Abstract
The main siderophores produced by streptomycetes are desferrioxamines. Here we show that Streptomyces sp. ATCC 700974 and several Streptomyces griseus strains, in addition, synthesize a hitherto unknown siderophore with a catechol-peptide structure, named griseobactin. The production is repressed by iron. We sequenced a 26-kb DNA region comprising a siderophore biosynthetic gene cluster encoding proteins similar to DhbABCEFG, which are involved in the biosynthesis of 2,3-dihydroxybenzoate (DHBA) and in the incorporation of DHBA into siderophores via a nonribosomal peptide synthetase. Adjacent to the biosynthesis genes are genes that encode proteins for the secretion, uptake, and degradation of siderophores. To correlate the gene cluster with griseobactin synthesis, the dhb genes in ATCC 700974 were disrupted. The resulting mutants no longer synthesized DHBA and griseobactin; production of both was restored by complementation with the dhb genes. Heterologous expression of the dhb genes or of the entire griseobactin biosynthesis gene cluster in the catechol-negative strain Streptomyces lividans TK23 resulted in the synthesis and secretion of DHBA or griseobactin, respectively, suggesting that these genes are sufficient for DHBA and griseobactin biosynthesis. Griseobactin was purified and characterized; its structure is consistent with a cyclic and, to a lesser extent, linear form of the trimeric ester of 2,3-dihydroxybenzoyl-arginyl-threonine complexed with aluminum under iron-limiting conditions. This is the first report identifying the gene cluster for the biosynthesis of DHBA and a catechol siderophore in Streptomyces.
Iron is an essential element for the growth and proliferation of nearly all microorganisms. In the presence of oxygen, the soluble ferrous iron is readily oxidized to its ferric form, which exists predominantly as a highly insoluble hydroxide complex at neutral pH. To overcome iron limitation, many bacteria synthesize and secrete low-molecular-weight, high-affinity ferric iron chelators, called siderophores (38, 53). Following the chelation of Fe3+ in the medium, the iron-siderophore complex is actively taken up by its cognate ABC transport system, and Fe3+ is subsequently released by reduction to Fe2+ and/or by hydrolysis of the siderophore (28, 32, 36). The three main classes of siderophores contain catecholates, hydroxamates, or (α-hydroxy-)carboxylates as iron-coordinating ligands, but mixed siderophores and siderophores containing other functional groups, such as diphenolates, imidazoles, and thiazolines, have also been found (16, 38).
Siderophores containing peptide moieties are synthesized by proteins belonging to the nonribosomal peptide synthetase (NRPS) family (16, 38). These multimodular enzymes function as enzymatic assembly lines in which the order of the modules usually determines the order of the amino acids incorporated into the peptide (19, 34). Each module contains the complete information for an elongation step combining the catalytic functions for the activation of the amino acid by the adenylation (A) domain, the tethering of the corresponding adenylate to the terminal thiol of the enzyme-bound 4′-phosphopantetheinyl (4′-PP) cofactor by the peptidyl carrier protein (PCP) domain, and the formation of the peptide bond by the condensation (C) domain (26, 34, 52). At the end, the product is released by the C-terminal thioesterase (TE) domain by hydrolysis or by cyclization via intramolecular condensation. Each adenylation domain recognizes a specific amino acid, and its substrate specificity can be predicted by its sequence. An NRPS specificity-conferring code consisting of 10 nonadjacent amino acid residues in the A domain has been proposed (49). Exceptions to the “colinearity-rule” (19) have been discovered. For example, in the biosynthesis of the siderophores enterobactin and bacillibactin, all the modules in the NRPS are used iteratively, and the TE domain stitches the chains together into a cyclic product (35, 45). Enterobactin is the trilactone of 2,3-dihydroxybenzoyl-serine, and bacillibactin is the lactone of 2,3-dihydroxybenzoyl-glycyl-threonine.
The typical siderophores produced by streptomycetes are desferrioxamines (24), and the genes encoding the enzymes for their biosynthesis have been identified (5). Recently, structurally different siderophores have been reported to be coproduced with desferrioxamines in some species, e.g., coelichelin in Streptomyces coelicolor (9, 30) and enterobactin in Streptomyces tendae (18). The genes encoding the proteins for the biosynthesis of enterobactin in S. tendae remain unknown.
Here we describe the gene cluster for the biosynthesis of a new siderophore, named griseobactin, produced by Streptomyces sp. strain ATCC 700974 and some strains of Streptomyces griseus. By sequencing two cosmids isolated from a Streptomyces sp. strain ATCC 700974 genomic library, we assigned the encoded proteins to enzymes that convert chorismate to 2,3-dihydroxybenzoate (DHBA), and to proteins involved in nonribosomal peptide biosynthesis and in the export, uptake, and utilization of siderophores. Knockout mutagenesis and heterologous expression confirmed the requirement of this gene cluster for the biosynthesis of griseobactin. This is the first report on the identification of the genes responsible for DHBA and catechol siderophore biosynthesis in Streptomyces.
MATERIALS AND METHODS
Bacterial strains, plasmids, cosmids, and culture conditions.
The Escherichia coli and Streptomyces strains, plasmids, and cosmids used are listed in Table 1. E. coli strains were cultured in liquid or on solid LB medium at 37°C (47). Streptomyces was routinely grown on solid mannitol soya flour medium at 30°C (27). For siderophore production, M9 minimal medium was used (47). When appropriate, the following supplements were added: apramycin (Apr), 50 μg/ml; chloramphenicol (Cam), 40 μg/ml; nalidixic acid (Nal), 25 μg/ml; neomycin (Neo), 50 μg/ml.
TABLE 1.
Strains, plasmids, and cosmids used in this study
| Strain, plasmid, or cosmid | Relevant genotype, description, or properties | Reference or sourcea |
|---|---|---|
| Strains | ||
| E. coli | ||
| MC4100 | araD139 Δ(lacIZYA-argF)U169 rpsL150 relA1 flhD5301 deoC1 fruA25 rbsR22 | 14 |
| ET12567 | dam-13::Tn9 dcm-6 hsdM hsdR zjj-202::Tn10 recF143 galK2 galT22 ara-14 lacY1 xyl-5 leuB6 thi-1 fhuA31 rpsL136 hisG4 tsx-78 mtl-1 glnV44 | 33 |
| DH5α | endA1 hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrA96 Δ(lacIZYA-argF)U169 relA1 (φ80lacZΔM15) | Stratagene (Heidelberg, Germany) |
| XL1-Blue MR | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac | Stratagene (Heidelberg, Germany) |
| Streptomyces | ||
| ATCC 700974 | Streptomyces sp.; Nalr | ATCC |
| Tü 6 | Streptomyces griseus | DSMZ |
| Tü 17 | S. griseus ETH 10073 | Strain collection, Tübingen |
| Tü 19 | S. griseus ETH 4289 | Strain collection, Tübingen |
| M145 | Streptomyces coelicolor A3(2) SLP2− SLP3− | 27 |
| SIP1192 | ATCC 700974 ΔdhbACE Neor with a possible polar effect on downstream dhbBG genes | This study |
| SIP1194 | ATCC 700974 ΔdhbCE Neor with a possible polar effect on downstream dhbBG genes | This study |
| TK23 | Streptomyces lividans 66 spc-1 SLP2− SLP3− | 27 |
| J1074 | Streptomyces albus G | 15 |
| Plasmids | ||
| pOJ436 | acc(3)IV (Aprr) λ cos oriTRK2; φC31 integration function, ColE1 replicon | 12 |
| pSET152ermE*p | acc(3)IV (Aprr) oriTRK2; pUC18 replicon, φC31 integration function, constitutive promoter ermE*p | Stefan Pelzer (Combinature, Berlin, Germany) |
| pKC1132 | acc(3)IV (Aprr) lacZα oriTRK2; pUC18 replicon | 12 |
| pSP126/1049 | dhbAEBG in pSET152ermE*p | This study |
| pSP126/1079 | dhbACEBG in pSET152ermE*p | This study |
| pSU18 | p15A replicon; Camr, high copy number | 6 |
| pUB307 | RP1 derivative; self-transmissible oriT-mobilizing plasmid | 8 |
| Cosmids | ||
| 679 | Integrative cosmid containing orf1, orf2, and dhbACEBG griABCD in pOJ436 | This study |
| 680 | Integrative cosmid comprising orf1, orf2, dhbACEBG griABCDEFGH, andorf3 in pOJ436 | This study |
ATCC, American Type Culture Collection, Manassas, VA; DSMZ, German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany.
Recombinant DNA techniques.
DNA was isolated and manipulated according to the methods of Sambrook and Russell (47) and Kieser et al. (27). DNA was purified on a silica matrix with NucleoSpin columns (Macherey-Nagel, Düren, Germany). Oligonucleotides were synthesized by VBC-Biotech (Vienna, Austria). PCRs were performed with a RoboCycler gradient 96 (Stratagene, Heidelberg, Germany) and the 3′-5′ proofreading polymerase ReproFast (Genaxxon, Biberach, Germany).
Construction of plasmids and strains.
Noncompatible ends were blunt ended prior to ligation by the removal of protruding 3′ termini or the filling in of recessed 3′ termini. E. coli DH5α was transformed with the ligation products. The constructs were confirmed by sequencing. Plasmids were introduced into Streptomyces by conjugation using the methylation-deficient E. coli strain ET12567, which harbored the intended plasmid as well as plasmid pUB307 (27), as a donor.
For construction of the plasmids used for gene inactivation, the EcoRI-EcoRI (nucleotides [nt] 4210 to 5715) and EcoRI-StuI (nt 5715 to 12053) fragments derived from cosmid 680 and comprising dhbE′, dhbC, dhbA, griA, and griB were adjacently cloned into the SmaI site of pSU18, thereby eliminating the EcoRI site at nt 5715 by ligation of the filled-in ends. From the resulting plasmid, the AspEI-SfiI fragment (dhbC, nt 5995 to 6912) or the DraIII fragment (dhbAC, nt 6113 to 7343) were replaced by the neomycin resistance cassette gained from transposon Tn5 by PCR with primers Tn5Neo1 (TTGCAGTGGGCTTACATGGCGATAGC) and Tn5Neo2 (GGCGAAGAACTCCAGCATGAGATCC). The inserts of these plasmids were first cut with EcoRI/HindIII and then cloned into the EcoRV-digested conjugative plasmid pKC1132. After conjugal transfer of these plasmids from E. coli ET12567(pUB307) into Streptomyces sp. strain ATCC 700974, potential double-crossover recombinants were selected with nalidixic acid and neomycin and, after several rounds of growth, by their sensitivity to apramycin. The gene replacements were confirmed by PCR and sequencing with various primers. The strain harboring the dhbC deletion with the neomycin resistance gene oriented in the same direction as the dhb genes was designated SIP1194, and the strain containing the dhbAC deletion with the neomycin resistance cassette oriented in the direction opposite to the dhb genes was designated SIP1192. The filled-in EcoRI site at nt 5715 caused a frameshift at the beginning of dhbE, resulting in dhbE inactivation. A polar effect on the downstream genes dhbBG cannot be excluded.
For complementation, the dhbACEBG genes were amplified by PCR with primers Dhb7 (CCTAACACCCGGAGGAGGTTGCTC) and Dhb5 (GACATACAGGTGGGCCAAGGTC), and the product was cloned into the EcoRV site of the pSET152ermE*p expression vector. Of five plasmids, only one, pSP126/1079, had the correct amino acid sequence, although it carried two silent mutations. Plasmid pSP126/1049 carried two mutations in the dhbC gene, resulting in the amino acid exchanges M148T and R235H, which render dhbC inactive, since, in contrast to pSP126/1079, it did not complement the ΔdhbACE and ΔdhbCE mutants SIP1192 and SIP1194, respectively. Thus, pSP126/1049 comprises only the functional genes dhbAEBG.
Cosmid library construction and sequencing.
Cosmids 679 and 680 were obtained from a genomic library of Streptomyces sp. strain ATCC 700974. For cosmid library construction, genomic DNA of Streptomyces sp. strain ATCC 700974 was partially digested with Sau3A1; fragments of about 40 kb were ligated to the BamHI/HpaI-digested, dephosphorylated cosmid vector pOJ436, packaged with the Gigapack III XL packaging extract (Stratagene, Heidelberg, Germany), and then used to transfect the restriction-minus E. coli strain XL1-Blue MR.
DNA sequencing.
Automated DNA sequencing was carried out on a 3730xl DNA analyzer (Applied Biosystems, Foster City, CA) by GATC Biotech (Konstanz, Germany) or on an ALFexpress II DNA sequencer (Amersham Pharmacia Biotech, Freiburg, Germany) using the SequiTherm Excel II long-read DNA sequencing kit-ALF (Epicentre, Madison, WI). Cosmids were subcloned prior to sequencing.
Determination of siderophore biosynthesis.
Catechol-type siderophores were detected spectrophotometrically at 515 nm using an assay developed by Arnow (3) with DHBA as a standard. Measurements were carried out in triplicate. For quantitative analysis of griseobactin composition and comparison with the amino acid analysis, the siderophore was hydrolyzed for 2 h in 5 N HCl at 100°C prior to the assay. This yielded 2- to 3-fold-higher values than the unhydrolyzed siderophore.
The peptide moiety of the siderophore was quantified according to the method of Bradford (13) with a Roti-Quant kit (Carl Roth, Karlsruhe, Germany) and bovine serum albumin as a standard. Measurements were carried out in triplicate.
Siderophore activity was measured using the chrome azurol S (CAS) liquid assay as described previously (48). This assay is based on the competition for Fe3+ ions between the CAS-Fe3+ complex and the excreted siderophore. The higher affinity constant of the siderophore for iron leads to a color change of the blue CAS-Fe3+ complex to the orange iron-free CAS, which is measured by the decrease in absorption at 630 nm. For quantification, the iron chelator desferrioxamine B mesylate (Sigma-Aldrich, Steinheim, Germany) was used as a standard.
Siderophore purification.
Griseobactin was obtained from the culture medium of Streptomyces sp. strain ATCC 700974. Cells were grown in iron-depleted M9 minimal medium (47) to the stationary phase with shaking at 30°C. The cells were sedimented by centrifugation for 20 min at 6,000 × g. The supernatant was passed through a 0.22-μm-pore-size filter (Schleicher & Schüll, Dassel, Germany) and applied to an XAD-2 (Supelco, Bellefonte, PA) column equilibrated with water. The column was washed with 5 volumes of water and then eluted with 50% methanol. The Arnow-positive fractions were pooled, evaporated to dryness, suspended in water, and subjected to gel filtration on a Superdex peptide HR 10/30 column (Amersham Bioscience, Freiburg, Germany) using water as the solvent. Griseobactin bound to XAD-2 resin but did not bind to XAD-4 resin, suggesting a mass above 1,000 Da.
Spectroscopy.
UV and visible (UV/Vis) spectra were recorded on an UltrospecIII spectrometer (Pharmacia Biosystems, Freiburg, Germany).
ESI-MS.
Electrospray ionization mass spectrometry (ESI-MS) and ESI-tandem MS (ESI-MS-MS) were performed on an HCT ultra ETD II mass spectrometer (Bruker-Daltonics, Bremen, Germany) operating in the positive ionization mode by Guido Sauer (this institute). Prior to analysis, samples were passed through a C18 cartridge. For fragmentation studies, ESI-MS-MS was performed by collision-induced dissociation with helium as the collision gas. High-resolution electrospray ionization-Fourier transform ion cyclotron resonance (FT-ICR) mass spectra were collected in the positive ionization mode on a 4.7-T Apex II FT-ICR mass spectrometer (Bruker-Daltonics) by Graeme Nicholson at the Institute of Organic Chemistry, University of Tübingen, Tübingen, Germany. Samples were dissolved in methanol containing 0.1% formic acid and were measured over the mass range of m/z 200 to 1,400.
Amino acid analysis.
Amino acid analysis was carried out by Norbert Tröndle (Genaxxon, Biberach, Germany). The sample was hydrolyzed for 24 h in 3 N HCl at 110°C and was then analyzed on an LC3000 amino acid analyzer.
Computer analyses.
Nucleotide and amino acid sequences were analyzed using the PC/GENE program package (IntelliGenetics, Mountain View, CA), Artemis (46), GenemarkS (10, 11), SignalP (7), LipoP (25), and the Clustal W program (51). Functional assignments were made by homology searches with the BLAST algorithm (1) and comparison with proteins of known functions in the database. The domain organization of the NRPS was analyzed according to references 2 and 34. The amino acid substrate specificities of the adenylation domains of NRPSs were predicted using NRPSpredictor (44).
Nucleotide sequence accession number.
The GenBank accession number for the complete 26-kb nucleotide sequence comprising the griseobactin biosynthesis gene cluster in Streptomyces sp. strain ATCC 700974 is FN545130.
RESULTS AND DISCUSSION
Sequencing and analysis of a gene cluster for a putative catecholate siderophore.
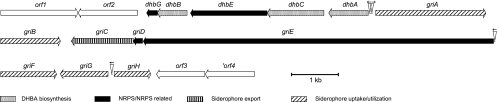
Cosmids 679 and 680 were prepared from the genome of Streptomyces sp. strain ATCC 700974 and then sequenced. A gene cluster with no nucleotide sequence similarity to sequences of the genomes of Streptomyces coelicolor and Streptomyces avermitilis or to any other entries in the most current database was found. The encoded proteins showed similarities to proteins involved in DHBA synthesis, NRPS-dependent siderophore biosynthesis, and siderophore-dependent iron transport systems from other organisms, which indicated their involvement in the biosynthesis of a catechol-peptide siderophore (Table 2; Fig. 1). Because this novel siderophore was synthesized mainly by Streptomyces griseus (see below), it was named griseobactin.
TABLE 2.
Proposed genes and encoded proteins of the griseobactin gene locus of Streptomyces sp. strain ATCC 700974 and comparison with proteins from databases
| Gene | Protein size (no. of amino acids) | Proposed function | Homologue (no. of amino acids)/organism (UniProt accession no.) | Identity, similarity (%) |
|---|---|---|---|---|
| orf1 | 547 | Phosphoglucomutase | Pgm (555)/E. coli (P36938) | 58, 72 |
| orf2 | 422 | Hypothetical protein | ||
| dhbG | 79 | Aryl carrier for DHBA | DhbB (312; C-terminal 88 amino acids)/B. subtilis (P45743) | 38, 52 |
| EntB (285; C-terminal 78 amino acids)/E. coli (P0ADI4) | 43, 58 | |||
| dhbB | 210 | Isochorismatase | DhbB (312; N-terminal 210 amino acids)/B. subtilis (P45743) | 54, 74 |
| EntB (285; N-terminal 210 amino acids)/E. coli (P0ADI4) | 44, 66 | |||
| dhbE | 545 | DHBA-AMP ligase | DhbE (539)/B. subtilis (P40871) | 65, 77 |
| EntE (536)/E. coli (P10378) | 46, 63 | |||
| dhbC | 401 | Isochorismate synthase | DhbC (398)/B. subtilis (P45744) | 52, 62 |
| EntC (391)/E. coli (P0AEJ2) | 37, 52 | |||
| dhbA | 276 | 2,3-Dihydro-DHBA dehydrogenase | DhbA (261)/B. subtilis (P39071) | 49, 64 |
| EntA (248)/E. coli (P15047) | 45, 56 | |||
| griA | 779 | Siderophore uptake membrane transporter | FepD, FepG (334, 330)/E. coli (P23876, P23877) | 33, 49 |
| FhuB (660)/E. coli (P06972) | 27, 42 | |||
| FhuB, FhuG (384, 336)/B. subtilis (P49936, P49937) | 29, 47 | |||
| griB | 424 | Esterase | Fes (374)/E. coli (P13039) | 30, 41 |
| griC | 439 | MFS exporter | HsrA (475)/E. coli (P31474) | |
| griD | 67 | MbtH-like protein | MbtH (71)/Mycobacterium tuberculosis (O05821) | 39, 49 |
| griE | 2,468 | NRPS | DhbF (2,378)/B. subtilis (P45745) | 47, 61 |
| griF | 397 | Amidohydrolase | AbgB (481)/E. coli (P76052) | 24, 34 |
| griG | 335 | Siderophore-binding lipoprotein | FhuD (315)/B. subtilis (P37580) | 27, 43 |
| griH | 263 | ATPase of siderophore uptake ABC transporter | FepC (271)/E. coli (P23878) | 46, 64 |
| orf3 | 339 | Lysine arginine ornithine transport system kinase | ArgK (331)/E. coli (P27254) | 41, 57 |
| orf4 | >349 | Methylmalonyl-CoA mutase large subunit | MutB (728; C-terminal 347 amino acids)/ Propionibacterium freudenreichii (P11653) | 68, 79 |
FIG. 1.
Organization of the griseobactin biosynthetic gene cluster from Streptomyces sp. strain ATCC 700974, as sequenced from two overlapping cosmids. The locations of putative iron boxes, the binding sites of Fe2+-loaded transcription repressors, are indicated by triangles, and their directions are indicated by arrows. The proposed functions of the individual genes are summarized in Table 2.
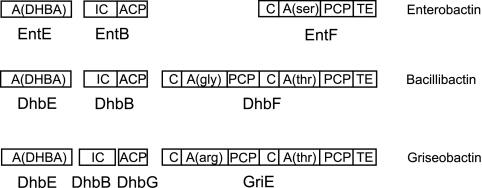
The five-cistron operon dhbACEBG (Fig. 1) encodes homologues of the DhbACEB proteins, required for the biosynthesis and activation of DHBA in the production of the Bacillus subtilis catechol siderophore bacillibactin (35). The isochorismate synthase DhbC first isomerizes chorismate to isochorismate, which is then hydrolyzed to 2,3-dihydro-DHBA through the cleavage of pyruvate by the isochorismatase DhbB (N-terminal part). The 2,3-dihydro-DHBA dehydrogenase DhbA oxidizes this product to the aromatic DHBA. The DHBA-AMP ligase DhbE activates DHBA and transfers it as a thioester onto the aryl carrier DhbB (C-terminal part), whereby ATP is hydrolyzed. Instead of having the isochorismatase and aryl carrier activities in a single bifunctional protein (DhbB), as in B. subtilis and E. coli, Streptomyces sp. strain ATCC 700974 has these activities in two separate proteins, DhbB (isochorismatase) and DhbG (aryl carrier) (Fig. 2). The phosphopantetheine binding site of DhbG, NLVDYGLDSVR, with the serine attachment site for phosphopantetheine, is consistent with the signature sequence of aryl carrier proteins: (N/D)LXXXGLDSXR.
FIG. 2.
Domain organization of the NRPS modules in griseobactin biosynthesis compared to those in bacillibactin and enterobactin biosynthesis. A, adenylation domain; ACP, aryl carrier protein; C, condensation domain; PCP, peptidyl carrier protein/thiolation domain; TE, thioesterase domain; IC, isochorismatase. The specificities of the adenylation domains are given in parentheses.
Sequence analysis of the NRPS-like 2,468-amino-acid protein encoded by griE suggests that it contains two NRPS modules, each comprising the core elongation domains C, A, and PCP, followed by one C-terminal TE domain (Fig. 2). The substrate specificity for the adenylation domains of GriE is predicted by the NRPSpredictor program to be threonine for module 1 and glutamine for module 2.
griD encodes a small (<100-amino-acid) MbtH-like protein that displays 39% sequence identity to the mbtH gene product in the gene cluster for the synthesis of mycobactin, a nonribosomally synthesized peptide siderophore in Mycobacterium tuberculosis (42). An MbtH-like protein is found in many of the gene clusters involved in the synthesis of siderophores and peptide and aminocoumarin antibiotics. Although the requirement of MbtH-like proteins for the formation or secretion of secondary metabolites in Streptomyces (31, 54) and for that of pyoverdine at wild-type levels in Pseudomonas aeruginosa (17) has been shown experimentally, the function of these proteins is still unknown, because different mbtH-like genes can functionally replace each other (31, 54).
The griC gene encodes a permease of the major facilitator superfamily (MFS). Analysis of GriC revealed the presence of at least 12 transmembrane helices. The sequence GWLGDRFGTKRVF, starting at amino acid 71, is consistent with a conserved 13-amino-acid motif, G(R/K/P/A/T/Y)L(G/A/S)(D/N)(R/K)(F/Y)GR(R/K)(R/K/P)(L/I/V/G/S/T)(L/I/M), present at a typical position between the second and third transmembrane domains, as found in MFS-type transporters (50), suggesting that GriC exports griseobactin from the cytoplasm into the medium.
The griA, griB, griF, griG, and griH genes are putatively involved in griseobactin uptake and utilization. GriA is homologous to siderophore uptake transporters. It consists of two halves that display 29% sequence identity to each other. It is predicted to form 20 transmembrane helices; hence, one protein is sufficient to constitute the whole transporter, like FhuB in E. coli, and no dimers of different proteins, such as FepDG or FecCD in E. coli, are necessary. GriG and GriH are similar to the siderophore-binding lipoprotein and the ATPase of iron-siderophore ABC transporters in other bacteria. In GriG, a cleavage site for signal peptidase II is predicted after amino acid 31, supporting its export and localization at the surface of the cytoplasmic membrane.
GriB belongs to the family of α,β-hydrolases (40) and possesses a conserved GXSXG serine esterase motif. It shows 30% identity to Fes, the enterochelin esterase that cleaves the ester bonds of the cyclic lactone and thereby releases iron from the imported enterobactin in E. coli (32). GriF belongs to the peptidase M20 superfamily. It is most similar to AbgB, the aminobenzoyl glutamate utilization protein B of E. coli, which is required but not essential for aminobenzoyl glutamate utilization (23). AbgB is proposed to hydrolyze aminobenzoyl glutamate to aminobenzoate and glutamate, either alone or in combination with AbgA (23). Therefore, GriF may contribute, together with GriB, to the release of iron by hydrolyzing the siderophore.
The boundaries of the griseobactin biosynthesis and utilization genes are delineated by the open reading frames orf1 and orf2 on one end and orf3 and orf4 on the other (Fig. 1). The predicted functions of these open reading frames are not related to those of griseobactin (Table 2). orf1 encodes an orthologue of the phosphoglucomutase Pgm in E. coli, and orf2 encodes a hypothetical protein with no similarity to known proteins. An extremely stable hairpin is found 22 nucleotides downstream of the orf2 stop codon. The stem-loop structure consists of a 20-nt GC-rich (G+C content, 90%) stem with a calculated ΔG (25°C) of −66.2 kcal/mol, followed by a U-rich region on the RNA, which may serve as a rho-independent terminator of transcription. orf3 and orf4 encode homologues of ArgK, the kinase that phosphorylates the periplasmic binding proteins for the transport of lysine, arginine, and ornithine, and MutB, the large subunit of methylmalonyl coenzyme A (CoA) mutase, respectively.
Four iron boxes are predicted in the griseobactin biosynthesis cluster.
The DmdR (divalent metal-dependent regulatory protein) family mediates iron regulation in Gram-positive bacteria and is also found in streptomycetes (22). A consensus iron box, TTAGGTTAGGCTCACCTAA, for DmdR binding in Streptomyces species has been deduced (20). The ferrous iron complex of DmdR binds to such iron boxes and prevents expression of the adjacent genes. A search of the griseobactin gene cluster for iron boxes revealed four well-conserved iron box sequences, all in regions upstream of putative genes (Fig. 1, triangles): TTAGGTTAccCTtACCTAA (84% identical to the consensus iron box), located 20 nt 5′ of dhbA; TTAGGcTAGcCTCACCTtA (84% identical), located 63 nt 5′ of griA; TTAacTTAGcCTtACCTAA (79% identical), located 12 nt 5′ of griE; and TaAaGTaAGGCTaACCTtA (74% identical), located 82 nt 5′ of griG and 68 nt 5′ of griH (capital letters indicate conservation of the nucleotide, and lowercase letters indicate mismatches.)
Catechol production by different Streptomyces strains and iron regulation.
To date, no gene cluster for a catechol siderophore has been identified in streptomycetes. Therefore, we examined whether Streptomyces sp. strain ATCC 700974 produces and secretes catechol compounds by using the Arnow assay (3), which is selective for aromatic vicinal diols devoid of substitution and steric hindrance at either the 3 or the 4 position, as found in catechol (4). We detected catechol in the culture supernatant of ATCC 700974 grown in M9 minimal medium (Fig. 3). Furthermore, the S. griseus strains Tü 6, Tü19, and, to a lesser extent, Tü 17 produced and secreted catechol compounds. In contrast, S. coelicolor M145, S. lividans TK23, and S. albus did not secrete catechol. The genome sequence of S. coelicolor M145 (9) lacks a dhbACEBG-griABCDEFGH gene cluster. In contrast to the culture supernatants, the methanol extracts of the mycelia of the Streptomyces strains mentioned above contained only negligible amounts of catechol (data not shown), indicating that there are low amounts of membrane-bound and intracellular catechol compounds. In contrast, enterobactin has been isolated from the methanol extracts of the mycelia of S. tendae Tü 901/8c and Streptomyces sp. strain Tü 6125 and only negligible amounts were found in the culture supernatants (18). Neither growth promotion experiments with an enterobactin-negative E. coli mutant, to prove the identity of enterobactin, nor studies of the iron dependence of enterobactin production have been carried out (18).
FIG. 3.
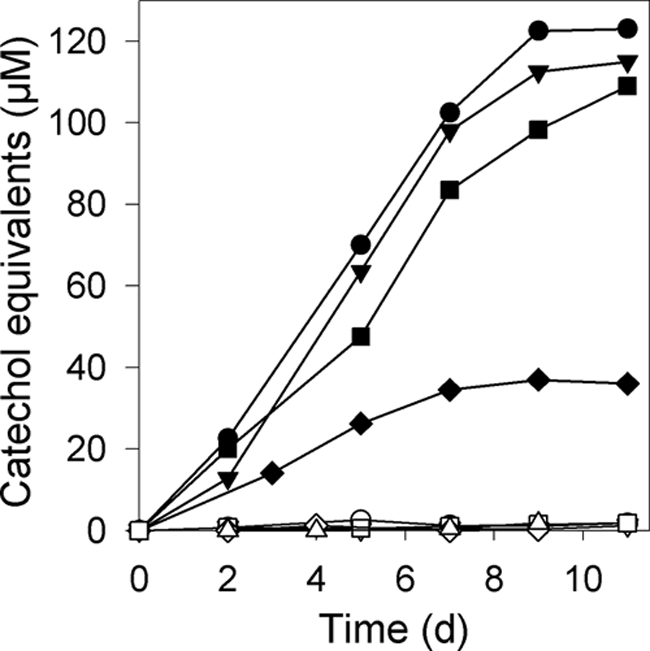
Catechol production by different Streptomyces strains. S. griseus Tü 19 (•, ▵), S. griseus Tü 6 (▾), Streptomyces sp. strain ATCC 700974 (▪, ⋄), S. griseus Tü 17 (♦), S. coelicolor M145 (○), S. lividans TK23 (▿), and S. albus G J1074 (□) were grown at 30°C in M9 minimal medium with (⋄, ▵) or without (•, ▾, ▪, ♦, ○, ▿, □) the addition of 50 μM Fe3+. The concentration of catechol in the supernatant was determined using the Arnow assay.
Since potential iron boxes were identified in the nucleotide sequence of the griseobactin gene cluster, the influence of iron on the biosynthesis of catechol was studied. Upon addition of 50 μM iron to the medium, catechol production ceased (Fig. 3), indicating that catechol biosynthesis is repressed by iron.
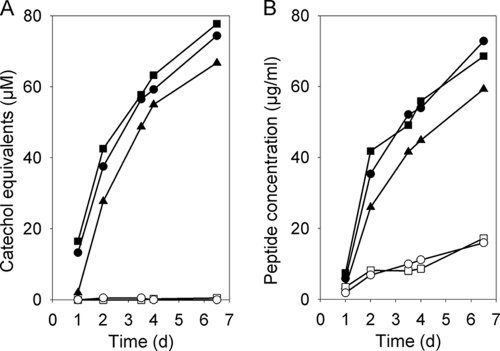
Disruption of the dhb genes abolishes griseobactin biosynthesis.
The data presented above strongly suggested that several Streptomyces strains produce a catechol-containing compound under low-iron conditions. To confirm that the dhbACEBG genes are responsible for griseobactin biosynthesis, the dhbC gene or the dhbAC genes of ATCC 700974 were replaced by a neomycin resistance cassette (see Materials and Methods). The resulting mutants, SIP1192 (ΔdhbACE) and SIP1194 (ΔdhbCE), were unable to synthesize any catechol compound (Fig. 4A). The supernatants of the dhb mutants contained very low peptide concentrations compared to that of the supernatant of the wild type (Fig. 4B), indicating that griseobactin contains not only a catechol but also a peptide moiety. In contrast to the Arnow assay, the peptide assay showed a certain background for the dhb mutants because of other proteinaceous compounds secreted by Streptomyces cells.
FIG. 4.
Catechol (A) and peptide (B) concentrations in the culture supernatants of dhb mutants and the same mutants complemented with dhb genes. Cells of the parental strain, Streptomyces sp. strain ATCC 700974 (▴), the ΔdhbACE mutant SIP1192 (○), the ΔdhbCE mutant SIP1194 (□), and the dhbACEBG-complemented strains SIP1192(pSP126/1079) (•) and SIP1194(pSP126/1079) (▪) were grown in M9 minimal medium at 30°C.
The siderophore activity of the culture supernatant was measured using the CAS liquid assay. The siderophore concentration in the parental strain, ATCC 700974, was about 100 μM desferrioxamine equivalents, whereas the concentrations in the dhb mutants were reduced to less than 50%. This showed that the dhb mutants are impaired in siderophore production but still synthesize at least one other siderophore. Strain ATCC 700974 possesses a gene cluster for desferrioxamine biosynthesis, as ascertained by nucleotide sequencing (data not shown).
Complementation of the dhb mutants with dhb genes.
To confirm that the loss of catechol siderophore biosynthesis was caused by the dhb mutation, the mutants SIP1194 (ΔdhbCE) and SIP1192 (ΔdhbACE) were complemented by integrative plasmids containing the dhbACEBG genes cloned under the control of the constitutive promoter ermE*p. Plasmid pSP129/1079 restored griseobactin production in both dhb mutants, as judged by the Arnow and peptide assays (Fig. 4). The mutants also produced the siderophore when complemented with cosmid 680 (data not shown). In this case, the catechol and peptide concentrations were 3- to 4-fold higher than those in the parental strain, ATCC 700974, probably because of a gene dosage effect (two copies of griD and griE).
The dhbC genotype is supplemented by the addition of DHBA.
The function of the dhbCBA genes was further examined by feeding the dhbC mutant with DHBA, which restored griseobactin synthesis. As shown in Fig. 5, the peptide content in the supernatant of the dhbC mutant SIP1192(pSP126/1049) increased more than 20-fold when DHBA was added to the medium. The same was true for the dhbC mutant SIP1194(pSP126/1049) (data not shown in Fig. 5 for clarity). Addition of DHBA to the media of the dhbACE and dhbCE mutants did not result in griseobactin production (Fig. 5), because these mutants lacked dhbE. This result corroborates the hypothesis that in griseobactin biosynthesis, DhbE catalyzes a step downstream of DHBA biosynthesis. Since the added DHBA causes a positive result in the Arnow assay, the Arnow assay could not be applied to determine the amount of the catechol moiety of griseobactin.
FIG. 5.
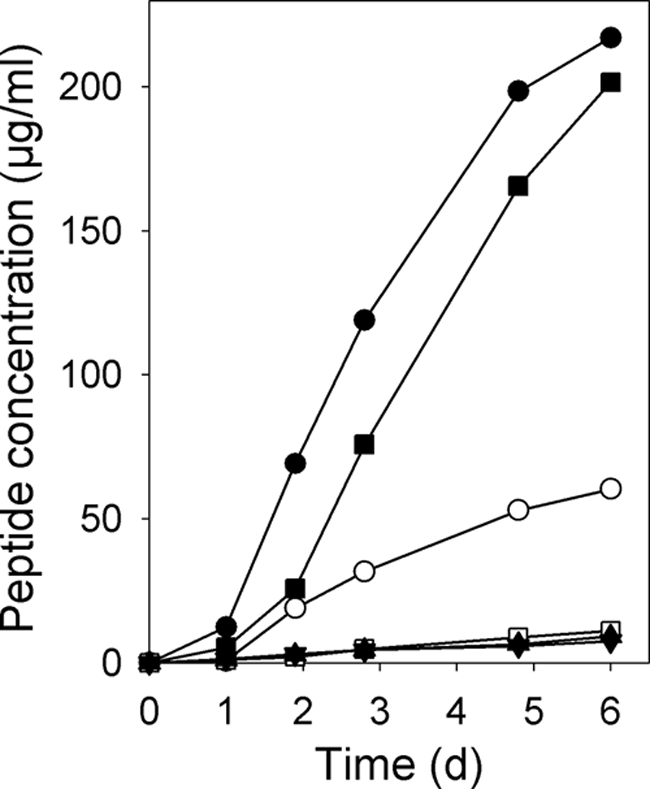
Supplementation of the dhbC mutant with DHBA. Peptide concentrations in the culture supernatants of the parental strain, Streptomyces sp. strain ATCC 700974 (circles), the dhbC mutant SIP1192(pSP126/1049) (squares), SIP1192 dhbACE (triangles), and SIP1194 dhbCE (inverted triangles) with (filled symbols) and without (open symbols) the addition of 2 mM DHBA are shown. Cells were grown in M9 minimal medium at 30°C.
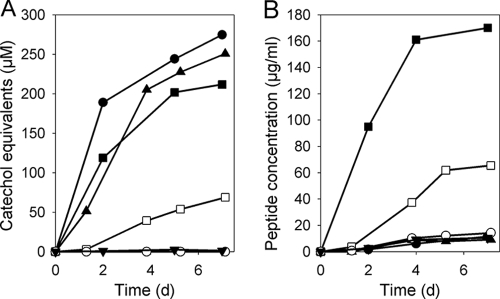
Heterologous expression of the dhb genes and of the entire griseobactin gene cluster in S. lividans TK23.
To investigate whether the dhbACEBG genes are sufficient for the production of DHBA, a plasmid containing the dhbACEBG genes under the control of the constitutive promoter ermE*p (pSP126/1079) was integrated into the genome of S. lividans TK23. No catechol was found in the supernatant of strain TK23, whereas the catechol concentration in the supernatant of TK23(pSP126/1079) was approximately 4-fold higher than that in the supernatant of ATCC 700974 (Fig. 6). The UV/Vis spectrum of the supernatant of TK23(pSP126/1079) displayed an absorption peak at 315 nm, which can be attributed to DHBA, whereas the spectrum of the supernatant of ATCC 700974 displayed an absorption peak at 330 nm, attributed to griseobactin (see below). The peptide concentration in the culture supernatant of TK23(pSP126/1079) was as low as that for TK23, indicating no siderophore production. The same held true for TK23 containing cosmid 679, which comprises dhbGBECA and griABCD but lacks the griE gene encoding the NRPS (Fig. 6). These data suggested that TK23(pSP126/1079) and TK23 containing cosmid 679 overproduce DHBA, which diffuses into the supernatant, but do not synthesize griseobactin. In contrast, ATCC 700974 secreted the complete siderophore but little or no free DHBA, because DHBA is incorporated into the siderophore. The secretion of DHBA in the dhbACEBG-complemented strain S. lividans TK23(pSP126/1079) is likely to be the consequence of intracellular DHBA overflow owing to a lack of the NRPS gene griE. In a B. subtilis dhbF mutant lacking a functional NRPS for bacillibactin biosynthesis, secretion of the bacillibactin precursor DHBA was 74-fold higher than that in the wild type (39).
FIG. 6.
Heterologous expression in S. lividans TK23. The catechol (A) and peptide (B) concentrations in the supernatants of S. lividans TK23 (○), the dhbACEBG-complemented strain TK23(pSP126/1079) (▴), TK23 containing cosmid 679 (•), TK23 containing cosmid 680 (▪, ▾), and Streptomyces sp. strain ATCC 700974 (□) were determined using the Arnow and peptide assays, respectively. Cells were grown at 30°C in M9 minimal medium with (▾) or without (•, ▪, ▴, □, ○) the addition of 50 μM Fe3+.
We expressed the entire griseobactin gene cluster in S. lividans to see whether it is sufficient for griseobactin production. Catechol, as well as the peptide moiety of griseobactin, was detected in the supernatant of TK23 containing cosmid 680 (Fig. 6). After subtraction of the background, the ratio of the catechol concentration to the peptide concentration was the same for TK23 containing cosmid 680 and ATCC 700974. This indicates that griseobactin was synthesized by TK23 containing cosmid 680. The higher levels of griseobactin synthesis in S. lividans TK23 containing cosmid 680 than in ATCC 700974 might be due to stronger gene expression. Biosynthesis of griseobactin in TK23 containing cosmid 680 was repressed upon the addition of iron (Fig. 6), which indicated negative iron regulation, as observed for ATCC 700974.
Chemical characterization of the siderophore.
We purified griseobactin on an XAD-2 column, followed by gel filtration. Purified griseobactin displayed a UV/Vis spectrum typical for catechol siderophores, with an absorption peak at 330 nm resulting from the π→π* transition of catechol (Fig. 7), in contrast to an absorption peak at 315 nm for free DHBA under these conditions. Upon addition of FeCl3, a broad absorption peak around 500 nm appeared (Fig. 7), and the solution turned from colorless to reddish brown. This absorption band is assigned to a ligand-to-metal charge transfer band characteristic for Fe3+ octahedrally coordinated by three catecholate units. This suggested that the siderophore was isolated predominantly in the iron-free form and formed a complex with ferric iron upon the addition of iron.
FIG. 7.
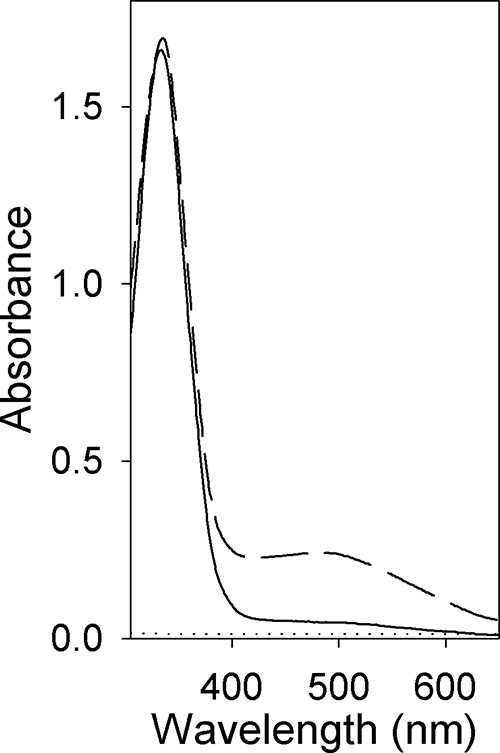
Absorption spectra of griseobactin (150 μM) in water with and without iron. Solid line, the purified siderophore without iron; dashed line, the siderophore after the addition of 150 μM FeCl3; dotted line, the same purification fraction of the dhbACE mutant SIP1192.
The NRPS activity in griseobactin synthesis is distributed among three proteins: DhbE, DhbG (aryl carrier for DHBA), and GriE (Fig. 2). The substrate specificity of the adenylation domains was predicted by the NRPSpredictor program to be DHBA for DhbE, threonine for GriE module 1, and glutamine for GriE module 2. Amino acid analysis of griseobactin revealed threonine (4.83 mM) but not glutamine. Instead, griseobactin contained arginine (4.87 mM) in amounts equimolar to those of threonine. For quantitative comparison of the amino acid concentration with the catechol content of the siderophore, the siderophore was hydrolyzed prior to the Arnow assay. This yielded catechol values 2- to 3-fold higher than those without hydrolysis, indicating that in the intact siderophore, only every second to third DHBA residue reacts in the Arnow assay, probably because of steric hindrance. Taking this into account, the DHBA concentration (4.5 mM) was as high as the concentrations of threonine or arginine.
Only a few nonribosomally synthesized peptides are known to contain arginine. The NRPSpredictor is based on only two peptides containing arginine: the toxins syringomycin (SyrE [UniProt accession no. O85168_m5]) of Pseudomonas syringae and microcystin (McyB [Q8RTG4_m1 and Q93LR1_m1] and McyC [Q9RNA9_m1 and Q9S1A7_m1]) of Microcystis aeruginosa (44). In contrast, the prediction of threonine is based on 24 threonine-incorporating modules. This may explain the prediction of glutamine instead of arginine by the NRPSpredictor. Thus, GriE may improve the prediction of NRPS modules incorporating arginine.
The ability of purified griseobactin to chelate iron was tested using the CAS liquid assay. The reaction was slower than the standard desferrioxamine B reaction. A griseobactin solution containing ∼200 μM catechol equivalents, as determined by the Arnow assay with prior hydrolysis of the siderophore, revealed ∼50 μM desferrioxamine B equivalents in the CAS assay. Taking into account the fact that three catechol equivalents are needed to bind one Fe3+ ion, this result indicated that approximately 75% of the isolated griseobactin was in the iron-free form and was capable of binding iron.
FT-ICR-MS of the siderophore revealed an m/z of 602.7284 for the double-negatively charged ion [M + 2H]2+. [M + 2H]+ was not detected, as often observed for samples with molecular masses greater than ≈1,200 Da. The monoisotopic molecular mass of the neutral molecular species, 1,203.4412 Da, agrees well with that of the aluminum complex of the cyclic trilactone of DHBA-arginine-threonine (C51H66N15O18Al; theoretical mass, 1,203.4525 Da). Aluminum siderophore complexes have been found previously, e.g., the iron-free siderophore pyoverdine, which contains aluminum in the cocrystal with the FpvA transporter (21). In addition to [M + 2H]2+ with an m/z of 602.7284, a smaller peak of [M + 2H]2+ with an m/z of 611.73411 was observed, corresponding to a monoisotopic molecular mass of 1,221.4526 Da for the neutral molecular species. The difference between these two species is 18.0114 Da and can be attributed to one H2O molecule with a relative error of 0.7 ppm. Thus, we propose that the molecule with an M of 1,221.4526 Da is the hydrolyzed linear form of the cyclic siderophore with an M of 1,203.4412 Da. In a sample purified in the same way but from the supernatant of SIP1192 (ΔdhbACE), both peaks were missing, which showed that this substance is not produced by the dhb mutant. However, these signals were present in the griseobactin fraction purified from the supernatants of the complemented dhbACE mutant SIP1192(pSP126/1079) and of the heterologously expressed griseobactin gene cluster in S. lividans TK23 containing cosmid 680, which indicated that the same siderophore is produced. In contrast to FT-ICR-MS, ESI-MS also detected the singly charged ion [M+H]+ with an m/z of 1,204.4 for the cyclic form and an m/z of 1,222.4 for the hydrated form, in addition to the double-positively charged ion [M + 2H]2+ in the hydrated form (m/z, 611.8) and, at a lower intensity, in the cyclic form (m/z, 602.8).
To date, no gene cluster for catechol siderophore biosynthesis in streptomycetes has been identified. The gene cluster of Streptomyces sp. strain ATCC 700974 sequenced in this study encodes proteins involved in the biosynthesis of DHBA and nonribosomal peptides, as well as proteins involved in the secretion, uptake, and utilization of siderophores. The culture supernatant of ATCC 700974 contained a catechol and a peptide moiety. Production of the siderophore was abolished by knocking out the dhbACE genes and was restored by complementation with the dhbACEBG genes. These results functionally link the synthesis of the catechol-peptide siderophore compound to the sequenced griseobactin biosynthesis gene cluster.
The entire gene cluster for griseobactin biosynthesis was heterologously expressed in S. lividans TK23. Griseobactin was identified by catechol and peptide quantification and by ESI-MS. To be functional, the aryl carrier protein DhbG and the PCP domains of the NRPS GriE have to be posttranslationally modified by the action of a dedicated 4′-phosphopantetheinyltransferase, which covalently links a phosphopantetheine moiety from coenzyme A to a highly conserved serine residue (29). Since no 4′-phosphopantetheinyltransferase gene was found within the griseobactin gene cluster, we propose that the function is taken over by a 4′-phosphopantetheinyltransferase encoded by the genome. 4′-Phosphopantetheinyltransferases have been shown to be rather nonselective, modifying all kinds of carrier protein domains (43).
The data suggest that griseobactin acts as a siderophore. Griseobactin biosynthesis is repressed by iron, and four putative iron boxes for repression by the DtxR-like iron regulator DmdR were identified in the nucleotide sequence. The iron-binding capacity of the purified griseobactin was shown by the CAS assay and by UV/Vis spectroscopy. Iron chelation was slow and incomplete, probably because the aluminum, which has to be displaced by iron, forms a kinetically stable complex. In addition, catechol compounds oxidize readily. Since ATCC 700974 synthesizes at least one other siderophore, dhb mutants were still able to grow under iron-limiting conditions. As in other bacteria, several streptomycetes do not rely only on one siderophore but satisfy their iron demand by using at least two siderophores, demonstrating the importance of iron.
The catechol-peptide siderophore griseobactin is structurally related to enterobactin, produced by the Gram-negative bacterium E. coli, and to bacillibactin, produced by the Gram-positive bacterium B. subtilis. These siderophores contain three DHBA residues for octahedral iron complexation that are linked to a cyclic amino acid scaffold synthesized by NRPSs. The structure of griseobactin is supported by the specificity of the two hydrolases homologous to GriB and GriF. Since the esterase Fes cleaves the ester bonds between serines in the trilactone enterobactin (32), GriB may cleave the ester bonds between threonines in the closely related trithreoninelactone in griseobactin. AbgB is believed to hydrolyze the amide bond of aminobenzoyl glutamate (23); thus, GriF may cleave the amide bond between the related aromatic DHBA and arginine in griseobactin. We predict a hydrolytic rather than a reductive release of iron, as observed for all other ester-linked bacterial siderophores, i.e., bacillibactin, enterobactin, and its glucosylated derivatives (32, 37).
In analogy with bacillibactin biosynthesis (35), we propose the following model for griseobactin biosynthesis. DHBA, arginine, and threonine are activated and transferred to their cognate aryl carrier/PCP domains by DhbE/DhbG, GriE module 1, and GriE module 2, respectively. The bound thioester PCP intermediates are subsequently condensed by the two C domains of GriE, leading to the attachment of DHBA-Arg-Thr as a thioester to the PCP domain of the second module of GriE (DHBA-Arg-Thr-S-PCP). The nucleophilic attack of the Ser hydroxyl group within the GXSXG motif of the TE domain of GriE leads to the attachment of DHBA-Arg-Thr as an ester to the TE domain (DHBA-Arg-Thr-O-TE). After a second round of DHBA-Arg-Thr-S-PCP synthesis, the thioester bond is attacked by the Thr hydroxyl group of DHB-Arg-Thr-O-TE, leading to (DHBA-Arg-Thr)2-O-TE. Iteration of this reaction results in the trimer (DHBA-Arg-Thr)3-O-TE, which is released by cyclization as a trilactone (DHBA-Arg-Thr)3. It will be interesting to elucidate the molecular mechanism that determines how in iterative NRPSs the TE domain counts the monomers stalled at the end of the assembly line and initiates release exactly when the desired length is achieved.
A catechol compound was found not only in the culture supernatant of Streptomyces sp. strain ATCC 700974 but also in those of the S. griseus strains Tü 6, Tü 17, and Tü 19. PCR of genomic DNAs of these strains with various primers, followed by partial sequencing, revealed the griseobactin gene cluster (data not shown). After the cloning and sequencing of cosmids 679 and 680 was finished and functions were assigned to the genes, the genome sequence of S. griseus IFO 13350 was published within the scope of genome projects (41), but no functional analyses had been performed. The IFO 13350 genome contains a gene cluster (SGR6730 to SGR6742) similar to the dhbGBECA and griABCDEFGH clusters. The sequence identity is approximately 90%, with minor deletions and insertions. Thus, we hypothesize that S. griseus IFO 13350 synthesizes griseobactin.
Acknowledgments
We gratefully acknowledge Graeme Nicholson (University of Tübingen) and Guido Sauer (this institute) for recording ESI-MS spectra, Norbert Tröndle (Genaxxon) for amino acid analysis, and Wolfgang Wohlleben and coworkers (University of Tübingen) for providing strains and plasmids. We thank Karen A. Brune for critical reading of the manuscript.
Footnotes
Published ahead of print on 13 November 2009.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansari, M. Z., G. Yadav, R. S. Gokhale, and D. Mohanty. 2004. NRPS-PKS: a knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res. 32:W405-W413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnow, L. E. 1937. Colorimetric determination of the components of 3,4-dihydroxyphenylalanine-tyrosine mixtures. J. Biol. Chem. 118:531-537. [Google Scholar]
- 4.Barnum, D. W. 1977. Spectrophotometric determination of catechol, epinephrine, dopa, dopamine and other aromatic vic-diols. Anal. Chim. Acta 89:157-166. [DOI] [PubMed] [Google Scholar]
- 5.Barona-Gómez, F., U. Wong, A. E. Giannakopulos, P. J. Derrick, and G. L. Challis. 2004. Identification of a cluster of genes that directs desferrioxamine biosynthesis in Streptomyces coelicolor M145. J. Am. Chem. Soc. 126:16282-16283. [DOI] [PubMed] [Google Scholar]
- 6.Bartolomé, B., Y. Jubete, E. Martínez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 7.Bendtsen, J. D., H. Nielsen, G. Von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 8.Bennett, P. M., J. Grinsted, and M. H. Richmond. 1977. Transposition of TnA does not generate deletions. Mol. Gen. Genet. 154:205-211. [DOI] [PubMed] [Google Scholar]
- 9.Bentley, S. D., K. F. Chater, A. M. Cerdeño-Tárraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 10.Besemer, J., and M. Borodovsky. 2005. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 33:W451-W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Besemer, J., A. Lomsadze, and M. Borodovsky. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 13.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 14.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 15.Chater, K. F., and L. C. Wilde. 1980. Streptomyces albus G mutants defective in the SalGI restriction-modification system. J. Gen. Microbiol. 116:323-334. [DOI] [PubMed] [Google Scholar]
- 16.Crosa, J. H., and C. T. Walsh. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66:223-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drake, E. J., J. Cao, J. Qu, M. B. Shah, R. M. Straubinger, and A. M. Gulick. 2007. The 1.8 Å crystal structure of PA2412, an MbtH-like protein from the pyoverdine cluster of Pseudomonas aeruginosa. J. Biol. Chem. 282:20425-20434. [DOI] [PubMed] [Google Scholar]
- 18.Fiedler, H. P., P. Krastel, J. Müller, K. Gebhardt, and A. Zeeck. 2001. Enterobactin: the characteristic catecholate siderophore of Enterobacteriaceae is produced by Streptomyces species. FEMS Microbiol. Lett. 196:147-151. [DOI] [PubMed] [Google Scholar]
- 19.Fischbach, M. A., and C. T. Walsh. 2006. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev. 106:3468-3496. [DOI] [PubMed] [Google Scholar]
- 20.Flores, F. J., and J. F. Martin. 2004. Iron-regulatory proteins DmdR1 and DmdR2 of Streptomyces coelicolor form two different DNA-protein complexes with iron boxes. Biochem. J. 380:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenwald, J., G. Zeder-Lutz, A. Hagege, H. Celia, and F. Pattus. 2008. The metal dependence of pyoverdine interactions with its outer membrane receptor FpvA. J. Bacteriol. 190:6548-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Günter-Seeboth, K., and T. Schupp. 1995. Cloning and sequence analysis of the Corynebacterium diphtheriae dtxR homologue from Streptomyces lividans and S. pilosus encoding a putative iron repressor protein. Gene 166:117-119. [DOI] [PubMed] [Google Scholar]
- 23.Hussein, M. J., J. M. Green, and B. P. Nichols. 1998. Characterization of mutations that allow p-aminobenzoyl-glutamate utilization by Escherichia coli. J. Bacteriol. 180:6260-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imbert, M., M. Béchet, and R. Blondeau. 1995. Comparison of the main siderophores produced by some species of Streptomyces. Curr. Microbiol. 31:129-133. [Google Scholar]
- 25.Juncker, A. S., H. Willenbrock, G. Von Heijne, S. Brunak, H. Nielsen, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keating, T. A., and C. T. Walsh. 1999. Initiation, elongation, and termination strategies in polyketide and polypeptide antibiotic biosynthesis. Curr. Opin. Struct. Biol. 3:598-606. [DOI] [PubMed] [Google Scholar]
- 27.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Charter, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 28.Köster, W. 2001. ABC transporter-mediated uptake of iron, siderophores, heme and vitamin B-12. Res. Microbiol. 152:291-301. [DOI] [PubMed] [Google Scholar]
- 29.Lambalot, R. H., A. M. Gehring, R. S. Flugel, P. Zuber, M. LaCelle, M. A. Marahiel, R. Reid, C. Khosla, and C. T. Walsh. 1996. A new enzyme superfamily—the phosphopantetheinyl transferases. Chem. Biol. 3:923-936. [DOI] [PubMed] [Google Scholar]
- 30.Lautru, S., R. J. Deeth, L. M. Bailey, and G. L. Challis. 2005. Discovery of a new peptide natural product by Streptomyces coelicolor genome mining. Nat. Chem. Biol. 1:265-269. [DOI] [PubMed] [Google Scholar]
- 31.Lautru, S., D. Oves-Costales, J. L. Pernodet, and G. L. Challis. 2007. MbtH-like protein-mediated cross-talk between non-ribosomal peptide antibiotic and siderophore biosynthetic pathways in Streptomyces coelicolor M145. Microbiology 153:1405-1412. [DOI] [PubMed] [Google Scholar]
- 32.Lin, H., M. A. Fischbach, D. R. Liu, and C. T. Walsh. 2005. In vitro characterization of salmochelin and enterobactin trilactone hydrolases IroD, IroE, and Fes. J. Am. Chem. Soc. 127:11075-11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacNeil, D. J., K. M. Gewain, C. L. Ruby, G. Dezeny, P. H. Gibbons, and T. MacNeil. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61-68. [DOI] [PubMed] [Google Scholar]
- 34.Marahiel, M. A., T. Stachelhaus, and H. D. Mootz. 1997. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97:2651-2673. [DOI] [PubMed] [Google Scholar]
- 35.May, J. J., T. M. Wendrich, and M. A. Marahiel. 2001. The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J. Biol. Chem. 276:7209-7217. [DOI] [PubMed] [Google Scholar]
- 36.Mies, K. A., J. I. Wirgau, and A. L. Crumbliss. 2006. Ternary complex formation facilitates a redox mechanism for iron release from a siderophore. Biometals 19:115-126. [DOI] [PubMed] [Google Scholar]
- 37.Miethke, M., O. Klotz, U. Linne, J. J. May, C. L. Beckering, and M. A. Marahiel. 2006. Ferri-bacillibactin uptake and hydrolysis in Bacillus subtilis. Mol. Microbiol. 61:1413-1427. [DOI] [PubMed] [Google Scholar]
- 38.Miethke, M., and M. A. Marahiel. 2007. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71:413-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miethke, M., S. Schmidt, and M. A. Marahiel. 2008. The major facilitator superfamily-type transporter YmfE and the multidrug-efflux activator Mta mediate bacillibactin secretion in Bacillus subtilis. J. Bacteriol. 190:5143-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nardini, M., and B. W. Dijkstra. 1999. α/β Hydrolase fold enzymes: the family keeps growing. Curr. Opin. Struct. Biol. 9:732-737. [DOI] [PubMed] [Google Scholar]
- 41.Ohnishi, Y., J. Ishikawa, H. Hara, H. Suzuki, M. Ikenoya, H. Ikeda, A. Yamashita, M. Hattori, and S. Horinouchi. 2008. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 190:4050-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quadri, L. E. N., J. Sello, T. A. Keating, P. H. Weinreb, and C. T. Walsh. 1998. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem. Biol. 5:631-645. [DOI] [PubMed] [Google Scholar]
- 43.Quadri, L. E. N., P. H. Weinreb, M. Lei, M. M. Nakano, P. Zuber, and C. T. Walsh. 1998. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry 37:1585-1595. [DOI] [PubMed] [Google Scholar]
- 44.Rausch, C., T. Weber, O. Kohlbacher, W. Wohlleben, and D. H. Huson. 2005. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs). Nucleic Acids Res. 33:5799-5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rusnak, F., M. Sakaitani, D. Drueckhammer, J. Reichert, and C. T. Walsh. 1991. Biosynthesis of the Escherichia coli siderophore enterobactin—sequence of the entF gene, expression and purification of EntF, and analysis of covalent phosphopantetheine. Biochemistry 30:2916-2927. [DOI] [PubMed] [Google Scholar]
- 46.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 48.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 49.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493-505. [DOI] [PubMed] [Google Scholar]
- 50.Stergiopoulos, I., L. H. Zwiers, and M. A. De Waard. 2002. Secretion of natural and synthetic toxic compounds from filamentous fungi by membrane transporters of the ATP-binding cassette and major facilitator superfamily. Eur. J. Plant Pathol. 108:719-734. [Google Scholar]
- 51.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.von Döhren, H., R. Dieckmann, and M. Pavela-Vrancic. 1999. The nonribosomal code. Chem. Biol. 6:R273-R279. [DOI] [PubMed] [Google Scholar]
- 53.Wandersman, C., and P. Delepelaire. 2004. Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58:611-647. [DOI] [PubMed] [Google Scholar]
- 54.Wolpert, M., B. Gust, B. Kammerer, and L. Heide. 2007. Effects of deletions of mbtH-like genes on clorobiocin biosynthesis in Streptomyces coelicolor. Microbiology 153:1413-1423. [DOI] [PubMed] [Google Scholar]