Abstract
Bacterial spores are encased in a multilayered proteinaceous shell known as the coat. In Bacillus subtilis, over 50 proteins are involved in spore coat assembly but the locations of these proteins in the spore coat are poorly understood. Here, we describe methods to estimate the positions of protein fusions to fluorescent proteins in the spore coat by using fluorescence microscopy. Our investigation suggested that CotD, CotF, CotT, GerQ, YaaH, YeeK, YmaG, YsnD, and YxeE are present in the inner coat and that CotA, CotB, CotC, and YtxO reside in the outer coat. In addition, CotZ and CgeA appeared in the outermost layer of the spore coat and were more abundant at the mother cell proximal pole of the forespore, whereas CotA and CotC were more abundant at the mother cell distal pole of the forespore. These polar localizations were observed both in sporangia prior to the release of the forespore from the mother cell and in mature spores after release. Moreover, CotB was observed at the middle of the spore as a ring- or spiral-like structure. Formation of this structure required cotG expression. Thus, we conclude not only that the spore coat is a multilayered assembly but also that it exhibits uneven spatial distribution of particular proteins.
Proper localization and assembly of proteins in cells and subcellular structures are essential features of living organisms. Complex protein assemblies, including ribosomes, flagella, and the cytokinetic machinery, play important roles in bacteria (26, 27, 40). Studying how these complex structures are formed is a fundamental theme in molecular biology. In this work, we developed a method to analyze one of the most complex bacterial protein assemblies: the spore coat of Bacillus subtilis.
Sporulation of B. subtilis is initiated in response to nutrient limitation, and it involves a highly ordered program of gene expression and morphological change (33, 42). The first morphological change of sporulation is the appearance of an asymmetrically positioned septum that divides the cell into a larger mother cell and a smaller forespore. Next, the mother cell membrane migrates around the forespore membrane during a phagocytosis-like process called engulfment. The completion of engulfment involves fusion of the mother cell membrane to pinch off the forespore within the mother cell. Compartment-specific gene expression brings about maturation of the spore and its release upon lysis of the mother cell (reviewed in reference 19). Mature spores remain viable during long periods of starvation and are resistant to heat, toxic chemicals, lytic enzymes, and other factors capable of damaging vegetative cells (30). Spores germinate and resume growth when nutrients become available (32).
The outer portions of Bacillus spores consist of a cortex, a spore coat layer, and in some cases, an exosporium. The cortex, a thick layer of peptidoglycan, is deposited between the inner and the outer membranes of the forespore, and it is responsible for maintaining the highly dehydrated state of the core, thereby contributing to the extreme dormancy and heat resistance of spores. Spore coat assembly involves the deposition of at least 50 protein species (12, 21, 24) into two major layers: an electron-dense outer layer, called the outer coat, and a less electron-dense inner layer with a lamellar appearance, called the inner coat (50). These layers provide a protective barrier against bactericidal enzymes and chemicals, such as lysozyme and organic solvents (30). Although disruption of any one gene encoding a spore coat protein typically has little or no effect on spore resistance, morphology, or germination, a few proteins, referred to as morphogenetic proteins, play central roles in the assembly of the spore coat (7, 10, 13). One of the morphogenetic proteins, CotE, is located between the inner and outer coats and directs the assembly of most or all of the outer coat proteins and also a few of the inner coat proteins (2, 9, 17, 25, 52). The locations of CotE, CotS, and SpoIVA in the spore coat were determined previously by immunoelectron microscopy (9, 43). CotA, CotB, CotC, and CotG were shown to be externally exposed on the surface of the spore by single-molecule recognition force spectroscopy or antibody accessibility (15, 18, 45, 28). However, the positions of most of the spore coat proteins in the coat have not been determined experimentally, although provisional assignments were made based largely on the control of assembly into the coat by CotE (17). In this study, we developed methods to estimate the positions of proteins in the spore coat layers by using fluorescence microscopy analysis of coat protein-fluorescent protein fusions, with resolution that allowed us to distinguish between the inner and outer coats. In addition, we discovered an asymmetric spatial distribution of four spore coat proteins and a ring- or spiral-like structure of CotB. These observations suggest that spore coat assembly is more intricate than previously appreciated.
MATERIALS AND METHODS
General methods and bacterial constructions.
B. subtilis was cultured in Luria-Bertani medium and induced to sporulate by exhaustion in Schaeffer's medium (31) at 37°C for 24 h. Plasmid DNA for transformation of B. subtilis was harvested from Escherichia coli strain JM109. All strain constructions were verified by the PCR. Bacterial strains, plasmids, and primers used in this study are listed in Tables S1 and S2 in the supplemental material. To fuse red fluorescent protein (RFP) to the C termini of CgeA, CotA, and CotB, each corresponding gene with its 5′ promoter region (36, 38, 53) was PCR amplified using genomic DNA from B. subtilis 168 as the template and primer pairs CGEAM490 and CGEA398R, COTAM430 and COTA1538R, and COTBM246 and COTB1139R, respectively (see Table S2 in the supplemental material). Fragments were digested with BamHI and XhoI and then cloned into BamHI/XhoI-digested pRFP3KP, resulting in the plasmids pCGEA3RP, pCOTA3RP, and pCOTB3RP (see Table S1 in the supplemental material). These plasmids were introduced into the aprE locus by a single-crossover event with selection for resistance to kanamycin (at 10 μg/ml), yielding strains CGEA3RP, COTA3RP, and COTB3RP (see Table S1 in the supplemental material). To fuse green fluorescent protein (GFP) to the C termini of CotB, CotC, CotD, CotF, CotZ, GerQ, YaaH, YmaG, YsnD, and YtxO, a segment of each corresponding gene was PCR amplified using genomic DNA from B. subtilis 168 as the template and primer pairs COTB8 and COTB1139R, COTC5 and COTC355R, COTD7 and COTD226R, COTF41 and COTF479R, COTZ20 and COTZ443R, GERQ20 and GERQ542R, YAAH636 and YAAH1380R, YMAG20 and YMAG368R, YSND20 and YSND532R, and YTXO23 and YTXO428R (see Table S1 in the supplemental material). Fragments were digested with BamHI and XhoI and then cloned into BamHI/XhoI-digested pGFP7C, resulting in the plasmids pCOTB8G, pCOTC8G, pCOTD8G, pCOTF8G, pCOTZ8G, pGERQ8G, pYAAH8G, pYMAG8G, pYSND8G, and pYTXO8G (see Table S1 in the supplemental material). These plasmids were introduced into the native locus by a single-crossover event with selection for resistance to chloramphenicol (at 5 μg/ml), yielding strains COTB8G, COTC8G, COTD8G, COTF8G, COTZ8G, GERQ8G, YAAH8G, YMAG8G, YSND8G, and YTXO8G (see Table S1 in the supplemental material). Although all GFP fusions used in this study have a six-histidine tag at the C-terminal end, we hereinafter refer to these constructs as GFP fusions.
Microscopy.
Following sporulation of the bacteria in Schaeffer's medium for 24 h, aliquots of cells were transferred onto microscope slides. The fluorescence of the GFP and RFP fusion proteins was observed using a BX51 fluorescent microscope fitted with mirror cube units for GFP (product no. U-MGFPHQ) and RFP (product no. U-MWG2; Olympus, Japan). A UPlanApo 100× objective lens (Olympus, Japan) and a U-TV1X-2 camera adapter were used. The images were captured using a cooled charge-coupled device camera (CoolSNAP ES/OL; Roper Scientific). Fluorescence intensities and the distance between two fluorescent peaks were measured using unadjusted merged images with RS Image Express processing software, version 4.5 (Roper Scientific). Autofluorescence of spores was observed after an exposure time of 5 or 3 s under our experimental conditions to capture the GFP or RFP fluorescence, respectively. The exposure time for image capture of GFP fusions was 0.5 to 2 s. The exposure time for image capture of RFP fusions was 0.5 to 1 s. One pixel corresponds to 64.5 nm in our detection system. Overlay images of GFP and RFP were generated by RS Image Express processing software, version 4.5. The contrast and tone balance of the overlay images of GFP and RFP were adjusted by using CANVAS 8 software (Deneba Systems Inc.).
RESULTS
Relative localization patterns of spore coat proteins.
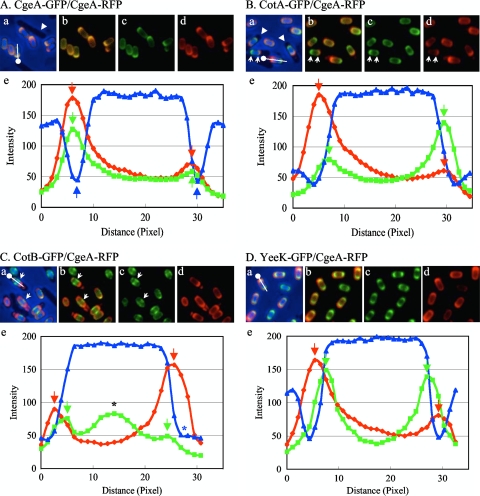
To localize spore coat proteins of B. subtilis by using fluorescent microscopy, we first fused GFP or RFP to the C termini of spore coat proteins CgeA, CotA, CotB, and YeeK. Although gfp was translationally fused to the spore coat protein gene at the native locus so that the fusion gene was the only copy expressed, genes encoding RFP-fused proteins were introduced at the aprE locus and the intact gene remained at the native locus. Strains capable of expressing both GFP-fused and RFP-fused coat proteins were induced to sporulate and observed by fluorescence microscopy. Up to 9% of the population exhibited only one color upon fluorescence microscopy analysis under our experimental conditions. These spores were not subjected to measurements. Both CgeA-GFP and CgeA-RFP surrounded the spore, and the fluorescence patterns matched very well (Fig. 1A, panels a to d). Intensities of the fluorescence of GFP and RFP along the long axis of the spore were measured (Fig. 1, line with dot in panels a), and the plot showed two peaks at the poles of a spore (Fig. 1A, panel e, green and red arrows). We defined the distance between the two peaks as the diameter of the protein layer. The diameters of the CgeA-GFP and CgeA-RFP layers were in very close agreement, as shown in Fig. 1A, panel e, suggesting that the two proteins assembled into the same layer of the spore coat. The diameters of the CotA-GFP, CotB-GFP, and YeeK-GFP layers were measured for strains engineered to also express CgeA-RFP, and the GFP diameters were shorter than the RFP diameter, suggesting that the CgeA-RFP layer is outside the CotA-GFP, CotB-GFP, and YeeK-GFP layers (Fig. 1B to D, panels e). Strains harboring a deletion of most of the cgeAB operon produced spores that had a tendency to clump, formed compact pellets (relative to those formed by wild-type spores) when centrifuged, and adhered to the surfaces of glass or plastic tubes (36). These observations suggest that the absence of CgeA and CgeB alters the surface of the spore, consistent with the idea that CgeA is in the outermost layer.
FIG. 1.
Fluorescence localization of coat protein-GFP and -RFP fusions and plots of fluorescence and phase-contrast intensities along the long axis of the spore. Strains possessing CgeA-RFP and CgeA-GFP (A), CotA-GFP (B), CotB-GFP (C), or YeeK-GFP (D) were induced to sporulate by nutrient exhaustion. Merged GFP and RFP fluorescence and blue phase-contrast images (a), merged GFP and RFP fluorescence images (b), GFP fluorescence images (c), and RFP fluorescence images (d) are shown. The dot with the line in each panel a indicates the long axis of the spore for which plots of fluorescence and phase-contrast intensities are shown in the corresponding panel e. The dot in panel a indicates the side of distance zero in panel e. Green and red arrows in panels e indicate peaks of GFP and RFP fluorescence intensities, respectively. (A) Blue arrows in panel e indicate negative peaks of phase-contrast intensity. The white arrowhead in panel a indicates a nonsporulating cell. (B) White arrowheads in panel a indicate mother cells. White arrows indicate opposite abundance patterns of CotA-GFP and CgeA-RFP. (C) White arrows indicate a ring- or spiral-like structure of CotB-GFP. The black asterisk in panel e indicates the peak of fluorescence intensity of the ring- or spiral-like structure of CotB-GFP. The blue asterisk in panel e indicates the absence of a negative peak of phase-contrast intensity.
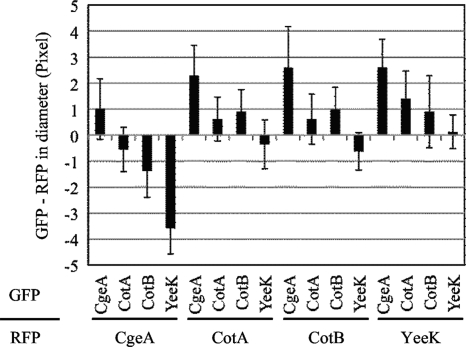
In order to determine the diameters of spore coat protein layers quantitatively, more than 20 free spores for each combination of GFP and RFP fusion proteins were subjected to measurements (Fig. 2). Unexpectedly, the diameters measured with GFP-fused proteins were typically slightly longer (up to 1 pixel) than those measured with the corresponding RFP-fused proteins. The differences may be due to effects of the fusion proteins on the assembly of the spore coat or to an optical property of green versus red fluorescence. Nevertheless, this approach clearly detected differences in the diameters of different protein layers. As shown in Fig. 2, CgeA fusions produced a larger diameter than all other proteins tested. CotA and CotB fusions were close in diameter. YeeK fusions produced a smaller diameter than the other proteins. These results are consistent with previous reports that CotA and CotB are located on the outer surfaces of spores and that YeeK is located in the inner coat (15, 44, 45). Since the diameters measured with GFP-fused proteins were typically slightly longer (up to 1 pixel) than those measured with the corresponding RFP-fused proteins, small differences in relative localizations are not very reliable. However, these data indicate that measurement of the fluorescence intensities from many spores is able to localize at least some proteins in the spore coat.
FIG. 2.
Difference in diameter between a GFP-fused protein layer and an RFP-fused protein layer. The diameter for the indicated RFP-fused protein (lower, horizontal label) was subtracted from that for the indicated GFP-fused protein (upper, vertical label). Bars indicate averages and error bars indicate standard deviations of results for 20 to 30 spores.
Absolute localization of spore coat proteins.
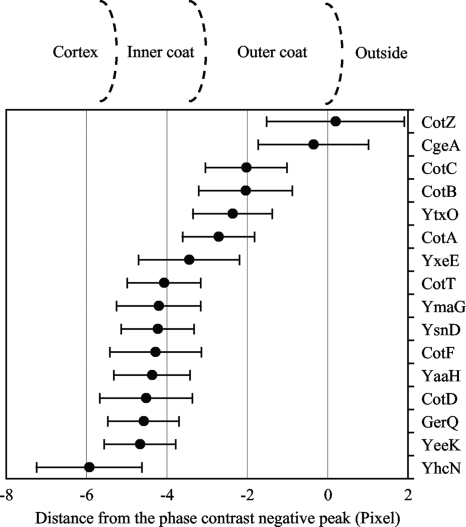
To determine the absolute locations of proteins in the spore coat, we utilized the observation that there is a negative peak of phase-contrast intensity at the boundary between the inside and the outside of the spore (Fig. 1A, panel e, blue arrows). Mature spores become bright as viewed by phase-contrast microscopy, whereas nonsporulating cells (Fig. 1A, panel a, arrowhead) and mother cells (Fig. 1B, panel a, arrowheads) are darker than the background. The negative peaks of phase-contrast intensity appeared to be near the spore surface since the diameter measured with these peaks was larger on average than the diameter measured with GFP-fused spore coat proteins, except in the case of CotZ-GFP (Fig. 3). Spores vary in length, but this variable was eliminated by comparing the GFP-fused spore coat protein diameter for each spore measured with the distance between the phase-contrast negative peaks by using merged images of GFP fluorescence and phase-contrast intensity. The strains did not have a gene encoding an RFP-fused protein, and only free spores were subjected to the measurements because a forespore surrounded by a dark mother cell did not have negative peaks of phase-contrast intensity (Fig. 1C, panel e).
FIG. 3.
Difference between the diameter of the GFP-fused protein layer and the distance between phase-contrast negative peaks. The distance between the two phase-contrast negative peaks was subtracted from the diameter of the GFP-fused protein layer. Points indicate averages and error bars indicate standard deviations of results for 60 spores. Estimated positions of the cortex, inner coat, and outer coat layers are shown above the graph.
As shown in Fig. 3, among the measured proteins, CotZ-GFP and CgeA-GFP were in the outermost layer. Since these measurements had large standard deviations, P values for the results were calculated to confirm this observation. The P value is the probability that the two samples have the same mean and that the observed difference is a coincidence of random sampling. If the P value was lower than 0.01, we considered the difference between the two samples to be statistically significant. The P value for CotZ-GFP and CgeA-GFP was 0.055, but that for CgeA-GFP and CotC-GFP was 7.8 × 10−12, suggesting that CotZ and CgeA are in a different layer from CotC, which is closer to the inside of the spore. CotE, which is sandwiched between the inner and outer coats, directs the assembly of most or all the outer coat proteins and some inner coat proteins (17). Assembly of CotZ onto the spore was shown previously to depend on CotE (17). We found that CgeA assembly also depends on CotE (see Fig. S1 in the supplemental material). We conclude that CotZ and CgeA are in the outermost layer of the spore coat.
CotC, CotB, YtxO, and CotA were in the outer coat layer based on the data in Fig. 3. CotB, YtxO, and CotA were previously shown to depend on CotE for assembly onto the spore (17, 52). We found that CotC assembly was also dependent on CotE (see Fig. S1 in the supplemental material). CotB, CotC, and CotA are believed to be exposed on the surface of the spore (15, 28, 45, 52). These data suggest that CotB, CotC, YtxO, and CotA are located in the outer spore coat, and presumably, although CgeA and CotZ are located farther out, these proteins do not cover the entire spore surface, so CotB, CotC, and CotA are also externally exposed.
YxeE appeared in a layer next to the outer coat (Fig. 3). CotE directs the assembly of the outer coat proteins and also a few of the inner coat proteins (17). Thus, dependence on CotE for assembly into the coat does not necessarily indicate that the protein is in the outer coat. However, CotE independence likely indicates that the protein is in the inner coat because cotE mutant spores lack the outer coat layer (52). YxeE assembly onto the spore was shown previously to be independent of CotE, suggesting that YxeE is an inner coat protein (17). Based on this report, we infer that YxeE and proteins farther in (with the exception of YhcN [see below]), including CotT, YmaG, YsnD, CotF, YaaH, CotD, GerQ, and YeeK, are located in the inner coat (Fig. 3). GerQ assembly was shown previously to depend on CotE (34), and we found that CotF assembly was also dependent on CotE (see Fig. S1 in the supplemental material). The assembly of YmaG, YsnD, and YeeK was shown previously to be independent of CotE (17, 44). The CotE independence of CotD assembly was demonstrated previously by SDS-PAGE (52). Using fluorescence microscopy, we found that CotT, YaaH, and CotD assembled onto the spore without CotE (see Fig. S1 in the supplemental material), although localization of CotT and YaaH was aberrant (see Fig. S1G and J in the supplemental material). Consistent with the idea that CotT is located in the inner coat, it has been reported previously that its absence or its overproduction causes alteration of the inner coat structure (4). YhcN is produced in the forespore in a σG-dependent manner, and it has a predicted lipoprotein signal peptide, suggesting that it is located in the inner membrane and the cortex (1, 46). All the other proteins tested were farther out than YhcN, supporting their assignment as spore coat proteins.
Uneven distributions of spore coat proteins.
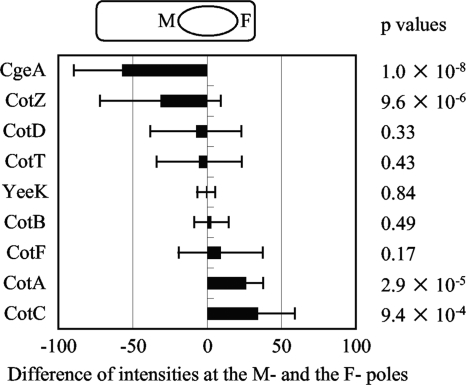
In the course of this study, we found that both CgeA-GFP and CgeA-RFP were more abundant at one pole of the spore than the other pole (Fig. 1A). On the other hand, CotA-GFP was more abundant at the pole opposite CgeA-RFP (Fig. 1B, white arrows). Although these opposite localizations were more apparent in sporangia, they were also observed in free spores. These opposite localizations were also observed in the strain possessing CotA-RFP and CgeA-GFP (data not shown). In sporangia, CgeA was abundant at the mother cell proximal pole of the forespore (M-pole) and CotA was abundant at the mother cell distal pole of the forespore (F-pole) (Fig. 1B, panels a to d, arrows). In order to investigate the polarity of the spore, differences between the fluorescence intensities of several spore coat proteins at the M- and F-poles were measured (Fig. 4). CotD, CotT, YeeK, CotB, and CotF did not show obvious differences between M- and F-pole intensities, and all P values were greater than 0.01, suggesting that these protein layers did not have polarity. However, CgeA and CotZ were more abundant at the M-pole and CotA and CotC were more abundant at the F-pole (Fig. 4). A few forespores had inverted CotZ-GFP intensity patterns, in which CotZ was more abundant at the F-pole, so the standard deviation for CotZ was greater than those for the other proteins, but CotZ was more abundant at the M-poles of most forespores in the population. Inverted fluorescence intensity patterns were not observed for CgeA-GFP, CotA-GFP, or CotC-GFP. The P values for the fluorescence intensities of CgeA-GFP, CotZ-GFP, CotA-GFP, and CotC-GFP at M- and F-poles were very low, indicating that these proteins were clearly more abundant at a particular pole. Therefore, we propose that the coats of B. subtilis spores have polarity, that CgeA and CotZ are M-pole proteins, and that CotA and CotC are F-pole proteins.
FIG. 4.
Difference between fluorescence intensities at forespore poles. The fluorescence intensity of the indicated GFP-fused protein at the M-pole peak was subtracted from that at the F-pole peak. Bars indicate averages and error bars indicate standard deviations of results for 20 to 30 forespores. The diagram at the top of the graph indicates that left and right bars represent higher intensity at the M-pole and the F-pole, respectively. Two-tail P values for fluorescence intensities at M- and F-poles of each strain are shown to the right.
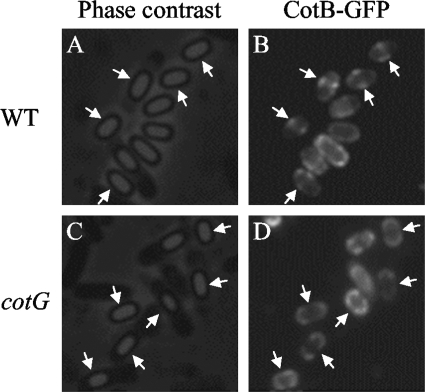
In contrast, CotB-GFP was observed to be abundant at the midspore position both before and after the release of the spore from the mother cell under our experimental conditions (Fig. 1C, black asterisk and white arrows). The ring- or spiral-like structure of CotB-GFP was also observed without CgeA-RFP (Fig. 5B, arrows), as well as for CotB-RFP (data not shown). CotB is synthesized as a 46-kDa protein and is converted into a 66-kDa form in a CotG-dependent manner (54). We tested whether CotB-GFP localization at midspore depends on cotG expression. Midspore localization of CotB-GFP was abolished in a cotG mutant, although CotB-GFP still surrounded the spore (Fig. 5D, arrows; also see Fig. S1C in the supplemental material for a merged color image). These results indicate that CotB-GFP is able to assemble on the spore but it needs cotG expression and/or conversion into the 66-kDa form to localize at midspore.
FIG. 5.
Midspore localization of CotB-GFP depends on cotG. Phase-contrast images (A and C) and fluorescence images (B and D) of CotB-GFP in the wild type (WT; A and B) and a cotG mutant (C and D) are shown. Arrows indicate midspore localization of CotB-GFP (A and B) and its absence (C and D).
Taken together, our results demonstrate not only that the spore coat of B. subtilis is a multilayered protein structure but also that it exhibits uneven spatial distribution of at least five proteins.
DISCUSSION
We have developed methods to estimate the positions of proteins in the coats of B. subtilis spores. These methods approach the limit of discrimination of our detection system, so the spore-to-spore variability was significant as shown by the error bars in Fig. 3. However, by measuring many spores (e.g., 60 for the results presented in Fig. 3), we could assign nine proteins to the inner coat and six proteins to the outer coat. Previously, the locations of CotE, CotS, and SpoIVA were determined by immunoelectron microscopy (9, 43). However, this method requires special training and equipment. CotA, CotB, CotC, and CotG were demonstrated previously to be exposed on the surface of the spore (15, 18, 28, 45). The methods used to obtain these findings rely on surface exposure and are unable to analyze the inner layer of the spore coat. In previous work, the locations of most proteins in the spore coat were provisionally assigned based largely on control by CotE (17). However, cotE disruption disrupts the outer coat layer and also affects the assembly of some inner coat proteins (17), so assignments based on CotE dependence are not definitive. Many recent studies of protein localization have exploited fluorescence microscopy with fluorescent proteins, but these methods have not been used to distinguish inner and outer spore coat proteins until recently because the resolution was insufficient (44). In this study, measurements of the diameters of the fluorescent spore coat protein layers of many spores achieved sufficient resolution to distinguish inner and outer spore coats. This approach may also be able to distinguish the inner spore membrane, the cortex, and the outer spore membrane.
In some cases, fusion to a fluorescent protein influences interaction with other molecules of the same or other proteins, with consequences for the assembly or precise localization of the protein. Indeed, GerQ-GFP was reported previously to be only partially functional (34), suggesting that the localization of GerQ-GFP observed in this study may not represent the native localization of GerQ. Webb et al. also reported that free spores released from the sporangia of a strain expressing the CotE-GFP fusion were somewhat abnormal (51), indicating that at least some spore coat proteins were affected by fusion with a fluorescent protein. Since disruption of any one gene encoding a spore coat protein typically has little or no effect on spore resistance, morphology, or germination, the effect of the fusion of a fluorescent protein to a spore coat protein cannot be assessed in most cases. Therefore, we cannot rule out the possibility that the assignment of proteins to the inner and outer spore coats and the uneven spatial localizations observed in this study were affected by the fluorescent protein fusions. However, in terms of assigning spore coat proteins to the inner or outer coat, our results agree with assignments made using other methods. Combining the other methods with immunofluorescence microscopy may be desirable. Together, the methods provide powerful approaches to investigate the structures of spores of many endospore-forming bacteria, including pathogens.
CotZ and CgeA appeared to be in the outermost layer of the spore coat, farther out than CotC, CotB, and CotA, which were previously localized to the outer coat, and farther out than YtxO, which our results indicate is in the outer coat. CotA, CotB, CotC, and CotG were demonstrated previously to be exposed on the surface of the spore by antibody accessibility (15, 18, 28, 45). There are several possible reasons why CotZ-GFP and CgeA-GFP appeared to be farther out than CotA-GFP, CotB-GFP, and CotC-GFP in this study. As noted above, fluorescent protein fusions may not localize properly, so CotZ-GFP and CgeA-GFP may have failed to assemble properly in the spore coat and therefore appeared to be in an outermost layer. On the other hand, if CotZ-GFP and CgeA-GFP are properly assembled, then given the antibody accessibility of CotA, CotB, and CotC in previous studies (15, 18, 28, 45) and our finding that these proteins are closer to the inside of the spore than CotZ and CgeA (Fig. 3), our results suggest that CgeA and CotZ do not cover the entire surface. Spores of several species of Bacillus and Clostridium, including B. anthracis, are surrounded by an exosporium, a loosely fitting outermost structure observed in thin sections by electron microscopy. However, an exosporium has not been observed surrounding B. subtilis spores, with the exception of spores of strains isolated from the human gastrointestinal tract (14). Sousa et al. observed an exosporium-like layer of B. subtilis after partial chemical extraction of the spore coats (39). More recently, an external glycoprotein layer surrounding spores of B. subtilis, which is less apparent with traditional staining methods used for electron microscopy, was found by ruthenium red staining (49). These observations suggest the existence of an outermost layer of the B. subtilis spore coat. Examination of CotZ-GFP and CgeA-GFP after partial chemical extraction of the spore coats or purification of the external glycoprotein layer is needed to determine whether the outermost layer observed in this study corresponds to that observed in other studies.
In this study, nine proteins were assigned to the inner coat (Fig. 3). Previously, YxeE, YmaG, YsnD, and CotD were provisionally assigned to the inner coat because their assembly was CotE independent (17), consistent with our results. YaaH and GerQ were previously assigned to the outer coat based on their CotE dependence (17, 25, 34). The CotE dependence of YaaH assembly onto the spore was tested by SDS-PAGE (25). Although YaaH-GFP localized on the spore without CotE, its localization was aberrant (see Fig. S1J in the supplemental material). Taken together, our results suggest that although YaaH and GerQ are in the inner coat, localization of these proteins is strongly affected by cotE disruption.
A novel finding of our study is that CgeA-GFP and CotZ-GFP are more abundant at the M-pole and that CotA-GFP and CotC-GFP are more abundant at the F-pole of the forespore (Fig. 4). Interestingly, the two M-pole proteins are the two proteins in the outermost layer of the spore coat. Since these proteins are synthesized in the mother cell and probably assemble onto the spore last, the forespore (enlarged by a thick cortex and previously assembled spore coat proteins) may impair the transfer of proteins synthesized in the larger space of the mother cell to the F-pole. Obviously, this would not explain the two F-pole proteins. There may be some sort of specific docking site for CotA and CotC on the F-pole of the spore. Asymmetric structures known as appendages are observed on the spores of several species (8, 29, 35, 48). However, B. subtilis spores do not exhibit an appendage, so the four polar proteins are a novel asymmetric structure of the spore coat. Spores of some species, including B. subtilis, split open at the midpoint during germination, but in other species, such as B. cereus and B. anthracis, the outgrowing cell emerges from one pole (11). The exosporium of B. anthracis spores contains alanine racemase, which is absent from one pole, called the cap-like region (41). The outgrowing cell always escapes from the cap-like region, suggesting that the cap is designed to facilitate the emergence of the outgrowing cell. Asymmetric distribution of spore coat proteins may also dictate where the coat splits during germination.
We observed CotB-GFP as a CotG-dependent ring- or spiral-like structure at the midpoints of B. subtilis spores (Fig. 5). Yeast two-hybrid analysis revealed that CotB directly interacts with CotG and also with itself, suggesting that CotB is capable of forming multimers (54). We speculate that CotG directs formation of a CotB filament, which was observed as a ring- or spiral-like structure at the midspore. Transient ring structures of the spore coat proteins YabP-GFP and YheD-GFP were observed previously (47). However, these structures were not observed with mature spores (47). Although three-dimensional analysis of CotB-GFP fluorescence is necessary to determine the exact nature of the CotB-GFP structure, if it is a ring or a spiral, it will resemble structures made by the cytoskeletal proteins MreB and Mbl or the cytokinetic protein FtsZ (3, 16). However, disruption of cotB did not alter the spore shape or impair germination (6) (data not shown). Further investigations are needed to reveal the role of the unevenly distributed spore coat proteins discovered in this study.
Supplementary Material
Acknowledgments
We are grateful to Lee Kroos for critical reading of the manuscript and helpful suggestions.
This work was supported by a grant-in-aid for scientific research (C; no. 20580089) and a grant-in-aid for young scientists (B; no. 19780066) from the Japanese Society for the Promotion of Science.
Footnotes
Published ahead of print on 20 November 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bagyan, I., M. Noback, S. Bron, M. Paidhungat, and P. Setlow. 1998. Characterization of yhcN, a new forespore-specific gene of Bacillus subtilis. Gene 212:179-188. [DOI] [PubMed] [Google Scholar]
- 2.Bauer, T., S. Little, A. G. Stöver, and A. Driks. 1999. Functional regions of the B. subtilis spore coat morphogenetic protein CotE. J. Bacteriol. 181:7043-7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben-Yehuda, S., and R. Losick. 2002. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109:257-266. [DOI] [PubMed] [Google Scholar]
- 4.Bourne, N., P. C. FitzJames, and A. I. Aronson. 1991. Structural and germination defects of Bacillus subtilis spores with altered contents of a spore coat protein. J. Bacteriol. 173:6618-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reference deleted.
- 6.Donovan, W., L. B. Zheng, K. Sandman, and R. Losick. 1987. Genes encoding spore coat polypeptides from Bacillus subtilis. J. Mol. Biol. 196:1-10. [DOI] [PubMed] [Google Scholar]
- 7.Driks, A. 2002. Proteins of the spore core and coat, p. 527-536. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives. American Society for Microbiology, Washington, DC.
- 8.Driks, A. 2007. Surface appendages of bacterial spores. Mol. Microbiol. 63:623-625. [DOI] [PubMed] [Google Scholar]
- 9.Driks, A., S. Roels, B. Beall, C. P. J. Moran, and R. Losick. 1994. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 8:234-244. [DOI] [PubMed] [Google Scholar]
- 10.Driks, A., and P. Setlow. 2000. Morphogenesis and properties of the bacterial spore, p. 191-218. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. American Society for Microbiology, Washington, DC.
- 11.Hamilton, W. A., and J. M. Stubbs. 1967. Comparison of the germination and outgrowth of spores of Bacillus cereus and Bacillus polymyxa. J. Gen. Microbiol. 47:121-129. [DOI] [PubMed] [Google Scholar]
- 12.Henriques, A. O., and C. P. J. Moran. 2007. Structure, assembly, and function of the spore surface layers. Annu. Rev. Microbiol. 61:555-588. [DOI] [PubMed] [Google Scholar]
- 13.Henriques, A. O., T. V. Costa, L. O. Martins, and R. Zilhao. 2004. The functional architecture and assembly of the coat, p. 65-86. In R. E. Ricca, A. O. Henriques, and S. M. Cutting (ed.), Bacterial spore formers: probiotics and emerging applications. Horizon Biosciences, Norfolk, United Kingdom.
- 14.Hong, H. A., R. Khaneja, N. M. Tam, A. Cazzato, S. Tan, M. Urdaci, A. Brisson, A. Gasbarrini, I. Barnes, and S. M. Cutting. 2009. Bacillus subtilis isolated from the human gastrointestinal tract. Res. Microbiol. 160:134-143. [DOI] [PubMed] [Google Scholar]
- 15.Isticato, R., G. Cangiano, H. T. Tran, A. Ciabattini, D. Medaglini, M. R. Oggioni, M. De Felice, G. Pozzi, and E. Ricca. 2001. Surface display of recombinant proteins on Bacillus subtilis spores. J. Bacteriol. 183:6294-6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, L. J., R. Carballido-López, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913-922. [DOI] [PubMed] [Google Scholar]
- 17.Kim, H., M. Hahn, P. Grabowski, D. C. McPherson, M. M. Otte, R. Wang, C. C. Ferguson, P. Eichenberger, and A. Driks. 2006. The Bacillus subtilis spore coat protein interaction network. Mol. Microbiol. 59:487-502. [DOI] [PubMed] [Google Scholar]
- 18.Kim, J. H., C. S. Lee, and B. G. Kim. 2005. Spore-displayed streptavidin: a live diagnostic tool in biotechnology. Biochem. Biophys. Res. Commun. 331:210-214. [DOI] [PubMed] [Google Scholar]
- 19.Kroos, L. 2007. The Bacillus and Myxococcus developmental networks and their transcriptional regulators. Annu. Rev. Genet. 41:13-39. [DOI] [PubMed] [Google Scholar]
- 20.Reference deleted.
- 21.Kuwana, R., Y. Kasahara, M. Fujibayashi, H. Takamatsu, N. Ogasawara, and K. Watabe. 2002. Proteomics characterization of novel spore proteins of Bacillus subtilis. Microbiology 148:3971-3982. [DOI] [PubMed] [Google Scholar]
- 22.Reference deleted.
- 23.Reference deleted.
- 24.Lai, E.-M., N. D. Phadke, M. T. Kachman, R. Giorno, S. Vazquez, J. A. Vazquez, J. R. Maddock, and A. Driks. 2003. Proteomic analysis of the spore coats of Bacillus subtilis and Bacillus anthracis. J. Bacteriol. 185:1443-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Little, S., and A. Driks. 2001. Functional analysis of the Bacillus subtilis morphogenetic spore coat protein CotE. Mol. Microbiol. 42:1107-1120. [DOI] [PubMed] [Google Scholar]
- 26.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77-100. [DOI] [PubMed] [Google Scholar]
- 27.Margolin, W. 2001. Spatial regulation of cytokinesis in bacteria. Curr. Opin. Microbiol. 4:647-652. [DOI] [PubMed] [Google Scholar]
- 28.Mauriello, E. M., L. H. Duc, R. Isticato, G. Cangiano, H. A. Hong, M. De Felice, E. Ricca, and S. M. Cutting. 2004. Display of heterologous antigens on the Bacillus subtilis spore coat using CotC as a fusion partner. Vaccine 22:1177-1187. [DOI] [PubMed] [Google Scholar]
- 29.Mizuki, E., M. Ohba, T. Ichimatsu, S. H. Hwang, K. Higuchi, H. Saitoh, and T. Akao. 1998. Unique appendages associated with spores of Bacillus cereus isolates. J. Basic Microbiol. 38:33-39. [DOI] [PubMed] [Google Scholar]
- 30.Nicholson, W. L., N. Munakata, G. Horneck, H. J. Melosh, and P. Setlow. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination, and outgrowth, p. 391-450. In C. R. Harding and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, United Kingdom.
- 32.Paidhungat, M., and P. Setlow. 2001. Spore germination and outgrowth, p. 537-548. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives. American Society for Microbiology, Washington, DC.
- 33.Piggot, P., and R. Losick. 2002. Sporulation genes and intercompartmental regulation, p. 483-517. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives. American Society for Microbiology, Washington, DC.
- 34.Ragkousi, K., P. Eichenberger, C. van Ooij, and P. Setlow. 2003. Identification of a new gene essential for germination of Bacillus subtilis spores with Ca2+-dipicolinate. J. Bacteriol. 185:2315-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rode, L. J., M. A. Crawford, and M. G. Williams. 1967. Clostridium spores with ribbon-like appendages. J. Bacteriol. 93:1160-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roels, S., and R. Losick. 1995. Adjacent and divergently oriented operons under the control of the sporulation regulatory protein GerE in Bacillus subtilis. J. Bacteriol. 177:6263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reference deleted.
- 38.Sandman, K., L. Kroos, S. Cutting, P. Youngman, and R. Losick. 1988. Identification of the promoter for a spore coat protein gene in Bacillus subtilis and studies on the regulation of its induction at a late stage of sporulation. J. Mol. Biol. 200:461-473. [DOI] [PubMed] [Google Scholar]
- 39.Sousa, J. C., M. T. Silva, and G. Balassa. 1976. An exosporium-like outer layer in Bacillus subtilis spores. Nature 263:53-54. [DOI] [PubMed] [Google Scholar]
- 40.Stark, H. 2002. Three-dimensional electron cryomicroscopy of ribosomes. Curr. Protein Pept. Sci. 3:79-91. [DOI] [PubMed] [Google Scholar]
- 41.Steichen, C. T., J. F. Kearney, and C. L. Turnbough, Jr. 2007. Non-uniform assembly of the Bacillus anthracis exosporium and a bottle cap model for spore germination and outgrowth. Mol. Microbiol. 64:359-367. [DOI] [PubMed] [Google Scholar]
- 42.Stragier, P., and R. Losick. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30:297-341. [DOI] [PubMed] [Google Scholar]
- 43.Takamatsu, H., Y. Chikahiro, T. Kodama, H. Koide, S. Kozuka, K. Tochikubo, and K. Watabe. 1998. A spore coat protein, CotS, of Bacillus subtilis is synthesized under the regulation of σK and GerE during development and is located in the inner coat layer of spores. J. Bacteriol. 180:2968-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takamatsu, H., D. Imamura, R. Kuwana, and K. Watabe. 2009. Expression of yeeK during Bacillus subtilis sporulation and localization of YeeK to the inner spore coat using fluorescence microscopy. J. Bacteriol. 191:1220-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang, J., D. Krajcikova, R. Zhu, A. Ebner, S. Cutting, H. J. Gruber, I. Barak, and P. Hinterdorfer. 2007. Atomic force microscopy imaging and single molecule recognition force spectroscopy of coat proteins on the surface of Bacillus subtilis spore. J. Mol. Recognit. 20:483-489. [DOI] [PubMed] [Google Scholar]
- 46.Tjalsma, H., A. Bolhuis, J. D. Jongbloed, S. Bron, and J. M. van Dijl. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64:515-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Ooij, C., P. Eichenberger, and R. Losick. 2004. Dynamic patterns of subcellular protein localization during spore coat morphogenesis in Bacillus subtilis. J. Bacteriol. 186:4441-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walker, J. R., A. J. Gnanam, A. L. Blinkova, M. J. Hermandson, M. A. Karymov, Y. L. Lyubchenko, P. R. Graves, T. A. Haystead, and K. D. Linse. 2007. Clostridium taeniosporum spore ribbon-like appendage structure, composition and genes. Mol. Microbiol. 63:629-643. [DOI] [PubMed] [Google Scholar]
- 49.Waller, L. N., N. Fox, K. F. Fox, A. Fox, and R. L. Price. 2004. Ruthenium red staining for ultrastructural visualization of a glycoprotein layer surrounding the spore of Bacillus anthracis and Bacillus subtilis. J. Microbiol. Methods 58:23-30. [DOI] [PubMed] [Google Scholar]
- 50.Warth, A. D., D. F. Ohye, and W. G. Murrell. 1963. The composition and structure of bacterial spores. J. Cell Biol. 16:579-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Webb, C. D., A. Decatur, A. Teleman, and R. Losick. 1995. Use of green fluorescent protein for visualization of cell-specific gene expression and subcellular protein localization during sporulation in Bacillus subtilis. J. Bacteriol. 177:5906-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng, L., W. P. Donovan, P. C. Fitz-James, and R. Losick. 1988. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 2:1047-1054. [DOI] [PubMed] [Google Scholar]
- 53.Zheng, L. B., and R. Losick. 1990. Cascade regulation of spore coat gene expression in Bacillus subtilis. J. Mol. Biol. 212:645-660. [DOI] [PubMed] [Google Scholar]
- 54.Zilhão, R., M. Serrano, R. Isticato, E. Ricca, C. P. J. Moran, and A. O. Henriques. 2004. Interactions among CotB, CotG, and CotH during assembly of the Bacillus subtilis spore coat. J. Bacteriol. 186:1110-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.