Abstract
In Saccharomyces cerevisiae, Snf1 protein kinase is important for growth on carbon sources that are less preferred than glucose. When glucose becomes limiting, Snf1 undergoes catalytic activation, which requires phosphorylation of its T-loop threonine (Thr210). Thr210 phosphorylation can be performed by any of three Snf1-activating kinases: Sak1, Tos3, and Elm1. These kinases are redundant in that all three must be eliminated to confer snf1Δ-like growth defects on nonpreferred carbon sources. We previously showed that in addition to glucose signaling, Snf1 also participates in nitrogen signaling and is required for diploid pseudohyphal differentiation, a filamentous-growth response to nitrogen limitation. Here, we addressed the roles of the Snf1-activating kinases in this process. Loss of Sak1 caused a defect in pseudohyphal differentiation, whereas Tos3 and Elm1 were dispensable. Sak1 was also required for increased Thr210 phosphorylation of Snf1 under nitrogen-limiting conditions. Expression of a catalytically hyperactive version of Snf1 restored pseudohyphal differentiation in the sak1Δ/sak1Δ mutant. Thus, while the Snf1-activating kinases exhibit redundancy for growth on nonpreferred carbon sources, the loss of Sak1 alone produced a significant defect in a nitrogen-regulated phenotype, and this defect resulted from deficient Snf1 activation rather than from disruption of another pathway. Our results suggest that Sak1 is involved in nitrogen signaling upstream of Snf1.
Snf1 protein kinase of the yeast Saccharomyces cerevisiae belongs to the conserved Snf1/AMP-activated protein kinase (AMPK) family; members of this family play central roles in responses to metabolic stress in eukaryotes (reviewed in references 17 and 18). Interest in Snf1/AMPK pathways is high due to their important functions. Deregulation of AMPK signaling in humans has been linked to type 2 diabetes, heart disease, and cancer (for a review, see reference 16). Snf1 homologs of pathogenic fungi have been implicated in virulence and drug resistance (23, 63, 64).
Yeast Snf1 (Cat1, Ccr1) was first identified by its requirement for growth on carbon sources that are less preferred than glucose (5, 7, 65). Subsequent evidence indicated that Snf1 protein kinase (6) is directly involved in glucose signaling, since its activity is stimulated in response to glucose limitation (62). Catalytic activation of Snf1 occurs through phosphorylation of its conserved T-loop threonine (Thr210) (12) by upstream kinases (40, 62). Three protein kinases—Sak1, Tos3, and Elm1—have been identified that can phosphorylate Thr210 of Snf1 (22, 41, 55). These kinases are related to the mammalian kinases that activate AMPK by phosphorylating the equivalent T-loop threonine (Thr172) (reviewed in references 17 and 18). We recently presented evidence that Snf1 homologs of two pathogenic Candida species, Candida albicans and C. glabrata, also undergo T-loop phosphorylation (42).
It is not entirely clear why S. cerevisiae has three different kinases that can activate Snf1. Judging by assays of Snf1 kinase activity, Sak1 makes the largest individual contribution to Snf1 activation in the cell (19, 22). However, deletion of SAK1 alone does not result in growth defects on alternative carbon sources, and all three Snf1-activating kinases must be eliminated to produce a phenotypic defect comparable to that of the snf1Δ mutant (22, 39, 55). Deletion of TOS3 was reported to moderately affect growth on nonfermentable carbon sources; this correlated with a reduction in Snf1 activity, although effects on another pathway(s) cannot be excluded (25). Mutation of ELM1 affects cell cycle progression and cell morphology, but this effect is unrelated to Elm1's role as a Snf1-activating kinase and pertains to its role in the activation of Nim1-related protein kinases involved in morphogenesis checkpoint control (1, 56).
While showing significant redundancy for growth on nonpreferred carbon sources, the Snf1-activating kinases could exhibit specialization in Snf1 signaling in response to stresses other than carbon stress. Evidence indicates that Snf1 is important for adaptation to a number of stress conditions (reviewed in reference 18). In some cases, such as genotoxic stress or exposure to hygromycin B, weak activity of unphosphorylated Snf1 appears to be sufficient for resistance (10, 48). In others, such as sodium ion stress and alkaline stress, Thr210 phosphorylation of Snf1 is required for adaptation, and Snf1 becomes activated upon stress exposure (21, 40). As with glucose limitation, however, in these latter cases Sak1 makes the largest contribution to Snf1 activation judging by biochemical assays, and yet it remains dispensable for wild-type levels of stress-resistant growth in phenotypic tests; loss of all three Snf1-activating kinases results in growth defects comparable to those of cells lacking Snf1 (21). Thus, investigation of these stresses provided no evidence for phenotypically relevant specialization of Sak1, Tos3, or Elm1 in Snf1 signaling.
Diploid pseudohyphal differentiation is a developmental response to nitrogen limitation (15). When nitrogen becomes limiting, diploid cells adopt elongated morphology, alter their budding pattern, and generate filaments (pseudohyphae) consisting of chains of cells attached to one another. One of the key events in this process is activation of the FLO11 (MUC1) gene, which encodes a cell surface glycoprotein involved in cell-cell adhesion (29, 33, 34). Following up on an observation that Snf1 is important for FLO11 expression on low glucose, we previously found that diploids lacking Snf1 fail to undergo pseudohyphal differentiation on low nitrogen (27, 28). The requirement of Snf1 for a nitrogen-regulated process raised the possibility that Snf1 is directly involved in nitrogen signaling. In support of this notion, we subsequently showed that weak activity of nonphosphorylatable Snf1-T210A is not sufficient for pseudohyphal differentiation and that Thr210 phosphorylation of Snf1 increases in response to nitrogen limitation (43).
Here, we have examined the roles of Sak1, Tos3, and Elm1 in pseudohyphal differentiation and Snf1 activation on low nitrogen. We show that elimination of Sak1 leads to a significant defect in nitrogen-regulated pseudohyphal differentiation, whereas Tos3 and Elm1 are dispensable. Sak1 is also required for normal Thr210 phosphorylation of Snf1 under nitrogen-limiting conditions. Our data strongly suggest that the loss of Sak1 affects pseudohyphal differentiation by affecting Snf1 activation and not by disruption of another pathway. Collectively, our findings implicate Sak1 in nitrogen signaling upstream of Snf1.
MATERIALS AND METHODS
Strains and genetic methods.
The S. cerevisiae strains used in the present study are listed in Table 1. The strains were in the Σ1278b genetic background and were descendants of strains MY1384 (MATa, prototroph), MY1401 (MATα ura3Δ leu2Δ his3Δ), and MY1402 (MATa ura3Δ leu2Δ trp1Δ) of the isogenic Sigma2000 series (Microbia, Cambridge, MA). Derivatives carrying snf1Δ::KanMX6 and sak1Δ::KanMX4 have been described (42, 43). To generate tos3Δ::His3MX6, elm1Δ::KanMX4, and swe1Δ::KanMX6 single mutants, wild-type strains were transformed with PCR fragments encompassing the marker sequences with flanks homologous to the flanks of the open reading frames to be deleted; all yeast transformations were performed by using standard methods (51). Gene deletions were confirmed by PCR analysis of genomic DNA. Strains carrying deletion combinations were constructed by genetic crossing and their genotypes were confirmed by PCR; homozygous mutant diploids were obtained by mating appropriate mutant haploids (51).
TABLE 1.
S. cerevisiae strainsa
| Strain | Genotype or description |
|---|---|
| KY70 | MATaleu2Δ |
| KY71 | MATα ura3Δ |
| KY72 | KY70 × KY71 |
| KY73 | MATaleu2Δ trp1Δ his3Δ sak1Δ::KanMX4 |
| KY74 | MATα ura3Δ sak1Δ::KanMX4 |
| KY75 | KY73 × KY74 |
| KY76 | MATaleu2Δ trp1Δ his3Δ tos3Δ::His3MX6 |
| KY77 | MATα ura3Δ his3Δ tos3Δ::His3MX6 |
| KY78 | KY76 × KY77 |
| KY79 | MATaleu2Δ trp1Δ his3Δ sak1Δ::KanMX4 tos3Δ::His3MX6 |
| KY80 | MATα ura3Δ his3Δ sak1Δ::KanMX4 tos3Δ::His3MX6 |
| KY81 | KY79 × KY80 |
| KY82 | MATaura3Δ trp1Δ |
| KY83 | MATα ura3Δ his3Δ |
| KY84 | KY82 × KY83 |
| KY85 | MATaura3Δ his3Δ sak1Δ::KanMX4 |
| KY86 | MATα ura3Δ trp1Δ sak1Δ::KanMX4 |
| KY87 | KY85 × KY86 |
| KY88 | MATaleu2Δ snf1Δ::KanMX6 |
| KY89 | MATα ura3Δ snf1Δ::KanMX6 |
| KY90 | KY88 × KY89 |
| KY91 | MATatrp1Δ elm1Δ::KanMX4 |
| KY92 | MATα ura3Δ leu2Δ elm1Δ::KanMX4 |
| KY93 | KY91 × KY92 |
| KY94 | MATaura3Δ trp1Δ elm1Δ::KanMX4 swe1Δ::KanMX6 |
| KY95 | MATα leu2Δ elm1Δ::KanMX4 swe1Δ::KanMX6 |
| KY96 | KY94 × KY95 |
| KY97 | MATaleu2Δ trp1Δ his3Δ swe1Δ::KanMX6 |
| KY98 | MATα ura3Δ swe1Δ::KanMX6 |
| KY99 | KY97 × KY98 |
| KY100 | MATatrp1Δ his3Δ sak1Δ::KanMX4 tos3Δ::His3MX6 elm1Δ::KanMX4 |
| KY101 | MATα ura3Δ his3Δ sak1Δ::KanMX4 tos3Δ::His3MX6 elm1Δ::KanMX4 |
| KY102 | KY100 × KY101 |
| KY103 | MATaura3Δ leu2Δ his3Δ sak1Δ::KanMX4 tos3Δ::His3MX6 elm1Δ::KanMX4 swe1Δ::KanMX6 |
| KY104 | MATα trp1Δ his3Δ sak1Δ::KanMX4 tos3Δ::His3MX6 elm1Δ::KanMX4 swe1Δ::KanMX6 |
| KY105 | KY103 × KY104 |
| KY106 | MATaura3Δ his3Δ tos3Δ::His3MX6 elm1Δ::KanMX4 swe1Δ::KanMX6 |
| KY107 | MATα leu2Δ trp1Δ his3Δ tos3Δ::His3MX6 elm1Δ::KanMX4 swe1Δ::KanMX6 |
| KY108 | KY106 × KY107 |
| KY115 | MATaura3Δ leu2Δ snf1Δ::KanMX6 |
| KY116 | MATα ura3Δ his3Δ snf1Δ::KanMX6 |
| KY117 | KY115 × KY116 |
The source for all of these strains was this study.
Plasmids.
pIT517 expresses N-terminal triple hemagglutinin (HA)-tagged Snf1-G53R from the yeast ADH1 promoter of vector pWS93 (43, 54). pM36 expresses N-terminal triple HA-tagged Sak1 and was constructed by introducing a PCR-amplified DNA fragment encompassing the SAK1 open reading frame into the XmaI site of vector pWS93. To construct pSM14, the wild-type SNF1 gene, including 1.1 kb of its upstream regulatory sequence, was amplified by PCR and cloned into the BamHI site of vector pRS316 (53). pMO16 was derived from pSM14 by introducing a mutation that changes the Thr210 codon of SNF1 to an Ala codon, using the QuikChange XL site-directed mutagenesis kit (Stratagene) and a pair of complementary mutagenic primers that also create a silent diagnostic HindIII site overlapping the Ala210 codon.
Pseudohyphal differentiation assays.
Solid synthetic low ammonia plus 2% dextrose (SLAD) medium containing 50 μM ammonium sulfate as the sole nitrogen source was used for standard pseudohyphal differentiation assays (15). Deviations from this medium composition were as specified in the text. Colonies were generated by micromanipulating single cells (at least a dozen per datapoint) to open areas of the plate, followed by incubation at 30°C for 4 days; when 1.0 mM ammonium sulfate was used as the nitrogen source, the cells were grown for 2 days. Representative colonies were photographed by using a Nikon Eclipse 80i microscope (10 × objective), a CoolSnap HQ2 camera (Photometrics), and NIS-Elements BR 3.01 software (Nikon Corp.).
Immunoblot assays of Thr210 phosphorylation.
For nitrogen-rich conditions, cells were grown at 30°C to mid-log phase (optical density at 600 nm [OD600] of 0.4 to 0.5) in synthetic complete medium (51). For nitrogen-limiting conditions, the above cells were shifted to liquid SLAD medium containing 50 μM ammonium sulfate as the sole nitrogen source and abundant (2%) glucose (15) for 20 min; alternatively, cells were inoculated directly in liquid SLAD at an OD600 of 0.02 and grown at 30°C for 15 h to an OD600 of 0.07 to 0.11 (i.e., approximately for two generations), which is still ∼2-fold below the maximum achievable on SLAD. Protein extracts were prepared by the heat inactivation/alkaline treatment method as recently described (42). Briefly, cultures or culture aliquots (3 to 5 ml) were placed in a boiling water bath for 3 min to arrest Snf1 in its culture-specific Thr210 phosphorylation state, followed by cooling, harvesting, mild alkaline treatment, and extraction by boiling in SDS-PAGE loading buffer. Proteins were separated by SDS-PAGE and analyzed by immunoblotting with anti-phospho-Thr172-AMPK (Cell Signaling Technology), which strongly recognizes phospho-Thr210-Snf1; total Snf1 protein levels were determined by reprobing the blots with anti-polyhistidine antibody H1029 (Sigma-Aldrich), which strongly recognizes a fortuitous stretch of 13 consecutive histidines (amino acids 18 to 30) present in Snf1 (42). The signals were detected by enhanced chemiluminescence using ECL Plus (Amersham Biosciences) or HyGlo (Denville Scientific).
RESULTS
Sak1 is required for pseudohyphal differentiation in response to low nitrogen, and Tos3 is dispensable.
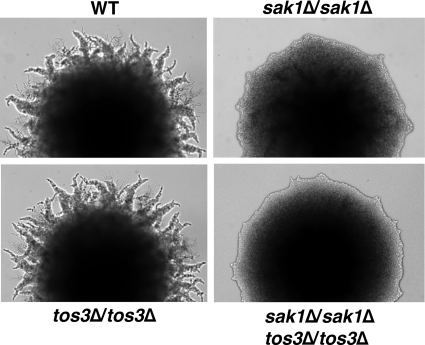
We first addressed the roles of Sak1 and Tos3; since the loss of Elm1 causes strong constitutive filamentation, its effects were analyzed in the absence of Swe1 (see below). Prototrophic wild-type, sak1Δ/sak1Δ, tos3Δ/tos3Δ, and sak1Δ/sak1Δ tos3Δ/tos3Δ diploids were constructed in the dimorphic Σ1278b genetic background and tested for pseudohyphal differentiation on low nitrogen SLAD medium containing 50 μM ammonium sulfate as the sole nitrogen source and abundant (2%) glucose (15). In this and other experiments, colonies were grown from single cells micromanipulated to open areas of the plate. The sak1Δ/sak1Δ mutant exhibited a significant phenotypic defect (Fig. 1). In contrast, the tos3Δ/tos3Δ mutant was indistinguishable from the wild type. The sak1Δ/sak1Δ tos3Δ/tos3Δ double mutant showed a defect similar to that of the sak1Δ/sak1Δ single mutant.
FIG. 1.
Pseudohyphal differentiation in diploids lacking Sak1 and/or Tos3. Prototrophic diploid strains KY72 (wild type, WT), KY75 (sak1Δ/sak1Δ), KY78 (tos3Δ/tos3Δ), and KY81 (sak1Δ/sak1Δ tos3Δ/tos3Δ) were assayed on low-nitrogen SLAD plates. Colonies were photographed after 4 days of growth at 30°C.
The mutant defects were not absolute, and filamentation could still be observed around dense cell patches. Such density dependence however suggested that this filamentation is facilitated by local colimitation for another nutrient, most likely glucose (8, 24, 60). As described further below, reducing the glucose content of the low nitrogen medium can restore filamentation in the absence of Sak1 (but not Snf1).
Elm1 is dispensable for nitrogen-regulated pseudohyphal differentiation.
Elm1 stands out among the Snf1-activating kinases in that its loss causes strong constitutive filamentation both in haploids and diploids (4), greatly complicating the analysis of its possible requirement for pseudohyphal differentiation as a response to low nitrogen. Lack of Elm1 does not lead to any unexpected increase in Snf1 activity (21) that could explain this constitutive phenotype. Evidence indicates that in addition to being a Snf1-activating kinase, Elm1 activates Nim1-related protein kinases involved in morphogenesis checkpoint control, which in turn negatively regulate Swe1, a protein kinase that phosphorylates and negatively regulates the cell cycle progression kinase Cdc28 (1, 2, 56). Previous studies done with haploids attributed the constitutive filamentation of elm1 mutants to the Swe1-dependent mitotic delay, as mutation of SWE1 suppresses elm1 (11, 44).
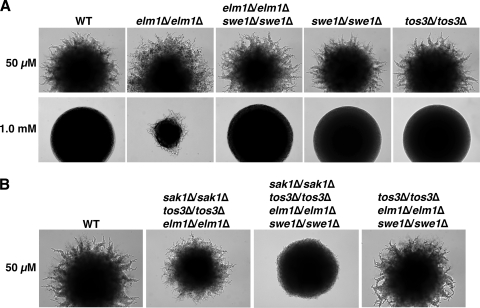
It seemed likely that, by analogy to haploids, elimination of Swe1 would suppress the constitutive filamentation of Elm1-less diploids and thus unmask any defects that the lack of Elm1 might produce in the nitrogen-regulated phenotype; importantly, Swe1 is dispensable for diploid pseudohyphal differentiation under standard nitrogen-limiting conditions (30). We constructed diploids lacking Elm1, Swe1, and both, and tested them on two media: standard SLAD medium containing 50 μM ammonium sulfate as the sole nitrogen source and an otherwise identical medium containing 1.0 mM ammonium sulfate, which inhibits pseudohyphal differentiation in the wild type (47) (Fig. 2A). The elm1Δ/elm1Δ mutant diploid was filamentous on both media. The elm1Δ/elm1 swe1Δ/swe1Δ double mutant lost the constitutive component but still exhibited pseudohyphal differentiation on standard SLAD. In control experiments, a triple mutant lacking all three Snf1-activating kinases (but retaining Swe1) was filamentous on standard SLAD, whereas the quadruple mutant lacking Sak1, Tos3, Elm1, and Swe1 exhibited a defect, a finding consistent with Thr210 phosphorylation being phenotypically relevant when the absence of Elm1 is suppressed by the absence of Swe1 (Fig. 2B). A triple mutant lacking Tos3, Elm1, and Swe1 (with Sak1 as the only Snf1-activating kinase) showed normal pseudohyphal differentiation. These results provided strong evidence that Elm1 is dispensable for nitrogen-regulated pseudohyphal differentiation, and that Sak1 alone is sufficient.
FIG. 2.
Elm1 is dispensable for nitrogen-regulated pseudohyphal differentiation. (A) Prototrophic diploid strains KY72 (wild type, WT), KY93 (elm1Δ/elm1Δ), KY96 (elm1Δ/elm1Δ swe1Δ/swe1Δ), KY99 (swe1Δ/swe1Δ), and KY78 (tos3Δ/tos3Δ) were assayed on standard pseudohyphal differentiation-inducing SLAD medium containing 50 μM ammonium sulfate as the sole nitrogen source (50 μM; upper panel) and on an otherwise identical medium containing 1.0 mM ammonium sulfate (1.0 mM; lower panel). Colonies were grown at 30°C for 4 days (upper panel) and for 2 days (lower panel). (B) Prototrophic diploid strains KY72 (WT), KY102 (sak1Δ/sak1Δ tos3Δ/tos3Δ elm1Δ/elm1Δ), KY105 (sak1Δ/sak1Δ tos3Δ/tos3Δ elm1Δ/elm1Δ swe1Δ/swe1Δ), and KY108 (tos3Δ/tos3Δ elm1Δ/elm1Δ swe1Δ/swe1Δ) were assayed on standard SLAD medium; colonies were photographed after 4 days of growth at 30°C.
Lack of Sak1 affects Thr210 phosphorylation of Snf1 on low nitrogen.
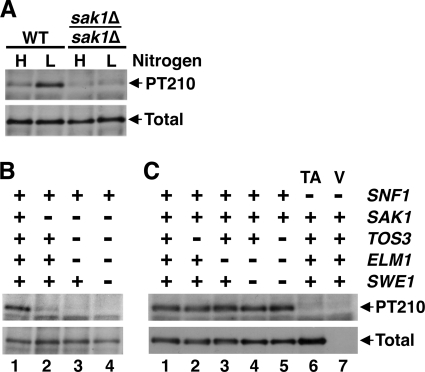
We addressed whether the pseudohyphal differentiation defect of the sak1Δ/sak1Δ mutant is associated with a defect in Snf1 activation on low nitrogen. We first performed immunoblot assays of Thr210 phosphorylation of Snf1 after growth in nitrogen-rich conditions and following a shift to nitrogen-limiting conditions. In the wild type, Thr210 phosphorylation increased in response to nitrogen limitation as expected (43); in contrast, no significant increase was observed in the sak1Δ/sak1Δ mutant (Fig. 3A). Since pseudohyphal differentiation is a response spanning more than one generation, we also assayed Thr210 phosphorylation after extended growth in liquid SLAD (approximately two generations; see Materials and Methods). Thr210 phosphorylation in SLAD-grown sak1Δ/sak1Δ cells was reduced relative to the wild type (Fig. 3B, lanes 1 and 2), which was consistent with the shift experiments. In control experiments, no appreciable signal was detected in SLAD-grown cells lacking all three Snf1-activating kinases, with or without Swe1 (Fig. 3B, lanes 3 and 4). Thr210 phosphorylation in cells lacking Tos3 alone, Elm1 alone, Elm1 and Swe1, as well as Tos3, Elm1, and Swe1, was similar to that in the wild type (Fig. 3C, lanes 1 to 5). These results suggested that Sak1 is necessary and sufficient for normal Thr210 phosphorylation of Snf1 under nitrogen-limiting conditions.
FIG. 3.
Loss of Sak1 affects Thr210 phosphorylation of Snf1 under nitrogen-limiting conditions. Cellular levels of phospho-Thr210-Snf1 (PT210) and total Snf1 protein (Total) were analyzed by immunoblotting as described in Materials and Methods. (A) Cells were grown to mid-log phase in nutrient-rich synthetic medium (high nitrogen, H) and shifted for 20 min to SLAD medium (low nitrogen, L). The strains were KY72 (wild type, WT) and KY75 (sak1Δ/sak1Δ). (B and C) Diploids homozygous for the indicated wild-type alleles (+) or homozygous for the corresponding knockout alleles (−) were grown in liquid SLAD for approximately two generations as described in Materials and Methods. (B) The strains were KY72, KY75, KY102, and KY105. (C) In lanes 1 to 5, the strains were KY72, KY78, KY93, KY96, and KY108, respectively; in lanes 6 and 7, strain KY117 carried plasmid pMO16 expressing Snf1-T210A (TA) or carried the corresponding empty vector pRS316 (V).
The Snf1 activation defect in the sak1Δ/sak1Δ mutant is phenotypically relevant.
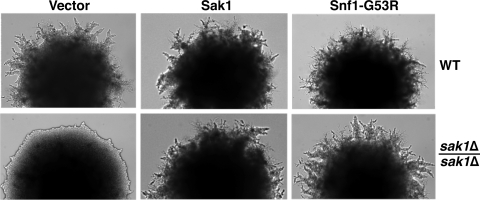
To assess the phenotypic relevance of the observed Thr210 phosphorylation defect in the sak1Δ/sak1Δ mutant, we used Snf1-G53R, a catalytically hyperactive Gly53-to-Arg mutant kinase (12); Snf1-G53R can stimulate pseudohyphal differentiation in the presence of elevated nitrogen levels (43). Expression of this hyperactive kinase restored normal pseudohyphal differentiation in a sak1Δ/sak1Δ mutant, as did expression of Sak1 itself from the same vector (Fig. 4), supporting the notion that the phenotypic defect in the absence of Sak1 results from deficient Snf1 activation.
FIG. 4.
Expression of hyperactive Snf1 restores pseudohyphal differentiation in the sak1Δ/sak1Δ mutant. Transformants of strains KY84 (WT) and KY87 (sak1Δ/sak1Δ) with vector pWS93 (vector) or with plasmids pM36 (Sak1) or pIT517 (Snf1-G53R) were assayed on low-nitrogen SLAD plates. Colonies were photographed after 4 days of growth at 30°C.
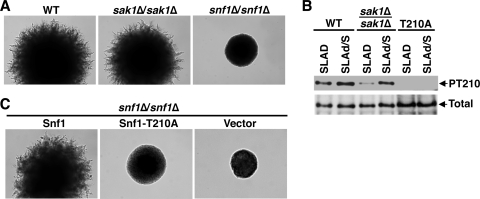
Furthermore, we reasoned that if Sak1 stimulates pseudohyphal differentiation by activating Snf1, then the requirement for Sak1 could be relaxed by engaging the other Snf1-activating kinases. Sak1, Tos3, and Elm1 all contribute to Snf1 activation during glucose limitation (22, 39, 55). We therefore tested whether reducing the glucose content of the low nitrogen medium would restore pseudohyphal differentiation in the absence of Sak1. Assays were performed on a medium identical to SLAD except that 2% glucose was replaced with 0.2% glucose plus 1% sucrose (designated SLAd/S medium); sucrose was added to satisfy the requirements of the Gpr1 sugar sensor pathway (36). Under these conditions, the sak1Δ/sak1Δ mutant exhibited vigorous pseudohyphal differentiation that was only slightly inferior to that of the wild type (Fig. 5A). Assays of Thr210 phosphorylation of Snf1 paralleled these phenotypic observations: upon shift from SLAD to SLAd/S, the level of Thr210 phosphorylation in the sak1Δ/sak1Δ mutant became similar to that observed in the SLAD-grown wild type (Fig. 5B). In control experiments, cells expressing no Snf1 or expressing nonphosphorylatable Snf1-T210A remained nonfilamentous on SLAd/S (Fig. 5A and C), as they are on standard SLAD (28, 43). Thus, these findings further supported Snf1 activation as the limiting step in the sak1Δ/sak1Δ mutant when nitrogen is the only limiting nutrient.
FIG. 5.
Reducing the glucose content of low nitrogen medium restores pseudohyphal differentiation in the absence of Sak1. (A) Strains KY72 (WT), KY75 (sak1Δ/sak1Δ), and KY90 (snf1Δ/snf1Δ) were analyzed for pseudohyphal differentiation on SLAd/S medium. Colonies were photographed after 4 days of growth at 30°C. (B) Cells were grown for approximately two generations in liquid SLAD medium (SLAD) and shifted for 20 min to SLAd/S medium (SLAd/S). The levels of phospho-Thr210-Snf1 (PT210) and total Snf1 protein (Total) were analyzed by immunoblotting. The strains were KY72 (WT), KY75 (sak1Δ/sak1Δ), and KY117 transformed with pMO16 expressing Snf1-T210A (T210A). (C) Transformants of strain KY117 (snf1Δ/snf1Δ) carrying plasmids pM14 (Snf1), pMO16 (Snf1-T210A), or the corresponding empty vector pRS316 (Vector) were analyzed for pseudohyphal differentiation on SLAd/S; colonies were photographed after 4 days of growth at 30°C.
Collectively, these results provided strong evidence that loss of Sak1 affects nitrogen-regulated pseudohyphal differentiation by affecting Snf1 activation.
DISCUSSION
The Snf1-activating kinases Sak1, Tos3, and Elm1 exhibit redundancy for growth on nonpreferred carbon sources and under conditions of alkaline and sodium stress. Here, we have addressed the roles of these kinases in diploid pseudohyphal differentiation in response to nitrogen limitation. We present evidence that loss of Sak1 confers a significant defect, whereas Tos3 and Elm1 are dispensable. Sak1 is also required for normal Thr210 phosphorylation of Snf1 under nitrogen-limiting conditions.
An important point to verify was that loss of Sak1 affects pseudohyphal differentiation primarily by affecting Snf1 activation and not by disrupting another pathway. Eukaryotic Snf1/AMPK-activating kinases can stimulate multiple protein kinase pathways. Mammalian LKB1, the principal upstream kinase for AMPK, phosphorylates and activates thirteen protein kinases (32). Recent studies in yeast showed that in addition to being a Snf1-activating kinase, Elm1 also phosphorylates and activates Gin4 and Hsl1, two Nim1-related, bud neck-associated protein kinases with roles in septin organization and morphogenesis checkpoint control (1, 56). Although Sak1 has not yet been shown to activate any other protein kinases besides Snf1, identification of additional physiologically relevant Sak1 targets may be a matter of time. To address the phenotypic relevance of the Snf1 activation defect in the sak1Δ/sak1Δ mutant, we conducted dominant epistasis tests using a hyperactive Snf1 mutant. A previous attempt to generate a constitutively activated version of Snf1 by replacing Thr210 with an aspartic acid was unsuccessful (12). We therefore used a catalytically hyperactive Snf1-G53R mutant identified by random mutagenesis and function-based screening (12). The exact mechanism by which the Gly53-to-Arg mutation confers catalytic hyperactivity is not well understood. Although the activity of Snf1-G53R still apparently benefits from Thr210 phosphorylation (26), the expression of Snf1-G53R can suppress the growth defect of the triple sak1Δ tos3Δ elm1Δ mutant on nonfermentable carbon sources (our unpublished result), suggesting that the mutation produces a conformational change resembling that induced by Thr210 phosphorylation. We show that expression of Snf1-G53R in a sak1Δ/sak1Δ diploid restores pseudohyphal differentiation to the wild-type level. This and other results provide strong evidence that the phenotypic defect caused by the absence of Sak1 is primarily due to deficient Snf1 activation. However, we cannot exclude that Sak1 at least partly contributes to pseudohyphal differentiation by additional mechanisms.
Because of its relevance to fungal pathogenesis, pseudohyphal differentiation in S. cerevisiae has been extensively studied, revealing the participation of a complex signaling network (reviewed in references 13, 14, 45, 46, 50, 58, and 61). Besides Snf1, this network includes prominent cellular regulators such as RAS, cAMP-dependent protein kinase (PKA), target-of-rapamycin (TOR) protein kinase, and others. RAS functions upstream of the Ste-Kss1 mitogen-activated protein kinase (MAPK) pathway, which positively regulates pseudohyphal differentiation (31, 37). RAS also stimulates PKA, which regulates pseudohyphal differentiation both positively and negatively via its distinct Tpk catalytic isoforms (47, 49). Pseudohyphal differentiation is both positively and negatively controlled by TOR, whose roles are mediated by multiple effectors, including PKA, a PKA-related protein kinase Sch9, and Npr1 protein kinase (9, 35, 36, 38, 52, 59). Critical signals regarding nitrogen and carbon availability also arrive from the Mep2 ammonium permease/sensor and the Gpr1/Gpa2 G protein-coupled receptor system, which affect the PKA pathway (35, 36, 57). Evidence within and outside the filamentous response suggests links of Snf1 to the Ste-MAPK, TOR, and PKA pathways (3, 8, 20, 43).
The molecular mechanisms by which various stresses, including glucose limitation, may regulate the Snf1-activating kinases remain unknown. Our results suggest that Sak1 is involved in nitrogen signaling upstream of Snf1. It will be interesting to determine whether Sak1 represents a direct regulatory substrate for one or more of the many protein kinases involved in controlling pseudohyphal differentiation. This could illuminate the molecular mechanisms of Snf1 regulation not only by nitrogen limitation but also by other stresses.
Acknowledgments
This study was supported by National Science Foundation grant MCB-0818837 (to S.K.) and by a UWM Research Growth Initiative grant (to S.K. and M.O.). L.B. was a UWM Advanced Opportunity Program Fellow.
We thank M. Carlson, S. Kron, and S. Palecek for reagents and D. Saffarini for helpful comments on the manuscript.
Footnotes
Published ahead of print on 30 October 2009.
REFERENCES
- 1.Asano, S., J. E. Park, L. R. Yu, M. Zhou, K. Sakchaisri, C. J. Park, Y. H. Kang, J. Thorner, T. D. Veenstra, and K. S. Lee. 2006. Direct phosphorylation and activation of a Nim1-related kinase Gin4 by Elm1 in budding yeast. J. Biol. Chem. 281:27090-27098. [DOI] [PubMed] [Google Scholar]
- 2.Barral, Y., M. Parra, S. Bidlingmaier, and M. Snyder. 1999. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 13:176-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertram, P. G., J. H. Choi, J. Carvalho, T. F. Chan, W. Ai, and X. F. Zheng. 2002. Convergence of TOR-nitrogen and Snf1-glucose signaling pathways onto Gln3. Mol. Cell. Biol. 22:1246-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blacketer, M. J., C. M. Koehler, S. G. Coats, A. M. Myers, and P. Madaule. 1993. Regulation of dimorphism in Saccharomyces cerevisiae: involvement of the novel protein kinase homolog Elm1p and protein phosphatase 2A. Mol. Cell. Biol. 13:5567-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson, M., B. C. Osmond, and D. Botstein. 1981. Mutants of yeast defective in sucrose utilization. Genetics 98:25-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Celenza, J. L., and M. Carlson. 1986. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science 233:1175-1180. [DOI] [PubMed] [Google Scholar]
- 7.Ciriacy, M. 1977. Isolation and characterization of yeast mutants defective in intermediary carbon metabolism and in carbon catabolite derepression. Mol. Gen. Genet. 154:213-220. [DOI] [PubMed] [Google Scholar]
- 8.Cullen, P. J., and G. F. Sprague, Jr. 2000. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. U. S. A. 97:13619-13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cutler, N. S., X. Pan, J. Heitman, and M. E. Cardenas. 2001. The TOR signal transduction cascade controls cellular differentiation in response to nutrients. Mol. Biol. Cell 12:4103-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubacq, C., A. Chevalier, and C. Mann. 2004. The protein kinase Snf1 is required for tolerance to the ribonucleotide reductase inhibitor hydroxyurea. Mol. Cell. Biol. 24:2560-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edgington, N. P., M. J. Blacketer, T. A. Bierwagen, and A. M. Myers. 1999. Control of Saccharomyces cerevisiae filamentous growth by cyclin-dependent kinase Cdc28. Mol. Cell. Biol. 19:1369-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estruch, F., M. A. Treitel, X. Yang, and M. Carlson. 1992. N-terminal mutations modulate yeast SNF1 protein kinase function. Genetics 132:639-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gagiano, M., F. F. Bauer, and I. S. Pretorius. 2002. The sensing of nutritional status and the relationship to filamentous growth in Saccharomyces cerevisiae. FEMS Yeast Res. 2:433-470. [DOI] [PubMed] [Google Scholar]
- 14.Gancedo, J. M. 2001. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25:107-123. [DOI] [PubMed] [Google Scholar]
- 15.Gimeno, C. J., P. O. Ljungdahl, C. A. Styles, and G. R. Fink. 1992. Unipolar cell divisions in the yeast Saccharomyces cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077-1090. [DOI] [PubMed] [Google Scholar]
- 16.Hardie, D. G. 2007. AMP-activated protein kinase as a drug target. Annu. Rev. Pharmacol. Toxicol. 47:185-210. [DOI] [PubMed] [Google Scholar]
- 17.Hardie, D. G. 2007. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell. Biol. 8:774-785. [DOI] [PubMed] [Google Scholar]
- 18.Hedbacker, K., and M. Carlson. 2008. SNF1/AMPK pathways in yeast. Front. Biosci. 13:2408-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hedbacker, K., S. P. Hong, and M. Carlson. 2004. Pak1 protein kinase regulates activation and nuclear localization of Snf1-Gal83 protein kinase. Mol. Cell. Biol. 24:8255-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hedbacker, K., R. Townley, and M. Carlson. 2004. Cyclic AMP-dependent protein kinase regulates the subcellular localization of Snf1-Sip1 protein kinase. Mol. Cell. Biol. 24:1836-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong, S. P., and M. Carlson. 2007. Regulation of snf1 protein kinase in response to environmental stress. J. Biol. Chem. 282:16838-16845. [DOI] [PubMed] [Google Scholar]
- 22.Hong, S. P., F. C. Leiper, A. Woods, D. Carling, and M. Carlson. 2003. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. U. S. A. 100:8839-8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu, G., P. Y. Cheng, A. Sham, J. R. Perfect, and J. W. Kronstad. 2008. Metabolic adaptation in Cryptococcus neoformans during early murine pulmonary infection. Mol. Microbiol. 69:1456-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iyer, R. S., M. Das, and P. J. Bhat. 2008. Pseudohyphal differentiation defect due to mutations in GPCR and ammonium signaling is suppressed by low glucose concentration: a possible integrated role for carbon and nitrogen limitation. Curr. Genet. 54:71-81. [DOI] [PubMed] [Google Scholar]
- 25.Kim, M. D., S. P. Hong, and M. Carlson. 2005. Role of Tos3, a Snf1 protein kinase kinase, during growth of Saccharomyces cerevisiae on nonfermentable carbon sources. Eukaryot. Cell 4:861-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuchin, S., I. Treich, and M. Carlson. 2000. A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. U. S. A. 97:7916-7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuchin, S., V. K. Vyas, and M. Carlson. 2003. Role of the yeast Snf1 protein kinase in invasive growth. Biochem. Soc. Trans. 31:175-177. [DOI] [PubMed] [Google Scholar]
- 28.Kuchin, S., V. K. Vyas, and M. Carlson. 2002. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol. Cell. Biol. 22:3994-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lambrechts, M. G., F. F. Bauer, J. Marmur, and I. S. Pretorius. 1996. Muc1, a mucin-like protein that is regulated by Mss10, is critical for pseudohyphal differentiation in yeast. Proc. Natl. Acad. Sci. USA 93:8419-8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.La Valle, R., and C. Wittenberg. 2001. A role for the Swe1 checkpoint kinase during filamentous growth of Saccharomyces cerevisiae. Genetics 158:549-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, H., C. A. Styles, and G. R. Fink. 1993. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262:1741-1744. [DOI] [PubMed] [Google Scholar]
- 32.Lizcano, J. M., O. Goransson, R. Toth, M. Deak, N. A. Morrice, J. Boudeau, S. A. Hawley, L. Udd, T. P. Makela, D. G. Hardie, and D. R. Alessi. 2004. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 23:833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lo, W.-S., and A. M. Dranginis. 1996. FLO11, a yeast gene related to the STA genes, encodes a novel cell surface flocculin. J. Bacteriol. 178:7144-7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lo, W. S., and A. M. Dranginis. 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9:161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lorenz, M. C., and J. Heitman. 1998. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 17:1236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lorenz, M. C., X. Pan, T. Harashima, M. E. Cardenas, Y. Xue, J. P. Hirsch, and J. Heitman. 2000. The G protein-coupled receptor Gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics 154:609-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madhani, H. D., C. A. Styles, and G. R. Fink. 1997. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 91:673-684. [DOI] [PubMed] [Google Scholar]
- 38.Martin, D. E., A. Soulard, and M. N. Hall. 2004. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119:969-979. [DOI] [PubMed] [Google Scholar]
- 39.McCartney, R. R., E. M. Rubenstein, and M. C. Schmidt. 2005. Snf1 kinase complexes with different beta subunits display stress-dependent preferences for the three Snf1-activating kinases. Curr. Genet. 47:335-344. [DOI] [PubMed] [Google Scholar]
- 40.McCartney, R. R., and M. C. Schmidt. 2001. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem. 276:36460-36466. [DOI] [PubMed] [Google Scholar]
- 41.Nath, N., R. R. McCartney, and M. C. Schmidt. 2003. Yeast Pak1 kinase associates with and activates Snf1. Mol. Cell. Biol. 23:3909-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orlova, M., L. Barrett, and S. Kuchin. 2008. Detection of endogenous Snf1 and its activation state: application to Saccharomyces and Candida species. Yeast 25:745-754. [DOI] [PubMed] [Google Scholar]
- 43.Orlova, M., E. Kanter, D. Krakovich, and S. Kuchin. 2006. Nitrogen availability and TOR regulate the Snf1 protein kinase in Saccharomyces cerevisiae. Eukaryot. Cell 5:1831-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palecek, S. P., A. S. Parikh, and S. J. Kron. 2000. Genetic analysis reveals that FLO11 upregulation and cell polarization independently regulate invasive growth in Saccharomyces cerevisiae. Genetics 156:1005-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palecek, S. P., A. S. Parikh, and S. J. Kron. 2002. Sensing, signalling and integrating physical processes during Saccharomyces cerevisiae invasive and filamentous growth. Microbiology 148:893-907. [DOI] [PubMed] [Google Scholar]
- 46.Pan, X., T. Harashima, and J. Heitman. 2000. Signal transduction cascades regulating pseudohyphal differentiation of Saccharomyces cerevisiae. Curr. Opin. Microbiol. 3:567-572. [DOI] [PubMed] [Google Scholar]
- 47.Pan, X., and J. Heitman. 1999. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4874-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Portillo, F., J. M. Mulet, and R. Serrano. 2005. A role for the non-phosphorylated form of yeast Snf1: tolerance to toxic cations and activation of potassium transport. FEBS Lett. 579:512-516. [DOI] [PubMed] [Google Scholar]
- 49.Robertson, L. S., and G. R. Fink. 1998. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc. Natl. Acad. Sci. USA 95:13783-13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rohde, J. R., and M. E. Cardenas. 2004. Nutrient signaling through TOR kinases controls gene expression and cellular differentiation in fungi. Curr. Top. Microbiol. Immunol. 279:53-72. [DOI] [PubMed] [Google Scholar]
- 51.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 52.Schmidt, A., T. Beck, A. Koller, J. Kunz, and M. N. Hall. 1998. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 17:6924-6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song, W., and M. Carlson. 1998. Srb/Mediator proteins interact functionally and physically with transcriptional repressor Sfl1. EMBO J. 17:5757-5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sutherland, C. M., S. A. Hawley, R. R. McCartney, A. Leech, M. J. Stark, M. C. Schmidt, and D. G. Hardie. 2003. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr. Biol. 13:1299-1305. [DOI] [PubMed] [Google Scholar]
- 56.Szkotnicki, L., J. M. Crutchley, T. R. Zyla, E. S. Bardes, and D. J. Lew. 2008. The checkpoint kinase Hsl1p is activated by Elm1p-dependent phosphorylation. Mol. Biol. Cell 19:4675-4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tamaki, H., T. Miwa, M. Shinozaki, M. Saito, C. W. Yun, K. Yamamoto, and H. Kumagai. 2000. GPR1 regulates filamentous growth through FLO11 in yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 267:164-168. [DOI] [PubMed] [Google Scholar]
- 58.Truckses, D. M., L. S. Garrenton, and J. Thorner. 2004. Jekyll and Hyde in the microbial world. Science 306:1509-1511. [DOI] [PubMed] [Google Scholar]
- 59.Urban, J., A. Soulard, A. Huber, S. Lippman, D. Mukhopadhyay, O. Deloche, V. Wanke, D. Anrather, G. Ammerer, H. Riezman, J. R. Broach, C. De Virgilio, M. N. Hall, and R. Loewith. 2007. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 26:663-674. [DOI] [PubMed] [Google Scholar]
- 60.Van de Velde, S., and J. M. Thevelein. 2008. Cyclic AMP-protein kinase A and Snf1 signaling mechanisms underlie the superior potency of sucrose for induction of filamentation in Saccharomyces cerevisiae. Eukaryot. Cell 7:286-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verstrepen, K. J., and F. M. Klis. 2006. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 60:5-15. [DOI] [PubMed] [Google Scholar]
- 62.Wilson, W. A., S. A. Hawley, and D. G. Hardie. 1996. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr. Biol. 6:1426-1434. [DOI] [PubMed] [Google Scholar]
- 63.Xu, Z., Y. B. Cao, J. D. Zhang, Y. Y. Cao, P. H. Gao, D. J. Wang, X. P. Fu, K. Ying, W. S. Chen, and Y. Y. Jiang. 2005. cDNA array analysis of the differential expression change in virulence-related genes during the development of resistance in Candida albicans. Acta Biochim. Biophys. Sin. 37:463-472. [DOI] [PubMed] [Google Scholar]
- 64.Yi, M., J. H. Park, J. H. Ahn, and Y. H. Lee. 2008. MoSNF1 regulates sporulation and pathogenicity in the rice blast fungus Magnaporthe oryzae. Fungal Genet. Biol. 45:1172-1181. [DOI] [PubMed] [Google Scholar]
- 65.Zimmermann, F. K., I. Kaufmann, H. Rasenberger, and P. Haubetamann. 1977. Genetics of carbon catabolite repression in Saccharomyces cerevisiae: genes involved in the derepression process. Mol. Gen. Genet. 151:95-103. [DOI] [PubMed] [Google Scholar]