Abstract
Tetrahydromonapterin is a major pterin in Escherichia coli and is hypothesized to be the cofactor for phenylalanine hydroxylase (PhhA) in Pseudomonas aeruginosa, but neither its biosynthetic origin nor its cofactor role has been clearly demonstrated. A comparative genomics analysis implicated the enigmatic folX and folM genes in tetrahydromonapterin synthesis via their phyletic distribution and chromosomal clustering patterns. folX encodes dihydroneopterin triphosphate epimerase, which interconverts dihydroneopterin triphosphate and dihydromonapterin triphosphate. folM encodes an unusual short-chain dehydrogenase/reductase known to have dihydrofolate and dihydrobiopterin reductase activity. The roles of FolX and FolM were tested experimentally first in E. coli, which lacks PhhA and in which the expression of P. aeruginosa PhhA plus the recycling enzyme pterin 4a-carbinolamine dehydratase, PhhB, rescues tyrosine auxotrophy. This rescue was abrogated by deleting folX or folM and restored by expressing the deleted gene from a plasmid. The folX deletion selectively eliminated tetrahydromonapterin production, which far exceeded folate production. Purified FolM showed high, NADPH-dependent dihydromonapterin reductase activity. These results were substantiated in P. aeruginosa by deleting tyrA (making PhhA the sole source of tyrosine) and folX. The ΔtyrA strain was, as expected, prototrophic for tyrosine, whereas the ΔtyrA ΔfolX strain was auxotrophic. As in E. coli, the folX deletant lacked tetrahydromonapterin. Collectively, these data establish that tetrahydromonapterin formation requires both FolX and FolM, that tetrahydromonapterin is the physiological cofactor for PhhA, and that tetrahydromonapterin can outrank folate as an end product of pterin biosynthesis.
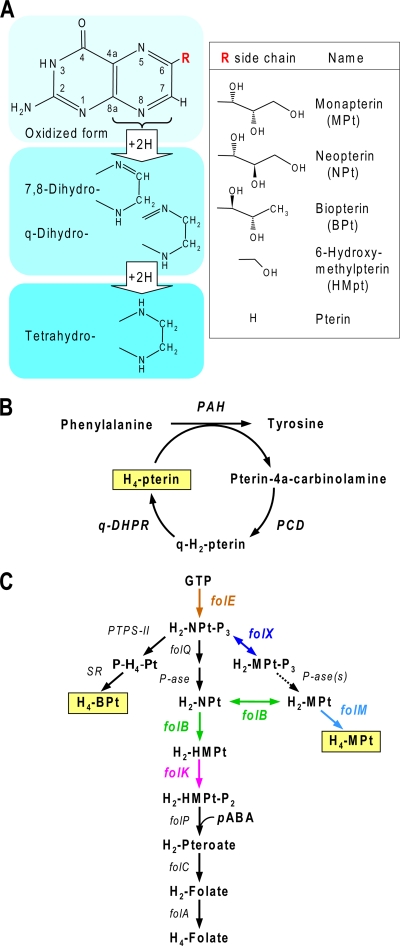
Pterins contain the bicyclic pteridine ring with an amino group in the 2-position and an oxo group in the 4-position; they can be reduced through the dihydro forms to the tetrahydro forms, which are active as cofactors (Fig. 1A). Tetrahydropterins are known to be the cofactors for phenylalanine hydroxylases from Pseudomonas and Chromatium species as well as for mammalian aromatic amino acid hydroxylases and other mammalian enzymes (13, 17, 38, 41) (Fig. 1B). Although the identity of the mammalian tetrahydropterin cofactor, tetrahydrobiopterin (H4-BPt), is firmly established (38), the same is not true for bacteria, and the biosynthesis of bacterial tetrahydropterins is not well understood.
FIG. 1.
Tetrahydropterin structure, cofactor role, and biosynthesis. (A) The pterin nucleus, its levels of reduction, and the structures of compounds relevant to this study. (B) The requirement for a tetrahydropterin (H4-pterin) cofactor for phenylalanine hydroxylase (PAH) and the cofactor regeneration cycle involving pterin-4a-carbinolamine dehydratase (PCD) and quinonoid dihydropterin (q-H2-pterin) reductase (q-DHPR; EC 1.5.1.34). (C) The established steps in tetrahydrobiopterin (H4-BPt) biosynthesis and possible routes for tetrahydromonapterin (H4-MPt) biosynthesis in relation to the folate pathway. H4-BPt is formed by the sequential action of 6-pyruvoyltetrahydropterin (P-H4-Pt) synthase (PTPS-II) and sepiapterin reductase (SR). H4-MPt could originate via the FolX-catalyzed epimerization of dihydroneopterin triphosphate (H2-NPt-P3) to dihydromonapterin triphosphate (H2-MPt-P3), followed by dephosphorylation to dihydromonapterin (H2-MPt) and reduction by a dihydropterin reductase (EC 1.5.1.33), putatively FolM. H4-MPt also could come from the FolB-mediated epimerization of dihydroneopterin (H2-NPt) followed by reduction. FolB also mediates the side chain cleavage of H2-NPt or H2-MPt to give 6-hydroxymethyldihydropterin (H2-HMPt); the H2-MPt cleavage is omitted for simplicity. Other abbreviations: P-ase, phosphatase; H2-HMPt-P2, 6-hydroxymethyldihydropterin diphosphate; pABA, p-aminobenzoate; H2-pteroate, dihydropteroate; H2-folate, dihydrofolate; H4-folate, tetrahydrofolate.
While a few bacterial taxa, such as Cyanobacteria and Chlorobia, produce H4-BPt, most do not, as judged directly from pterin analysis and indirectly from the rarity of H4-BPt biosynthesis genes 6-pyruvoyltetrahydropterin synthase II (PTPS-II) and sepiapterin reductase (SR) (Fig. 1C) among sequenced genomes (12, 25). As bacteria lacking H4-BPt include Pseudomonas and many others with phenylalanine hydroxylase genes, it is clear that bacterial phenylalanine hydroxylases generally must use a cofactor other than H4-BPt. The most prominent candidate is tetrahydromonapterin (H4-MPt), which occurs in Escherichia coli (21) and almost certainly also in Pseudomonas species (11, 17). H4-MPt could be derived from the dihydropterin intermediates of folate biosynthesis via two different routes (Fig. 1C). These are (i) the conversion of dihydroneopterin triphosphate (H2-NPt-P3) to dihydromonapterin triphosphate (H2-MPt-P3) by H2-NPt-P3 epimerase (FolX) followed by dephosphorylation and reduction to the tetrahydro level, and (ii) the conversion of dihydroneopterin (H2-NPt) to dihydromonapterin (H2-MPt) by the epimerase action of dihydroneopterin aldolase (FolB) and then reduction. FolB is a fairly well-understood enzyme of folate synthesis (9), but FolX has no known biological role and a folX deletant has no obvious phenotype (19). folX genes apparently are confined to Gammaproteobacteria (9).
Although the epimerase activities of FolX and FolB have been demonstrated amply in vitro (1, 5, 19), no genetic evidence links either enzyme to H4-MPt formation in vivo. The situation with the reduction of H2-MPt to H4-MPt is even less clear, because this activity has not been investigated experimentally. A candidate enzyme for this step nevertheless can be proposed on bioinformatic grounds: the somewhat mysterious FolM protein (9). FolM belongs to a subset of the short-chain dehydrogenase/reductase (SDR) family having the characteristic motif TGX3RXG (in place of TGX3GXG, which typifies other SDRs). The archetype of this subset is Leishmania pteridine reductase 1 (PTR1), which reduces various dihydropterins to the tetrahydro state (15). E. coli FolM has low dihydrofolate (H2-folate) and dihydrobiopterin (H2-BPt) reductase activities in vitro (14), but neither of these is likely to be its physiological function, since H2-folate reduction normally is mediated by FolA and E. coli lacks H4-BPt. folM genes occur in many Gram-negative organisms, including Chlamdiae, Chloroflexi, Cyanobacteria, Acidobacteria, Planctomycetes, Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and Deltaproteobacteria (9).
We report here comparative genomic and genetic evidence that FolX and FolM are required for H4-MPt synthesis in E. coli and P. aeruginosa, the bacteria in which H4-MPt has been most studied, and biochemical evidence that FolM has high H2-MPt reductase activity. We also point out gaps in the understanding of pterin metabolism that our data bring sharply into focus.
MATERIALS AND METHODS
Bioinformatics.
Bacterial genomes were analyzed using the SEED database and its tools (31). Full results of the analysis are available at the public SEED server (theseed.uchicago.edu) in the subsystem pterin metabolism 3.
Pterins.
7,8-Dihydromonapterin, 7,8-dihydroneopterin, 6-hydroxymethyl-7,8-dihydropterin hydrochloride, l-monapterin, 5,6,7,8-tetrahydromonapterin, and 7,8-dihydrofolate were obtained from Schircks Laboratories (www.schircks.com). Near-saturated solutions were freshly prepared in N2-sparged potassium phosphate (10 mM, pH 7.5), excluding light, and titers were determined spectrophotometrically using published extinction coefficients (4, 32). Quinonoid dihydromonapterin was prepared in situ in enzyme assays using H2O2 and horseradish peroxidase (26). Assay mixtures (100 μl) contained 100 mM potassium phosphate (pH 6.0), 1 μg of horseradish peroxidase, 0.5 mM H2O2, 50 μM H4-MPt, 100 μM NADPH, and 5 μg of purified FolM.
Bacterial strains, plasmids, and media.
E. coli K-12 MG1655 and P. aeruginosa PAO1 were grown at 37°C on Luria-Bertani medium (LB) or minimal medium containing M9 salts (35), 0.1 mM CaCl2, 0.5 mM MgSO4, and 0.4% (wt/vol) glucose as the carbon source unless otherwise noted. Media were solidified with 15 g of agar/liter. Kanamycin (50 μg/ml), ampicillin (100 μg/ml), spectinomycin (100 μg/ml), thiamine (17 μg/ml), l-phenylalanine (50 μg/ml), l-tyrosine (54 μg/ml), l-tryptophan (50 μg/ml), isopropyl-β-d-thiogalactopyranoside (IPTG; 1 mM), p-aminobenzoic acid (10 μM), 4-hydroxybenzoate (10 μM), and 2,3-dihydroxybenzoate (10 μM) were added as required. Plasmids used in this study are listed in Table S1 in the supplemental material.
E. coli deletions and functional complementation tests.
E. coli strains MG1655 folX::Kn, MG1655 folM::Kn, JP2255 folX::Kn, and JP2255 folM::Kn were created by the P1 transduction of the respective mutant allele from the Keio collection into strain MG1655 or JP2255 (2). Where necessary, the kanamycin cassette was recombined out using pCP20 (8). Deletions were confirmed by PCR and sequencing (see Table S2 in the supplemental material). The plasmids pJS11 (37), which carries P. aeruginosa phhA, and pBluescript, carrying P. aeruginosa phhB (27), were transformed simultaneously by electroporation into strains JP2255, JP2255 folM, and JP2255 folX. Complementation was assessed by plating transformants onto minimal medium with 0.4% (vol/vol) glycerol as the carbon source, plus appropriate antibiotics, thiamine, and l-phenylalanine, with or without l-tyrosine. Plates were incubated at 37°C for 2 to 3 days. To restore the phenotype by complementation, folM and folX were PCR amplified and cloned into pEXT21 (10). These plasmids and the empty vector then were transformed by electroporation into their respective host strains expressing phhA and phhB and were plated on LB containing ampicillin, kanamycin, and spectinomycin. Transformants then were plated on minimal medium as described above.
P. aeruginosa deletions and functional complementation tests.
Genes were deleted in P. aeruginosa using pEX18Gm and pEX18Tet (20), both of which contain the sacB gene for the negative selection of the suicide marker. Approximately 1-kbp regions flanking each target gene were amplified by PCR. To delete folX, the gent gene from pEX18Gm was amplified by PCR. In a three-way ligation, the resistance gene was cloned between the flanking regions into pEX18Tet. In a similar manner, to generate the deletion construct for tyrA, the tet gene from pEX18Tet was amplified and cloned between the flanking regions into pEX18Gm. The deletion constructs were transformed by electroporation as described previously (7), and after being plated on LB containing the appropriate antibiotic, resistant colonies capable of growth on LB containing 6% (wt/vol) sucrose were identified and verified by PCR. For deletions in tyrA, the following also were added to media: p-aminobenzoic acid, 4-hydroxybenzoate, 2,3-dihydroxybenzoate, l-tryptophan, l-phenylalanine, and l-tyrosine at the concentrations specified above. Deletants were confirmed by PCR and sequencing (see Table S2 in the supplemental material). To negate the potentially lethal polar effects of deleting tyrA on the immediately downstream gene cmk (encoding cytidylate kinase), cmk was cloned by PCR into pHERD20T (33) and transformed into the ΔtyrA and ΔfolX ΔtyrA strains. PhhA function was assessed by being streaked on minimal medium plates (as described above) containing appropriate antibiotics and p-aminobenzoic acid, 4-hydroxybenzoate, 2,3-dihydroxybenzoate, and l-tryptophan, with or without l-phenylalanine and l-tyrosine. The plates were incubated at 37°C for 2 to 3 days.
Pterin and folate analysis.
Wild-type and ΔfolX E. coli strains were grown at 37°C in 50 ml of minimal medium until the A600 reached 1.0 ± 0.1. Cells were harvested by centrifugation (6,000 × g, 20 min, 4°C) and washed three times with cold 25 mM potassium phosphate, pH 7.5 (wash buffer). Washed pellets and the corresponding culture medium were stored at −80°C until use. For pterin analysis, pellets were resuspended in 1 ml of wash buffer and broken in a Mini-Bead-Beater using 0.1-mm zirconia-silica beads. The extract was removed, and the beads were washed twice with 0.75 ml of wash buffer. The extract and washes were pooled and centrifuged (10,000 × g, 10 min, 4°C). Aliquots (100 μl) of supernatants (intracellular fraction) and culture media (extracellular fraction) were oxidized for 1 h in the dark at 4°C by adding 10 μl of 1% (wt/vol) I2-2% (wt/vol) KI in 0.1 or 1 M HCl, after which the excess iodine was oxidized by adding 10 μl of 10% (wt/vol) sodium ascorbate. Pterins (50- or 100-μl injections) were separated on a 4-μm, 250- by 4.6-mm Synergi Fusion-RP 80 column (Phenomenex) that was eluted isocratically with 10 mM sodium phosphate (pH 6.0) at 1.5 ml/min. Peaks were detected by fluorescence (350 nm excitation, 450 nm emission) and identified relative to standards. Pterins in P. aeruginosa wild-type and ΔfolX strains were analyzed the same way, except that the cells were harvested when the A600 reached 0.6 ± 0.1. For folate analysis, cell pellets were suspended in 50 mM HEPES-CHES [2-(N-cyclohexylamino)ethanesulfonic acid] (pH 7.85, 10 ml final volume) containing 2% (wt/vol) ascorbic acid and 10 mM β-mercaptoethanol (extraction buffer), sonicated, boiled for 10 min, and centrifuged (13,000 × g, 10 min). The samples were reextracted the same way, and the combined extracts were treated for 2 h at 37°C with 2 ml of dialyzed rat plasma to deglutamylate folates to the monoglutamate level. Samples then were boiled for 10 min, centrifuged as described above, and filtered. Culture medium samples (4 ml) were processed similarly after being mixed with 6 ml of ×1.67 extraction buffer. Folates were isolated using 2-ml folate affinity columns (16). Samples of the eluate were analyzed by high-performance liquid chromatography (HPLC) with electrochemical detection (3). The detector response was calibrated with folate standards from Merck Eprova (www.merckeprova.com).
Expression and purification of recombinant FolM.
E. coli BL21-CodonPlus (DE3)-RIPL cells (Stratagene) were transformed with pFolM (14), which encodes E. coli FolM with an N-terminal His tag. Cells were grown at 37°C in LB containing appropriate antibiotics. When the A600 reached 0.6, IPTG was added to a final concentration of 1 mM and growth was continued for 4 h. Subsequent steps were carried out at 4°C. Cells from a 200-ml culture were harvested by centrifugation, resuspended in 50 mM sodium phosphate (pH 8.0), 300 mM NaCl, 10% (vol/vol) glycerol, 10 mM imidazole, and disrupted by sonication. The cleared supernatant was loaded onto a 0.35-ml Ni2+-nitrilotriacetic acid agarose resin column (Qiagen) equilibrated with the sonication buffer. After the column was washed with 16 ml of 50 mM sodium phosphate, 300 mM NaCl, 10% (vol/vol) glycerol, 20 mM imidazole (pH 8.0), the recombinant protein was eluted with 3 ml of the same buffer containing 250 mM imidazole and was desalted on PD-10 columns equilibrated in 50 mM sodium phosphate (pH 8.0), 100 mM NaCl, 10% (vol/vol) glycerol. The purified protein was stable to freezing in liquid N2 and storage at −80°C. The protein concentration was estimated by the Bradford method (6) using bovine serum albumin as a standard.
Biochemical analysis of FolM.
FolM activity was measured spectrophotometrically. Assays (100 μl) were run at 22 to 23°C in 100 mM potassium phosphate (pH 6.0) and contained 100 μM NADPH, purified enzyme (2 to 20 μg), a pterin, and (except when quinonoid dihydromonapterin was the substrate) 10 mM β-mercaptoethanol. All components except NADPH were incubated together for 30 s before starting the reaction by adding NADPH. Reductase activities using various substrates (50 μM) were measured by continuous assay, obtaining initial velocities from the rate of decrease of absorbance at 340 nm. Blank rates measured in the absence of enzyme were used to correct for the nonenzymatic breakdown of the substrate and the oxidation of NADPH. As pterins exhibit absorbance changes when reduced, the following molar extinction coefficients (ɛ340, M−1 cm−1; from the literature or determined empirically) were used for the coupled oxidation/reduction of NADPH-pterin at pH 6.0: H2-MPt, 6,028; H2-folate, 12,300 (14); and H2-HMPt, 10,295. The Km and Vmax values for H2-MPt were measured in a discontinuous assay because of the high absorbance of H2-MPt at 340 nm. Reactions were stopped after 30 s by adding 400 μl of 0.5 M Tris-HCl (pH 8.0) (FolM is inactive at this pH), and rates were determined as described above (ɛ340 for NADPH/H2-MPt at pH 8.0 is 11,040 M−1 cm−1). Data were fitted to a hyperbolic curve with the MicroCal Origin 3.0 software (Microcal Software, Inc., Northampton, MA).
RESULTS
Genomic context connects folX and folM with tetrahydropterin production.
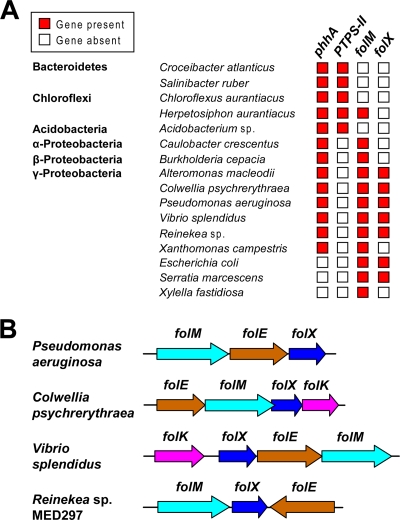
The analysis of 724 bacterial genomes using the SEED database and its tools (31) revealed 184 that encode phenylalanine hydroxylase (PhhA), only 19 of which also encode the characteristic H4-BPt synthesis enzyme PTPS-II (25). Of the remaining 165 genomes with PhhA, the majority (108 genomes) encode FolM and, in Gammaproteobacteria, frequently also FolX. Conversely, genomes with PTPS-II very rarely encode FolM and never encode FolX. Thus, there is a certain reciprocity between the phyletic distribution of PTPS-II and that of FolM and FolX (Fig. 2A), suggesting that the latter produces an alternative to H4-BPt. Gene-clustering evidence (Fig. 2B) supports this possibility, for it shows that folX and folM often are associated with each other and with folE (the first gene of pterin synthesis), and sometimes also with folK (encoding hydroxymethyldihydropterin pyrophosphokinase, which converts H2-HMPt to its pyrophosphate) (Fig. 1C). The folM-folE association was noted previously (9). Taken together, the comparative genomic data suggest that FolX and FolM have related functions in tetrahydropterin biosynthesis. A simple hypothesis is that they mediate the epimerization and reduction steps in H4-MPt synthesis (Fig. 1C).
FIG. 2.
Comparative genomic evidence implicating folX and folM in tetrahydromonapterin synthesis. (A) Phyletic distribution of phenylalanine hydroxylase (phhA), PTPS-II, folM, and folX genes among representative genomes. Note the anticorrelation between PTPS-II and folM or folX. Note also that folM sometimes is unaccompanied by folX, and that folM and folX sometimes occur without phhA. (B) Clustering of folX and folM with each other and with folE in diverse Gammaproteobacteria. In two genomes these genes also cluster with folK. Genes are colored to match Fig. 1C. Arrows indicate the transcriptional direction; overlaps between arrows indicate translational coupling.
Deleting E. coli folX selectively eliminates monapterin production and secretion.
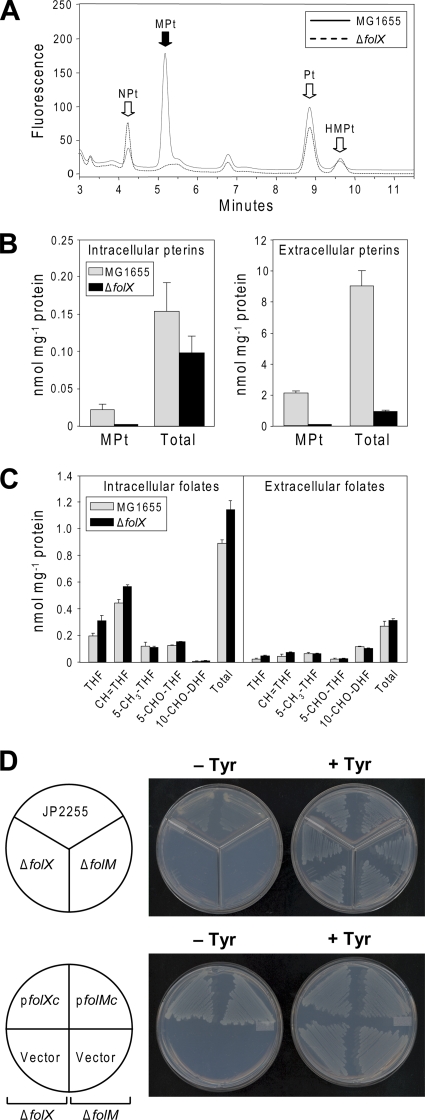
The hypothesis described above was tested first in E. coli, because E. coli has H4-MPt as a prominent intracellular pterin (21) and secretes large amounts of monapterin in some form (40). The reduction state of the secreted form is uncertain, because H4-MPt and H2-MPt can spontaneously oxidize to MPt and are expected to do so once released to the medium (29, 34). A folX deletant was constructed, and its intra- and extracellular pterin and folate profiles were compared to those of the parent wild-type strain. Pterins were analyzed by fluorometric HPLC after acidic iodine oxidation to their fluorescent aromatic forms. The ΔfolX strain showed the selective loss of the MPt peak from the intracellular profile (Fig. 3A) and the complete loss of extracellular MPt and most other extracellular pterins (which appear to be MPt breakdown products) (Fig. 3B). The intra- and extracellular folate profiles of the ΔfolX strain were, however, essentially identical to those of the wild type (Fig. 3C). These results establish that FolX is the only source of MPt in E. coli, and it has no role in folate synthesis. The combined intra- plus extracellular pterin production of wild-type cells was 9.1 nmol/mg protein; it was 1 nmol/mg protein in the deletant. Since folates contain a pterin moiety, both values should be expanded by 1.3 nmol/mg protein (the approximate sum of intra- plus extracellular folate production) to obtain total pterin production. Total pterin values thus are 10.4 and 2.3 nmol/mg protein for the wild type and deletant, respectively. The difference between these values (8.1 nmol/mg protein) provides an estimate of the normal FolX-mediated flux to MPt. This flux is sixfold larger than the flux to folates (1.3 nmol/mg protein). It is noteworthy that 99% of the MPt produced by wild-type E. coli cells was extracellular, as was 98% of total pterin (Fig. 3B).
FIG. 3.
Genetic evidence implicating folX and folM in tetrahydromonapterin synthesis in E. coli and tetrahydromonapterin as the cofactor for P. aeruginosa PhhA. (A) Fluorometric HPLC profile of pterins extracted from wild-type (MG1655) and ΔfolX E. coli strains grown in liquid M9 medium plus 0.4% glucose to an A600 of 1.5. Samples were oxidized before analysis to convert di- and tetrahydropterins to their fluorescent aromatic forms. Note the selective loss of monapterin (MPt) from the folX deletant. NPt, neopterin; Pt, pterin; HMPt, 6-hydroxymethylpterin. (B) Quantitation of intra- and extracellular pterins (as MPt equivalents per mg cellular protein) in MG1655 and ΔfolX cultures grown to an A600 of 1.0 in the medium described for panel A. Data are means and standard errors from three replicates. Besides MPt, the most prominent extracellular species was pterin, a breakdown product of MPt (34). (C) Quantitation of intra- and extracellular folates (per mg cellular protein) from MG1655 and ΔfolX strains grown as described for panel B. Data are means and standard errors from three replicates. THF, tetrahydrofolate; CH = THF, 5,10-methenyltetrahydrofolate; 5-CH3-THF, 5-methyltetrahydrofolate; 5-CHO-THF, 5-formyltetrahydrofolate; 10-CHO-DHF, 10-formydihydrofolate. Note that the analytical procedures convert 5,10-methylenetetrahydrofolate to THF and 10-formyltetrahydrofolate to CH = THF. 10-CHO-DHF is an oxidation product of 10-formyltetrahydrofolate. (D) The upper frames show the abrogation of tyrosine prototrophy by deleting folX or folM from the tyrA strain JP2255 harboring plasmids carrying P. aeruginosa phhA and phhB. Duplicate strains were tested. The lower frames show the restoration of tyrosine prototrophy to folX or folM deletants in the system described in the upper frames by introducing plasmid-borne folX (pfolXc) or folM (pfolMc), respectively. Controls with the plasmid alone (Vector) were included. Plates contained M9 minimal medium supplemented with 0.4% glycerol, 17 μg/ml thiamine, 50 μg/ml l-phenylalanine, 1 mM IPTG, and appropriate antibiotics, with or without 54 μg/ml l-tyrosine.
folX and folM are essential for P. aeruginosa phenylalanine hydroxylase function in E. coli.
To further explore the roles of folX and folM in H4-MPt formation and function, we turned to a convenient heterologous expression system in which P. aeruginosa PhhA is expressed in E. coli together with the pterin recycling enzyme pterin-4a-carbinolamine dehydratase (PhhB) (27, 37, 41). As E. coli has neither PhhA nor PhhB, their expression confers the ability to form tyrosine from phenylalanine, thereby allowing the rescue of the tyrosine auxotrophy caused by a tyrA mutation. This rescue was abrogated by deleting either folX or folM and restored by expressing the deleted gene from a plasmid (Fig. 3D). Because the ΔfolX strain lacks MPt and only MPt (Fig. 3A), these results establish that H4-MPt is the physiological cofactor for PhhA, at least in E. coli. They also show that FolM is required for PhhA function, most probably to reduce H2-MPt to H4-MPt. An alternative possibility, that FolM is the quinonoid dihydropterin reductase that recycles oxidized H4-MPt (Fig. 1B), is unlikely, because this activity is reported to reside in the E. coli NfnB protein (39) and was not detected in recombinant FolM (see below).
Recombinant FolM has high H2-MPt reductase activity.
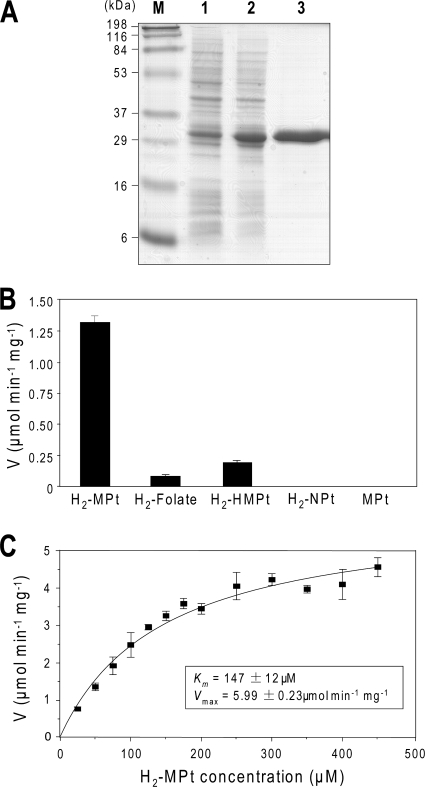
To corroborate the inference that FolM is the source of H2-MPt reductase activity, recombinant FolM was purified (Fig. 4A) and assayed using H2-MPt or other pterins (50 μM) as substrates and NADPH as a cofactor (Fig. 4B). Assays were made at pH 6.0 because we found that FolM is most active at acidic pH, as previously reported (14). FolM activity with H2-MPt was 16-fold higher than that with H2-folate, previously the best known substrate (14). FolM also showed some activity with the folate pathway intermediate 6-hydroxymethyldihydropterin (H2-HMPt) but not with dihydroneopterin (H2-NPt), the epimer of H2-MPt. Monapterin was not a substrate, and neither, as far as could be judged, was the quinonoid form of dihydromonapterin. This form (prepared in situ by oxidizing H4-MPt) rearranges very fast to H2-MPt (23), and no activity beyond that attributable to H2-MPt thus formed was detected. NADH could not replace NADPH as the cofactor. The kinetic characterization of the H2-MPt reductase activity (Fig. 4C) indicated a higher Vmax than that reported for H2-folate (5.99 versus 0.083 μmol min−1 mg−1) and also a higher Km (147 versus 9.5 μM) (14).
FIG. 4.
Purification and characterization of E. coli FolM. (A) SDS-polyacrylamide gel electrophoresis of extracts of cells (∼17 μg protein) harboring pFolM before and after 4 h IPTG induction (lanes 1 and 2, respectively) and 10 μg of Ni2+ affinity-purified FolM protein (lane 3). Staining was done with Coomassie blue. The positions of molecular markers (M; in kDa) are indicated. (B) Reductase activities measured at pH 6.0 using various substrates (50 μM) and 100 μM NADPH. Values are means and standard errors from three replicates. Abbreviations are defined in the legend to Fig. 1. (C) Dihydromonapterin reductase activity as a function of dihydromonapterin concentration. Assays were made at pH 6.0 and contained 100 μM NADPH. The Km and Vmax values estimated by hyperbolic curve fitting are shown in the inset.
FolX is essential for PhhA function in P. aeruginosa.
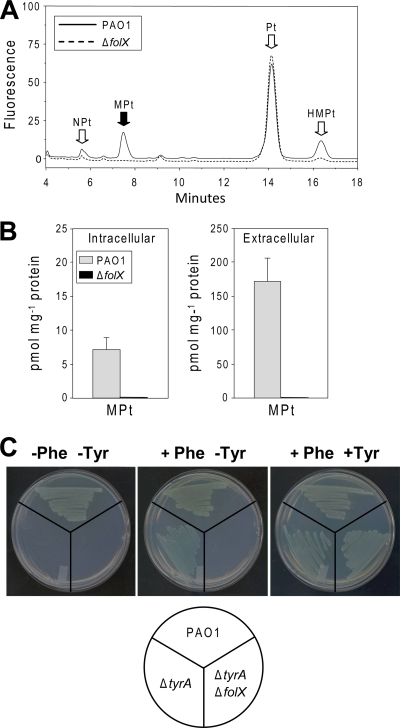
To extend the findings described above to P. aeruginosa, we first constructed a ΔfolX strain and demonstrated that the deletion abolished intra- and extracellular MPt production (Fig. 5A and B). As in E. coli, >95% of the MPt made by wild-type cells was extracellular (Fig. 5B). A tyrA deletant then was constructed as well as a ΔtyrA ΔfolX strain. The ΔtyrA strain was, as expected, prototrophic for tyrosine, since PhhA allows tyrosine formation from phenylalanine (Fig. 4C). The ΔtyrA ΔfolX strain was auxotrophic for tyrosine (Fig. 4C), showing that folX is required for PhhA function in situ as well as when expressed heterologously in E. coli.
FIG. 5.
Genetic evidence implicating folX in tetrahydromonapterin synthesis in P. aeruginosa and supporting the tetrahydromonapterin requirement of P. aeruginosa PhhA in situ. (A) Fluorometric HPLC analysis of pterins extracted from wild-type (PAO1) and ΔfolX strains grown to an A600 of 0.6 in liquid M9 medium supplemented with 0.4% glucose. Note the loss of the monapterin peak in the ΔfolX strain. Abbreviations are the same as those in the legend to Fig. 3A. (B) Quantitation of intra- and extracellular monapterin (per mg cellular protein) in wild-type and ΔfolX strains grown to an A600 of 0.6 in the medium described for panel A. Data are means and standard errors from three replicates. (C) Abolition of the tyrosine prototrophy of a ΔtyrA strain by the deletion of folX. Wild-type P. aeruginosa was included as a control. Plates contained M9 medium supplemented with 0.4% glucose, 0.1% arabinose, 50 μg/ml l-tryptophan, 10 μM p-aminobenzoate, 10 μM 4-hydroxybenzoate, and 10 μM 2,3-dihydroxybenzoate, with or without 50 μg/ml l-phenylalanine and 54 μg/ml l-tyrosine. Note that the ΔtyrA strain requires l-phenylalanine as well as l-tryptophan and p-aminobenzoate, because P. aeruginosa TyrA is a bifunctional cyclohexadienyl dehydrogenase/5-enolpyruvylshikimate-3-phosphate synthase whose ablation eliminates the synthesis of chorismate as well as that of tyrosine.
DISCUSSION
Our data confirm the hypothesis (1) that FolX is the sole source of MPt in E. coli, and they show that this is the case in P. aeruginosa as well. They also establish a physiological function for FolM as an H2-MPt reductase, again confirming an earlier hypothesis (9). In this connection, the lack of activity against H2-NPt is significant, because, as an intermediate of folate synthesis (Fig. 1C), this pterin must remain in the dihydro form. Although the Km for H2-MPt is higher than that for H2-folate, the latter would not be expected to displace H2-MPt in vivo, because H2-folate concentrations are kept extremely low by FolA, whose Km for H2-folate is close to 1 μM (30, 36).
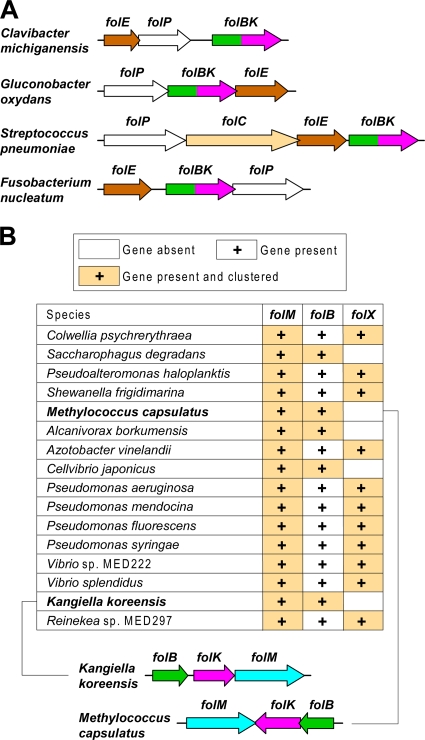
Our results also raise three questions that reveal gaps in the present understanding of bacterial pterin metabolism. First, since FolB has high H2-NPt epimerase activity in vitro (16% of its H2-NPt→H2-HMPt cleavage activity) (19), why can FolB not give rise to H2-MPt when FolX is ablated? The answer may lie in the occurrence in diverse bacteria of genes encoding fusions of FolB with FolK, the next enzyme in the folate synthesis pathway (Fig. 6A). Such a fusion implies the possibility of the metabolic channeling of H2-NPt straight through to H2-HMPt diphosphate (Fig. 1C), with any H2-MPt formed via the epimerase activity of FolB being inaccessible to other enzymes. It therefore is conceivable that FolB-FolK fusions are covalently linked versions of a noncovalent FolB-FolK complex in which channeling also occurs. A related possibility is that the FolB-FolK complex lacks epimerase activity.
FIG. 6.
Comparative genomic evidence bearing on possible metabolic channeling between FolB and FolK and on the possible substrate for FolM in bacteria without FolX. (A) The widespread occurrence in folate synthesis gene clusters of folB-folK fusion genes. The species shown represent four diverse phyla (Actinobacteria, Alphaproteobacteria, Firmicutes, and Fusobacteria). (B) Clustering patterns of folM genes in Gammaproteobacteria with or without folX genes. Note that folM-folB clusters occur only if folX is absent. The folM-folB clusters also include folK, as illustrated by two representative examples (Kangiella koreensis and Methylococcus capsulatus).
The second question is about the role of FolM in bacteria whose genomes lack a folX gene. Such bacteria are numerous, and some of them (e.g., Burkholderia and Xanthomonas) contain a phhA gene and, hence, almost surely produce a tetrahydropterin (Fig. 2A). Perhaps in such cases, FolM has access to H2-MPt produced by FolB because FolB is not tightly complexed with FolK. In this connection, it is interesting that, in Gammaproteobacteria that lack folX, folM clusters on the chromosome with folB instead of folX (Fig. 6B). Alternatively, FolM might reduce the folate synthesis intermediate H2-HMPt, which is a substrate for E. coli FolM (Fig. 4B), but this would set up competition between tetrahydropterin and folate synthesis.
The third question concerns the biological role of H4-MPt in E. coli and other bacteria (e.g., Serratia and Shigella) that have folX and folM but no phenylalanine hydroxylase gene (Fig. 2A). It is probable that, like E. coli, the others produce and secrete H4-MPt, because high levels of MPt were found in the culture medium of Serratia marcescens (synonym, S. indica)(22, 24). The scale of pterin production in E. coli and S. marcescens far exceeds that of folates, although the latter usually are considered the main end products of pterin biosynthesis (Fig. 3B) (22). In E. coli at least, pterin production peaks as log-phase growth ends (40), and folX expression is 39-fold higher in stationary-phase cells than in mid-log-phase cells (18). It therefore seems likely that a major tetrahydropterin-dependent process remains to be discovered in these bacteria, that this process is extracellular, and that it is most active when growth ceases. In this connection, it may be relevant that the FolM homolog PTR1 of the parasitic protist Leishmania major has an important (but biochemically undefined) role in oxidative stress resistance that depends upon the reduction of H2-BPt to H4-BPt (28).
Supplementary Material
Acknowledgments
This work was supported by the National Research Initiative of the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service (grant no. 2008-35318-04589 to A.D.H.), by the National Institutes of Health (grant no. R01 GM70641-01 to V.C.-L.), and by an endowment from the C. V. Griffin Sr. Foundation.
We thank M. Mevarech (Tel Aviv University, Israel) for pFolM, S. Jin (University of Florida) for P. aeruginosa PAO1 and the pEX18 vectors, and H. Yu (Marshall University, Huntington, WV) for pHERD20T.
Footnotes
Published ahead of print on 6 November 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ahn, C., J. Byun, and J. Yim. 1997. Purification, cloning, and functional expression of dihydroneopterin triphosphate 2′-epimerase from Escherichia coli. J. Biol. Chem. 272:15323-15328. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagley, P. J., and J. Selhub. 2000. Analysis of folate form distribution by affinity followed by reversed-phase chromatography with electrochemical detection. Clin. Chem. 46:404-411. [PubMed] [Google Scholar]
- 4.Blakley, R. L. 1969. Chemical and physical properties of pterins and folate derivatives, p. 58-105. In R. L. Blakley (ed.), The biochemistry of folic acid and related pteridines. Wiley, New York, NY.
- 5.Blaszczyk, J., Y. Li, J. Gan, H. Yan, and X. Ji. 2007. Structural basis for the aldolase and epimerase activities of Staphylococcus aureus dihydroneopterin aldolase. J. Mol. Biol. 368:161-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Choi, K.-H., A. Kumar, and H. P. Schweizer. 2006. A 10 min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64:391-397. [DOI] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Crécy-Lagard, V., B. El Yacoubi, R. D. de la Garza, A. Noiriel, and A. D. Hanson. 2007. Comparative genomics of bacterial and plant folate synthesis and salvage: predictions and validations. BMC Genomics 8:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dykxhoorn, D. M., R. St. Pierre, and T. Linn. 1996. A set of compatible tac promoter expression vectors. Gene 177:133-136. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez, R. F., E. Dolghih, and D. A. Kunz. 2004. Enzymatic assimilation of cyanide via pterin-dependent oxygenolytic cleavage to ammonia and formate in Pseudomonas fluorescens NCIMB 11764. Appl. Environ. Microbiol. 70:121-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forrest, H. S., and C. Van Baalen. 1970. Microbiology of unconjugated pteridines. Annu. Rev. Microbiol. 24:91-108. [DOI] [PubMed] [Google Scholar]
- 13.Fujisawa, H., and H. Nakata. 1987. Phenylalanine 4-monooxygenase from Chromobacterium violaceum. Methods Enzymol. 142:44-49. [DOI] [PubMed] [Google Scholar]
- 14.Giladi, M., N. Altman-Price, I. Levin, L. Levy, and M. Mevarech. 2003. FolM, a new chromosomally encoded dihydrofolate reductase in Escherichia coli. J. Bacteriol. 185:7015-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gourley, D. G., A. W. Schüttelkopf, G. A. Leonard, J. Luba, L. W. Hardy, S. M. Beverley, and W. N. Hunter. 2001. Pteridine reductase mechanism correlates pterin metabolism with drug resistance in trypanosomatid parasites. Nat. Struct. Biol. 8:521-525. [DOI] [PubMed] [Google Scholar]
- 16.Gregory, J. F., and J. P. Toth. 1988. Chemical synthesis of deuterated folate monoglutamate and in vivo assessment of urinary excretion of deuterated folates in man. Anal. Biochem. 170:94-104. [DOI] [PubMed] [Google Scholar]
- 17.Guroff, G., and C. A. Rhoads. 1969. Phenylalanine hydroxylation by Pseudomonas species (ATCC 11299a). Nature of the cofactor. J. Biol. Chem. 244:142-146. [PubMed] [Google Scholar]
- 18.Gutiérrez-Ríos, R. M., D. A. Rosenblueth, J. A. Loza, A. M. Huerta, J. D. Glasner, F. R. Blattner, and J. Collado-Vides. 2003. Regulatory network of Escherichia coli: consistency between literature knowledge and microarray profiles. Genome Res. 13:2435-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haussmann, C., F. Rohdich, E. Schmidt, A. Bacher, and G. Richter. 1998. Biosynthesis of pteridines in Escherichia coli. Structural and mechanistic similarity of dihydroneopterin-triphosphate epimerase and dihydroneopterin aldolase. J. Biol. Chem. 273:17418-17424. [DOI] [PubMed] [Google Scholar]
- 20.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 21.Ikemoto, K., T. Sugimoto, S. Murata, M. Tazawa, T. Nomura, H. Ichinose, and T. Nagatsu. 2002. (6R)-5,6,7,8-Tetrahydro-l-monapterin from Escherichia coli, a novel natural unconjugated tetrahydropterin. Biol. Chem. 383:325-330. [DOI] [PubMed] [Google Scholar]
- 22.Iwai, K., M. Kobashi, and H. Fujisawa. 1970. Occurrence of Crithidia factors and folic acid in various bacteria. J. Bacteriol. 104:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaufman, S. 1961. The nature of the primary oxidation product formed from tetrahydropteridines during phenylalanine hydroxylation. J. Biol. Chem. 236:804-810. [PubMed] [Google Scholar]
- 24.Kobashi, M., and K. Iwai. 1971. Biochemical studies on naturally occurring pteridines. II. Isolation and characterization of l-threoneopterin as the Crithidia factor, and on isoxanthopterin produced by Serratia indica. Agric. Biol. Chem. 35:47-52. [Google Scholar]
- 25.Kong, J. S., J. Y. Kang, H. L. Kim, O. S. Kwon, K. H. Lee, and Y. S. Park. 2006. 6-Pyruvoyltetrahydropterin synthase orthologs of either a single or dual domain structure are responsible for tetrahydrobiopterin synthesis in bacteria. FEBS Lett. 580:4900-4904. [DOI] [PubMed] [Google Scholar]
- 26.Lye, L. F., M. L. Cunningham, and S. M. Beverley. 2002. Characterization of quinonoid-dihydropteridine reductase (QDPR) from the lower eukaryote Leishmania major. J. Biol. Chem. 277:38245-38253. [DOI] [PubMed] [Google Scholar]
- 27.Naponelli, V., A. Noiriel, M. J. Ziemak, S. M. Beverley, L. F. Lye, A. M. Plume, J. R. Botella, K. Loizeau, S. Ravanel, F. Rébeillé, V. de Crécy-Lagard, and A. D. Hanson. 2008. Phylogenomic and functional analysis of pterin-4a-carbinolamine dehydratase family (COG2154) proteins in plants and microorganisms. Plant Physiol. 146:1515-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nare, B., L. A. Garraway, T. J. Vickers, and S. M. Beverley. 2009. PTR1-dependent synthesis of tetrahydrobiopterin contributes to oxidant susceptibility in the trypanosomatid protozoan parasite Leishmania major. Curr. Genet. 55:287-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noiriel, A., V. Naponelli, J. F. Gregory III, and A. D. Hanson. 2007. Pterin and folate salvage. Plants and Escherichia coli lack capacity to reduce oxidized pterins. Plant Physiol. 143:1101-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohmae, E., Y. Fukumizu, M. Iwakura, and K. Gekko. 2005. Effects of mutation at methionine-42 of Escherichia coli dihydrofolate reductase on stability and function: implication of hydrophobic interactions. J. Biochem. 137:643-652. [DOI] [PubMed] [Google Scholar]
- 31.Overbeek, R., T. Begley, R. M. Butler, J. V. Choudhuri, H. Y. Chuang, M. Cohoon, V. de Crécy-Lagard, N. Diaz, T. Disz, R. Edwards, M. Fonstein, E. D. Frank, S. Gerdes, E. M. Glass, A. Goesmann, A. Hanson, D. Iwata-Reuyl, R. Jensen, N. Jamshidi, L. Krause, M. Kubal, N. Larsen, B. Linke, A. C. McHardy, F. Meyer, H. Neuweger, G. Olsen, R. Olson, A. Osterman, V. Portnoy, G. D. Pusch, D. A. Rodionov, C. Ruckert, J. Steiner, R. Stevens, I. Thiele, O. Vassieva, Y. Ye, O. Zagnitko, and V. Vonstein. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1,000 genomes. Nucleic Acids Res. 33:5691-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfleiderer, W. 1985. Chemistry of naturally occurring pteridines, p. 43-114. In R. L. Blakley, and S. J. Benkovic (ed.), Folates and pterins, vol. 2. Wiley, New York, NY. [Google Scholar]
- 33.Qiu, D., F. H. Damron, T. Mima, H. P. Schweizer, and H. D. Yu. 2008. PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl. Environ. Microbiol. 74:7422-7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rembold, H., and R. Gyure. 1972. Biochemistry of the pteridines. Angew. Chem. Int. Ed. Engl. 11:1061-1072. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 36.Shaw, D., J. D. Odom, and R. B. Dunlap. 1999. High expression and steady-state kinetic characterization of methionine site-directed mutants of Escherichia coli methionyl- and selenomethionyl-dihydrofolate reductase. Biochim. Biophys. Acta 1429:401-410. [DOI] [PubMed] [Google Scholar]
- 37.Song, J., T. Xia, and R. A. Jensen. 1999. PhhB, a Pseudomonas aeruginosa homolog of mammalian pterin 4a-carbinolamine dehydratase/DCoH, does not regulate expression of phenylalanine hydroxylase at the transcriptional level. J. Bacteriol. 181:2789-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thöny, B., G. Auerbach, and N. Blau. 2000. Tetrahydrobiopterin biosynthesis, regeneration and functions. Biochem. J. 347:1-16. [PMC free article] [PubMed] [Google Scholar]
- 39.Vasudevan, S. G., D. C. Shaw, and W. L. Armarego. 1988. Dihydropteridine reductase from Escherichia coli. Biochem. J. 255:581-588. [PMC free article] [PubMed] [Google Scholar]
- 40.Wachter, H., A. Hausen, E. Reider, and M. Schweiger. 1980. Pteridine excretion from cells as indicator of cell proliferation. Naturwissenschaften 67:610-611. [DOI] [PubMed] [Google Scholar]
- 41.Zhao, G., T. Xia, J. Song, and R. A. Jensen. 1994. Pseudomonas aeruginosa possesses homologues of mammalian phenylalanine hydroxylase and 4 alpha-carbinolamine dehydratase/DCoH as part of a three-component gene cluster. Proc. Natl. Acad. Sci. USA 91:1366-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.