Abstract
Sexual reproduction in fungi is governed by a specialized genomic region, the mating type (MAT) locus, whose gene identity, organization, and complexity are diverse. We identified the MAT locus of five dermatophyte fungal pathogens (Microsporum gypseum, Microsporum canis, Trichophyton equinum, Trichophyton rubrum, and Trichophyton tonsurans) and a dimorphic fungus, Paracoccidioides brasiliensis, and performed phylogenetic analyses. The identified MAT locus idiomorphs of M. gypseum control cell type identity in mating assays, and recombinant progeny were produced. Virulence tests in Galleria mellonella larvae suggest the two mating types of M. gypseum may have equivalent virulence. Synteny analysis revealed common features of the MAT locus shared among these five dermatophytes: namely, a small size (∼3 kb) and a novel gene arrangement. The SLA2, COX13, and APN2 genes, which flank the MAT locus in other Ascomycota are instead linked on one side of the dermatophyte MAT locus. In addition, the transcriptional orientations of the APN2 and COX13 genes are reversed compared to the dimorphic fungi Histoplasma capsulatum, Coccidioides immitis, and Coccidioides posadasii. A putative transposable element, pogo, was found to have inserted in the MAT1-2 idiomorph of one P. brasiliensis strain but not others. In conclusion, the evolution of the MAT locus of the dermatophytes and dimorphic fungi from the last common ancestor has been punctuated by both gene acquisition and expansion, and asymmetric gene loss. These studies further support a foundation to develop molecular and genetic tools for dermatophyte and dimorphic human fungal pathogens.
In the fungal kingdom, sexual reproduction is pervasive and produces offspring in which the parental genetic material has been recombined. Meiotic recombination during sexual reproduction is considered a major driving force for diversification in natural populations of fungi (6, 24, 41). One advantage of sexual reproduction is the generation of genetic variation, which may produce individuals with altered virulence or increased fitness in new ecological niches (24, 41).
Many fungi are only known to undergo clonal, asexual reproduction. In some fungi, such as Aspergillus fumigatus (12), Coccidioides immitis (5), and Candida glabrata (10), although there was strong evidence for genetic recombination and gene flow based on population genetic studies, no known sexual cycle had been observed in nature or the laboratory. Recent genomic evidence disclosed cryptic mating potential in these fungi by identification of a MAT locus and machinery for sex in the genome (16, 38, 42, 58), and a complete sexual cycle for A. fumigatus was recently discovered and found to require 6 months of incubation in the dark on oatmeal agar (42). Compared to a sexual cycle, asexual reproduction is economical (7, 11, 49). Many fungi have the ability to reproduce both asexually or sexually (9). It has been hypothesized that fungi may preferentially clonally expand by asexual reproduction in stable environmental niches but also undergo genetic exchange via sexual reproduction in response to stressful conditions such as new environments and changes in the human host such as antimicrobial therapy (19, 41).
Mating in ascomycetous fungi is governed by a single mating type (MAT) locus with two idiomorphs of highly divergent sequences (6, 9). In the euascomycetes, each MAT idiomorph encodes an alpha domain or HMG domain transcription factor (6, 9, 52). Heterothallic MAT1-1 and MAT1-2 strains undergo sexual reproduction when cocultured under appropriate conditions. MAT1-1 and MAT1-2 fusions, or unlinked integrations on different chromosomes, enable homothallic, self-fertile sexual reproduction (13, 36, 46, 61).
Although alpha domain and HMG domain genes are essential elements of MAT, the MAT locus structure of each fungal species is unique in terms of locus number, gene number and arrangement, and transcriptional orientation (6, 9, 52). In addition, both homothallic and heterothallic mating systems coexist and are closely aligned throughout the Ascomycota, but heterothallism is more prevalent in some lineages. By comparative analyses, some studies have hypothesized that the ancestral mating system in the Ascomycota was heterothallic (16, 25, 46, 61), but other studies have derived the opposite conclusion, namely, that the ancestral mating system was homothallic (17, 59). This controversy may in part result from a limited number of fungal species with identified MAT loci and defined sexual cycles. In addition, as mentioned above, the MAT locus may be associated with virulence, ecology, and even pathogenicity (24, 41). Thus, identification of the MAT locus in pathogenic fungi and evolutionary studies are important, not only to understand their pathogenicity but also to elucidate both the ancestral and evolving organization of mating systems.
The dermatophytes are a group of closely related fungi that can invade skin, hair, and nails in humans and other animals and are the most common cause of fungal infections worldwide (56, 57). The dermatophytes are divided into three ecological groups depending on host preference and natural habitat: anthropophiles, zoophiles, and geophiles (18). Geophiles are primarily soil-dwelling organisms, but some taxa are pathogenic to humans. Zoophiles are essentially animal pathogens, although they may also cause human infections in some circumstances. Anthropophiles are largely restricted to humans, but some species can in some cases also infect other animals. Most dermatophytes are thought to be capable of reproducing sexually based on direct observation of mating structures or indirect results derived from population genetic studies. The sexual (teleomorphic or perfect) states of dermatophyte species align with the family Arthrodermataceae, the order Onygenales, and the class Eurotiomycetes (56). The anamorphic (asexual or imperfect) dermatophytes comprise three genera with diverse morphologies: Microsporum, Trichophyton, and Epidermophyton (56). According to genotypic and phylogenetic studies, dermatophytes can be considered a homogeneous group with recent evolutionary divergence, despite their diverse morphological and host characteristics (20, 21, 27). By direct mating assays and indirect population genetic studies, it has been suggested that geophilic dermatophytes typically have an extant sexual cycle, the zoophilic dermatophytes also frequently retain the ability to sexually reproduce, whereas anthropophilic dermatophytes are frequently infertile (57). Dermatophyte fungi therefore represent an ideal model to study how different environmental niches have influenced evolution of the MAT locus and sexual reproduction. The dermatophytes are phylogenetically closely related to dimorphic fungi such as Histoplasma capsulatum and Coccidioides spp. in which the MAT locus has been characterized, and Paracoccidioides brasiliensis in which mating and the MAT locus are unknown (16, 34, 38). In the present study, we identified and characterized the MAT locus in five dermatophytes: Microsporum gypseum (geophilic), Microsporum canis (zoophilic), Trichophyton equinum (zoophilic), Trichophyton rubrum (anthropophilic), and Trichophyton tonsurans (anthropophilic) and P. brasiliensis, by bioinformatics, direct sequencing, mating assays, and phylogeny.
MATERIALS AND METHODS
Strains and cultivation.
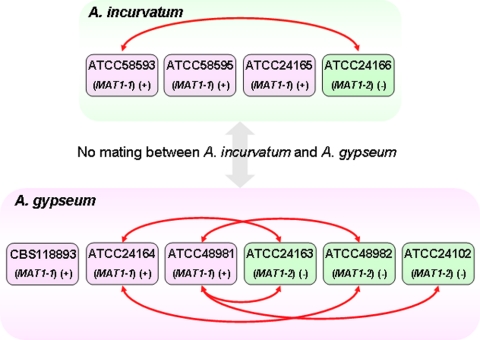
Three M. gypseum strains that were reported to mate and form the teleomorph Arthroderma gypseum (CBS118893, ATCC 24163, and ATCC 24164) and four M. gypseum strains reported to form the A. incurvatum teleomorph (ATCC 24165, ATCC 24166, ATCC 58593 and ATCC 58595) were purchased from the CBS (Centraalbureau voor Schimmelcultures) and the ATCC (American Type Culture Collection) (see Fig. 5). Strain ATCC 24102 was kindly provided by Wiley Schell and Jonathan Benton at Duke University Medical Center. All of the strains of M. gypseum were cultivated on Sabouraud dextrose agar (Difco) at room temperature.
FIG. 5.
M. gypseum mating types determined by MAT locus sequencing and mating assays. The MAT locus idiomorph in all strains was determined by PCR (Fig. 4). The MAT locus in strains CBS118893, ATCC 24163, ATCC 24164, ATCC 48981, ATCC 48982, ATCC 58593, and ATCC 58595 was confirmed by MAT locus sequencing (GenBank accession numbers FJ798794 to FJ798800). Pairs of strains connected by red lines with double arrows are fertile and produced sexual structures in mating assays (Fig. 6).
Identification of the MAT locus and flanking regions in the genomic sequences and gene prediction.
The genomic sequences of 1 M. gypseum, 1 M. canis, 1 T. equinum, 1 T. rubrum, 1 T. tonsurans, 4 H. capsulatum, 4 C. immitis, 10 C. posadasii, and 3 P. brasiliensis strains were downloaded from the Broad Institute (http://www.broad.mit.edu/science/data#), and a database containing these 26 genomic sequences was constructed with the Linux operating system (Table 1). The protein sequences of the alpha domain gene of H. capsulatum strain ATCC 22636 (GenBank accession no. EF472256) and the protein sequences of the HMG domain gene of H. capsulatum strain ATCC 22635 (GenBank accession no. EF472255) (16) were used as queries to search for orthologs in the 26 genomic sequences by tBlastn (matrix, Blosum62; E-value, 1e−3; gap cost, existence 11; extension 1; and low-complexity-region filter) (1). Once there was a significant hit, the corresponding genomic and flanking sequences were obtained for further analysis. Putative genes in the MAT locus and flanking regions were predicted by using a combination of primary annotation from the Broad Institute, GeneMark (37), BLAST (1), ORF Finder (http://www.ncbi.nlm.nih.gov/projects/gorf/), and FGENESH (http://linux1.softberry.com/berry.phtml), which predicts open reading frames (genes) using gene sequences of H. capsulatum and Coccidioides as models.
TABLE 1.
Fungal strains and genomes analyzed to identify and characterize the MAT locus
| Species | Strain |
MAT |
%GC of flanking region (∼10 kb) | Genome |
|||
|---|---|---|---|---|---|---|---|
| Locus | Size (bp)a | %GC | Size (Mb) | %GC | |||
| M. gypseum | CBS118893 | MAT1-1 | 2,941 | 45.77 | 47.38 | 23.25 | 48.48 |
| M. canis | CBS113480 | MAT1-1 | 3,383 | 43.54 | 47.85 | 23.24 | 47.52 |
| M. gypseum | ATCC48982 | MAT1-2 | 3,184 | 44.44 | 47.46 | NAb | NA |
| T. equinum | CBS127.97 | MAT1-2 | 3,161 | 44.86 | 48.29 | 24.13 | 47.39 |
| T. rubrum | CBS118892 | MAT1-1 | 3,005 | 45.26 | 47.34 | 22.51 | 48.3 |
| T. tonsurans | CBS112818 | MAT1-1 | 2,972 | 45.86 | 47.78 | 22.96 | 48.15 |
| H. capsulatum | H143 | MAT1-1 | 5,706 | 44.34 | 47.16 | 38.92 | 41.73 |
| H. capsulatum | NAm1 | MAT1-1 | 5,554* | 43.62 | 46.89 | 32.99 | 46.16 |
| H. capsulatum | H88 | MAT1-2 | 5,829 | 43.82 | 46.99 | 37.9 | 42.02 |
| H. capsulatum | G186AR | MAT1-2 | 5,371 | 44.37 | 46.37 | 30.44 | 44.57 |
| C. immitis | H538.4 | MAT1-1 | 8,167 | 44.1 | 49.04 | 27.73 | 46.96 |
| C. immitis | RMSCC 3703 | MAT1-1 | NA | 44.68 | 47.12 | 27.65 | 47.08 |
| C. posadasii | RMSCC 3488 | MAT1-1 | 8,149 | 44.15 | 47.56 | 28.15 | 46.33 |
| C. posadasii | RMSCC 2133 | MAT1-1 | 8,151 | 44.11 | 47.34 | 27.85 | 47.07 |
| C. posadasii | CPA 0001 | MAT1-1 | 8,176 | 44.04 | 48.12 | 28.62 | 46.09 |
| C. immitis | RS | MAT1-2 | 9,142 | 44.33 | 48.77 | 28.89 | 46.02 |
| C. immitis | RMSCC 2394 | MAT1-2 | 9,142 | 44.32 | 47.59 | 28.82 | 46.22 |
| C. posadasii | Silveira | MAT1-2 | 9,140* | 43.88 | 48.56 | 27.47 | 46.86 |
| C. posadasii | RMSCC 3700 | MAT1-2 | 9,250* | 44.26 | 47.34 | 25.45 | 48.11 |
| C. posadasii | CPA 0020 | MAT1-2 | 9,152* | 44.06 | 49.56 | 27.29 | 46.84 |
| C. posadasii | CPA 0066 | MAT1-2 | 9,128* | 44.17 | 47.87 | 27.71 | 46.32 |
| C. posadasii | RMSCC 1037 | MAT1-2 | 9,010 | 44.16 | 46.79 | 26.59 | 46.9 |
| C. posadasii | RMSCC 1038 | MAT1-2 | 9,136* | 44.13 | 47.63 | 26.12 | 47.19 |
| C. posadasii | C735 (TIGR) | MAT1-2 | 9,153 | 44.14 | 45.98 | 26.69 | 46.74 |
| P. brasiliensis | P01 | MAT1-1 | 6,851 | 42.55 | 46.82 | 32.94 | 42.82 |
| P. brasiliensis | P03 | MAT1-2 | 6,736 | 42.74 | 47.18 | 29.06 | 44.5 |
| P. brasiliensis | P18 | MAT1-2 | 9,558* | 43.12 | 47.01 | 29.95 | 44.36 |
*, a gap is present in the MAT locus, and the %GC was calculated without this gap.
NA, not available.
Isolation of genomic DNA: amplification and sequencing of the MAT locus and flanking genes.
After 2 weeks of growth on Sabouraud dextrose agar, M. gypseum strains were collected directly for genomic DNA isolation by using a MasterPure yeast DNA purification kit (Epicentre Biotechnologies) with minor modifications. Briefly, 500-μl glass beads (425 to 600 nm) were added into the mixture of strains and 300 μl of cell lysis solution to break the cells. Other steps followed the protocol provided by the manufacturer.
Four sets of primers were chosen for flanking genomic regions of the APN2, SLA2, and COX13 genes in the genome sequence of M. gypseum strain CBS118893. The primer sequences are listed in Table 2. PCR assays were conducted in a PTC-200 automated thermal cycler (MJ Research, Waltham, MA). A total of 300 ng of each DNA preparation was amplified in a 20-μl reaction mixture containing 10 pM concentrations of each primer, 2 mM concentrations of each nucleotide (dATP, dCTP, dGTP, and dTTP), 4 μl of 5× Iproof HF buffer, 0.2 μl of iProof High-Fidelity Taq polymerase (Bio-Rad), and 12.4 μl of distilled water. The following conditions were used for the amplification: an initial 30 s of denaturation at 98°C, followed by 35 cycles of denaturation for 10 s at 98°C, an annealing time of 30 s at 54°C, and an extension cycle for 5 min at 72°C. The amplification was completed by an extension period of 5 min at 72°C. Sterile water was used as a negative control in each assay. PCR products were analyzed on 1.5% agarose gels. Amplicons were purified by using a QIAquick PCR purification kit (Qiagen) as recommended by the manufacturer. PCR products were then sequenced by direct sequencing and/or primer walking in both directions using a BigDye terminator (version 3.1) cycle sequencing ready reaction mix (Applied Biosystems, Foster City, CA). Sequencing products were resolved by using an ABI 3130 automated sequencer (Applied Biosystems). Sequences were assembled by using Sequencher 4.8 (Gene Codes). The sequences of the MAT1-1 and MAT1-2 idiomorphs of M. gypseum were deposited in GenBank under accession numbers FJ798794 to FJ798800 for strains CBS118893, ATCC 24163, ATCC 24164, ATCC 48981, ATCC 48982, ATCC 58593, and ATCC 58595, respectively.
TABLE 2.
Primer pairs used for amplification of MAT and flanking regions and mating type determination of M. gypseum
| Primer 1 | Primer 1 sequence (5′-3′) | Primer 2 | Primer 2 sequence (5′-3′) | Amplicon size (bp)a | Gene amplified | Aim |
|---|---|---|---|---|---|---|
| JOHE20209 | CGAGAGCAACGACGTTCATA | JOHE20213 | AACAACCGTTGGACAGAAGG | 11,048 | Unknown gene, APN2, COX13, and SLA2 | Amplification of MAT locus and flanking region |
| JOHE20210 | GATTGCCCGTTCTCTCTCAG | JOHE20211 | GCATAACTGGCACCCTTTGT | 5,910 | Alpha box and APN2 | Amplification of MAT locus and flanking region |
| JOHE20463 | CACTGGCGAAAGTTATGCAA | JOHE20464 | GCCAGCCAACCTTTCACTTA | 5,549 | COX13, and SLA2 | Amplification of MAT locus and flanking region |
| JOHE20465 | CACTCGGGCTTCGAGTAAAC | JOHE20466 | TCTGGTCTGGCATGAAAGTG | 5,658 | APN2, COX13, and SLA2 | Amplification of MAT locus and flanking region |
| JOHE20887 | CCTCTTCTACTGCCATGACA | JOHE20888 | CCATGGGATTGATGTGTGCA | 452 | MAT1-1 alpha domain | Mating type assignment |
| JOHE20893 | GGATGAGTCCTGATATGTCA | JOHE20894 | CCTATGGGTTTAGCTTCTGA | 502 | MAT1-2 HMG domain | Mating type assignment |
| JOHE21325 | GCTATTGGTCGAGATAACGTCTTT | JOHE21326 | TTAATCAATAACTGACCTGCTCCA | 624 | Polysaccharide export protein | Recombination test |
| JOHE21327 | TGCTGTCCGTAAGTGTAGGTATGT | JOHE21328 | CCAGCCTTGCTCTCAAGTAGTTAT | 889 | DNJ-oxidoreductase intergenic spacer | Recombination test |
Amplicon sizes were deduced from the genome sequence of strain CBS118893 (Broad Institute) and the MAT sequence of strain ATCC 48982 (this study).
Sequence and structure comparison of MAT.
Syntenic comparison of the opposite MAT locus and flanking regions within a species was performed by using CLUSTAL W (51), which was also used to identity the highly diverse DNA regions as MAT idiomorphs. Because CLUSTAL W is used for pairwise alignment of highly similar sequences, we used Blastn to identify the MAT loci of M. canis, T. rubrum, T. tonsurans, and T. equinum, which are different species from M. gypseum and have only one genome sequence available, and to search for the homologs of the M. gypseum MAT locus in the genome. A Dotpath program in the EMBOSS package (45) was used to generate the dot plot comparisons of the MAT1-1 and MAT1-2 of M. gypseum. Artemis was used for synteny analysis of the MAT locus among the five dermatophyte species (8).
Mating type determination for M. gypseum by PCR.
The available MAT1-1 and MAT1-2 sequences were aligned by using CLUSTAL W to choose primers for mating type determination for M. gypseum by PCR. In addition, the selected primers were further used as query sequences in BLAST searches of the M. gypseum genome to avoid potential nonspecific amplification. The primers used for mating type determination PCR are listed in Table 2. PCR assays were conducted in a PTC-200 automated thermal cycler (MJ Research). A total of 300 ng of each DNA preparation was amplified in a 25-μl reaction mixture containing a 10 pM concentration of each primer, a 2 mM concentration of each nucleotide (dATP, dCTP, dGTP, and dTTP), 2.5 μl of 10× Ex Taq buffer, 0.125 μl of Ex Taq polymerase (Takara), and an appropriate volume of distilled water. The following conditions were used for the amplification: an initial 3 min of denaturation at 95°C, followed by 35 cycles of denaturation for 30 s at 94°C, an annealing time of 20 s at 54°C, and an extension cycle for 1 min at 72°C. The amplification was completed by an extension period of 5 min at 72°C. Sterile water served as a negative control in each assay. PCR products were analyzed on 1.5% agarose gels.
Mating assays and observation of sexual structures by light microscopy and scanning electron microscopy.
Medium E (12 g of oatmeal agar [Difco], 1 g of MgSO4·7H2O, 1 g of KH2PO4, 1 g of NaNO3, and 16 g of agar/liter) was used for mating assays as described previously (55). Small cubes of a growth patch were cut from cultures that had been grown on Sabouraud dextrose agar for 1 week and then placed in the center of a mating plate side by side. The plates were incubated at room temperature in the dark for 1 month without parafilm and then examined for cleistothecium production.
To observe asci and ascospore formation, a cleistothecium was placed on the microscope slide, mounted with a coverslip, and examined with an Axioskop 2 Plus upright microscope (Zeiss). Images were captured by using an AxioCam MRm camera and AxioVision application software (Zeiss). Cleistothecia and ascospores were also examined by scanning digital electronic microscopy. The cleistothecium was broken and rolled on a 3% agarose plate using a fine needle to expose the ascospores. The agar cubes containing cleistothecia and ascosporea were fixed in 3% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 6.8) for several days at 4°C. They were then rinsed in three 30-min changes of cold 0.1 M sodium cacodylate buffer (pH 6.8), followed by a graded dehydration series of 30-min changes in cold 30 and 50% ethanol (EtOH) and then held overnight in 70% EtOH. Dehydration was completed with 1-h changes of cold 95% and 100% EtOH at 4°C with warming to room temperature in the 100% EtOH. Two additional 1-h changes of room temperature 100% EtOH completed the dehydration series. The samples were then critical point dried in liquid CO2 (Samdri-795; Tousimis Research Corp., Rockville, MD) for 15 min at critical point. The samples were mounted on stubs with either double-stick tape or silver paint to ensure good conductivity, followed by sputter coating the samples with 50 Å of Au/Pd (Hummer 6.2; Anatech U.S.A., Hayward, CA). Samples were held in the vacuum desiccator until viewed at a JEOL JSM 5900LV SEM at 10 kV.
Evaluation of genetic recombination in progeny of M. gypseum.
Genetic recombination in progeny of strains ATCC 24164 and ATCC 48982 was evaluated by a combination of genotype of two unlinked genetic markers and the mating type. A cleistothecium was taken from the mating plate containing M. gypseum strains ATCC 24164 and ATCC 48982 and cleaned by rolling and tearing on a 3% agarose plate. The cleaned and disrupted cleistothecium was dispersed in 1 ml of double-distilled H2O, and spores were plated at a limiting dilution on Sabouraud dextrose agar. After 1 week's growth, 10 single isolated clones were colony purified and subcultured on Sabouraud dextrose agar for 10 days before genomic DNA isolation.
Two unlinked genetic markers, one encoding a polysaccharide export protein (CAP59) and one intergenic spacer flanked by the DNJ gene and a gene encoding an oxidoreductase, were selected from the genomic sequences of M. gypseum CBS118893. These two genetic markers were amplified and sequenced from parental strains and the 10 F1 progeny to designate sequence types. Primers used for PCR amplification and sequencing are listed in Table 2. PCR assays were carried out in a PTC-200 automated thermal cycler (MJ Research). A total of 300 ng of each DNA preparation was amplified in a 25-μl reaction mixture containing a 10 pM concentration of each primer, a 2 mM concentration of each nucleotide (dATP, dCTP, dGTP, and dTTP), 2.5 μl of 10× Ex Taq buffer, 0.125 μl of Ex Taq polymerase (Takara), and an appropriate volume of distilled water. The following conditions were used for the amplification: an initial 3 min of denaturation at 95°C, followed by 35 cycles of denaturation for 30 s at 94°C, an annealing time of 20 s at 54°C, and an extension cycle for 1 min at 72°C. The amplification was completed by an extension period of 5 min at 72°C. Sterile water was used as a negative control in each assay. PCR products were analyzed on 1.5% agarose gels. Amplicon sequencing was performed with the same primers for PCR as described above for sequencing the MAT locus. The genotype of each genetic marker was designated as a unique DNA sequence. We also incorporated the mating type (MAT1-1/MAT1-2) that was determined by PCR for genetic recombination. The progeny genotype and mating type were compared to the parental strains for genetic recombination analysis.
Virulence tests of the two mating types of M. gypseum in Galleria mellonella.
G. mellonella has been increasingly used as a model to test virulence of several pathogenic fungi such as Cryptococcus (40), Aspergillus (26), and Candida (3) but had not been applied for the virulence assays of dermatophytes. We tested M. gypseum virulence in the heterologous host G. mellonella (greater wax moth). G. mellonella larvae were infected with M. gypseum by placing them on M. gypseum culture plates and allowing surface exposure and infection (see Fig. 6A). All G. mellonella larvae were purchased in bulk and a single shipment used for each replicate virulence test (Van der Horst Wholesale, St. Marys, OH). Twelve G. mellonella larvae were placed on each Sabouraud dextrose agar plate where an M. gypseum strain had been growing for 10 days. After this, the plate was covered and stored at room temperature for 12 days. The insects that turned dark in pigmentation and were nonresponsive to touch were scored as dead and were then counted and removed from the plate each day. All 10 M. gypseum strains included in the present study were tested for virulence in the wax moth. Prism software was used to plot the survival curves and for statistical analysis.
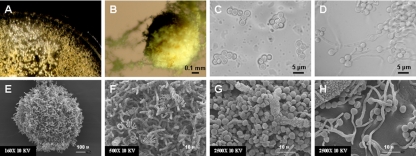
FIG. 6.
Sexual reproduction of M. gypseum involves MAT1-1 and MAT1-2 heterothallic isolates. After incubation at room temperature in the dark for 1 month, cleistothecium formation and ascospore production were observed on Medium E plates when MAT1-1 and MAT1-2 M. gypseum strains were coincubated. (A) The cleistothecium is a small white structure formed on mating plates and visible with the naked eye. (B) Under light microscopy, the mature cleistothecium is yellowish. (E and F) The cleistothecium monitored with scanning electron microscopy (SEM) was enveloped by mature peridial hyphae, many of which are coiled and spiral. The ascospores are round cells with a diameter of ∼3 μm (as observed by light microscopy [C] and SEM [G]). (C) Asci containing eight ascospores were visible by light microscopy. Ascospores germinated to produce hyphae when spread on Sabouraud dextrose agar and were incubated for 5 days at room temperature (as observed by light microscopy [D] and SEM [H]).
Phylogenetic analysis.
The DNA sequences of the alpha domain and HMG domain genes in MAT1-1 and MAT1-2, respectively, were extracted from the 24 genomic sequences. FGENESH was used to generate the predicted amino acid sequences encoded by the alpha domain and HMG domain genes. The amino acid sequences of the alpha domain and HMG domain of other related fungi were downloaded from GenBank. The alpha domain and HMG domain protein were aligned with CLUSTAL W (51) and subsequently imported into Mega 3.1 (28) for phylogenetic analysis using the neighbor-joining and maximum-parsimony methods.
RESULTS
Identification of the MAT idiomorphs, MAT1-1 and MAT1-2, of M. gypseum.
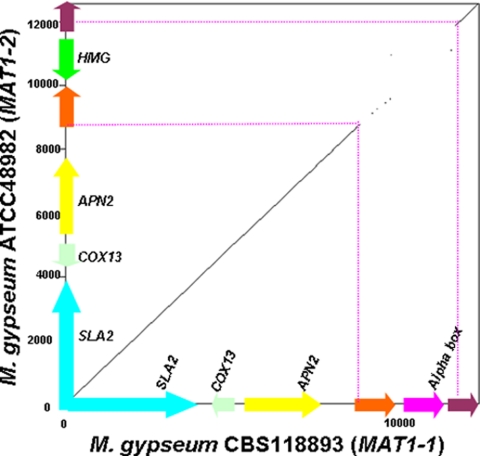
A homolog of the MAT1-1 alpha domain gene of H. capsulatum ATCC 22636 was identified in the genomic sequence of M. gypseum CBS118893 by BLAST search. The deduced amino acid sequences exhibit 45.3% identity, 18.6% strong similarity, and 11.6% weak similarity to the H. capsulatum ortholog (see Fig. S1 in the supplemental material). An alpha domain gene and flanking sequences were obtained by direct sequencing in M. gypseum strains CBS118893, ATCC 24164, ATCC 48981, ATCC 58593, and ATCC 58595, respectively (see Fig. S1 in the supplemental material). The sequence obtained from strain CBS118893 is identical to the corresponding sequence from the M. gypseum CBS118893 genome sequence obtained by the Broad Institute. The opposite MAT idiomorph expressing a conserved HMG domain gene and flanking sequences was identified from strains ATCC 24163 and ATCC 48982 by direct sequencing and primer walking, respectively. The deduced amino acid sequences exhibit 44.6% identity, 17.5% strong similarity, and 11.0% weak similarity with the HMG domain of H. capsulatum ATCC 22635 (see Fig. S1 in the supplemental material). By sequence alignment, the MAT idiomorphs of M. gypseum were identified as a 2,941-bp MAT1-1 idiomorph containing an alpha domain gene and a 3,184-bp MAT1-2 idiomorph containing an HMG domain gene (Fig. 1 and 2; Table 1). Compared to the high sequence identity (99.4%) of the flanking genes (SLA2, COX13, ANP2, and two predicted genes) in a total of 9,423 bp from strains CBS118893 and ATCC 48982, the sequence identity of the MAT1-1 and MAT1-2 idiomorphs was significantly lower (42.8%). Similar to other fungi in the Ascomycota, the M. gypseum MAT locus is closely linked to the SLA2, COX13, and APN2 genes.
FIG. 1.
Synteny of MAT1-1 and MAT1-2 mating type locus idiomorphs of M. gypseum. Pairwise comparison of MAT1-1 of M. gypseum CBS118893 and MAT1-2 of M. gypseum ATCC 48982 was performed by the Dotpath program in the EMBOSS package. The highly diverse MAT locus flanked by highly similar sequences comprises an alpha domain/HMG gene and genes encoding two hypothetical proteins. The corresponding MAT loci in both strains are highlighted with broken red lines.
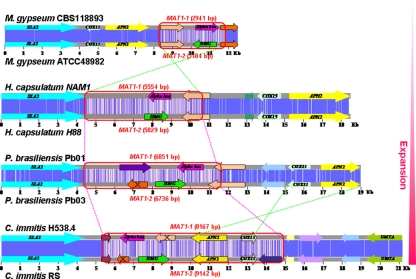
FIG. 2.
Comparing the MAT locus of dermatophyte and dimorphic fungi. A DNA sequence alignment of the opposite MAT loci and flanking genes of M. gypseum, H. capsulatum, C. immitis, C. posadasii, and P. brasiliensis was prepared by using CLUSTAL W. MAT locus genes are represented by arrows outlined with black lines. Homologous genes are represented by the same color. The DNA sequence similarity between two opposite MAT loci is represented in blue, and darker colors indicate greater sequence similarity. The main events during the MAT locus evolution of these fungal species are indicated by broken red lines (expansion) and crossed broken green lines (inversion). Genes present in only one idiomorph are marked with black crosses.
Strains ATCC 24163 and ATCC 48982 are designated as minus (−) mating type due to the presence of the MAT1-2 locus. Strains CBS118893, ATCC 24164, ATCC 48981, ATCC 58593, and ATCC 58595 are designated as plus (+) mating type due to the presence of the MAT1-1 locus in the genome. We note that our molecular designation of mating types does not in all cases match previous designations of (+) and (−) mating type based on mating assays (23, 43, 54) (http://www.atcc.org/) (see Table S1 in the supplemental material). Historically, the two teleomorphic forms were randomly assigned (+) and (−) and were assigned opposite to each other. However, this discord in the designation of mating types cannot be completely explained by this inversion of previous designations. Thus, previous assignments of mating type were in some cases incorrect, and correlating both the molecular and the genetic mating type is essential. CBS118893 is used here as the (+) reference standard.
MAT locus of M. canis, T. rubrum, T. tonsurans, and T. equinum.
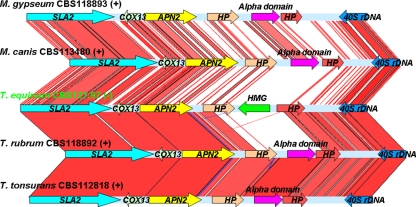
Homologs of the MAT1-1 gene of H. capsulatum ATCC 22636 were identified in the genomic sequences of M. canis CBS113480, T. rubrum CBS118892, and T. tonsurans CBS112818, respectively, and a homolog of the MAT1-2 gene of H. capsulatum ATCC 22635 was identified in the genomic sequence of T. equinum CBS127.97. The deduced amino acid sequences of the alpha domain of M. canis CBS113480, T. rubrum CBS118892, T. tonsurans CBS112818, and H. capsulatum ATCC 22636 exhibit 41% identity, 14.2% strong similarity, and 11.7% weak similarity (see Fig. S1 in the supplemental material). The deduced amino acid sequences of the HMG domain of T. equinum exhibits 42.4% identity, 19.4% strong similarity, and 10.6% weak similarity with the HMG domain of H. capsulatum ATCC 22635 (see Fig. S1 in the supplemental material). Next, the alpha domain, HMG domain, and flanking genes (SLA2, COX13, ANP2, and two predicted genes) were extracted from the four genomic sequences. By comparison with the M. gypseum MAT locus using Blastn, an ∼3-kb MAT1-1 or MAT1-2 idiomorph was determined in the genomic sequences of M. canis (MAT1-1), T. rubrum (MAT1-1), T. tonsurans (MAT1-1), and T. equinum (MAT1-2), and they exhibit a highly similar organization (Fig. 3).
FIG. 3.
Genomic synteny of dermatophyte MAT locus. The sequence and structural comparison of the MAT loci of the five dermatophyte species M. gypseum (MAT1-1), M. canis (MAT1-1), T. equinum (MAT1-2), T. rubrum (MAT1-1), and T. tonsurans (MAT1-1) was performed using Artemis. The genomic regions linked with red and blue lines are highly conserved DNA sequences with forward and reverse orientations, respectively. This reveals a similar MAT locus structure among the five dermatophyte species that exhibit identical gene orders and transcriptional orientations of the SLA2, COX13, APN2, hypothetical proteins (HP), and alpha domain/HMG genes.
Determination of M. gypseum mating type by PCR.
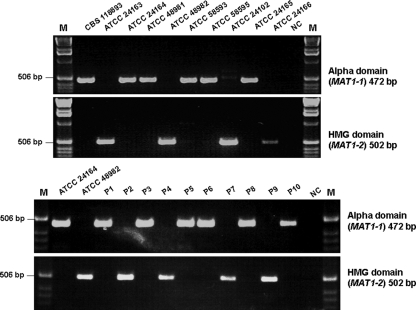
We next developed a simple PCR based assay to rapidly assign mating type molecularly for M. gypseum. A single product was generated by PCR from each M. gypseum strain using primer pairs complementary to the alpha domain or HMG domain genes, respectively (Fig. 4). Amplicon sequencing confirmed these products result from specific amplification of portions of the alpha domain gene of MAT1-1 (472 bp) and the HMG domain gene of MAT1-2 (502 bp) of M. gypseum. Diagnostic PCR demonstrated that strains CBS118893, ATCC 24164, ATCC 48981, ATCC 58593, ATCC 58595, and ATCC 24165 contain the MAT1-1 idiomorph (alpha domain gene), and strains ATCC 24163, ATCC 48982, ATCC 24102, and ATCC 24166 contain the MAT1-2 idiomorph. This PCR-based mating type determination is in accord with MAT locus sequencing, genome analysis, and mating assays (Table 1 and Fig. 5).
FIG. 4.
PCR-based determination of MAT locus idiomorphs of M. gypseum. A 472-bp portion of the alpha domain MAT1-1 gene was obtained from strains CBS118893, ATCC 24164, ATCC 48981, ATCC 58593, ATCC 58595, and ATCC 24165 and progeny P1, P3, P5, P6, P8, and P10 of strains ATCC 24164 and ATCC 48982. A 502-bp portion of the HMG domain MAT1-2 gene was obtained from strains ATCC 24163, ATCC 48982, ATCC 24102, ATCC 24166 and the progeny P2, P4, P7, and P9 of strains ATCC 24164 and ATCC 48982. NC, negative control; M, DNA ladder marker.
Mating ability of M. gypseum.
M. gypseum has a defined sexual cycle (50, 54), and mating assays were conducted to examine the control of mating type identity by the MAT locus. Cleistothecium formation and production of asci and ascospores (Fig. 6) were observed in mating assays on Medium E for several strain pairs (Fig. 5) with opposite MAT locus configurations as identified by genome analysis (Table 1), MAT locus sequencing, and PCR (Fig. 4). The cleistothecia appear as small white dots on the mating plate when observed by the naked eye (Fig. 6A). By light microscopy, the mature cleistothecium has a yellow hue (Fig. 6B). As observed by scanning electron microscopy, the cleistothecium was enveloped by mature peridial hyphae, many of which are coiled and spiral (Fig. 6E and F). Ascospores released from the cleistothecium were round with a diameter of ∼3 μm (Fig. 6C and G). Asci containing eight ascospores were observed by light microscopy (Fig. 6C). Ascospores germinated to produce hyphae when spread on Sabouraud dextrose agar and incubated for 5 days at room temperature (Fig. 6D and H). No sexual structures were produced by any isolate when cultured alone. Although these sexual structures were not produced by every cocultured potentially compatible strain pair combination, the positive mating results are in accord with the MAT locus idiomorphs identified by PCR, sequencing, and genome analysis and productive mating always involved coincubation of MAT1-1 and MAT1-2 strains (Fig. 5). No sexual structures were observed for any mating with strains CBS118893, ATCC 24165, and ATCC 58595, even after incubation of the mating assay plates at room temperature for up to 2 months, indicating that these isolates may be sterile or require different conditions. Our observations of cleistothecium formation, production of asci and ascospores, and ascospore germination support the conclusion that the identified MAT locus idiomorphs control cell type identity of M. gypseum.
Genetic recombination in the progeny of M. gypseum.
Based on PCR and sequence analysis, the two parental strains, ATCC 24164 and ATCC 48982, have different genotypes at CAP59 and the DNJ-oxidoreductase intergenic spacer (Table 3). Three SNPs were found between these two CAP59 genotypes, and one SNP and an 8-bp indel were found between the two genotypes of the DNJ-oxidoreductase intergenic spacer (see Fig. S2 in the supplemental material). Ten F1 progeny of a cross between strains ATCC 24164 and ATCC 48982 exhibited one or the other of these genotypes at CAP59 and the DNJ-oxidoreductase intergenic spacer (Table 3). The PCR-based assay was then used to assign mating type for these F1 progeny. Among the 10 progeny, six (P1, P3, P5, P6, P8, and P10) are the “(+)” mating type due to amplification of the alpha domain gene, and four (P2, P4, P7, and P9) are the “(−)” mating type due to amplification of the HMG domain gene (Fig. 4). Based on the combinations of genotypes at these two genetic markers and mating type, 8 of 10 progeny exhibit genetic recombination compared to the parental isolates (Table 3). No genetic recombination was observed at these loci in progeny P2 and P4, which may be attributable to the limited number of genetic markers analyzed. The evidence of genetic recombination in F1 progeny further confirms that mating occurs between M. gypseum strains with opposite mating types and that the identified MAT locus idiomorphs control cell type identity.
TABLE 3.
Mating of M. gypseum produces recombinant F1 progenya
| Strain | CAP59 genotype | DNJ-oxidoreductase intergenic spacer genotype | Mating typeb | Recombination |
|---|---|---|---|---|
| Parental strain | ||||
| ATCC 24164 | 1 | 1 | MAT1-1 | |
| ATCC 48982 | 2 | 2 | MAT1-2 | |
| F1 progeny | ||||
| P1 | 1 | 2 | MAT1-1 | Yes |
| P2 | 2 | 2 | MAT1-2 | No (ATCC 48982) |
| P3 | 2 | 2 | MAT1-1 | Yes |
| P4 | 2 | 2 | MAT1-2 | No (ATCC 48982) |
| P5 | 1 | 2 | MAT1-1 | Yes |
| P6 | 1 | 2 | MAT1-1 | Yes |
| P7 | 1 | 1 | MAT1-2 | Yes |
| P8 | 2 | 1 | MAT1-1 | Yes |
| P9 | 1 | 2 | MAT1-2 | Yes |
| P10 | 2 | 2 | MAT1-1 | Yes |
Genetic recombination of the F1 progeny of ATCC 24164 × ATCC 48982 was determined by sequencing two genetic markers, CAP59 and the DNJ-oxidoreductase intergenic spacer and by mating type PCR tests.
The mating types were identified by PCR (see Fig. 5).
M. gypseum is virulent in G. mellonella.
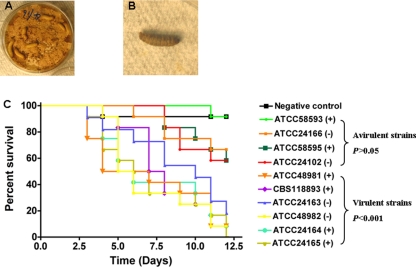
The virulences of M. gypseum strains of the (+) and (−) mating types were evaluated in the heterologous host, G. mellonella (greater wax moth) (Fig. 7A). Compared to noninfected G. mellonella larvae on Sabouraud dextrose agar (negative control), the exposed insects initially became dark several days postinfection but were still alive (Fig. 7B and see Video S1 in the supplemental material). At 2 to 4 h after the larvae turned dark, they became nonresponsive to touch and were therefore inviable (Fig. 7B). Based on survival curves and statistical analysis, four M. gypseum strains ATCC 24102 (−), ATCC 24166 (−), ATCC 58593 (+) and ATCC 58595 (+) were not virulent (P > 0.05) (Fig. 7C). Four other mating type (+) strains (CBS118893, ATCC 48981, ATCC 24164, and ATCC 24165) and two mating type (−) strains (ATCC 24163 and ATCC 48982) showed significant virulence (P < 0.001) (Fig. 7C). Among the eight known clinical strains, five (ATCC 24163, ATCC 24164, ATCC 24165, ATCC 48981, and ATCC 48982) are virulent, but two others (ATCC 24102 and ATCC 24166) exhibited low virulence in G. mellonella (Fig. 7C and see Table S1 in the supplemental material). No significant virulence difference was found between the mating type (+) and mating type (−) strains in the virulent group (P > 0.05). Our results suggest the two mating types strains of M. gypseum may have equivalent virulence, in accord with previous reports that the two mating types are recovered at similar frequencies from infected patients (23, 54).
FIG. 7.
Virulence tests of M. gypseum in G. mellonella. (A) Twelve wax moth larvae were placed on Sabouraud dextrose agar plates on which M. gypseum strains were cultivated and surface infected by contact with the fungus. A Sabouraud dextrose agar plate without any M. gypseum served as the negative control. (B) G. mellonella larvae that turned dark and were nonresponsive to touch were scored as dead and removed. (C) The curves show survival of larvae during a 12-day period. By statistical analysis, M. gypseum strains ATCC 24102 (−), ATCC 24166 (−), ATCC 58593 (+), and ATCC 58595 (+) are not significantly virulent (P > 0.05). The remaining six strains CBS118893 (+), ATCC 48981 (+), ATCC 24164 (+), ATCC 24165 (+), ATCC 24163 (−), and ATCC 48982 (−) showed significant virulence (P < 0.001). No significant virulence difference was found between the mating type (+) and mating type (−) strains in the virulent group (P > 0.05). Our results suggest the two mating type strains of M. gypseum may have equivalent virulence in G. mellonella.
Comparing the MAT locus of dermatophytes with closely related dimorphic fungi.
H. capsulatum, C. immitis, and C. posadasii are heterothallic dimorphic fungal pathogens whose MAT locus has previously been characterized (4, 16, 38). Here, we further identified MAT1-1 or MAT1-2 idiomorphs in the genomic sequences of 4 H. capsulatum, 4 C. immitis, and 10 C. posadasii strains by Blastn searches (Table 1). They share highly similar gene organization and sequence identity (Table 1).
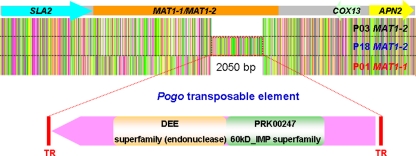
We also identified two MAT idiomorphs, MAT1-1 containing an alpha domain gene and MAT1-2 containing an HMG domain gene, in the genomic sequences of three strains of the related dimorphic fungal pathogen P. brasiliensis (Table 1 and Fig. 2) (51a). The MAT1-1 idiomorph of P. brasiliensis strain P01 is 6,851 bp and the MAT1-2 idiomorph in P. brasiliensis strain P03 is 6,736 bp (Fig. 2). Strain P18 also has a MAT1-2 idiomorph containing an HMG domain gene, but a 2,850-bp insertion is located between the HMG domain gene and the 3′ predicted gene (Fig. 8). The deduced amino acid sequence of this insertion exhibits 50% sequence identity with the Pogo transposable element (TE; GenBank accession no. EED21380) of Talaromyces stipitatus, an ascomycete fungus, and 34% sequence identity with the Pogo TE previously identified from A. fumigatus (22). This insertion shares significant identity with 18, 12, and 17 sequences in the genomes of strains P01, P03, and P18, respectively (data not shown). The assignment of this insertion as a Pogo-related TE is supported by its presence in multiple copies in the genome, the high similarity shared with known Pogo TEs, flanking terminal repeats (TRs), and an open reading frame encoding a DEE superfamily endonuclease domain (Fig. 8). The presence of a TE in one but not both MAT alleles is in accord with the known features of this genomic region involving a sheltering of recombination and accumulation of repetitive elements.
FIG. 8.
Identification of the P. brasiliensis MAT locus. The DNA sequence alignment of the MAT loci of P. brasiliensis strains P01 (MAT1-1), P03 (MAT1-2), and P18 (MAT1-2) was determined by using CLUSTAL W. A putative Pogo transposable element (2,850 bp) flanked by two terminal repeats (TRs) was found in the MAT1-2 locus of P. brasiliensis isolate P18. Two superfamily domains, DEE superfamily endonuclease and the PRK00247 60kD_IMP superfamily, were identified in this Pogo TE.
H. capsulatum, C. immitis, C. posadasii, and P. brasiliensis share a similar MAT locus organization with the five dermatophytes M. gypseum, M. canis, T. equinum, T. rubrum, and T. tonsurans (Fig. 2). All of the MAT loci described here comprise a putative alpha domain or HMG domain gene, which are tightly linked with the SLA2, APN2, and COX13 genes (Fig. 2). However, the arrangement of these genes differed significantly. In the dermatophytes, H. capsulatum, and P. brasiliensis, the orientation of the SLA2, APN2, and COX13 genes are the same with respect to each other. The HMG and alpha domain genes are in opposite orientation in all species, and both of their orientations are reversed in the dermatophytes compared to the other species. The directions of APN2 and COX13 in C. immitis and C. posadasii are reversed compared to the dermatophytes, H. capsulatum, and P. brasiliensis (Fig. 2).
Each defined MAT locus idiomorph also contains one or more predicted genes; however, these genes share very low sequence similarity and have different orientations (Fig. 2). Some of these predicted genes are completely contained within the MAT locus, whereas others are only partially included and span the junction of the MAT locus with the flanking genomic DNA (Fig. 2). In addition, the size of MAT1-1 and MAT1-2 in C. immitis and C. posadasii is larger than those in the dermatophytes, H. capsulatum, and P. brasiliensis due to incorporation of the APN2 and COX13 genes into the MAT locus (Fig. 2) (16, 38).
We also compared the GC content of the MAT locus with that of the flanking sequences (10 kb) and the GC content of the genome. Our results revealed a similar GC content between MAT1-1 and MAT1-2 regardless of variation in genomic GC content in different species and strains (Table 1).
Phylogenetic relationships of dermatophyte and dimorphic fungi based on alpha domain and HMG domain genes.
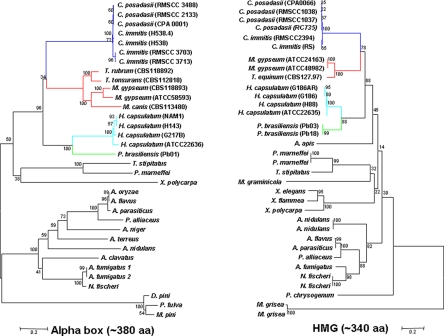
Based on the amino acid sequences of the alpha domain and HMG domain genes, dendrograms of dermatophytes, dimorphic fungi, and other fungi in the order Onygenales, as deduced by neighbor-joining and maximum-parsimony methods, exhibited a similar phylogenetic organization (Fig. 9). The dermatophytes are closely related to dimorphic fungi phylogenetically. P. brasiliensis has a closer relationship to H. capsulatum than to C. immitis and C. posadasii compared to previous studies based on 18S rDNA and 25S rDNA sequences (2, 34). The rDNA sequence deduced phylogenetic organization had previously suggested that the dimorphic fungal species were significantly separated from the dermatophytes (2, 34). On the basis of the amino acid sequences of the alpha domain and HMG domain genes elucidated in the present study, however, the dermatophytes group together with the dimorphic fungi and share a closer relationship with C. immitis and C. posadasii (Fig. 9).
FIG. 9.
Dermatophyte and dimorphic fungal pathogens are phylogenetically related based on alpha domain and HMG genes. The phylogenetic organization of M. gypseum, M. canis, T. equinum, T. tonsurans, T. rubrum, H. capsulatum, C. immitis, C. posadasii, P. brasiliensis, and closely related species in the Ascomycota based on the amino acid sequences encoded by the alpha domain gene (left) and HMG domain gene (right) was generated by using Mega 4.1 software. The phylogenetic classification of these fungal species based on the protein sequences of alpha domain and HMG domain genes is highly similar but differs from those based on 18S and 25S rDNA sequences in which the dermatophytes were clearly separated from the dimorphic fungi (2, 34).
DISCUSSION
In this study we identified and characterized the MAT locus that orchestrates the sexual cycle of M. gypseum. We established genetic crosses and characterized mating both morphologically and molecularly and also demonstrated that genetic recombination had occurred in the F1 progeny. By identification of the MAT locus in other dermatophytes and dimorphic fungi and by comparative analysis, we further present an evolutionary trajectory of the MAT locus in these closely related fungal species. In summary, this represents the first systematic study of sex and virulence in the dermatophytes.
The MAT locus was identified through a combined bioinformatic and direct sequencing approach. Our identification of MAT idiomorphs and meiotic genes (data not shown) in different strains of M. gypseum is the first genomic evidence for sexual reproduction in this dermatophyte species and is well correlated with its known ability to sexually reproduce under laboratory conditions (23, 43, 50, 54). The MAT locus of M. gypseum is similar to those in other Ascomycota: two idiomorphs in which one encodes an alpha domain factor and the other encodes an HMG domain factor, both of which are associated with the APN2, SLA2, and COX13 genes.
The ∼3-kb MAT locus (3,161 bp for MAT1-1 and 2,941 bp for MAT1-2) of M. gypseum appears to be a reduced version in the phylum of Ascomycota, especially compared to related species: H. capsulatum (5,554 bp for MAT1-1 and 5,829 bp for MAT1-2), C. immitis and C. posadasii (8,167 bp for MAT1-1 and 9,142 bp for MAT1-2), and P. brasiliensis (6,851 bp for MAT1-1 and 6,736 bp for MAT1-2) (Table 1 and Fig. 2). Although there is no direct association between the size of MAT and mating ability, a larger MAT locus may facilitate suppression of recombination. In humans, other animals, and plants, the sex genomic region independently evolved as chromosomes, with suppressed recombination during meiosis. Notably, mating ability is still retained in M. gypseum even though the MAT locus size spans only 3 kb. This raises several questions: what is the minimum size of a MAT locus sufficient to drive sexual reproduction in fungi? For example, could a single nucleotide difference in a key regulatory factor suffice? An ancestral MAT locus may have contained only an alpha domain or only an HMG gene and may be extant in some taxa. Characterization of the MAT locus in additional fungi will be useful for addressing this question.
Although a high similarity of MAT locus structure has been observed between M. gypseum and other Ascomycota, the MAT locus of M. gypseum exhibits a unique arrangement. In M. gypseum, the alpha domain and HMG gene orientations are reversed with respect to the SLA2 gene, and the APN2 and COX13 genes are still linked to the MAT locus but lie on the 5′ side linked to SLA2 rather than flanking the locus at the 3′ side (SLA2-COX13-APN2-alpha domain/HMG rather than SLA2-alpha domain/HMG-COX13-APN2) (Fig. 2). As a result of this atypical arrangement, it would not be possible to amplify this configuration of MAT locus using degenerate PCR with APN2 and SLA2 gene primers. This may provide clues to identify MAT in other Ascomycota.
A laboratory-defined sexual cycle is known in M. gypseum, and this is also supported by population genetic studies (23, 44, 54). Two teleomorphic forms, A. gypseum and A. incurvatum, have been classified into the M. gypseum complex. The MAT1-1 sequence identity assessed here between A. gypseum (CBS118893, ATCC 24164, and ATCC 48981) and A. incurvatum (ATCC 58593 and ATCC 58595) was 82.5%. The sequence variation between A. gypseum and A. incurvatum is mainly the result of divergence in the genes flanking the alpha domain (data not shown). Our MAT locus identification and mating type designation of M. gypseum are in accord with mating assays (Fig. 6).
Sexual structures were only observed in crosses between MAT1-1 and MAT1-2 strains. Taken together with identification of only one or the other MAT locus idiomorph in each isolate, this further supports the assignment of M. gypseum as a heterothallic fungal species. In addition, strains CBS118893, ATCC 24165, and ATCC 58593 appear sterile. Other unknown genetic (or epigenetic) factors may contribute to control sexual reproduction of M. gypseum. In addition, some potentially compatible (MAT1-1 and MAT1-2) strain pairs failed to mate (Fig. 5). A loss of mating ability has also been observed in H. capsulatum with repeated passage, but the mechanism is not known (4, 29).
While mating tests to determine teleomorphic states previously served as an important approach to characterizing M. gypseum clinical strains (50, 54), they are not sufficient in all cases. As mentioned above, no sexual structures were observed for some MAT1-1/MAT1-2 strain pairs based on mating assays in the present study (Fig. 5). The mating types of some isolates previously detected by mating assays are also not in accord with the MAT locus idiomorphs identified by PCR and DNA sequencing in the present study (see Table S1 in the supplemental material). In addition, mating tests are time-consuming and usually require more than 1 month to observe sexual structures. Based on the available sequences of MAT1-1 and MAT1-2, we designed two sets of primers to rapidly determine the mating type of the M. gypseum strains by PCR (Fig. 4). This simple PCR assay can serve as a rapid, sensitive, and reliable method for molecular determination of the mating type of M. gypseum isolates, possibly even involving environmental samples.
Sexual reproduction had also been proposed for a large number of dermatophytes, mostly for geophilic species, although the genomic basis of the mating type had not been studied (57). Based on the characterization of the MAT locus in M. gypseum, we were able to infer the MAT locus in four additional dermatophyte species. By syntenic analysis between M. gypseum and M. canis, T. equinum, T. rubrum, and T. tonsurans, we identified a similar MAT structure in M. canis and T. equinum (Fig. 3). We speculate that other dermatophytes may share a similar MAT locus organization, as observed in these five dermatophyte species with only sequence diversity of the genes.
M. gypseum is geophilic, M. canis and T. equinum are zoophilic, and T. rubrum and T. tonsurans are anthropophilic. However, all of the five species are pathogenic to humans. Sexual reproduction has often been observed in geophilic dermatophytes but has not often been observed in zoophilic and anthropophilic dermatophytes and is especially rare in anthropophilic dermatophytes. Identification of MAT loci in geophilic (M. gypseum), zoophilic (M. canis and T. equinum), and anthropophilic (T. rubrum and T. tonsurans) dermatophytes suggests that most anthropophilic dermatophytes may retain a MAT locus and extant sexual or parasexual cycles, like C. albicans and A. fumigatus.
In previous studies, the MAT locus has been identified in the closely related dimorphic fungal pathogen species H. capsulatum, C. immitis, and C. posadasii (4, 16, 38). An expansion model of MAT locus evolution in Coccidioides species, including C. immitis and C. posadasii, has been proposed based on the observation that the SLA2 and COX13 genes have been captured into the MAT loci in these two species (16, 38). In the present study, we provide further insights regarding the MAT locus from 4 genomic sequences of H. capsulatum and 14 genomic sequences of C. immitis and C. posadasii (Table 1). Our results confirm the previous observation that the MAT locus in C. immitis and C. posadasii has expanded relative to H. capsulatum (4, 16, 38).
We also extended knowledge of MAT locus evolution by identification of two MAT idiomorphs in P. brasiliensis (51a). P. brasiliensis is a dimorphic human pathogenic fungus and the causative agent of paracoccidioidomycosis, an important systemic mycosis in Latin America (47). Although no sexual cycle has been described for P. brasiliensis, phylogenetic studies strongly suggest speciation and sexual reproduction of at least one species of the complex (39). Recent transcriptional analysis provided further evidence for sexual reproduction of this dimorphic fungus by detecting the expression of meiotic and MAT locus genes (14). We also found a corresponding expressed sequence tag (EST) (GenBank accession no. CN253714) for the alpha domain gene of P. brasiliensis strain P01 and a corresponding EST (GenBank accession no. BQ498044) for the HMG domain gene of P. brasiliensis strains P03 and P18 in GenBank by BLAST search. A recent study performed by Torres et al. reveals an ∼1:1 distribution of MAT1-1 and MAT1-2 strains of P. brasiliensis in South America (51a). Taken together, we hypothesize that P. brasiliensis is a heterothallic fungus that could have an extant sexual cycle.
The P. brasiliensis MAT locus has an organization similar to closely related dimorphic fungi and is more similar to H. capsulatum than to C. immitis or C. posadasii (Fig. 2). A Pogo TE has translocated into the MAT1-2 locus of P. brasiliensis P18; however, no similar TE was found in the MAT1-1 idiomorph of strain P01 or MAT1-2 of strain P03 (Fig. 8). Homologs of this TE are widely distributed in the genomic sequences of M. gypseum, H. capsulatum, C. immitis, and C. posadasii. In A. fumigatus, insertion of this Pogo TE inactivated a G-protein-coupled receptor gene (22). Previously, TEs have been found within or flanking MAT loci, such as the homothallic fungus Neosartorya fischeri (46) and in the MAT locus of C. neoformans (35), but the influence of TE transposition on MAT locus genes or mating ability is unknown. It is also currently unknown whether the TE insertion in the MAT locus of P. brasiliensis is a common feature in this species. Furthermore, the role of this TE insertion as it relates to the sexual cycle, if any, is still unknown. However, transposition of TE into or around the MAT locus may significantly increase the sequence diversity between two idiomorphs and decrease recombination of this genomic region if sexual reproduction is still possible. Insertion and local transposition likely also provides homologous sequences driving recombination events at the MAT locus.
Compared to the closely related dimorphic fungal species H. capsulatum, C. immitis, C. posadasii, and P. brasiliensis, the dermatophytes have the smallest MAT locus size and a unique MAT organization (Fig. 2 and 3). In C. immitis and C. posadasii, the size of MAT is significantly increased by capture of the APN2 and COX13 genes, and the orientation of the APN2 and COX13 genes within the MAT locus was reversed (16, 38) (Fig. 2). Based on the comparison of size and structure of the MAT locus of the dermatophyte and dimorphic fungal pathogens and phylogenetic analysis, an expansion rather than a contraction model for MAT evolution is proposed (Fig. 2). Expansion of MAT mostly derives from changes in gene orientation or gene order, and gene acquisition but gene loss can also occur. These three types of events may promote each other. with an increasing number of whole-genome sequences available, gene arrangement or gene order resulting from gene transposition, duplication, inversion, or loss has been considered an important genetic polymorphism as well as gene content. Changes in gene order can alter gene expression (48). There may be functional, and adaptive, relationships between gene order and phenotype mediated by gene order effects on transcriptional regulation (53). Transposition of TEs can remarkably change gene order and in some cases may result in inactivation of neighboring genes (22). The influence of the dynamic gene order of the MAT locus and flanking regions on gene function or mating ability is unknown. Identification of the MAT locus in other closely related fungi such as Blastomyces dermatitidis and Lacazia loboi, as well as functional studies, will begin to address this. Despite acquisition of the APN2 and COX13 genes as a route to MAT locus expansion in C. immitis and C. posadasii, diversity between MAT1-1 and MAT1-2 idiomorphs is also significantly increased by gene loss. Only one of the two unknown genes present in the M. gypseum MAT locus is found in the MAT1-1 and MAT1-2 idiomorphs of C. immitis and C. posadasii (Fig. 2). In addition, by searching the genomic sequences of C. immitis and C. posadasii, there was no evidence that the lost genes transposed to other genomic regions. Thus, gene loss from only one idiomorph also contributes to MAT locus remodeling. A unilateral loss of genes of unknown function from the MAT locus was also found in H. capsulatum and P. brasiliensis (Fig. 2).
A similar phylogenetic relationship was found among the species analyzed based on the alpha domain in MAT1-1 (∼380 amino acids) or the HMG domain in MAT1-2 (∼340 amino acids) (Fig. 9). This suggests that the MAT locus alpha and HMG domain genes are conserved and evolved coordinately, whereas the MAT locus organization is remarkably dynamic. The close phylogenetic relationship of dermatophytes with dimorphic human pathogenic fungi based on MAT locus genes further supports their close evolutionary association. However, the dermatophytes were grouped into the dimorphic fungi with an even closer relationship with C. immitis and C. posadasii based on the MAT locus genes. This phylogenetic organization differs from previous reports that separated the dermatophytes from dimorphic fungi based on 18S and 25S rDNA sequences (2, 34).
An unequal prevalence of the two opposite mating types has been observed in clinical isolates of several pathogenic fungi, such as C. neoformans (30), C. gattii (15), and H. capsulatum (32). Thus, mating type identification of these fungal species has important potential clinical ramifications because of potential associations of virulence with specific mating types (31). For M. gypseum, however, the two mating types have almost equal prevalence among clinical isolates (23, 54). Our virulence tests in the G. mellonella larvae heterologous host further support that the two mating type strains of M. gypseum may have similar virulence (Fig. 8). Previously, mating types of M. gypseum strains were determined by crosses on defined medium, which is a time-consuming and labor-intensive method. In addition, not all strains are fertile, and in some examples their reported mating type may not be accurate (see Table S1 in the supplemental material). The PCR assay developed here for M. gypseum will be useful for reevaluating the distribution of the two mating types among clinical isolates. Thus, characterization of the MAT locus of dermatophytes will not only facilitate classical genetic studies involving mating but also provide information potentially relevant to virulence by rapid determination of the mating types by PCR.
Some environmental or commensal fungi can complete sexual cycles on human or animal hosts, including mating on skin. For example, C. albicans mates on the skin of nude mice (33). The causative agents of dandruff and other skin disorders, Malassezia globosa and M. restricta, have been speculated to sexually reproduce on skin based on genomic evidence for the MAT locus, mating, and meiotic genes (60). Given that sexual reproduction can alter the virulence of fungi (24, 41), mating of environmental or commensal fungi on the human host may be associated with antifungal resistance or pathogenicity. It will therefore be of considerable interest to test whether M. gypseum mates on human or animal skin and to also characterize progeny that are produced to explore pathogenesis of the dermatophytes. In conclusion, molecular identification and characterization of the MAT locus of the dermatophytes has opened a door enabling further studies of their mating, virulence, and pathogenicity.
Supplementary Material
Acknowledgments
We thank Wiley A. Schell and Jonathan Benton for M. gypseum strain ATCC 24102; Yonathan Lewit, Xuying Wang, Soo Chan Lee, and Min Ni for technical assistance; and Edmond Byrnes, Yen-Ping Hsueh, Keisha Findley, and Robert Bastidas for valuable discussions and reading the manuscript. We thank Angela Restrepo's group and Axel Brakhage's group for sharing unpublished data. We thank the Broad Institute for access to the genomic sequences included in this study and Christina Cuomo for her leadership for the dermatophyte genome project.
This study was supported by NIH/NIAID R01 grant AI50113 to J.H. and NIH/NIAID R21 grant AI081235 to T.C.W.
W.L., B.M., T.C.W., and J.H. conceived and designed the experiments. W.L. performed the bioinformatic analyses, genetic studies, and virulence tests, and W.L. and B.M. performed the mating assays. W.L. and J.H. analyzed the results and wrote the manuscript, and B.M. and T.C.W. provided edits and comments on the manuscript.
Footnotes
Published ahead of print on 30 October 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bialek, R., A. Ibricevic, A. Fothergill, and D. Begerow. 2000. Small subunit ribosomal DNA sequence shows Paracoccidioides brasiliensis closely related to Blastomyces dermatitidis. J. Clin. Microbiol. 38:3190-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brennan, M., D. Y. Thomas, M. Whiteway, and K. Kavanagh. 2002. Correlation between virulence of Candida albicans mutants in mice and Galleria mellonella larvae. FEMS Immunol. Med. Microbiol. 34:153-157. [DOI] [PubMed] [Google Scholar]
- 4.Bubnick, M., and A. G. Smulian. 2007. The MAT1 locus of Histoplasma capsulatum is responsive in a mating type-specific manner. Eukaryot. Cell 6:616-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burt, A., D. A. Carter, G. L. Koenig, T. J. White, and J. W. Taylor. 1996. Molecular markers reveal cryptic sex in the human pathogen Coccidioides immitis. Proc. Natl. Acad. Sci. USA 93:770-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler, G. 2007. The evolution of MAT: the ascomycetes, p. 3-18. In J. Heitman, J. W. Kronstad, J. Taylor, and L. Casselton (ed.), Sex in fungi: molecular determination and evolutionary implications. ASM Press, Washington, DC.
- 7.Butlin, R. 2002. Evolution of sex: the costs and benefits of sex: new insights from old asexual lineages. Nat. Rev. Genet. 3:311-317. [DOI] [PubMed] [Google Scholar]
- 8.Carver, T., M. Berriman, A. Tivey, C. Patel, U. Bohme, B. G. Barrell, J. Parkhill, and M. A. Rajandream. 2008. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24:2672-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coppin, E., R. Debuchy, S. Arnaise, and M. Picard. 1997. Mating types and sexual development in filamentous ascomycetes. Microbiol. Mol. Biol. Rev. 61:411-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dodgson, A. R., C. Pujol, M. A. Pfaller, D. W. Denning, and D. R. Soll. 2005. Evidence for recombination in Candida glabrata. Fungal Genet. Biol. 42:233-243. [DOI] [PubMed] [Google Scholar]
- 11.Doncaster, C. P., G. E. Pound, and S. J. Cox. 2000. The ecological cost of sex. Nature 404:281-285. [DOI] [PubMed] [Google Scholar]
- 12.Dyer, P. S., and M. Paoletti. 2005. Reproduction in Aspergillus fumigatus: sexuality in a supposedly asexual species? Med. Mycol. 43(Suppl. 1):S7-S14. [DOI] [PubMed] [Google Scholar]
- 13.Dyer, P. S., M. Paoletti, and D. B. Archer. 2003. Genomics reveals sexual secrets of Aspergillus. Microbiology 149:2301-2303. [DOI] [PubMed] [Google Scholar]
- 14.Felipe, M. S., R. V. Andrade, F. B. Arraes, A. M. Nicola, A. Q. Maranhao, F. A. Torres, I. Silva-Pereira, M. J. Pocas-Fonseca, E. G. Campos, L. M. Moraes, P. A. Andrade, A. H. Tavares, S. S. Silva, C. M. Kyaw, D. P. Souza, M. Pereira, R. S. Jesuino, E. V. Andrade, J. A. Parente, G. S. Oliveira, M. S. Barbosa, N. F. Martins, A. L. Fachin, R. S. Cardoso, G. A. Passos, N. F. Almeida, M. E. Walter, C. M. Soares, M. J. Carvalho, and M. M. Brigido. 2005. Transcriptional profiles of the human pathogenic fungus Paracoccidioides brasiliensis in mycelium and yeast cells. J. Biol. Chem. 280:24706-24714. [DOI] [PubMed] [Google Scholar]
- 15.Fraser, J. A., S. S. Giles, E. C. Wenink, S. G. Geunes-Boyer, J. R. Wright, S. Diezmann, A. Allen, J. E. Stajich, F. S. Dietrich, J. R. Perfect, and J. Heitman. 2005. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature 437:1360-1364. [DOI] [PubMed] [Google Scholar]
- 16.Fraser, J. A., J. E. Stajich, E. J. Tarcha, G. T. Cole, D. O. Inglis, A. Sil, and J. Heitman. 2007. Evolution of the mating type locus: insights gained from the dimorphic primary fungal pathogens Histoplasma capsulatum, Coccidioides immitis, and Coccidioides posadasii. Eukaryot. Cell 6:622-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galagan, J. E., S. E. Calvo, C. Cuomo, L. J. Ma, J. R. Wortman, S. Batzoglou, S. I. Lee, M. Basturkmen, C. C. Spevak, J. Clutterbuck, V. Kapitonov, J. Jurka, C. Scazzocchio, M. Farman, J. Butler, S. Purcell, S. Harris, G. H. Braus, O. Draht, S. Busch, C. D'Enfert, C. Bouchier, G. H. Goldman, D. Bell-Pedersen, S. Griffiths-Jones, J. H. Doonan, J. Yu, K. Vienken, A. Pain, M. Freitag, E. U. Selker, D. B. Archer, M. A. Penalva, B. R. Oakley, M. Momany, T. Tanaka, T. Kumagai, K. Asai, M. Machida, W. C. Nierman, D. W. Denning, M. Caddick, M. Hynes, M. Paoletti, R. Fischer, B. Miller, P. Dyer, M. S. Sachs, S. A. Osmani, and B. W. Birren. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105-1115. [DOI] [PubMed] [Google Scholar]
- 18.Georg, L. K. 1960. Epidemiology of the dermatophytoses sources of infection, modes of transmission and epidemicity. Ann. N. Y. Acad. Sci. 89:69-77. [DOI] [PubMed] [Google Scholar]
- 19.Goddard, M. R., H. C. Godfray, and A. Burt. 2005. Sex increases the efficacy of natural selection in experimental yeast populations. Nature 434:636-640. [DOI] [PubMed] [Google Scholar]
- 20.Graser, Y., S. De Hoog, and R. C. Summerbell. 2006. Dermatophytes: recognizing species of clonal fungi. Med. Mycol. 44:199-209. [DOI] [PubMed] [Google Scholar]
- 21.Graser, Y., J. Scott, and R. Summerbell. 2008. The new species concept in dermatophytes-a polyphasic approach. Mycopathologia 166:239-256. [DOI] [PubMed] [Google Scholar]
- 22.Hey, P., G. Robson, M. Birch, and M. Bromley. 2008. Characterisation of Aft1 a Fot1/Pogo type transposon of Aspergillus fumigatus. Fungal Genet. Biol. 45:117-126. [DOI] [PubMed] [Google Scholar]
- 23.Hironaga, M., S. Tanaka, and S. Watanabe. 1982. Distribution of mating types among clinical isolates of the Microsporum gypseum complex. Mycopathologia 77:31-35. [DOI] [PubMed] [Google Scholar]
- 24.Hsueh, Y. P., and J. Heitman. 2008. Orchestration of sexual reproduction and virulence by the fungal mating-type locus. Curr. Opin. Microbiol. 11:517-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inderbitzin, P., J. Harkness, B. G. Turgeon, and M. L. Berbee. 2005. Lateral transfer of mating system in Stemphylium. Proc. Natl. Acad. Sci. USA 102:11390-11395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson, J. C., L. A. Higgins, and X. Lin. 2009. Conidiation color mutants of Aspergillus fumigatus are highly pathogenic to the heterologous insect host Galleria mellonella. PLoS One 4:e4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kac, G. 2000. Molecular approaches to the study of dermatophytes. Med. Mycol. 38:329-336. [DOI] [PubMed] [Google Scholar]
- 28.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 29.Kwon-Chung, K. J. 1972. Sexual stage of Histoplasma capsulatum. Science 175:326. [DOI] [PubMed] [Google Scholar]
- 30.Kwon-Chung, K. J., and J. E. Bennett. 1978. Distribution of alpha and a mating types of Cryptococcus neoformans among natural and clinical isolates. Am. J. Epidemiol. 108:337-340. [DOI] [PubMed] [Google Scholar]
- 31.Kwon-Chung, K. J., J. C. Edman, and B. L. Wickes. 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60:602-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwon-Chung, K. J., R. J. Weeks, and H. W. Larsh. 1974. Studies on Emmonsiella capsulata (Histoplasma capsulatum). II. Distribution of the two mating types in 13 endemic states of the United States. Am. J. Epidemiol. 99:44-49. [DOI] [PubMed] [Google Scholar]
- 33.Lachke, S. A., S. R. Lockhart, K. J. Daniels, and D. R. Soll. 2003. Skin facilitates Candida albicans mating. Infect. Immun. 71:4970-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leclerc, M. C., H. Philippe, and E. Gueho. 1994. Phylogeny of dermatophytes and dimorphic fungi based on large subunit rRNA sequence comparisons. J. Med. Vet. Mycol. 32:331-341. [DOI] [PubMed] [Google Scholar]
- 35.Lengeler, K. B., D. S. Fox, J. A. Fraser, A. Allen, K. Forrester, F. S. Dietrich, and J. Heitman. 2002. Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot. Cell 1:704-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin, X., and J. Heitman. 2007. Mechanisms of homothallism in fungi and transitions between heterothallism and homothallism, p. 35-57. In J. Heitman, J. W. Kronstad, J. Taylor, and L. Casselton (ed.),. Sex in fungi: molecular determination and evolutionary implications. ASM Press, Washington, DC.
- 37.Lomsadze, A., V. Ter-Hovhannisyan, Y. O. Chernoff, and M. Borodovsky. 2005. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 33:6494-6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandel, M. A., B. M. Barker, S. Kroken, S. D. Rounsley, and M. J. Orbach. 2007. Genomic and population analyses of the mating type loci in Coccidioides species reveal evidence for sexual reproduction and gene acquisition. Eukaryot. Cell 6:1189-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matute, D. R., J. G. McEwen, R. Puccia, B. A. Montes, G. San-Blas, E. Bagagli, J. T. Rauscher, A. Restrepo, F. Morais, G. Nino-Vega, and J. W. Taylor. 2006. Cryptic speciation and recombination in the fungus Paracoccidioides brasiliensis as revealed by gene genealogies. Mol. Biol. Evol. 23:65-73. [DOI] [PubMed] [Google Scholar]
- 40.Mylonakis, E., R. Moreno, J. B. El Khoury, A. Idnurm, J. Heitman, S. B. Calderwood, F. M. Ausubel, and A. Diener. 2005. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect. Immun. 73:3842-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nielsen, K., and J. Heitman. 2007. Sex and virulence of human pathogenic fungi. Adv. Genet. 57:143-173. [DOI] [PubMed] [Google Scholar]
- 42.O'Gorman, C. M., H. T. Fuller, and P. S. Dyer. 2009. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature 457:471-474. [DOI] [PubMed] [Google Scholar]
- 43.Punsola, L., and J. Guarro. 1984. Distribution of mating types of the “Microsporum gypseum complex” in Spanish soils. Mykosen 27:191-193. [DOI] [PubMed] [Google Scholar]
- 44.Ranganathan, S., S. A. Balajee, and T. Menon. 1996. Mating patterns of dermatophytes of diverse origin in India. Mycopathologia 136:91-94. [DOI] [PubMed] [Google Scholar]
- 45.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 46.Rydholm, C., P. S. Dyer, and F. Lutzoni. 2007. DNA sequence characterization and molecular evolution of MAT1 and MAT2 mating-type loci of the self-compatible ascomycete mold Neosartorya fischeri. Eukaryot. Cell 6:868-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.San-Blas, G. 1993. Paracoccidioidomycosis and its etiologic agent Paracoccidioides brasiliensis. J. Med. Vet. Mycol. 31:99-113. [PubMed] [Google Scholar]
- 48.Shapiro, J. A. 1982. Changes in gene order and gene expression. Natl. Cancer Inst. Monogr. 60:87-110. [PubMed] [Google Scholar]
- 49.Siller, S. 2001. Sexual selection and the maintenance of sex. Nature 411:689-692. [DOI] [PubMed] [Google Scholar]
- 50.Stockdale, P. M. 1963. The Microsporum gypseum complex (Nannizzia incurvata Stockd., N. gypsea (Nann.) comb. nov., N. fulva sp. nov.). Sabouraudia 3:114-126. [PubMed] [Google Scholar]
- 51.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51a.Torres, I., A. M. García, O. Hernández, A. González, J. G. McEwen, A. Restrepo, and M. Arango. 20 November 2009, posting date. Presence and expression of the mating type locus in Paracoccidioides brasiliensis isolates. Fungal Genet. Biol. doi: 10.1016/j.fgb.2009.11.005. [DOI] [PubMed]
- 52.Turgeon, B. G., and O. C. Yoder. 2000. Proposed nomenclature for mating type genes of filamentous ascomycetes. Fungal Genet. Biol. 31:1-5. [DOI] [PubMed] [Google Scholar]
- 53.Vision, T. J. 2005. Gene order in plants: a slow but sure shuffle. New Phytol. 168:51-60. [DOI] [PubMed] [Google Scholar]
- 54.Weitzman, I., M. A. Gordon, and S. A. Rosenthal. 1971. Determination of the perfect state, mating type and elastase activity in clinical isolates of the Microsporum gypseum complex. J. Investig. Dermatol. 57:278-282. [DOI] [PubMed] [Google Scholar]
- 55.Weitzman, I., and M. Silva-Hutner. 1967. Non-keratinous agar media as substrates for the ascigerous state in certain members of the Gymnoascaceae pathogenic for man and animals. Sabouraudia 5:335-340. [DOI] [PubMed] [Google Scholar]
- 56.Weitzman, I., and R. C. Summerbell. 1995. The dermatophytes. Clin. Microbiol. Rev. 8:240-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.White, T. C., B. G. Oliver, Y. Graser, and M. R. Henn. 2008. Generating and testing molecular hypotheses in the dermatophytes. Eukaryot. Cell 7:1238-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wong, S., M. A. Fares, W. Zimmermann, G. Butler, and K. H. Wolfe. 2003. Evidence from comparative genomics for a complete sexual cycle in the ‘asexual’ pathogenic yeast Candida glabrata. Genome Biol. 4:R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Woo, P. C., K. T. Chong, H. Tse, J. J. Cai, C. C. Lau, A. C. Zhou, S. K. Lau, and K. Y. Yuen. 2006. Genomic and experimental evidence for a potential sexual cycle in the pathogenic thermal dimorphic fungus Penicillium marneffei. FEBS Lett. 580:3409-3416. [DOI] [PubMed] [Google Scholar]
- 60.Xu, J., C. W. Saunders, P. Hu, R. A. Grant, T. Boekhout, E. E. Kuramae, J. W. Kronstad, Y. M. Deangelis, N. L. Reeder, K. R. Johnstone, M. Leland, A. M. Fieno, W. M. Begley, Y. Sun, M. P. Lacey, T. Chaudhary, T. Keough, L. Chu, R. Sears, B. Yuan, and T. L. Dawson, Jr. 2007. Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proc. Natl. Acad. Sci. USA 104:18730-18735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yun, S. H., M. L. Berbee, O. C. Yoder, and B. G. Turgeon. 1999. Evolution of the fungal self-fertile reproductive lifestyle from self-sterile ancestors. Proc. Natl. Acad. Sci. U.S.A. 96:5592-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.