Abstract
The genetic context, phylogeny, and biochemistry of a gene flanking the H2-forming methylene-H4-methanopterin dehydrogenase gene (hmdA), here designated hmdB, indicate that it is a new member of the radical S-adenosylmethionine enzyme superfamily. In contrast to the characteristic CX3CX2C or CX2CX4C motif defining this family, HmdB contains a unique CX5CX2C motif.
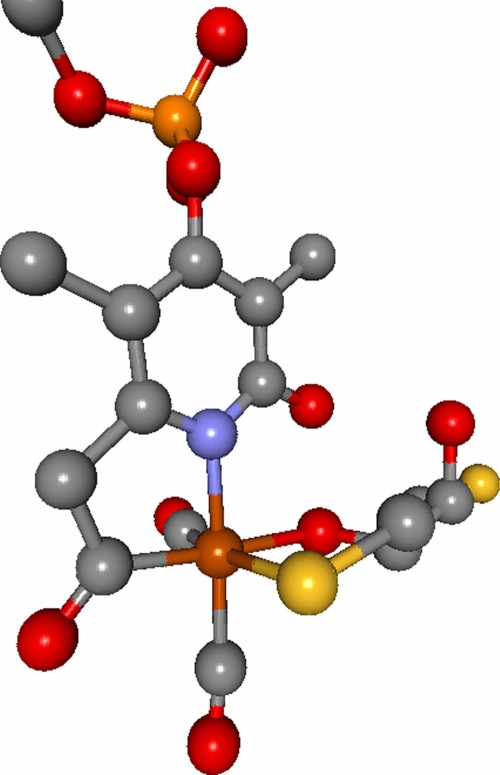
The HmdA enzyme is found in hydrogenotrophic methanogens, where it functions in the reversible reduction of methenyl-tetrahydromethanopterin (H4MPT+) to methylene-H4MPT and H+, an intermediary step in CO2 reduction to CH4 using H2 as an electron donor (33). Biochemical characterization of the enzyme has revealed the presence of a unique active site metal cofactor which consists of an Fe ion ligated by two CO molecules, a cysteine side chain, a guanylyl pyridinol cofactor (GP cofactor), and an unknown ligand suggested from crystallographic data to be an acyl group (Fig. 1) (8, 9, 12, 22, 28, 29). The active site features and activity of the HmdA protein, namely, the CO-coordinated Fe and the ability to react with H2, bear similarity to the [NiFe] and [FeFe] hydrogenases. These three hydrogenase classes are evolutionarily unrelated (17, 36, 34, 35), but all have CO-coordinated Fe at the active site (21, 29, 37), indicating that active site and functional similarities are a result of convergent evolution (14).
FIG. 1.
Active site of the Hmd hydrogenase as determined by X-ray crystallography (8), ligated by DTT and an acyl group from the pyridinol ring. Oxygen, red; nitrogen, blue; carbon, gray; iron, rust; sulfure, gold; phosphorus, orange. The figure was generated using the BALLView 1.3.0 software program (18).
The synthesis of the active site cluster of [NiFe] hydrogenase requires at least seven proteins (see reference 1 and references contained therein), whereas three are required for the synthesis of the active site cluster of the [FeFe] hydrogenases (15, 16, 24). Two oxygen-sensitive (10) radical S-adenosylmethionine (AdoMet) enzymes are involved in [FeFe] hydrogenase maturation (23, 24), whereas none are involved in the maturation of the [NiFe] hydrogenases. This observation may reflect the fact that [FeFe] hydrogenase is present only in anaerobic bacteria and several lower eukaryotes, whereas [NiFe] hydrogenases are widely distributed among aerobic and anaerobic members of both the archaea and bacteria (17, 34-36). None of the proteins thought to be involved in the assembly of the [NiFe] hydrogenase share homology with proteins involved in the assembly of the active site of [FeFe] hydrogenases, indicating that the maturation genes, like the structural genes, evolved independently. The presence of CO-ligated Fe and the GP cofactor suggests the involvement of a number of gene products in HmdA cofactor biosynthesis; however, such gene products have yet to be identified.
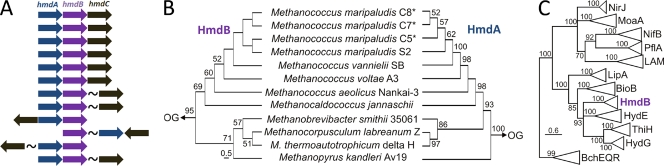
The genes involved in active site cluster biosynthesis in [FeFe] and [NiFe] hydrogenase are often colocalized with structural genes on the chromosome (17). We screened all available genome sequences for the presence of hmdA and examined the flanking genes. Two protein-encoding genes were colocalized with hmdA (Fig. 2A)(27), whose products are referred to herein as HmdB and HmdC. HmdB and HmdC exhibit sequence homology with radical AdoMet enzymes and eukaryotic fibrillarin, respectively. Importantly, these genes were not found associated with the hmdA homologs encoding HmdAII and HmdAIII, which have been proposed to act as scaffolds or cellular reservoirs of GP cofactor given the ability of HmdAII from Methanocaldococcus jannaschii to bind cofactor in vitro and the similarity of Hmd active sites (7, 27, 30).
FIG. 2.
(A) Gene context of hmdA, hmdB, and hmdC. hmdA, blue; hmdB, purple; hmdC, black. (B) Chronogram of HmdB rooted with a homolog identified in the genome of Desulfitobacterium hafniense Y51 and HmdAI rooted with HmdAII from Methanococcus maripaludis S2. The scale bar represents 0.5 substitutions per site. (C) Phylogram of representative members of the radical AdoMet superfamily of enzymes. Descriptions of the functions of the various AdoMet enzymes can be found in the work of Sofia et al. (31). The depth of the collapsed clades is proportional to the diversity of the individual enzyme class. The scale bar represents 0.6 substitutions per site.
As a first step toward investigating the role of the HmdB and HmdC proteins, we reconstructed the evolutionary history of HmdA, HmdB, and HmdC (see Methods in the supplemental material). The HmdA topology is nearly mirrored in the HmdB topology (Fig. 2B), with the primary exception being the sequence from Methanopyrus kandleri, which forms a lineage at the base of the Methanobacteriales/Methanomicrobiales lineage. In the HmdC phylogram, the deeply branching lineages were not statistically well supported, thereby precluding a detailed examination of the branching order among early-emerging taxa (data not shown). Nonetheless, two distinct lineages comprising sequences from Methanobacteriales/Methanomicrobiales/Methanopyrales and Methanococcales were apparent. Thus, the topology of the HmdC phylogram is similar to that observed in the HmdB phylogram. Similar tree topology for each of the individual HmdA, HmdB, and HmdC phylograms (27) suggests that these protein-encoding genes have coevolved.
The evolutionary relationship of HmdB to several members of the radical AdoMet superfamily of enzymes was determined by evolutionary model prediction using the ProtTest software program and maximum-likelihood phylogenetic reconstruction (see the supplemental material) and as previously described (2). HmdB clustered within a well-supported sequence lineage that contained ThiH, HydE, and HydG (Fig. 2C). HydE and HydG are required for the synthesis of the active site cluster of the [FeFe] hydrogenase (24). Thus, these proteins recently diverged from a common ancestor and therefore may catalyze similar chemistry. Sequence alignment of HmdB proteins indicates that they universally harbor a unique CX5CX2C motif, rather than the CX3CX2C or CX4CX2C motif characteristic of radical AdoMet enzymes (4, 6, 13, 31) (see Fig. S1 in the supplemental material).
To examine the biochemical characteristics of HmdB, the protein from Methanococcus maripaludis S2 was heterologously expressed in Escherichia coli. Cloning was accomplished as described in the supplemental material to allow for the expression of a 6×-His tag variant of HmdB. Ni2+ affinity chromatography yielded protein estimated to be ∼99% pure (data not shown). Gel filtration chromatography indicated that the protein existed as a monomer (data not shown). Iron analysis via inductively coupled plasma mass spectrometry (ICP-MS) on an Agilent 7500ce ICP-MS (see the supplemental material) revealed the protein bound 2.2 ± 0.3 Fe atoms/protein as isolated. Reconstitution of the protein with iron and sulfide by incubation with 25 equivalents dithiothreitol (DTT), 6.5 equivalents FeCl3, and 6.5 equivalents Na2S showed that the protein was capable of binding 3.91 ± 0.4 Fe atoms/protein.
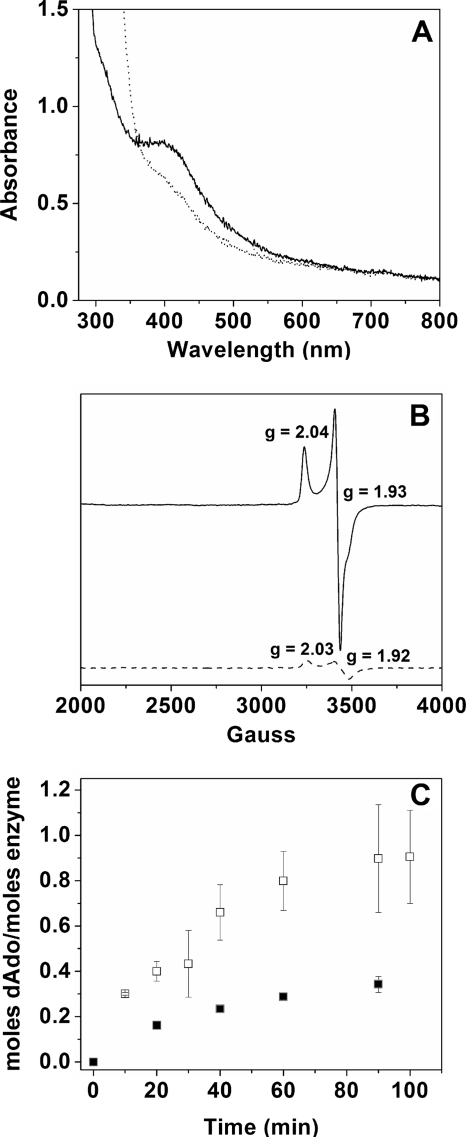
Room temperature UV/visible spectroscopic characterization of the protein as isolated, acquired on a Cary 6000i UV/visible/near-infrared spectrophotometer (Varian), showed an absorbance peak centered at ∼410 nm (Fig. 3A), indicating the presence of an iron-sulfur cluster. Reduction of the protein by addition of dithionite (DT) to a final concentration of 2 mM showed a decreased intensity of the ∼410-nm peak, indicating reduction of an iron-sulfur cluster. Low-temperature electron paramagnetic resonance (EPR) analysis of the as-isolated protein performed at 12 K (see the supplemental material for parameters) revealed a small isotropic signal characteristic of [3Fe-4S]1+ clusters, although spin integration versus a copper standard indicated this signal accounted for less than 1% of bound iron (data not shown). Upon addition of DT (8 mM, final concentration), the isotropic signal disappeared and a strong axial signal corresponding to spin = 1/2 [4Fe-4S]1+ cluster appeared which accounted for approximately 31% of bound iron as calculated from spin integration (Fig. 3B). Power and temperature dependence indicated the presence of a mixture of [4Fe-4S] and [2Fe-2S] clusters bound by the protein (see Fig. S2 in the supplemental material). To investigate possible interaction of the cluster with AdoMet, AdoMet was added to the reduced protein (8 mM, final concentration) immediately prior to freezing it for EPR analysis. The resulting EPR spectrum revealed a markedly decreased signal intensity (approximately 5% of total iron), as well as slightly altered G values (Fig. 3B). The decrease in signal, which is similar to that observed previously when the radical AdoMet enzyme spore photoproduct lyase was incubated in the presence of AdoMet (3, 25), could be the result of reductive cleavage of AdoMet (which would be accompanied by cluster oxidation); alternatively, a change in the spin state of the [4Fe-4S]1+ cluster could give rise to a decrease in the spin = 1/2 EPR signal. In addition to the decrease in signal intensity, the change in G values observed with the addition of AdoMet may be indicative of interaction with the cluster. From these studies, it is clear that HmdB binds an iron sulfur cluster and the cluster likely interacts with AdoMet.
FIG. 3.
(A) UV-visible spectra of HmdB obtained in the presence (dotted) or absence (solid) of dithionite; the protein concentration was 95 μM. (B) EPR spectra of dithionite-reduced HmdB in the presence (dashed) or absence (solid) of SAM; the protein concentration was 395 μM. (C) Production of dAdo by HmdB (20 μM) in the presence of 2 mM SAM and 1 mM DT as determined by high-performance liquid chromatography. Time points correspond to assays performed in triplicate which contained (open squares) or lacked (closed squares) 5 mM DTT.
Enzymatic assays performed in sealed and anaerobic vials (see the supplemental material for details) containing purified HmdB protein were monitored for the production of 5′-deoxyadenosine (dAdo) at time points of 10, 20, 40, 60, and 90 min. Experiments were performed in triplicate at 37°C. In the presence of HmdB, the concentration of dAdo increased over time. Deoxyadenosine was not observed in incubations performed in the absence of HmdB, demonstrating that dAdo production due to cleavage of AdoMet was catalyzed by HmdB, presumably through the formation of a 5′-dAdo radical (Fig. 3C). An approximate 2-fold increase in the reaction rate was observed in reaction mixtures containing DTT, a result consistent with the observed activities of the AdoMet enzymes HydE and HydG, where an increase in the reaction rate has been attributed to DTT acting as a radical acceptor, shifting the reaction equilibrium (26). Under these conditions, maximal dAdo production of 1 mol of product per mol HmdB enzyme was observed at a rate estimated to be 0.87 mol dAdo/mol HmdB/h, similar to that observed for HydE and HydG from Thermatoga maritima (26).
The possible involvement of HmdB in the synthesis of the iron-carbonyl linkage in the Hmd cofactor is suggested by the collective evolutionary and biochemical data presented herein. The AdoMet phylogram indicates that HmdB is evolutionarily related to HydE and HydG, which have been proposed to be involved in the synthesis of the CO ligand in the active site of HydA (20). The colocalization of hmdB and hmdC with hmdA in the genomes of hydrogenotrophic methanogens, coupled with the similar evolutionary histories observed for deduced amino acid sequences for hmdA, hmdB, and hmdC (Fig. 2), suggests that the genes are likely involved in a common process. This argument is bolstered by the observation that homologs of HmdB and HmdC were identified only in the genomes of hydrogenotrophic methanogens that contained hmdA. The fibrillarin homolog HmdC bears homology to members of a class of enzymes involved in RNA maturation, including enzymes which catalyze nucleoside modification, such as the methylation and methoxycarboxylation of target molecules, using AdoMet as a substrate (5, 11, 19, 32). Thus, HmdC may be involved in the methylation of the GP cofactor on the pyridinol ring; however, the demonstration of this activity is beyond the scope of the current study.
The combination of bioinformatics and biochemical approaches presented here represents a powerful approach for developing hypotheses regarding the functions of uncharacterized protein-encoding genes in genomic databases. Further spectroscopic and biochemical characterization of the HmdA cofactor synthesized in a background lacking hmdB and/or hmdC, in addition to further characterization of the HmdB protein in vitro, will continue to provide insight into the specific role of these enzymes in the maturation of HmdA.
Supplementary Material
Acknowledgments
This work was supported by the NASA Astrobiology Institute (NAI)-Montana State University Astrobiology Biogeocatalysis Research Center (NNA08CN85A) to J.B.B. and J.W.P. We acknowledge funding for the establishment of the Environmental and Biofilm Mass Spectrometry Facility through the Defense University Research Instrumentation Program (DURIP), contract number W911NF0510255. S.E.M. is supported by an NSF IGERT fellowship from the MSU program in Geobiological Systems (DGE 0654336). E.S.B. was supported by an NAI postdoctoral fellowship.
We thank David Schwab of Montana State University for assistance in running EPR samples.
Footnotes
Published ahead of print on 6 November 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bock, A., P. W. King, M. Blokesch, and M. C. Posewitz. 2006. Maturation of hydrogenases. Adv. Microb. Physiol. 51:1-71. [DOI] [PubMed] [Google Scholar]
- 2.Boyd, E. S., J. R. Spear, and J. W. Peters. 2009. [FeFe]-hydrogenase genetic diversity provides insight into molecular adaptation in a saline microbial mat community. Appl. Environ. Microbiol. 13:4620-4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buis, J. M., J. Cheek, E. Kalliri, and J. B. Broderick. 2006. Characterization of an active spore photoproduct lyase, a DNA repair enzyme in the radical S-adenosylmethionine superfamily. J. Biol. Chem. 281:25994-26003. [DOI] [PubMed] [Google Scholar]
- 4.Chatterjee, A., Y. Li, Y. Zhang, T. L. Grove, M. Lee, C. Krebs, S. J. Booker, T. P. Begley, and S. E. Ealick. 2008. Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily. Nat. Chem. Biol. 4:758-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chow, C. S., T. N. Lamichhane, and S. K. Mahto. 2007. Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. ACS Chem. Biol. 2:610-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frey, P. A., A. D. Hegeman, and F. Ruzicka. 2008. The radical SAM superfamily. Crit. Rev. Biochem. Mol. Biol. 43:63-88. [DOI] [PubMed] [Google Scholar]
- 7.Goldman, A. D., J. A. Leigh, and R. Samudrala. 2009. Comprehensive computational analysis of Hmd enzymes and paralogs in methanogenic Archaea. BMC Evol. Biol. 9:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiromoto, T., K. Ataka, O. Pilak, S. Vogt, M. S. Stagni, W. Meyer-Klaucke, E. Warkentin, R. K. Thauer, S. Shima, and U. Ermler. 2009. The crystal structure of C176A mutated [Fe]-hydrogenase suggests an acyl-iron ligation in the active site iron complex. FEBS Lett. 583:585-590. [DOI] [PubMed] [Google Scholar]
- 9.Hiromoto, T., E. Warkentin, J. Moll, U. Ermler, and S. Shima. 2009. The crystal structure of an [Fe]-hydrogenase-substrate complex reveals the framework for H2 activation. Angew. Chem. Int. Ed. Engl. 48:6457-6460. [DOI] [PubMed] [Google Scholar]
- 10.Imlay, J. A. 2006. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 59:1073-1082. [DOI] [PubMed] [Google Scholar]
- 11.Kozbial, P. Z., and A. R. Mushegian. 2005. Natural history of S-adenosylmethionine-binding proteins. BMC Struct. Biol. 5:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyon, E. J., S. Shima, R. Boecher, R. K. Thauer, F. W. Grevels, E. Bill, W. Roseboom, and S. P. Albracht. 2004. Carbon monoxide as an intrinsic ligand to iron in the active site of the iron-sulfur-cluster-free hydrogenase H2-forming methylenetetrahydromethanopterin dehydrogenase as revealed by infrared spectroscopy. J. Am. Chem. Soc. 126:14239-14248. [DOI] [PubMed] [Google Scholar]
- 13.Martinez-Gomez, N. C., and D. M. Downs. 2008. ThiC is an [Fe-S] cluster protein that requires AdoMet to generate the 4-amino-5-hydroxymethyl-2-methylpyrimidine moiety in thiamin synthesis. Biochemistry 47:9054-9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGlynn, S. E., D. W. Mulder, E. M. Shepard, J. B. Broderick, and J. W. Peters. 2009. Hydrogenase cluster biosynthesis: organometallic chemistry nature's way. Dalton Trans. 2009:4274-4285. [DOI] [PubMed] [Google Scholar]
- 15.McGlynn, S. E., S. S. Ruebush, A. Naumov, L. E. Nagy, A. Dubini, P. W. King, J. B. Broderick, M. C. Posewitz, and J. W. Peters. 2007. In vitro activation of [FeFe] hydrogenase: new insights into hydrogenase maturation. J. Biol. Inorg. Chem. 12:443-447. [DOI] [PubMed] [Google Scholar]
- 16.McGlynn, S. E., E. M. Shepard, M. A. Winslow, A. V. Naumov, K. S. Duschene, M. C. Posewitz, W. E. Broderick, J. B. Broderick, and J. W. Peters. 2008. HydF as a scaffold protein in [FeFe] hydrogenase H-cluster biosynthesis. FEBS Lett. 582:2183-2187. [DOI] [PubMed] [Google Scholar]
- 17.Meyer, J. 2007. [FeFe] hydrogenases and their evolution: a genomic perspective. Cell. Mol. Life Sci. 64:1063-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moll, A., A. Hildebrandt, H. P. Lenhof, and O. Kohlbacher. 2005. BALLView: an object-oriented molecular visualization and modeling framework. J. Comput. Aided Mol. Des. 19:791-800. [DOI] [PubMed] [Google Scholar]
- 19.Noma, A., Y. Kirino, Y. Ikeuchi, and T. Suzuki. 2006. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 25:2142-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters, J. W., R. K. Szilagyi, A. V. Naumov, and T. Douglas. 2006. A radical solution for the biosynthesis of the H-cluster of hydrogenase. FEBS Lett. 580:363-367. [DOI] [PubMed] [Google Scholar]
- 21.Peters, J. W., W. N. Lanzilotta, B. J. Lemon, and L. C. Seefeldt. 1998. X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution. Science 282:1853-1858. [DOI] [PubMed] [Google Scholar]
- 22.Pilak, O., B. Mamat, S. Vogt, C. H. Hagemeier, R. K. Thauer, S. Shima, C. Vonrhein, E. Warkentin, and U. Ermler. 2006. The crystal structure of the apoenzyme of the iron-sulphur cluster-free hydrogenase. J. Mol. Biol. 358:798-809. [DOI] [PubMed] [Google Scholar]
- 23.Posewitz, M. C., D. W. Mulder, and J. W. Peters. 2008. New frontiers in hydrogenase structure and biosynthesis. Curr. Chem. Biol. 2:178-199. [Google Scholar]
- 24.Posewitz, M. C., P. W. King, S. L. Smolinski, L. Zhang, M. Seibert, and M. L. Ghirardi. 2004. Discovery of two novel radical S-adenosylmethionine proteins required for the assembly of an active [Fe] hydrogenase. J. Biol. Chem. 279:25711-25720. [DOI] [PubMed] [Google Scholar]
- 25.Rebeil, R., and W. L. Nicholson. 2001. The subunit structure and catalytic mechanism of the Bacillus subtilis DNA repair enzyme spore photoproduct lyase. Proc. Natl. Acad. Sci. U. S. A. 98:9038-9043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubach, J. K., X. Brazzolotto, J. Gaillard, and M. Fontecave. 2005. Biochemical characterization of the HydE and HydG iron-only hydrogenase maturation enzymes from Thermatoga maritima. FEBS Lett. 579:5055-5060. [DOI] [PubMed] [Google Scholar]
- 27.Schick, M. 2008. Methanogenen Archaea ohne Gene für die vermuteten FeGP-Kofaktor Scaffold-Proteine Hmd II und Hmd III. Philipps Universität Marburg, Marburg, Germany.
- 28.Shima, S., E. J. Lyon, M. Sordel-Klippert, M. Kauss, J. Kahnt, R. K. Thauer, K. Steinbach, X. Xie, L. Verdier, and C. Griesinger. 2004. The cofactor of the iron-sulfur cluster free hydrogenase Hmd: structure of the light-inactivation product. Angew. Chem. Int. Ed. Engl. 43:2547-2551. [DOI] [PubMed] [Google Scholar]
- 29.Shima, S., O. Pilak, S. Vogt, M. Schick, M. Stagni, W. Meyer-Klaucke, E. Warkentin, R. K. Thauer, and U. Ermler. 2008. The crystal structure of [Fe]-hydrogenase reveals the geometry of the active site. Science 321:572-575. [DOI] [PubMed] [Google Scholar]
- 30.Shima, S., and R. K. Thauer. 2007. A third type of hydrogenase catalyzing H2 activation. Chem. Rec. 7:37-46. [DOI] [PubMed] [Google Scholar]
- 31.Sofia, H. J., G. Chen, B. G. Hetzler, J. F. Reyes-Spindola, and N. E. Miller. 2001. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 29:1097-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki, Y., A. Noma, T. Suzuki, R. Ishitani, and O. Nureki. 2009. Structural basis of tRNA modification with CO2 fixation and methylation by wybutosine synthesizing enzyme TYW4. Nucleic Acids Res. 37:2910-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thauer, R. K. 1998. Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Microbiology 144:2377-2406. [DOI] [PubMed] [Google Scholar]
- 34.Vignais, P. M., and B. Billoud. 2007. Occurrence, classification, and biological function of hydrogenases: an overview. Chem. Rev. 107:4206-4272. [DOI] [PubMed] [Google Scholar]
- 35.Vignais, P. M., B. Billoud, and J. Meyer. 2001. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 25:455-501. [DOI] [PubMed] [Google Scholar]
- 36.Vignais, P. M. 2008. Hydrogenases and H±-reduction in primary energy conservation. Results Probl. Cell Differ. 45:223-252. [DOI] [PubMed] [Google Scholar]
- 37.Volbeda, A., M. H. Charon, C. Piras, E. C. Hatchikian, M. Frey, and J. C. Fontecilla-Camps. 1995. Crystal-structure of the nickel-iron hydrogenase from Desulfovibrio-Gigas. Nature 373:580-587. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.