of each mRNA, thereby making an uncapped Pol I-derived transcript available for translation (34, 39, 42, 47). In order to provide the large amount of SL RNA necessary for trans-splicing, the trypanosome has up to 200 SL RNA transcription units which are highly transcribed by Pol II. The SL RNA promoter is the only Pol II promoter in trypanosomes which has been well characterized (14, 23, 70).
In bloodstream form T. brucei antigenic variation of a VSG coat is used to escape host antibodies. This is accomplished through monoallelic exclusion of VSG expression, whereby within a single cell only one of about 15 telomeric VSG blood stream form expression sites (BESs) is transcribed at a time by Pol I (7, 9, 58). The nature of the chromatin structure of VSG BESs has been a controversial issue. Earlier studies have argued that there are no detectable differences in nucleosomal organization between active and silent VSG BESs, although active VSG BESs are sensitive to digestion by endonucleases including single-strand-specific endonucleases (20, 51, 57). Studies using exogenous T7 RNA polymerase as a probe for chromatin accessibility also did not find evidence that active VSG BESs in bloodstream form T. brucei were more accessible for transcription than inactive ones (52). Recently, however, the role of chromatin in the downregulation of silent BESs is being reevaluated as chromatin remodeling proteins have been shown to be important for VSG BES control (15). First, the chromatin remodeling protein T. brucei ISWI (TbISWI) has been shown to be important for VSG BES silencing in both bloodstream and insect forms of T. brucei (28). In addition, downregulation of the histone modification protein DOT1B affects the kinetics of VSG BES switching, arguing that histone modification plays a role in monoallelic transcription (16).
The way in which African trypanosomes regulate transcription is unique compared with most eukaryotes, and little is known about the chromatin structure of the trypanosome genome. Here, we investigated nucleosome distribution within the genome of both bloodstream and insect forms of T. brucei. In general, we find that nucleosomes are enriched on the nontranscribed repeat arrays and are depleted from the promoter regions of a variety of different transcription units (including examples transcribed by Pol I, II, and III) in both bloodstream and insect forms of T. brucei. We used two isogenic bloodstream form T. brucei lines containing marker genes in silent and active VSG BESs to investigate whether differences in chromatin structure exist between VSG BESs when they are either transcriptionally active or silent. Strikingly, we find that the active VSG BES in bloodstream form T. brucei is particularly depleted of nucleosomes compared with all other regions analyzed and shows a more open chromatin structure than silent VSG BESs. This difference may provide a level of epigenetic control whereby bloodstream form trypanosomes are able to efficiently pass on the transcriptional states of active and silent BESs to daughter cells.
MATERIALS AND METHODS
Trypanosome strains and culturing.
Bloodstream form and procyclic-form T. brucei subsp. brucei strain 427 was used for all experiments. Wild-type procyclic cells were grown at 27°C in SDM-79 medium containing 10% fetal bovine serum (8). Bloodstream form trypanosomes were grown at 37°C in HMI-9 medium containing 10% fetal bovine serum and 10% SerumPlus (SAFC Biosciences) (26). Two isogenic bloodstream form T. brucei cell lines containing a hygromycin resistance gene downstream of the promoter of the VSG221 BES and a neomycin resistance gene downstream of the promoter of the VSGVO2 BES were used (T. brucei HNI). In T. brucei HNI (221+) the VSG221 BES was active, and the VSGVO2 BES was silent, while in T. brucei HNI (VO2+) the VSGVO2 BES was active, and the VSG221 BES was silent (66).
ChIP.
Chromatin immunoprecipitation (ChIP) was performed essentially as described previously (43). In brief, cultures of T. brucei were fixed in 1% formaldehyde for 20 min at room temperature. The reaction was stopped by the addition of glycine to a final concentration of 125 mM. Chromatin was sonicated to an average DNA size of about 200 to 500 bp using a BioRuptor (Wolf Laboratories) and clarified by centrifugation at 15,000 × g for 5 min at 4°C. The sonicated extract was precleared using protein A-Sepharose CL-4B beads (GE Healthcare) and incubated for 16 h either with or without the relevant anti-histone antibody. Chromatin from 7 × 107 cell equivalents was used per immunoprecipitation (IP). Antibodies used for the ChIP experiments include anti-histone H3 (ab1791; AbCam) at a concentration of 2 μg per IP, anti-T. brucei histone H4 (kind gift of the George Cross laboratory) at a concentration of 10 μg per IP (75), and anti-histone H2A (07-146; Upstate) at a final dilution of 1:58. The protein-DNA complexes were incubated with protein A-Sepharose CL-4B beads for 2 h and eluted from the beads after a wash in 1% SDS-0.1 M NaHCO3 preheated to 65°C. The cross-links were reversed by adding NaCl to a final concentration of 325 mM and incubated at 65°C overnight. Following RNase A and proteinase K treatment, DNA was purified using a QIAquick PCR purification kit (Qiagen). Each ChIP experiment was performed two or three times, as indicated in the figure legends, and the results were analyzed by hybridization of slot blots or quantitative PCR (qPCR).
Slot blot analysis.
ChIP material was analyzed on slot blots using radiolabeled probes. Twenty microliters of input chromatin (10% of total) and immunoprecipitated DNA was diluted in TE (Tris-EDTA) buffer (pH 8.0) and denatured by heating to 65°C for 30 min in a final concentration of 0.3 M NaOH. The denatured DNA were neutralized by the addition of ammonium acetate, pH 7.0, to a final concentration of 1.0 M and immediately transferred to Hybond XL membrane (GE Healthcare) using a Schleicher and Schuell Minifold II slot blot apparatus. Membranes were hybridized with a probe for the 50-bp repeats directly flanking VSG BESs (84) or the 177-bp repeats comprising the bulk of the T. brucei minichromosomes (81). The radioactive probes were labeled with [α-32P]dCTP using Ready To-Go DNA Labeling Beads and Illustra ProbeQuant G-50 Micro Columns according to the manufacturer's instructions (GE Healthcare). Membranes were hybridized with radiolabeled probes at 42°C overnight and washed to an end stringency of 0.3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Blots were exposed to a PhosphorImager screen (Bio-Rad), and the percentage of DNA immunoprecipitated was determined using QuantityOne software (Bio-Rad).
Quantitative PCR.
The ChIP material obtained was analyzed using qPCR. Input chromatin (10% of total) and immunoprecipitated material were analyzed using the primers listed in Table S1 in the supplemental material. All qPCR samples were amplified in triplicate in a 20-μl total reaction volume containing 19 μl of qPCR reaction mix (LightCycler480 SYBR green Master Mix; Roche), 0.5 μM (each) forward and reverse primers, and 1 μl of the DNA template. Reaction mixtures were amplified using a LightCycler480 Real-Time PCR System (Roche) according to the manufacturer's instructions. All PCR primers and conditions were optimized to produce a single amplicon of the correct size.
Micrococcal nuclease digestion of T. brucei chromatin.
Digitonin was used to permeabilize bloodstream form T. brucei prior to micrococcal nuclease (MNase) treatment, as previously described (43, 51). In brief, 3 × 108 T. brucei HNI (221+) or T. brucei HNI (VO2+) cells were incubated in permeabilization buffer (100 mM KCl, 10 mM Tris [pH 8.0], 25 mM EDTA, 1 mM dithiothreitol [DTT]) containing digitonin at a final concentration of 40 μM for 5 min at room temperature. Cells were pelleted at 1,200 × g for 5 min at 4°C, and unincorporated digitonin was removed by washing the cells in isotonic buffer (100 mM KCl, 10 mM Tris [pH 8.0], 10 mM CaCl2, 5% glycerol, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 1 μg/ml each of pepstatin A, leupeptin, and aprotinin). Cells were then centrifuged at 1,200 × g for 5 min at 4°C and resuspended in MNase buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.25 M sucrose, 3 mM CaCl2, 100 μM PMSF) to a concentration of 5 × 105 cells μl−1. Six units of MNase (Worthington Biochemicals) was added to the cell suspension (two units per 1 × 108 cells) and incubated for 5 min at 37°C. The reaction was stopped by the addition of EDTA and EGTA to a final concentration of 10 mM. Nuclei were pelleted at 10,000 × g for 10 min at 4°C, and the supernatant was discarded. Chromatin was solubilized by resuspending the nuclear pellet in 200 μl of RSB buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 5 mM EDTA, 5 mM EGTA, 0.05% NP-40, 200 mM NaCl). Cell debris was removed by centrifuging at 10,000 × g for 10 min at 4°C. A 15-μl sample of the supernatant was removed as an input control, and the remaining supernatant was loaded immediately on a preprepared 5 to 30% sucrose gradient, as described below.
Sucrose density gradient fractionation of multinucleosomes.
MNase-digested DNA was separated into mono-, di-, tri-, and tetranucleosome fractions using 5 to 30% sucrose gradients, essentially as described previously (24). Discontinuous sucrose gradients were prepared containing 2 ml each of 5, 10, 15, 20, 25, and 30% sucrose solution in gradient buffer (10 mM Tris-HCl [pH 7.5], 0.25 mM EDTA, 100 mM NaCl, 0.1 mM PMSF, and 0.5 μg/ml each of pepstatin A, leupeptin, and aprotinin) 24 h prior to use and stored at 4°C. The supernatant from the MNase treatment was carefully loaded on the 5 to 30% sucrose gradient and centrifuged at 40,000 rpm (Beckman SW41 Ti rotor) for 16 h at 4°C. Twenty-four 500-μl fractions were isolated. Proteins were removed by treating fractions with proteinase K for 30 min at 37°C, followed by phenol-chloroform and chloroform extractions and ethanol precipitation. Aliquots from each of the MNase-digested DNA fractions were analyzed on a 2% agarose gel for the presence of mono- and multinucleosomes. Fractions containing di-/trinucleosomes (fractions 18 to 21) and tri-/tetranucleosomes (fractions 22 and 23) were pooled for analysis by qPCR using the primer pairs indicated in the figures, sequences of which can be found in Table S1 in the supplemental material. The data were plotted as the fold difference between T. brucei HNI (221+) and T. brucei HNI (VO2+). Error bars in the figures reflect standard deviations of the average signal obtained from three independent experiments.
RESULTS
Histone H3 is enriched in nontranscribed repeat sequences compared with regions highly transcribed by Pol II or Pol III.
Most of the trypanosome genome is transcribed as extensive polycistronic transcription units extending for many tens of kilobases (10, 75), with relatively few regions remaining nontranscribed. Notable exceptions which are kept transcriptionally silent include the 50-bp and 177-bp repeat arrays. The 50-bp repeats form the upstream flanks of all known VSG BESs and typically extend for 40 to 50 kb (66, 73, 84). The 177-bp repeat arrays comprise the bulk of the transcriptionally silent T. brucei minichromosomes (81). This highly abundant class of about 100 small chromosomes (each ranging between 50 to 150 kb) makes up about 10% of the T. brucei nuclear genome (81).
To determine the distribution of histone H3 over different types of genomic regions in T. brucei, we first looked at histone H3 occupancy over the nontranscribed 50-bp and 177-bp repeat arrays. Histones are extremely well conserved within eukaryotes, and an antibody against the C terminus of human histone H3 specifically recognizes T. brucei histone H3 (46). Chromatin immunoprecipitation (ChIP) using formaldehyde cross-linked T. brucei chromatin was performed with the anti-histone H3 antibody or no antibody as a negative control. Input chromatin (10% of total) and immunoprecipitated material were analyzed by slot blot using radiolabeled probes for either the 50-bp repeats or the 177-bp repeats (Fig. 1A). The signal on the blots was quantitated, and values obtained from the no-antibody control were subtracted from the values obtained using the anti-histone H3 antibody. In both bloodstream form and procyclic-form T. brucei, about 8% of the input DNA was immunoprecipitated with anti-histone H3 antibody, showing that histone H3 is very abundant in these nontranscribed regions (Fig. 1D).
FIG. 1.
Histone H3 is relatively abundant on nontranscribed repeat regions in T. brucei chromatin compared with the SL and 5S RNA transcription units. (A) Histone H3 distribution analyzed by ChIP performed using an anti-histone H3 antibody (α-H3) or no antibody (No Ab) in bloodstream form (BF) or procyclic form (PF) T. brucei, as described previously (43). Input material (10% of total [10% In]) was compared with immunoprecipitated samples using slot blot analysis. The slot blots were hybridized with radiolabeled probes for the transcriptionally silent 50-bp repeat regions flanking VSG BESs (Fig. 4A) or the 177-bp repeats, which make up the bulk of the transcriptionally silent minichromosomes. Three independent ChIP experiments were performed, and a representative slot blot is shown. (B) Schematic of the genomic region containing the SL RNA transcription units transcribed by RNA Pol II. The SL promoters are indicated with flags, and the SL genes, as well as characteristic transposable elements found within the spliced leader arrays (SLACS) (1) are indicated with black boxes. Regions analyzed by qPCR are indicated with lettered brackets. (C) Schematic of the 5S rDNA transcription units, transcribed by RNA Pol III. The 5S rDNA promoters are indicated with flags, the 5S rDNA genes are indicated with black boxes, and regions amplified using qPCR are indicated by lettered brackets. (D) Quantification of histone H3 distribution in bloodstream form and procyclic-form T. brucei analyzed either by hybridization of slot blots (panel A) using a Phosphorimager for quantitation of the 50-bp or 177-bp simple sequence repeats or qPCR for the Pol II- or Pol III-transcribed transcription units. Regions analyzed include the gene encoding the large subunit of RNA Pol I, γ-tubulin (γTub), regions around the SL RNA (a to d), or 5S rRNA transcription units (e to g) (panels B and C show the location of the amplified regions). Signals are expressed as a percentage of input immunoprecipitated after subtraction of signal from the no-antibody control. The data shown are the average of three independent ChIP experiments, with the standard deviations indicated with error bars. ψ, pseudogene.
We next looked at the distribution of histone H3 on actively transcribed regions of the T. brucei genome. As with the majority of genes in T. brucei, the genes encoding the RNA polymerase I large subunit (Pol I) and γ-tubulin are present in large polycistronic transcription units constitutively transcribed by Pol II in both bloodstream form and procyclic form T. brucei. Quantitative PCR (qPCR) was used to determine the relative abundance of these two genes in the material immunoprecipitated with the anti-histone H3 antibody. The primers used for the qPCR experiments are shown in Table S1 in the supplemental material. In both life cycle stages, about 5 to 6% of the input DNA was immunoprecipitated (Fig. 1D), indicating that these Pol II-transcribed genes contain less histone H3 and are in a more open chromatin conformation than the silent repeats.
In trypanosomes, maturation of the precursor mRNAs requires the addition of a 39-nucleotide spliced leader (SL) RNA onto the 5′ end of every mRNA (39). In order to provide the large amount of SL RNA necessary for trans-splicing, the trypanosome has up to 200 SL RNA transcription units which are transcribed by RNA Pol II (19, 70). The repetitive SL genes are highly transcribed regions in the T. brucei genome, and are the only Pol II-transcribed genes with a well-characterized promoter (14, 23, 70). Here, we used ChIP followed by qPCR to determine the distribution of histone H3 in the genomic region containing the SL RNA transcription unit repeats. Figure 1B depicts a portion of a genomic region containing 10 to 11 tandem SL RNA transcription units, as well as a retrotransposable element called the SL-associated conserved sequence (SLACS), which has integrated exclusively in the SL RNA genes (1, 55). Using ChIP-qPCR, we found that the SLACS elements contain amounts of histone H3 similar to those of the genes encoding the Pol I large subunit or γ-tubulin, which are also transcribed in both life cycle stages by Pol II (Fig. 1D). However, approximately 3-fold less histone H3 was found on the SL gene promoters, and about 2-fold less histone H3 was present on the SL RNA genes themselves or the SL RNA spacer region (Fig. 1D). This indicates that the SL RNA transcription units are more transcriptionally open than those areas containing SLACSs.
We next determined the distribution of histone H3 across the 5S rDNA transcription units transcribed by RNA Pol III. 5S rRNA is an integral component of the large subunit of ribosomes, and a schematic depicting part of a region of eight tandem 5S rRNA transcription units is shown in Fig. 1C. The histone H3 distribution across the 5S rDNA region was slightly lower than that found within the Pol II-transcribed polycistronic transcription units and was constant between both T. brucei life cycle stages (Fig. 1D).
Histone H3 is particularly depleted around the promoters of the Pol I-transcribed rDNA transcription units.
It has been shown in a variety of eukaryotes that the ribosomal DNA transcription units are transcribed at a very high rate by Pol I. In T. brucei there are about 11 to 12 rDNA transcription units arranged over six to seven chromosomes (4). In order to determine the distribution of histone H3 across these highly transcribed rDNA transcription units, we designed qPCR primers spanning the length of an rDNA transcription unit as well as in the nontranscribed rDNA spacer (80) (Fig. 2A). We performed ChIP-qPCR in both bloodstream form and procyclic-form T. brucei and found that the rDNA promoter region contains approximately 3-fold less histone H3 (approximately 1 to 2% input precipitated) than the nontranscribed rDNA spacer (5 to 6% of the input was immunoprecipitated) (Fig. 2B). This is compatible with an open chromatin structure that is depleted of nucleosomes immediately around the transcriptionally highly active Pol I promoters. Additionally, a gradient of histone H3 distribution was found along the rDNA transcription unit, with levels that were lowest immediately downstream of the promoter and gradually increasing up to those observed in the spacer region (Fig. 2B).
FIG. 2.
T. brucei ribosomal DNA transcription units show relative depletion of histone H3 around the promoter regions. (A) Schematic depicting the region around a T. brucei rDNA transcription unit according to White et al. (80); the rDNA promoter is indicated by a flag, the rDNA genes are shown as black boxes, and the regions analyzed by qPCR are indicated by lettered brackets. (B) Distribution of histone H3 over the rDNA as determined by ChIP using a histone H3 antibody (or no antibody as a negative control) in bloodstream form (BF) or procyclic form (PF) T. brucei. qPCR was used to amplify regions indicated in panel A. Signals are expressed as the percentage of input immunoprecipitated after subtraction of signal from the no-antibody control. Results show the average from three independent experiments, with the standard deviations indicated with error bars.
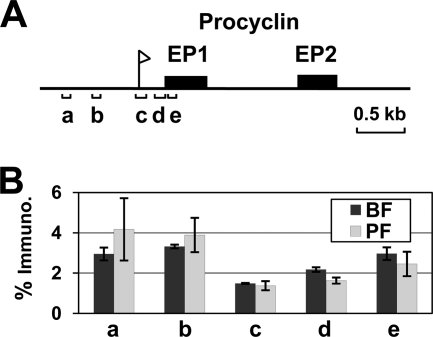
Histone H3 is depleted from the procyclin gene promoters in both bloodstream form and procyclic form T. brucei.
In most eukaryotes RNA Pol I exclusively transcribes rDNA transcription units. However, T. brucei is unique in its ability to use Pol I to transcribe the major cell surface proteins of both procyclic form and bloodstream form trypanosomes (22, 32, 64). The surface coat of procyclic form trypanosomes is composed of members of the procyclin family of glycoproteins including the EP and GPEET procyclin variants (62, 63), of which one EP procyclin transcription unit is depicted in Fig. 3A. Using ChIP-qPCR analysis, we found that histone H3 is more than 3-fold depleted on the procyclin EP promoter in both bloodstream and procyclic T. brucei (less than 1.5% ChIP material immunoprecipitated) compared with regions upstream of the EP procyclin transcription unit (Fig. 3B). The EP procyclin genes contain more histone H3 than the promoter region (about 3% of input immunoprecipitated) (Fig. 3B); however, this is 2-fold less than that observed in genes present in the Pol II-transcribed polycistronic arrays, indicating a more open chromatin structure (Fig. 1D).
FIG. 3.
Histone H3 is less abundant on procyclin promoters in both bloodstream form and procyclic form T. brucei. (A) Schematic depicting part of an EP procyclin locus (PARP B1) (see references 62 and 67). The EP procyclin promoter is indicated with a flag, the EP procyclin genes are indicated by black boxes, and the genomic regions analyzed using qPCR are indicated by lettered brackets. (B) Histone H3 distribution over the procyclin locus was investigated using ChIP with an anti-histone H3 antibody (or no antibody as a negative control) in bloodstream form (BF) and procyclic form (PF) T. brucei. Quantitative PCR was used to amplify the procyclin genomic regions indicated in panel A. The data are expressed as the percentage of input immunoprecipitated after subtraction of signal from the no-antibody control. The results shown are the average signal from three independent ChIP experiments, with the standard deviations indicated with error bars.
Analysis of these two Pol I transcription units (rDNA and procyclin) is complicated by the fact that the genes within them are present in multiple, nearly identical copies. In a number of other eukaryotes, it has been shown that about half of the rDNA transcription units are transcriptionally active (11). In T. brucei this is also likely to be the case, although this issue has not yet been resolved. Multiple procyclin transcription units are also present, and within a single cell multiple units can be simultaneously transcriptionally active (reviewed in reference 62). In order to better study the chromatin structure of a single Pol I transcription unit, we investigated the chromatin structure of VSG BESs since in bloodstream form T. brucei a single VSG BES is transcribed by Pol I in a monoallelic fashion (9, 22).
Active VSG expression sites are dramatically depleted of histones compared with silent VSG expression sites.
VSG BESs are extensive telomeric transcription units typically varying between 40 to 60 kb and containing a variety of expression site-associated genes (ESAGs) in addition to the telomeric VSG gene (Fig. 4A) (25). Although different members of individual ESAG gene families are polymorphic, they remain highly similar to each other in sequence (25), which greatly complicates the analysis of BES transcriptional states. Here, we use two isogenic bloodstream form T. brucei cell lines containing different single-copy marker genes in both the active and silent VSG BES (Fig. 4B) to investigate the distribution of histone H3 along the length of VSG BESs as well as between silent and active BESs. T. brucei HNI (221+) has an active VSG221 BES with a hygromycin resistance gene inserted behind the promoter and a neomycin resistance gene inserted behind the promoter of the silent VSGVO2 BES. In T. brucei HNI (VO2+) the VSG221 BES is silent, and the VSGVO2 BES is active.
FIG. 4.
Active VSG expression sites (BESs) are depleted of histone H3 compared with silent VSG BESs in bloodstream form T. brucei. (A) A typical VSG BES (modified from AnTat1.3A) (5, 41) is indicated in the schematic shown, with the promoter indicated by a flag, the expression site-associated genes (ESAGs) are indicated with numbered boxes, and characteristic BES repeat arrays are indicated with striped boxes. The area immediately around the BES promoter is shown expanded below. The locations of BES regions used for qPCR analysis that are common to all BESs are indicated with lettered brackets. (B) The two isogenic bloodstream form T. brucei 427 cell lines used for investigating the distribution of histone H3 along active versus silent VSG BESs are shown as large red or blue boxes. T. brucei HNI (221+) expresses VSG221 from the active VSG221 BES, and T. brucei HNI (VO2+) expresses VSGVO2 from the active VSGVO2 BES (66). BESs are shown with the promoters indicated with flags. Relevant single copy genes, including hygromycin resistance (Hyg), a VSG pseudogene (ψ), VSG221, neomycin resistance (Neo), and VSGVO2 are indicated. Transcription of the active VSG BES is indicated with arrows. The BES regions analyzed by qPCR are indicated with lettered bars. (C) Histone H3 distribution over active or silent VSG BESs. ChIP analysis was carried out on isogenic bloodstream form T. brucei cell lines containing either an active VSG221 BES [HNI (221+)] or an active VSGVO2 BES [HNI (VO2+)] (panel B). Chromatin from each cell line was immunoprecipitated using an anti-histone H3 antibody or no antibody as a negative control. Quantitative PCR was used to amplify conserved regions shared by all BESs indicated in panel A or the single-copy marker genes specific for the individual BESs shown in panel B. After subtraction of signal from the no-antibody control, signals are expressed as a percentage of input precipitated. ChIP results from the HNI (221+) cell line are shown with red bars, and results from the HNI (VO2+) cell line are shown with blue bars. The results presented are the average signal obtained from three independent experiments, with the standard deviations indicated with error bars.
Using ChIP-qPCR, we first used these cell lines to investigate the distribution of histone H3 along the length of VSG BESs. In T. brucei 427 there are approximately 15 BESs (25), all of which are silent except one. Due to the high degree of sequence similarity between BESs, most of the primer pairs used for amplifying genes within the BES would be expected to recognize most if not all BESs (BES Fig. 4C) (25). The distribution of histone H3 over BESs was found to be particularly low over the “core” BES promoter (29) (Fig. 4C, qPCR primer pair c) in isogenic T. brucei strains expressing either the VSG221 or VSGVO2 BES (Fig. 4C, red or blue bars, respectively). Levels of histone H3 over the BES core promoter were about 2-fold lower than in the region upstream of the BES promoter (primer pair a) and about 3-fold lower than levels in ESAG6/7 (primer pair e). The low level of histone H3 present at the BES core promoter is consistent with data from Figueiredo and Cross (17) and suggests that the BES promoter region is in a more open chromatin conformation in multiple BESs. These results are compatible with the observation that there is transcription extending from most if not all silent BES promoters in bloodstream form T. brucei, but this transcription attenuates relatively rapidly downstream of the BES promoter in all except for the active BES (2, 65, 79).
We next investigated the distribution of histone H3 between silent and active BESs by performing ChIP-qPCR analysis on the single-copy marker genes present in our two isogenic T. brucei cell lines (Fig. 4C). We found that there is a striking 11- to 40-fold difference in distribution of histone H3 on active versus silent BESs. Less than 0.5% of input was immunoprecipitated from the single-copy genes present in the actively transcribed VSG221 BES (Fig. 4C, primer pairs i to k) and VSGVO2 BES (Fig. 4C, primer pairs l and m) compared to over 4% when the same genes were silent. This indicates that the active VSG BES is in a far more open chromatin conformation than the silent BESs. As expected, in the isogenic T. brucei strain where the VSG221 BES has been silenced and the VSGVO2 BES activated, the distribution of histone H3 over the two BESs is reversed (Fig. 4C).
To determine whether other histones exhibit a similar distribution pattern between silent and active BESs, we performed additional ChIP experiments using antibodies against histone H4 and histone H2A. qPCR analysis was performed using a subset of the BES primer pairs and γ-tubulin as a control. As observed with the histone H3 ChIP experiments, histone H4 was found to be significantly depleted (5- to 10-fold) on the active BES compared with the silent BES (Fig. 5A) while histone H2A was 2- to 4-fold depleted (Fig. 5B). Taken together, these data strongly suggest that the active BES in bloodstream form T. brucei is depleted of nucleosomes.
FIG. 5.
Histone H4 and histone H2A are relatively depleted from active compared with silent VSG BESs. (A) Histone distribution was determined in T. brucei cell lines containing either an active VSG221 [HNI (221+)] or an active VSGVO2 [HNI (VO2+)] BES. Diagrams for these cell lines are shown in Fig. 4B. ChIP experiments were performed using an antibody against histone H4 or no antibody as a negative control. Quantitative PCR was used to analyze two regions common to all BESs: the BES core promoter (primer c in Fig. 4A) and ESAG7/6 (ESAG6 and primer e in Fig. 4A). Additionally, the five single-copy sequences, shown schematically in Fig. 4B, specific for either the VSG221 or VSGVO2 BES were analyzed: i, hygromycin; j, ψ pseudogene; k, VSG221; l, neomycin; and m, VSGVO2. ChIP data from the VSG221-expressing cell line are indicated with red bars, and data from the VSGVO2-expressing line are shown with blue bars. As a control, histone H4 presence in the γ-tubulin locus (γTub) was also analyzed. Signals are expressed as a percentage of input immunoprecipitated, with the results shown being the average of three independent experiments; the standard deviations are indicated with error bars. (B) ChIP was performed as described for panel A, except that an anti-histone H2A antibody was used. Signals from T. brucei cell lines containing an active VSG221 (red bars) or an active VSGVO2 (blue bars) gene are expressed as a percentage of input immunoprecipitated and are the average signal obtained from two independent experiments. The primers used for qPCR analysis are as indicated in panel A.
In insect form T. brucei, all BESs are downregulated as VSG is not necessary in this life cycle stage. BES transcriptional silencing appears to be more “leaky” in procyclic form than in bloodstream form T. brucei, and significant levels of transcription extending from most, if not all, BES promoters has been reported (2, 65, 79). Using a subset of the BES qPCR primer pairs, we next investigated the distribution of histone H3 along BESs in procyclic-form T. brucei. Less histone H3 was present on the BES core promoter than the rest of the BES, compatible with the transcriptional activity observed in this life cycle stage (Fig. 6).
FIG. 6.
Histone H3 distribution over VSG expression sites in procyclic form T. brucei. ChIP was performed in procyclic form T. brucei using either an anti-histone H3 antibody or no antibody. Quantitative PCR was used to analyze three regions common to all BESs with the primers indicated in Fig. 4A: the BES core promoter (ESProm, primer c in Fig. 4A), the area immediately downstream of the BES promoter (ESProm Dn; primer d), and ESAG7/6 (ESAG6; primer e). Additionally, the single-copy VSG221 gene (primer k in Fig. 4B) was amplified. The results are expressed as the percentage of input immunoprecipitated after the signal obtained from the no-antibody control was subtracted. Three independent experiments were performed, and the standard deviations are indicated with error bars.
Active VSG expression sites are depleted of nucleosomes.
To ensure that the results obtained from the ChIP experiments were not an artifact of cross-linking, we investigated BES chromatin structure using an alternative experimental approach. In order to confirm that the relative depletion of histones over the active BES observed in the ChIP experiments indeed reflected reduced nucleosome occupancy, we examined the relative sensitivity of the active and silent BES to digestion by micrococcal nuclease (MNase). Rather than using the traditional MNase experimental procedure requiring large numbers of cells and Southern blotting, we used an approach which involves fractionating MNase-digested chromatin with sucrose gradients, followed by qPCR. This newly developed approach has been used to establish nucleosome depletion over mammalian promoters and provides a more sensitive and quantitative way of investigating chromatin structure than older Southern blotting-based methods (18, 40). Genomic areas which are depleted for nucleosomes are in a more open chromatin structure and are therefore hypersensitive to digestion by MNase. Using this reasoning, less qPCR product would be expected to be amplified from oligonucleosomal DNA template in regions of the genome that are in a relatively more open chromatin state than in those genomic regions with a more closed chromatin structure.
Chromatin from permeabilized T. brucei HNI (221+) and HNI (VO2+) cell lines was partially digested with MNase to yield oligonucleosomes. This preparation was then separated on 5 to 30% sucrose gradients, and DNA was isolated from the collected fractions (Fig. 7A). Fractions enriched in di- and trinucleosomes were pooled to form nucleosomal fraction I (Fig. 7A, NI), as were fractions enriched in tri- and tetranucleosomes (Fig. 7A, NII). These two pools of oligonucleosomal DNA were then subjected to qPCR using a subset of the primer pairs used in the ChIP experiments (Fig. 7B and C) and plotted as the fold difference in the amount of DNA amplified between the two isogenic T. brucei lines. For the housekeeping genes (Pol I, γ-tubulin, and α-tubulin) and two regions common to all BESs (BES promoter and ESAG6/7), equivalent levels of DNA were amplified from the NI and NII fractions using both T. brucei HNI (221+) and HNI (VO2+), indicating that the chromatin structure within these regions does not differ between these two cell lines.
FIG. 7.
The active VSG expression site in T. brucei is relatively depleted of nucleosomes. (A) Chromatin from the T. brucei HNI (221+) or HNI (VO2+) cell lines was partially digested by MNase and fractionated through sucrose gradients to obtain mono- and oligonucleosomes. DNA was extracted from the input material as well as fractions 9 to 24 (Frac. nr.) and electrophoresed on a 2% agarose gel stained with ethidium bromide. Size markers are indicated on the left, and the locations of mono-, di-, tri-, and tetranucleosomes are indicated on the right. Oligonucleosomal fractions comprised primarily of di- and trinucleosomes were pooled to serve as the template DNA in subsequent qPCR analysis (NI). In addition, fractions containing primarily tri- and tetranucleosomes were pooled (NII). Three independent experiments were performed, and a representative gel is shown. (B) qPCR was used to amplify oligonucleosomal DNA from the NI pool of fractions containing primarily di- and trinucleosomes. These data are plotted as the fold difference in amount of DNA amplified (indicating nucleosome occupancy) between the two isogenic T. brucei lines expressing either VSG221 or VSGVO2. Red bars indicate the values obtained from the VSG221-expressing cells divided by the values obtained from the VSGVO2-expressing cell line (221/VO2). Blue bars represent the reciprocal calculation (VO2/221). Three housekeeping genes were monitored as a control and include γ-tubulin (γtub), α-tubulin (αtub), and the large subunit of RNA Pol I. Two primer pairs common to all BESs were also used and include the BES core promoter (ESPro) and ESAG7/6 (represented in Fig. 4A as primers c and e, respectively). The five single-copy sequences specific for either the VSG221 or VSGVO2 BES were analyzed (shown schematically in Fig. 4B as primers i to m). The results presented are the average from three independent experiments, with the standard deviations indicated with error bars. (C) qPCR was used to amplify DNA from the NII pool containing primarily tri- and tetranucleosomes, as described in panel B. Average values are shown from three independent experiments, with standard deviations indicated with error bars.
Using primers for the five single-copy genes (Fig. 4B), we found that DNA from silent BESs was about 3-fold more enriched in the di-/trinucleosomal fraction than DNA from the active BES (Fig. 7B, NI), indicating that the active BES is indeed more sensitive to digestion by MNase. Comparable results were obtained using the tri-/tetranucleosomal fraction (Fig. 7C, NII). This result is similar to that observed in the ChIP experiments using anti-histone antibodies and confirms that the active BES is depleted of nucleosomes. As a control, qPCR analysis was performed on “naked” DNA that was stripped of nucleosomes and then partially digested with MNase (see Fig. S1A in the supplemental material). As expected, no relative enrichment between the different BESs was found, providing further support that nucleosome positioning is responsible for the differences observed between silent and active expression sites (see Fig. S1B in the supplemental material).
DISCUSSION
Here, we examined the distribution of nucleosomes throughout the genome of T. brucei using histone H3 as a marker. We find that histone H3 is enriched at the extensive nontranscribed 50-bp and 177-bp simple sequence repeats (8 to 10% of input precipitated) in both bloodstream form and procyclic-form T. brucei, compatible with the idea that these nontranscribed regions are present in a more closed chromatin state. In contrast, histone H3 is relatively depleted in the Pol II transcription units (γ-tubulin or Pol I large-subunit genes with 5 to 6% of input precipitated) or Pol III-transcribed 5S rRNA transcription units (approximately 4% input precipitated), which is compatible with the regions being present in a relatively open chromatin state. Histone H3 is particularly depleted in the spliced leader (SL) arrays which are highly transcribed by Pol II, with only 2% of the input precipitated. These results are in agreement with what has been found in yeast, where histone occupancy has in general been shown to be inversely correlated with rates of transcription (3, 35).
Within the SL RNA genomic loci we observed differences in chromatin structure and found about 3-fold more histone H3 present within characteristic SLACS retroposons (about 6% input precipitated) than within the SL transcription units themselves (about 2% input precipitated). This is in agreement with the observed reduced density of Pol II over the SLACS elements, indicating that transcription through the SLACS open reading frames (ORFs) is relatively inefficient compared with the SL RNA (55). Recently, similar observations have been reported for other trypanosomatids. In Trypanosoma cruzi relatively few nucleosomes are detected at the SL promoter (61), and in Leishmania tarentolae the SL RNA promoters and transcribed SL RNA genes are nucleosome free (27).
In the Pol I-transcribed rDNA we find that histone H3 occupancy is greatly reduced in the area immediately downstream of the rDNA promoter (approximately 1% input precipitated) compared with the nontranscribed rDNA spacer (5 to 6% input precipitated). Therefore, the promoter regions of T. brucei transcription units transcribed by all three RNA polymerases (Pol I, II, and III) appear to be particularly depleted of histone H3. This relative depletion of histone H3 at promoter regions is in agreement with previous studies in yeast which provide evidence for nucleosome depletion at active regulatory regions (3, 35, 83). Within the rDNA in T. brucei, we find evidence for a gradient of histone H3 distribution extending along the length of the rDNA transcription unit, ranging from relatively little histone H3 in the neighborhood of the rDNA promoter, up to higher levels of histone H3 at the end of the transcription unit and approaching levels found in the nontranscribed spacer. A gradient of histone H3 distribution over the rDNA transcription unit has also been shown in S. cerevisiae (30) although these data appear to show relatively less histone H3 in the nontranscribed spacer region than we found using T. brucei.
Procyclin is another Pol I transcription unit in T. brucei which encodes the major surface protein in insect form trypanosomes. We found approximately 3-fold less histone H3 on the procyclin EP promoter in both bloodstream form and procyclic form T. brucei in comparison with sequences upstream of these transcription units. These data are compatible with previous studies which found significant transcriptional activity from the procyclin promoter even in bloodstream form T. brucei (56, 78), where the elongation of transcription appears to be developmentally controlled (78).
However, a complicating factor in the analysis of rDNA Pol I transcription units has been the repetitive nature of the nearly identical genes contained within them. This complication applies to the analysis of rDNA as well as procyclin transcription in T. brucei. Bloodstream form T. brucei is unique, however, in transcribing a single VSG BES by Pol I using strict monoallelic exclusion (7, 9, 53). We have been able to investigate the chromatin structure of different single-copy sequences within VSG BESs using isogenic T. brucei strains with two different VSG BESs in either “on” or “off” states. Here, we show that there is a striking depletion of histone H3 along the entire length of an active VSG BES, with an 11- to 40-fold difference between silent and active sites. Using antibodies against histone H4 and H2A in additional ChIP experiments, we identified a similar trend, albeit less pronounced, with 5- to 10-fold and 2- to 4-fold depletion, respectively, compared with the same BES in its silent state. The fact that VSG BESs are monoallelically expressed and contain, or can be modified to contain, single-copy sequences greatly facilitates the analysis of the chromatin architecture of these Pol I transcription units. Our results showing the dramatic depletion of histones within the active VSG BES are consistent with those from Figueiredo and Cross (17) and are in agreement with studies which argue that actively transcribed rRNA genes are largely devoid of histones (12, 50).
We find more striking differences in the distribution of histone H3 than in histone H2A distribution in active versus silent VSG BESs. It is important to note, however, that the anti-histone antibodies used here differ in how efficiently they are able to immunoprecipitate their target. Using the Pol II-transcribed γ-tubulin genes as a control, the anti-histone H3 antibody immunoprecipitated ∼5% of the input (Fig. 1D), and anti-histone H4 immunoprecipitated ∼2% of the input (Fig. 5A) while anti-histone H2A immunoprecipitated only ∼0.5% of the input (Fig. 5B). The observed variations in abundance of these histones could therefore be a consequence of different antibody affinities. However, in addition these results could be explained by nucleosome dynamics. Within the nucleosome, DNA is first wrapped around a core histone H3/H4 tetramer before two histone H2A/H2B dimers are added (44). Consistent with this, H2A/H2B dimers have been shown to be more dynamic in chromatin, and there is a relatively rapid exchange of histone H2B compared with the histone H3/H4 tetramers, which are more tightly and stably associated in chromatin (31). Our results showing relatively less enrichment of histone H2A in silent BESs is compatible with a scenario whereby histone H2A is less stably associated with the nucleosomes present in silent BESs than histone H3.
Histone H3 was found to be depleted from the multicopy VSG BES promoters in both bloodstream and insect form T. brucei, indicating a more open chromatin conformation. In bloodstream form T. brucei this depletion is about 2-fold greater than in the procyclic form T. brucei, with ∼2% rather than 4% of the input immunoprecipitated. These results agree with the previous observation that a prominent nuclease-hypersensitive site is present within the core promoter of both active and inactive VSG BESs in bloodstream form T. brucei (51). Furthermore, it agrees with the extensive evidence that there is significant nonprocessive transcription from multiple “inactive” VSG BESs in both bloodstream and insect form T. brucei (2, 65, 79), which could explain the open chromatin structure in this region that we observe.
An inherent problem with ChIP experiments is the fact that formaldehyde cross-linking can potentially inhibit antibody access to more interior targets. In order to ensure that we were not observing an artifact related to this, we used an additional experimental approach that did not require cross-linking to investigate VSG BES chromatin structure. Micrococcal nuclease digestion followed by nucleosome fractionation provides a powerful tool to investigate chromatin structure in a quantitative fashion using limited material. Using this approach, we confirm our histone ChIP results and show that the active VSG BESs are in a more open chromatin structure than the silent VSG BESs. This observation that active VSG BESs are highly sensitive to digestion by micrococcal nuclease is in agreement with results obtained using Southern blot analysis of MNase-digested chromatin (17). These results in some ways contradict the findings from previous studies, which concluded that no difference exists in the nucleosome structure between active and silent VSG BESs.
Why has this difference in nucleosomal structure between the two different VSG BES activation states been missed in the past? Early studies comparing the nucleosomal structure of active versus silent BESs have argued that there is no detectably altered nucleosomal organization in active versus silent VSG BESs (20). However, this study did not compare the same VSG BES sequence (in either an active or silent state) with a single probe to allow effective comparison. In our study, using isogenic T. brucei strains, we have been able to compare the same set of single-copy sequences present within an BES in either a transcriptionally active or silent state, allowing us to directly compare differences. A later study concentrated on investigating the chromatin structure immediately around the BES core promoter in both an active and silent state in bloodstream form T. brucei and found a prominent DNase I-hypersensitive site in both cases (51). Our data are in agreement with the observation that most, if not all, VSG BES promoters are present in an open chromatin state (51). However, using two different methods, we find that the active VSG BES is depleted of nucleosomes along its entire length. An additional study using exogenous T7 RNA polymerase to probe for chromatin structure in T. brucei found that the silent VSG BESs in insect form but not bloodstream form T. brucei were refractory to transcription by T7 RNA polymerase (52). The authors argued from these data that chromatin remodeling plays a role in VSG BES silencing specifically in insect form T. brucei but did not find evidence for this being the case in bloodstream form T. brucei. However, it is unclear exactly which aspects of eukaryotic chromatin can impede transcription by T7 RNA polymerase.
We do not know if the relative depletion of nucleosomes on the active VSG BES is a direct consequence of transcriptional activity or if this open chromatin structure is uncoupled from transcription itself. It has been shown in other experimental systems that nucleosome loss is not necessarily a consequence of transcription. For example, after the induction of a heat shock, initial loss of nucleosomes from the Drosophila heat shock locus occurs extremely rapidly and before the first RNA polymerases reach the corresponding region of the gene (59). This initial rapid loss of nucleosomes after heat shock was shown to be independent of transcription and to occur over a larger region than a single transcription unit (59). In these experiments, a second and later wave of nucleosome loss was dependent on transcription. Although we have found that the active VSG BES is significantly depleted of nucleosomes, we do not know the order of events and whether nucleosome depletion precedes VSG BES activation.
In summary, we show that highly transcribed loci within the T. brucei genome have reduced nucleosomes, with the active VSG BES showing the lowest nucleosomal occupancy of all regions investigated. The observed depletion of nucleosomes within the active VSG BES indicates that chromatin remodeling may play a critical role in the monoallelic exclusion necessary for VSG BES control in bloodstream T. brucei. The transcriptional states of active and repressed BESs are efficiently inherited in bloodstream form trypanosomes, suggesting the presence of epigenetic marks. The lack of nucleosomes on the active BES may be one of the factors involved in marking and propagating the epigenetic state of the BES from one generation to the next. Future studies will be necessary to allow us to determine exactly how this chromatin remodeling proceeds during VSG BES switching.
Supplementary Material
Acknowledgments
We are very grateful to Luisa Figueiredo, Nicolai Siegel, and George Cross (Rockefeller University, New York, NY) for providing the T. brucei anti-histone H4 antibody, suggesting the use of the specific anti-histone H2A antibody, and communicating unpublished results on the chromatin structure of VSG expression sites. We thank Jane Mellor and David Clynes (Department of Biochemistry, University of Oxford) for discussions on chromatin structure, for advice on experimental procedures, and for generously allowing us to use the BioRuptor sonicator. We thank Mani Narayanan, Megan Lindsay, Manish Kushwaha, Viola Denninger, Nadina Vasileva, and Alexander Fullbrook for comments on the manuscript.
G.R. is a Wellcome Senior Fellow in the Basic Biomedical Sciences. This research was funded by the Wellcome Trust.
Footnotes
Published ahead of print on 13 November 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Aksoy, S., S. Williams, S. Chang, and F. F. Richards. 1990. SLACS retrotransposon from Trypanosoma brucei gambiense is similar to mammalian LINEs. Nucleic Acids Res. 18:785-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansorge, I., D. Steverding, S. Melville, C. Hartmann, and C. Clayton. 1999. Transcription of “inactive” expression sites in African trypanosomes leads to expression of multiple transferrin receptor RNAs in bloodstream forms. Mol. Biochem. Parasitol. 101:81-94. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, B. E., C. L. Liu, E. L. Humphrey, E. O. Perlstein, and S. L. Schreiber. 2004. Global nucleosome occupancy in yeast. Genome Biol. 5:R62. doi: 10.1186/gb-2004-5-9-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berriman, M., E. Ghedin, C. Hertz-Fowler, G. Blandin, H. Renauld, D. C. Bartholomeu, N. J. Lennard, E. Caler, N. E. Hamlin, B. Haas, U. Bohme, L. Hannick, M. A. Aslett, J. Shallom, L. Marcello, L. Hou, B. Wickstead, U. C. Alsmark, C. Arrowsmith, R. J. Atkin, A. J. Barron, F. Bringaud, K. Brooks, M. Carrington, I. Cherevach, T. J. Chillingworth, C. Churcher, L. N. Clark, C. H. Corton, A. Cronin, R. M. Davies, J. Doggett, A. Djikeng, T. Feldblyum, M. C. Field, A. Fraser, I. Goodhead, Z. Hance, D. Harper, B. R. Harris, H. Hauser, J. Hostetler, A. Ivens, K. Jagels, D. Johnson, J. Johnson, K. Jones, A. X. Kerhornou, H. Koo, N. Larke, S. Landfear, C. Larkin, V. Leech, A. Line, A. Lord, A. Macleod, P. J. Mooney, S. Moule, D. M. Martin, G. W. Morgan, K. Mungall, H. Norbertczak, D. Ormond, G. Pai, C. S. Peacock, J. Peterson, M. A. Quail, E. Rabbinowitsch, M. A. Rajandream, C. Reitter, S. L. Salzberg, M. Sanders, S. Schobel, S. Sharp, M. Simmonds, A. J. Simpson, L. Tallon, C. M. Turner, A. Tait, A. R. Tivey, S. Van Aken, D. Walker, D. Wanless, S. Wang, B. White, O. White, S. Whitehead, J. Woodward, J. Wortman, M. D. Adams, T. M. Embley, K. Gull, E. Ullu, J. D. Barry, A. H. Fairlamb, F. Opperdoes, B. G. Barrell, J. E. Donelson, N. Hall, C. M. Fraser, S. E. Melville, and N. M. El-Sayed. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309:416-422. [DOI] [PubMed] [Google Scholar]
- 5.Berriman, M., N. Hall, K. Sheader, F. Bringaud, B. Tiwari, T. Isobe, S. Bowman, C. Corton, L. Clark, G. A. Cross, M. Hoek, T. Zanders, M. Berberof, P. Borst, and G. Rudenko. 2002. The architecture of variant surface glycoprotein gene expression sites in Trypanosoma brucei. Mol. Biochem. Parasitol. 122:131-140. [DOI] [PubMed] [Google Scholar]
- 6.Birch, J. L., and J. C. Zomerdijk. 2008. Structure and function of ribosomal RNA gene chromatin. Biochem. Soc. Trans. 36:619-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borst, P. 2002. Antigenic variation and allelic exclusion. Cell 109:5-8. [DOI] [PubMed] [Google Scholar]
- 8.Brun, R., and Schonenberger. 1979. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop. 36:289-292. [PubMed] [Google Scholar]
- 9.Chaves, I., G. Rudenko, A. Dirks-Mulder, M. Cross, and P. Borst. 1999. Control of variant surface glycoprotein gene-expression sites in Trypanosoma brucei. EMBO J. 18:4846-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clayton, C. E. 2002. Life without transcriptional control? From fly to man and back again. EMBO J. 21:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conconi, A., R. M. Widmer, T. Koller, and J. M. Sogo. 1989. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell 57:753-761. [DOI] [PubMed] [Google Scholar]
- 12.Dammann, R., R. Lucchini, T. Koller, and J. M. Sogo. 1993. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 21:2331-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dammann, R., R. Lucchini, T. Koller, and J. M. Sogo. 1995. Transcription in the yeast rRNA gene locus: distribution of the active gene copies and chromatin structure of their flanking regulatory sequences. Mol. Cell. Biol. 15:5294-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das, A., Q. Zhang, J. B. Palenchar, B. Chatterjee, G. A. Cross, and V. Bellofatto. 2005. Trypanosomal TBP functions with the multisubunit transcription factor tSNAP to direct spliced-leader RNA gene expression. Mol. Cell. Biol. 25:7314-7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Figueiredo, L. M., G. A. Cross, and C. J. Janzen. 2009. Epigenetic regulation in African trypanosomes: a new kid on the block. Nat. Rev. Microbiol. 7:504-513. [DOI] [PubMed] [Google Scholar]
- 16.Figueiredo, L. M., C. J. Janzen, and G. A. Cross. 2008. A histone methyltransferase modulates antigenic variation in African trypanosomes. PLoS Biol. 6:e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Figueiredo, L. M., and G. A. M. Cross. 2010. Nucleosomes are depleted at the VSG expression site transcribed by RNA polymerase I in African trypanosomes. Eukaryot. Cell 9:148-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gal-Yam, E. N., S. Jeong, A. Tanay, G. Egger, A. S. Lee, and P. A. Jones. 2006. Constitutive nucleosome depletion and ordered factor assembly at the GRP78 promoter revealed by single molecule footprinting. PLoS Genet. 2:e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilinger, G., and V. Bellofatto. 2001. Trypanosome spliced leader RNA genes contain the first identified RNA polymerase II gene promoter in these organisms. Nucleic Acids Res. 29:1556-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greaves, D. R., and P. Borst. 1987. Trypanosoma brucei variant-specific glycoprotein gene chromatin is sensitive to single-strand-specific endonuclease digestion. J. Mol. Biol. 197:471-483. [DOI] [PubMed] [Google Scholar]
- 21.Grummt, I. 2007. Different epigenetic layers engage in complex crosstalk to define the epigenetic state of mammalian rRNA genes. Hum. Mol. Genet. 16(R1):R21-R27. [DOI] [PubMed] [Google Scholar]
- 22.Gunzl, A., T. Bruderer, G. Laufer, B. Schimanski, L. C. Tu, H. M. Chung, P. T. Lee, and M. G. Lee. 2003. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot. Cell 2:542-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunzl, A., E. Ullu, M. Dorner, S. P. Fragoso, K. F. Hoffmann, J. D. Milner, Y. Morita, E. K. Nguu, S. Vanacova, S. Wunsch, A. O. Dare, H. Kwon, and C. Tschudi. 1997. Transcription of the Trypanosoma brucei spliced leader RNA gene is dependent only on the presence of upstream regulatory elements. Mol. Biochem. Parasitol. 85:67-76. [DOI] [PubMed] [Google Scholar]
- 24.Hebbes, T. R., A. L. Clayton, A. W. Thorne, and C. Crane-Robinson. 1994. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 13:1823-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hertz-Fowler, C., L. M. Figueiredo, M. A. Quail, M. Becker, A. Jackson, N. Bason, K. Brooks, C. Churcher, S. Fahkro, I. Goodhead, P. Heath, M. Kartvelishvili, K. Mungall, D. Harris, H. Hauser, M. Sanders, D. Saunders, K. Seeger, S. Sharp, J. E. Taylor, D. Walker, B. White, R. Young, G. A. Cross, G. Rudenko, J. D. Barry, E. J. Louis, and M. Berriman. 2008. Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS One 3:e3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirumi, H., and K. Hirumi. 1989. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 75:985-989. [PubMed] [Google Scholar]
- 27.Hitchcock, R. A., S. Thomas, D. A. Campbell, and N. R. Sturm. 2007. The promoter and transcribed regions of the Leishmania tarentolae spliced leader RNA gene array are devoid of nucleosomes. BMC Microbiol. 7:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes, K., M. Wand, L. Foulston, R. Young, K. Harley, S. Terry, K. Ersfeld, and G. Rudenko. 2007. A novel ISWI is involved in VSG expression site downregulation in African trypanosomes. EMBO J. 26:2400-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jefferies, D., P. Tebabi, and E. Pays. 1991. Transient activity assays of the Trypanosoma brucei variant surface glycoprotein gene promoter: control of gene expression at the posttranscriptional level. Mol. Cell. Biol. 11:338-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones, H. S., J. Kawauchi, P. Braglia, C. M. Alen, N. A. Kent, and N. J. Proudfoot. 2007. RNA polymerase I in yeast transcribes dynamic nucleosomal rDNA. Nat. Struct. Mol. Biol. 14:123-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimura, H., and P. R. Cook. 2001. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153:1341-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kooter, J. M., and P. Borst. 1984. Alpha-amanitin-insensitive transcription of variant surface glycoprotein genes provides further evidence for discontinuous transcription in trypanosomes. Nucleic Acids Res. 12:9457-9472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lam, F. H., D. J. Steger, and E. K. O'Shea. 2008. Chromatin decouples promoter threshold from dynamic range. Nature 453:246-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LeBowitz, J. H., H. Q. Smith, L. Rusche, and S. M. Beverley. 1993. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev. 7:996-1007. [DOI] [PubMed] [Google Scholar]
- 35.Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl, and J. D. Lieb. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36:900-905. [DOI] [PubMed] [Google Scholar]
- 36.Lee, W., D. Tillo, N. Bray, R. H. Morse, R. W. Davis, T. R. Hughes, and C. Nislow. 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 39:1235-1244. [DOI] [PubMed] [Google Scholar]
- 37.Li, B., M. Carey, and J. L. Workman. 2007. The role of chromatin during transcription. Cell 128:707-719. [DOI] [PubMed] [Google Scholar]
- 38.Li, J., G. Langst, and I. Grummt. 2006. NoRC-dependent nucleosome positioning silences rRNA genes. EMBO J. 25:5735-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang, X. H., A. Haritan, S. Uliel, and S. Michaeli. 2003. trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell 2:830-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin, J. C., S. Jeong, G. Liang, D. Takai, M. Fatemi, Y. C. Tsai, G. Egger, E. N. Gal-Yam, and P. A. Jones. 2007. Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell 12:432-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lips, S., P. Revelard, and E. Pays. 1993. Identification of a new expression site-associated gene in the complete 30.5 kb sequence from the AnTat 1.3A variant surface protein gene expression site of Trypanosoma brucei. Mol. Biochem. Parasitol. 62:135-137. [DOI] [PubMed] [Google Scholar]
- 42.Lo, H. J., H. K. Huang, and T. F. Donahue. 1998. RNA polymerase I-promoted HIS4 expression yields uncapped, polyadenylated mRNA that is unstable and inefficiently translated in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:665-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lowell, J. E., and G. A. Cross. 2004. A variant histone H3 is enriched at telomeres in Trypanosoma brucei. J. Cell Sci. 117:5937-5947. [DOI] [PubMed] [Google Scholar]
- 44.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 45.Reference deleted.
- 46.Mandava, V., C. J. Janzen, and G. A. Cross. 2008. Trypanosome H2Bv replaces H2B in nucleosomes enriched for H3 K4 and K76 trimethylation. Biochem. Biophys. Res. Commun. 368:846-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matthews, K. R., C. Tschudi, and E. Ullu. 1994. A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev. 8:491-501. [DOI] [PubMed] [Google Scholar]
- 48.Mavrich, T. N., C. Jiang, I. P. Ioshikhes, X. Li, B. J. Venters, S. J. Zanton, L. P. Tomsho, J. Qi, R. L. Glaser, S. C. Schuster, D. S. Gilmour, I. Albert, and B. F. Pugh. 2008. Nucleosome organization in the Drosophila genome. Nature 453:358-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McStay, B., and I. Grummt. 2008. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 24:131-157. [DOI] [PubMed] [Google Scholar]
- 50.Merz, K., M. Hondele, H. Goetze, K. Gmelch, U. Stoeckl, and J. Griesenbeck. 2008. Actively transcribed rRNA genes in S. cerevisiae are organized in a specialized chromatin associated with the high-mobility group protein Hmo1 and are largely devoid of histone molecules. Genes Dev. 22:1190-1204. [DOI] [PMC free article] [PubMed]
- 51.Navarro, M., and G. A. Cross. 1998. In situ analysis of a variant surface glycoprotein expression-site promoter region in Trypanosoma brucei. Mol. Biochem. Parasitol. 94:53-66. [DOI] [PubMed] [Google Scholar]
- 52.Navarro, M., G. A. Cross, and E. Wirtz. 1999. Trypanosoma brucei variant surface glycoprotein regulation involves coupled activation/inactivation and chromatin remodeling of expression sites. EMBO J. 18:2265-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Navarro, M., and K. Gull. 2001. A Pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature 414:759-763. [DOI] [PubMed] [Google Scholar]
- 54.Nogi, Y., R. Yano, and M. Nomura. 1991. Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc. Natl. Acad. Sci. U. S. A. 88:3962-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patrick, K. L., P. M. Luz, J. P. Ruan, H. Shi, E. Ullu, and C. Tschudi. 2008. Genomic rearrangements and transcriptional analysis of the spliced leader-associated retrotransposon in RNA interference-deficient Trypanosoma brucei. Mol. Microbiol. 67:435-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pays, E., H. Coquelet, P. Tebabi, A. Pays, D. Jefferies, M. Steinert, E. Koenig, R. O. Williams, and I. Roditi. 1990. Trypanosoma brucei: constitutive activity of the VSG and procyclin gene promoters. EMBO J. 9:3145-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pays, E., M. Lheureux, and M. Steinert. 1981. The expression-linked copy of a surface antigen gene in Trypanosoma is probably the one transcribed. Nature 292:265-267. [DOI] [PubMed] [Google Scholar]
- 58.Pays, E., L. Vanhamme, and D. Perez-Morga. 2004. Antigenic variation in Trypanosoma brucei: facts, challenges and mysteries. Curr. Opin. Microbiol. 7:369-374. [DOI] [PubMed] [Google Scholar]
- 59.Petesch, S. J., and J. T. Lis. 2008. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134:74-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Radman-Livaja, M., and O. J. Rando. 13 June 2009, posting date. Nucleosome positioning: how is it established, and why does it matter? Dev. Biol. doi: 10.1016/j.ydbio.2009.06.012. [DOI] [PMC free article] [PubMed]
- 61.Respuela, P., M. Ferella, A. Rada-Iglesias, and L. Aslund. 2008. Histone acetylation and methylation at sites initiating divergent polycistronic transcription in Trypanosoma cruzi. J. Biol. Chem. 283:15884-15892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roditi, I., A. Furger, S. Ruepp, N. Schurch, and P. Butikofer. 1998. Unravelling the procyclin coat of Trypanosoma brucei. Mol. Biochem. Parasitol. 91:117-130. [DOI] [PubMed] [Google Scholar]
- 63.Roditi, I., and M. Liniger. 2002. Dressed for success: the surface coats of insect-borne protozoan parasites. Trends Microbiol. 10:128-134. [DOI] [PubMed] [Google Scholar]
- 64.Rudenko, G., D. Bishop, K. Gottesdiener, and L. H. Van der Ploeg. 1989. Alpha-amanitin resistant transcription of protein coding genes in insect and bloodstream form Trypanosoma brucei. EMBO J. 8:4259-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rudenko, G., P. A. Blundell, M. C. Taylor, R. Kieft, and P. Borst. 1994. VSG gene expression site control in insect form Trypanosoma brucei. EMBO J. 13:5470-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rudenko, G., I. Chaves, A. Dirks-Mulder, and P. Borst. 1998. Selection for activation of a new variant surface glycoprotein gene expression site in Trypanosoma brucei can result in deletion of the old one. Mol. Biochem. Parasitol. 95:97-109. [DOI] [PubMed] [Google Scholar]
- 67.Rudenko, G., S. Le Blancq, J. Smith, M. G. Lee, A. Rattray, and L. H. Van der Ploeg. 1990. Procyclic acidic repetitive protein (PARP) genes located in an unusually small alpha-amanitin-resistant transcription unit: PARP promoter activity assayed by transient DNA transfection of Trypanosoma brucei. Mol. Cell. Biol. 10:3492-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saha, A., J. Wittmeyer, and B. R. Cairns. 2006. Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 7:437-447. [DOI] [PubMed] [Google Scholar]
- 69.Santoro, R., J. Li, and I. Grummt. 2002. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 32:393-396. [DOI] [PubMed] [Google Scholar]
- 70.Schimanski, B., T. N. Nguyen, and A. Gunzl. 2005. Characterization of a multisubunit transcription factor complex essential for spliced-leader RNA gene transcription in Trypanosoma brucei. Mol. Cell. Biol. 25:7303-7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schones, D. E., K. Cui, S. Cuddapah, T. Y. Roh, A. Barski, Z. Wang, G. Wei, and K. Zhao. 2008. Dynamic regulation of nucleosome positioning in the human genome. Cell 132:887-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schones, D. E., and K. Zhao. 2008. Genome-wide approaches to studying chromatin modifications. Nat. Rev. Genet. 9:179-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sheader, K., M. Berberof, T. Isobe, P. Borst, and G. Rudenko. 2003. Delineation of the regulated Variant Surface Glycoprotein gene expression site domain of Trypanosoma brucei. Mol. Biochem. Parasitol. 128:147-156. [DOI] [PubMed] [Google Scholar]
- 74.Shivaswamy, S., A. Bhinge, Y. Zhao, S. Jones, M. Hirst, and V. R. Iyer. 2008. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 6:e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Siegel, T. N., D. R. Hekstra, L. E. Kemp, L. M. Figueiredo, J. E. Lowell, D. Fenyo, X. Wang, S. Dewell, and G. A. Cross. 2009. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 23:1063-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reference deleted.
- 77.Strohner, R., A. Nemeth, K. P. Nightingale, I. Grummt, P. B. Becker, and G. Langst. 2004. Recruitment of the nucleolar remodeling complex NoRC establishes ribosomal DNA silencing in chromatin. Mol. Cell. Biol. 24:1791-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vanhamme, L., M. Berberof, D. Le Ray, and E. Pays. 1995. Stimuli of differentiation regulate RNA elongation in the transcription units for the major stage-specific antigens of Trypanosoma brucei. Nucleic Acids Res. 23:1862-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vanhamme, L., P. Poelvoorde, A. Pays, P. Tebabi, H. Van Xong, and E. Pays. 2000. Differential RNA elongation controls the variant surface glycoprotein gene expression sites of Trypanosoma brucei. Mol. Microbiol. 36:328-340. [DOI] [PubMed] [Google Scholar]
- 80.White, T. C., G. Rudenko, and P. Borst. 1986. Three small RNAs within the 10 kb trypanosome rRNA transcription unit are analogous to domain VII of other eukaryotic 28S rRNAs. Nucleic Acids Res. 14:9471-9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wickstead, B., K. Ersfeld, and K. Gull. 2004. The small chromosomes of Trypanosoma brucei involved in antigenic variation are constructed around repetitive palindromes. Genome Res. 14:1014-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Workman, J. L. 2006. Nucleosome displacement in transcription. Genes Dev. 20:2009-2017. [DOI] [PubMed] [Google Scholar]
- 83.Yuan, G. C., Y. J. Liu, M. F. Dion, M. D. Slack, L. F. Wu, S. J. Altschuler, and O. J. Rando. 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309:626-630. [DOI] [PubMed]
- 84.Zomerdijk, J. C., M. Ouellette, A. L. ten Asbroek, R. Kieft, A. M. Bommer, C. E. Clayton, and P. Borst. 1990. The promoter for a variant surface glycoprotein gene expression site in Trypanosoma brucei. EMBO J. 9:2791-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.