Abstract
This paper describes the cloning, purification, and characterization of thioredoxin (Trx) and thioredoxin reductase (TrxR) and the structure determination of TrxR from the ionizing radiation-tolerant bacterium Deinococcus radiodurans strain R1. The genes from D. radiodurans encoding Trx and TrxR were amplified by PCR, inserted into a pET expression vector, and overexpressed in Escherichia coli. The overexpressed proteins were purified by metal affinity chromatography, and their activity was demonstrated using well-established assays of insulin precipitation (for Trx), 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) reduction, and insulin reduction (for TrxR). In addition, the crystal structure of oxidized TrxR was determined at 1.9-Å resolution. The overall structure was found to be very similar to that of E. coli TrxR and homodimeric with both NADPH- and flavin adenine dinucleotide (FAD)-binding domains containing variants of the canonical nucleotide binding fold, the Rossmann fold. The Km (5.7 μM) of D. radiodurans TrxR for D. radiodurans Trx was determined and is about twofold higher than that of the E. coli thioredoxin system. However, D. radiodurans TrxR has a much lower affinity for E. coli Trx (Km, 44.4 μM). Subtle differences in the surface charge and shape of the Trx binding site on TrxR may account for the differences in recognition. Because it has been suggested that TrxR from D. radiodurans may have dual cofactor specificity (can utilize both NADH and NADPH), D. radiodurans TrxR was tested for its ability to utilize NADH as well. Our results show that D. radiodurans TrxR can utilize only NADPH for activity.
Deinococcus radiodurans is a gram-positive bacterium capable of withstanding exposure to extreme gamma ray and UV radiation, oxidants, and desiccation (6, 10, 26). The mechanism behind the ability of D. radiodurans to survive exposure to extreme conditions has been a subject of intense research (10, 43). Its ability to survive exposure to extreme conditions has been attributed a number of factors, as follows: a high number of genome copies (8), ring-like nucleoid organization (22), high manganese content (8), and a higher ability to scavenge reactive oxygen species (ROS) (43). However, the mechanism responsible for its extremophilic nature is not clearly understood (25).
Efforts to understand the mechanism behind the capability of D. radiodurans to tolerate extreme conditions have focused on understanding its ability to prevent or repair genomic damage, because if unrepaired, genomic damage is lethal to the cell (7). The ability of D. radiodurans to repair genomic damage is likely due to its ability to prevent proteome damage, i.e., its ability to maintain sufficient enzymatic activity for genome repair after irradiation. Therefore, genome repair probably plays a bigger role than prevention of genome damage in making D. radiodurans radiation tolerant (7, 8). Indeed, some experimental evidence suggests that efficient DNA repair is solely responsible for the ability of D. radiodurans to withstand ionizing radiation. D. radiodurans DNA sustains the same amount of genome damage at high radiation doses as other bacteria, but unlike other bacteria, its damage is mended within hours (25). However, some recent evidence suggests that it is likely that prevention of DNA damage (reactive oxygen species [ROS] scavenging) supplements DNA repair to make D. radiodurans ionizing radiation tolerant. It is worth noting that only about 20% of radiation-induced damage to the genome is due to the direct effect of irradiation (the rest is due to radiation-induced ROS) and that cellular extracts of D. radiodurans are more effective in scavenging ROS than Escherichia coli extracts when subjected to oxidative stress (43). Moreover, D. radiodurans has higher basal levels of some antioxidant enzymatic systems (catalase and superoxide dismutase), and disruption of superoxide dismutase (sodA) and catalase (katA) genes results in increased sensitivity of D. radiodurans to ionizing radiation. In addition D. radiodurans catalase is more resistant to inhibition by substrate H2O2 than bovine or Aspergillus niger catalase (17). Taken together, these experimental results suggest a significant contribution of antioxidant systems to the ability of D. radiodurans to withstand extreme ionizing radiation.
While the contribution of some antioxidant enzymatic systems to the extremophilic nature of D. radiodurans has been extensively studied, the role of the thioredoxin system has not been investigated (40, 43). The thioredoxin system is composed of thioredoxin reductase (TrxR), thioredoxin (Trx), and various cellular targets. The system is found in both prokaryotes and eukaryotes, and homologues of both TrxR and Trx have been isolated from many species. Trx proteins are low-molecular-mass proteins (12 kDa) that possess a highly conserved active site motif, WCGPC (27, 41). TrxR is a homodimeric enzyme and is a member of the family of pyridine nucleotide-disulfide oxidoreductase flavoenzymes. Each monomer possesses a flavin adenine dinucleotide (FAD) prosthetic group, a NADPH-binding site, and an active site comprising a redox-active disulfide. There are two distinct forms of this enzyme, as follows: low-molecular-mass TrxR (35 kDa), found in prokaryotes and some eukaryotes, and high-molecular-mass TrxR (55 kDa), found in eukaryotes (41). The two types of TrxR proteins have some differences in structure and mechanism. However, in both cases, reducing equivalents are transferred from NADPH to TrxR, from TrxR to Trx, and finally, from Trx to various cellular proteins (29, 41). Trx targets include proteins which take part in the scavenging of ROS-like thioredoxin-dependent thiol peroxidase (29). The thioredoxin system is thus an important antioxidant enzymatic system.
In this study we report the expression, purification, and biochemical characterization of the main components of the D. radiodurans thioredoxin system. In addition, the structural characterization of D. radiodurans TrxR is reported.
MATERIALS AND METHODS
Bacterial strains.
Genomic DNA of wild-type D. radiodurans strain R1 and wild-type E. coli strain K-12 was obtained from the ATCC. E. coli expression strain Rosetta 2(DE3) and cloning strain NovaBlue were obtained from Novagen. E. coli expression strain BL21-Gold(DE3) was obtained from Stratagene.
Cloning of D. radiodurans TrxR and Trx1 and E. coli Trx1.
D. radiodurans TrxR was cloned into an expression vector as previously described (26). D. radiodurans Trx1 and E. coli Trx1 coding sequences were obtained by PCR using D. radiodurans strain R1 and E. coli strain K-12 genomic DNA templates, respectively. The sequences used as forward and reverse oligonucleotide primers for PCR in D. radiodurans Trx1 cloning were 5′-GCG CCA TGG GTA TGA GTG ACA TCC TGA CCTG-3′ and 5′-GCG GGA TCC TCA GGA AAG CTG GTT GAG-3′, respectively. For E. coli Trx1 cloning, the sequences used as forward and reverse oligonucleotide primers were 5′-GCG CCA TGG GTA TGA TGA GCG ATA AAA TT-3′ and 5′-GCG GGA TCC CGC CAG GTT AG-3′, respectively.
The resulting amplified 0.5-kbp fragments were purified by gel extraction, digested with NcoI and BamHI, and cloned into a modified pET-30b vector (thrombin cleavage site replaced with a TEV protease site) kindly provided by H. Liu (23). The resulting constructs were transformed into E. coli bacterial strain NovaBlue and plated on Luria-Bertani (LB) agar containing kanamycin (50 μg/ml). Positive clones were screened by restriction analysis using NcoI and BamHI and analyzed by agarose gel electrophoresis. Positive clones were further verified by DNA sequencing (Plant Biotechnology Institute, Saskatoon, Canada).
E. coli TrxR in the ampicillin-resistant vector PROK-1 was kindly provided by Leslie Poole (Wake Forest University, Winston-Salem, NC).
Protein production.
Confirmed constructs were transformed into Rosetta 2 cells. D. radiodurans TrxR, E. coli TrxR, and E. coli Trx1 were overexpressed and harvested as previously described for D. radiodurans TrxR (31). D. radiodurans Trx1 was also overexpressed and harvested as previously described for D. radiodurans TrxR, but with some modifications (31). Briefly, BL21-Gold cells were transformed with the modified pET-30b vector containing the D. radiodurans Trx1 gene, plated on LB-kanamycin plates containing 50 μg/ml kanamycin, and incubated at 37°C overnight. A single colony was isolated from an LB-kanamycin plate and used to grow 100 ml of inoculum culture on LB media with 50 μg/ml kanamycin. After overnight incubation at 37°C, 10 ml of the inoculum culture was added to 1 liter of LB media containing 50 μg/ml kanamycin. The cells were grown in Pyrex Fernbach flasks at 37°C until the optical density at 600 nm (OD600) reached 0.6. D. radiodurans Trx1 expression was then induced by adding isopropyl-β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 0.25 mM. The cells were grown at 30°C for 4 h after induction and harvested by centrifugation (20 min, 8,000 × g, 4°C), and the pellets were stored at −80°C.
Purification of D. radiodurans Trx1.
The frozen cell pellets were thawed on ice and resuspended in 5 mM EDTA, 50 mM Tris-HCl at pH 8.0, 0.5% Triton X-100, 1 mM AEBSF [4-(2-aminoethyl)-benzenesulfonyl fluoride], 20 μg ml−1 DNase, and 20 μg ml−1 lysozyme. The thawed cells were mechanically disrupted by sonication and centrifuged (20 min, 8,000 × g, 4°C). The protein was found in inclusion bodies by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of the resulting pellet and supernatant. It was then solubilized from the pellet by resuspending the pellet in 5 mM EDTA, 50 mM Tris-HCl at pH 8.0, 0.5% deoxycholic acid, and sodium salt (deoxycholate [DOC]), followed by sonication and centrifugation (26,500 × g). The resulting supernatant and pellet were separated by decanting. The solubilization procedure was repeated for the pellet. The two supernatants were pooled and loaded onto a POROS MC50 metal chelation column (Applied Biosystems) preequilibrated with a buffer containing 1 mM imidazole, 0.5 M NaCl, and 50 mM Tris-HCl at pH 8.0. The column was washed with the same buffer to remove nonspecific binding, and the protein was eluted with a 0 to 0.5 M imidazole gradient. Peak fractions (5 ml) were collected, and the purity of the protein was checked using Coomassie blue-stained SDS-PAGE. Those fractions showing high levels of purity were pooled and concentrated. The concentrated protein was then dialyzed against 50 mM Tris-HCl at pH 8.
Purification of E. coli TrxR.
E. coli TrxR was purified essentially as previously reported using ion-exchange and affinity chromatography, with small modifications (33). Instead of a DE52 column, an HQ 20 anion-exchange column (Applied Biosystems) preequilibrated with 25 mM K2HPO4/KHPO4 buffer at pH 8.0 was used. The protein was eluted with a linear gradient of 0 to 500 mM NaCl.
Purification of D. radiodurans TrxR and E. coli Trx1.
D. radiodurans TrxR and E. coli Trx1 were purified as previously reported for D. radiodurans TrxR using metal affinity chromatography (31).
D. radiodurans Trx1 activity.
The activity of D. radiodurans Trx1 was tested using the insulin precipitation assay by monitoring the increase in turbidity at 650 nm (14). The reaction was conducted at room temperature in 50 mM Tris-HCl at pH 8. The assay mixture contained insulin (0.85 mM), dithiothreitol (1 mM), and D. radiodurans Trx1 (5 to 75 μM).
Insulin reduction assay.
D. radiodurans TrxR activity was measured by using D. radiodurans Trx1 as a substrate and by monitoring the consumption of NADPH by measuring the decrease in absorbance at 340 nm. The assay mixture (1 ml) contained 100 mM HEPES buffer (pH 7.4), 2 mM EDTA, 30 μM bovine insulin (Sigma), 0.1 mM NADPH (Calbiochem), 0.1 μM D. radiodurans TrxR, and D. radiodurans Trx1 (0 to 25 μM). All the measurements were carried out at room temperature using a Cary 50 spectrophotometer (Varian). The reaction was initiated by adding D. radiodurans Trx1, and the resulting change in absorbance was recorded. The data were fit to a straight line over a ∼1-min range to calculate the change in absorbance per minute. Initial velocities (Vo) were calculated as μM of NADPH oxidized/min in accordance with the following relationship: Vo = ΔA340/0.0062 (15).
DTNB reduction assay.
D. radiodurans TrxR activity was also measured using the 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) reduction assay. The reaction mixture contained 20 mM HEPES (pH 7.4), 2 mM EDTA, 0.24 mM NADPH, 0.1 μM D. radiodurans TrxR, 1.6 mM DTNB, and 0.1 to 20 μM D. radiodurans Trx1 in a final volume of 1 ml. The reaction was initiated by adding DTNB and monitored by noting the increase in absorbance at 412 nm in a Cary 50 spectrophotometer (Varian). Initial velocities were calculated as μM of DTNB reduced/min in accordance with the following relationship: Vo = ΔA412/0.0136 (24, 30).
D. radiodurans TrxR structure determination and refinement.
D. radiodurans TrxR was purified and crystallized, and data were collected as previously described. The details of data processing and structure solution have also been reported previously (31). Briefly, the structure was determined by molecular replacement, using the Mycobacterium tuberculosis TrxR structure (Protein Data Bank [PDB] accession no. 2A87) as the search model. The solution was obtained by MOLREP (44) using MrBUMP (16), an automated scheme for molecular replacement. The correctness of the resulting solution was confirmed from the electron density of FAD (which appeared in the expected positions, although it was omitted from the initial search model). Initial restrained refinement carried out using REFMAC5 (28) as a part of MrBUMP resulted in an Rfree value of 0.431, with an initial Rfree value of 0.547. Further refinement was performed using REFMAC5 with manual rebuilding using Coot (11). Tight noncrystallographic symmetry restraints for the main chain atoms and loose restraints for the side chain atoms were applied in the late stages of refinement. FAD was easily modeled into the positive difference electron density at the expected sites. The program ARP/wARP (32) was used to add water molecules, and the resulting model was manually inspected for correctness using Coot. The stereochemical quality of the resulting model was analyzed using PROCHECK (20). The refinement and final model statistics for D. radiodurans TrxR are shown in Table 1.
TABLE 1.
Refinement statistics for D. radiodurans TrxRa
| Statistics | Value(s) |
|---|---|
| Resolution (Å) | 80-1.9 |
| R factor (%) | 19.2 |
| Rfree (%) | 24.2 |
| Overall B factor | 22.0 |
| Average B values | |
| Main chain | 19.6 |
| Side chain and water | 24.0 |
| No. of cofactors | 2 |
| No. of water molecules | 521 |
| No. of atoms | |
| Protein | 4,728 |
| Ligand (FAD) | 106 |
| RMSD bonds (Å) | 0.016 |
| RMSD angles (°) | 1.715 |
| Ramachandran plot—nonglycine residues in: | |
| Most favorable region (%) | 89.5 |
| Additionally allowed region (%) | 10.2 |
| Generously allowed region (%) | 0.20 |
| Disallowed region (%) | 0.20 |
Data collection parameters and crystallographic data statistics have been previously reported (31).
Homology modeling of D. radiodurans Trx1 and FR conformation of D. radiodurans TrxR.
Models of D. radiodurans Trx1 and the FR conformation of D. radiodurans TrxR were generated by MODELLER 9v1 (with default parameters) (37). Coordinates of the E. coli Trx crystal structure (PDB accession no. 2trx) and the E. coli TrxR crystal structure (PDB accession no. 1F6M) were used as templates to build the D. radiodurans Trx1 model and the D. radiodurans TrxR model (FR conformation), respectively. Side chains of the D. radiodurans TrxR model (FR conformation) were manually adjusted to match those of the D. radiodurans TrxR structure (PDB accession no. 2Q7V) using Coot. The overall quality of the two models was checked by PROCHECK.
Rigid-body docking experiments.
Rigid-body docking simulations (of the four complexes that were kinetically characterized in Table 2) were done using the ZDOCK server (5; http://zdock.bu.edu/), with constraints based on the E. coli TrxR/Trx complex structure (PDB accession no. 1F6M). E. coli TrxR/Trx structures (PDB accession no. 1F6M) and D. radiodurans TrxR (FR conformation)/Trx homology models were used for the docking studies. The top complex (best ZDOCK score) was compared to the E. coli TrxR/Trx complex structure for correctness and then used for shape and complementarity evaluation. One way to evaluate shape and complementarity is to calculate a gap volume (GV) index using the following equation: GV (Å) = Gap volume between molecules (Å3)/interface area (Å2) (per complex) (19). The gap volume index was calculated using the PROTORP server (36; http://www.bioinformatics.sussex.ac.uk/protorp/).
TABLE 2.
Kinetic constants for the D. radiodurans thioredoxin system using both D. radiodurans Trx1 and E. coli Trx1 as substratesa
| Assay | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|
| D. radiodurans TrxR/D. radiodurans Trx1 | 5.7 ± 1.9 (2.75 ± 0.40) | 1.1 ± 0.1 (9.4 ± 1.0) | 1.9 × 105 (3.4 × 106) |
| D. radiodurans TrxR/E. coli Trx1 | 32.4 ± 8.5 | 18.0 ± 2.6 | 5.5 × 105 |
| E. coli TrxR/D. radiodurans Trx1 | 44.4 ± 5.5 | 4.6 ± 0.2 | 1.0 × 105 |
| E. coli TrxR/E. coli Trx1 | 2.7 ± 0.7 (0.7 ± 0.003) | 3.6 ± 0.01 (4.4 ± 0.04) | 1.3 × 106 (6.3 × 106) |
Protein structure accession number.
The D. radiodurans TrxR coordinates have been deposited in the Brookhaven Protein Data Bank under accession no. 2Q7V.
RESULTS AND DISCUSSION
Kinetic characterization of D. radiodurans TrxR and Trx1.
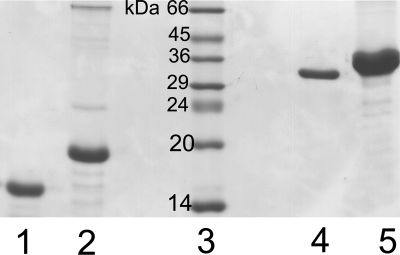
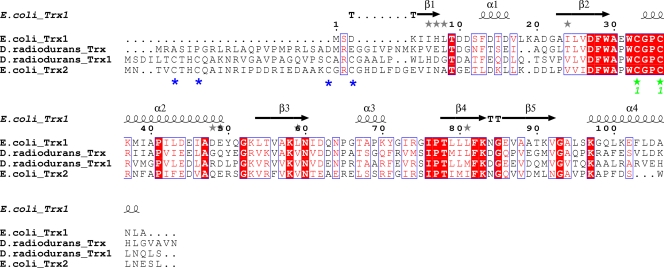
Recombinant D. radiodurans TrxR was expressed from a modified pET-30b vector in E. coli strain Rosetta and purified to homogeneity, resulting in a high yield of soluble protein. Similarly, D. radiodurans Trx1 was expressed in E. coli strain Rosetta; however, it resulted in the protein being found in the pellet after sonication and centrifugation. D. radiodurans Trx1 was subsequently resolubilized from the pellet using DOC and purified to homogeneity. Both Trx1 and TrxR from D. radiodurans resulted in the expected molecular masses of ∼12 kDa and ∼35 kDa, respectively, when analyzed by SDS-PAGE (Fig. 1). All thioredoxins reported so far effectively reduce insulin disulfides. Upon reduction, the A and B chains of insulin dissociate, resulting in aggregation and precipitation of the B chain. The resulting turbidity can be used to measure the activity of thioredoxin (14). We performed the standard insulin precipitation assay, as described in Materials and Methods, to confirm the identity of D. radiodurans Trx1. The addition of 5 μM D. radiodurans Trx1 to the reaction mixture described above resulted in rapid precipitation of insulin after 16 min, confirming the identity of the protein. The results also suggest that D. radiodurans Trx1 reduces insulin in a manner similar to that of E. coli Trx1 (7.8 μM E. coli Trx1 resulted in rapid precipitation of insulin after 9 min) (14). Examination of the completed D. radiodurans genome revealed two thioredoxins, annotated as Trx and Trx1 (46). Several other bacteria also contain at least two thioredoxins, usually designated Trx1 and Trx2. In a classic thioredoxin system, Trx2 has slightly lower redox activity than Trx1 and contains extra N-terminal cysteines (Fig. 2), which have been suggested to play a role in regulating thioredoxin activity (27). Efforts are under way to clone D. radiodurans Trx in order to compare its activity with that of D. radiodurans Trx1. D. radiodurans Trx1 contains extra N-terminal cysteines, whereas D. radiodurans Trx does not have any cysteines on the N terminus, suggesting that D. radiodurans Trx1 is more similar to classic Trx2 than to Trx1.
FIG. 1.
SDS-PAGE of purified samples of various TrxR and Trx proteins, as follows: (1) E. coli thioredoxin (Trx), (2) D. radiodurans thioredoxin (Trx1), (3) low-molecular-mass protein marker, (4) E. coli thioredoxin reductase (TrxR), and (5) D. radiodurans thioredoxin reductase (TrxR).
FIG. 2.
Sequence alignment of thioredoxins from E. coli and D. radiodurans. The N terminus of D. radiodurans Trx1 is more similar to that of E. coli Trx2 than that of E. coli Trx1. Absolutely conserved active-site cysteines are indicated with green asterisks, whereas N terminus-conserved cysteines of D. radiodurans Trx1 and E. coli Trx2 are indicated with blue asterisks. The figure was produced using the program ClustalW version 2.0 (18) and the ESPript server (12).
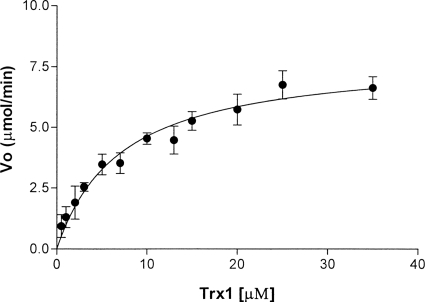
The redox activity of the D. radiodurans thioredoxin system was measured using the insulin reduction assay and the DTNB reduction assay (15, 24, 30). The results are shown in Fig. 3, and the kinetic data are summarized in Table 2. The catalytic efficiency (kcat/Km) of the D. radiodurans thioredoxin system is similar to that of the E. coli thioredoxin system (about 6-fold lower using the insulin reduction assay). These results suggest that abundance rather than efficiency of some ROS-scavenging enzymatic systems may be responsible for the higher ROS-scavenging ability seen in D. radiodurans cell extracts than in E. coli cell extracts (43).
FIG. 3.
Michaelis-Menten plot of D. radiodurans TrxR, determined using D. radiodurans Trx1 as a substrate. The reaction was monitored by consumption of NADPH via a decrease in absorbance at 340 nm. Initial velocity (Vo) was measured as μmoles of NADPH consumed per minute. Reaction mixtures contained 100 mM HEPES buffer (pH 7.4), 2 mM EDTA, 30 μM solution of bovine insulin (Sigma), 0.1 mM NADPH (Calbiochem), 0.1 μM D. radiodurans TrxR, and D. radiodurans Trx1 (0 to 35 μM).
Cross-reactivity with the E. coli thioredoxin system.
D. radiodurans TrxR and D. radiodurans Trx1 were tested for their ability to cross-react with E. coli Trx1 and E. coli TrxR, respectively, using the insulin reduction assay described above, with small modifications. For the D. radiodurans TrxR/E. coli Trx1 assay, E. coli Trx1 (0 to 75 μM) was used instead of D. radiodurans Trx1, and for the E. coli TrxR/D. radiodurans Trx1 assay, E. coli TrxR (0.1 μM) was used instead of D. radiodurans TrxR. The results are summarized in Table 2. Consistent with results from other bacterial thioredoxin systems, D. radiodurans TrxR prefers its cognate thioredoxin (Km = 5.7 μM) as a substrate rather than thioredoxin from different species (E. coli thioredoxin, Km = 44.4 μM) (3).
Cofactor specificity.
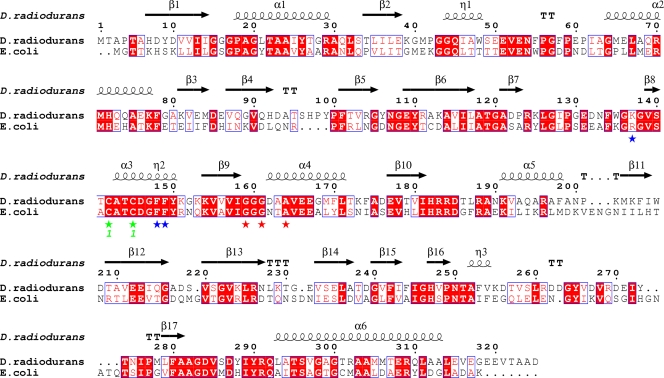
It has been reported that D. radiodurans TrxR may have dual cofactor specificity, i.e., it can utilize NADPH or NADH for its redox activity (39). To test this possibility, we performed the insulin reduction assay under standard assay conditions, with either NADPH or NADH as a source of reducing equivalents. The concentrations of NADPH and NADH varied from 0 to 0.6 mM, with a constant D. radiodurans Trx1 concentration of 10 μM. We determined that D. radiodurans TrxR can utilize only NADPH as a source of reducing equivalents. No activity was seen when NADH was used as a reductant for the insulin reduction assay (some trace activity was seen with the DTNB reduction assay). Sequence analysis of D. radiodurans TrxR also suggests NADPH dependence (Fig. 4); D. radiodurans TrxR has the GXGXXA motif (common in NADPH-dependent enzymes) instead of the GXGXXG motif (common in NADH-dependent enzymes) (2). Our results are consistent with previous studies which showed that bacterial TrxR proteins had no activity (3) or very low activity (42) when NADH is used as a reductant. To the best of our knowledge, all bacterial thioredoxin reductases utilize only NADPH as a source of reducing equivalents; only thioredoxin reductase-related flavoenzymes (NADH:peroxiredoxin oxidoreductases) are capable of utilizing NADH as a reductant (34, 35).
FIG. 4.
Sequence alignment of TrxR proteins from E. coli and D. radiodurans. The conserved active-site cysteine residues are indicated with green asterisks, the key residues involved in interaction with Trx are indicated with blue asterisks, and the GXGXXA motifs are indicated with red asterisks. The figure was produced using the program ClustalW version 2.0 (18) and the ESPript server (12).
D. radiodurans TrxR structure.
The structure of oxidized D. radiodurans TrxR was refined to a 1.9-Å resolution. The final restrained refinement resulted in R and Rfree values of 0.19 and 0.24, respectively. All the residues of the polypeptide chain of both subunits (A and B) were well defined in the electron density maps, except for the terminal residues of both the C terminus and the N terminus. One residue from the N terminus and 11 residues from the C terminus were not observed in the electron density for subunit A, and five residues from the N terminus and 11 residues from the C terminus were not observed. The FAD cofactors were also well defined in the electron density for both chains A and B. The stereochemistry of the model is good, as judged by the Ramachandran plot. About 90% of the residues lie within the most favorable region of the Ramachandran plot, with the rest of the residues in the additionally allowed regions. One residue (E271) is well defined in the electron density maps, but it is in the disallowed region of the Ramachandran plot. This residue is part of a tight turn (γ-turn) involving three residues, D270, E271, and I272. The asymmetric unit contained two monomers, forming a homodimer, as found in solution.
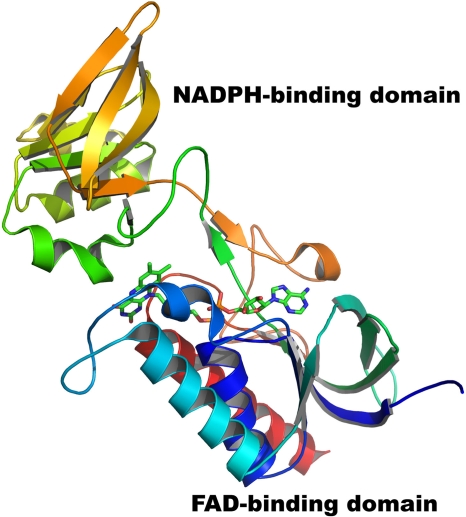
The overall structure of D. radiodurans TrxR is very similar to other bacterial thioredoxin reductase structures (1, 13, 45). Each subunit consists of an FAD-binding domain (residues 1 to 123 and 249 to 325) and an NADPH-binding domain (residues 124 to 248), as shown in Fig. 5. Both the NADPH-binding and the FAD-binding domains contain variants of the canonical nucleotide binding fold that was first seen in glutathione reductase (38). Each domain contains two β-sheets (one parallel and the other antiparallel β-sheet) and three α-helices. The two domains are connected by two β-strands and loops but otherwise are separated by a large cleft which is filled mostly with water molecules. Three hundred four Cα atoms of D. radiodurans TrxR of the A chain can be superposed to the E. coli TrxR structure (PDB accession no. 1tde), with a root mean square deviation (RMSD) of 1.57 Å. Like other bacterial thioredoxin reductases, the active-site cysteines (residues 142 and 145) are located on the NADPH domain, but the active-site disulfide is buried and unavailable for reaction with thioredoxin in the FO conformation.
FIG. 5.
Overall structure of the A chain of D. radiodurans TrxR in the FO conformation. In this conformation, the FAD-binding domain is too far from the NADPH-binding domain for electron transfer to occur from NADPH to FAD. A 67° rotation by the NADPH-binding domain brings NADPH close to FAD, allowing electron transfer to occur from NADPH to FAD (21). The figure was generated by PYMOL (9).
Trx-binding site.
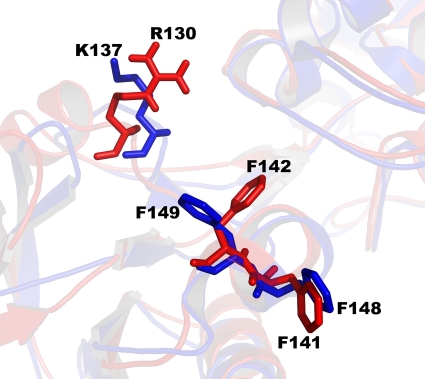
Our kinetic results show that D. radiodurans TrxR has a lower affinity (8-fold-higher Km) for E. coli Trx1 than for its own Trx. The structural basis of binding E. coli Trx1 to E. coli TrxR has been elucidated (21). Eight residues (G129, R130, G131, V132, S133, F141, F142, and Y143) on the surface of the NADPH domain of E. coli TrxR form a groove to which a complementary thioredoxin loop binds. Two of these residues (F141 and F142) fit into a hydrophobic pocket on E. coli Trx1, making additional interactions with E. coli Trx1 (21). Shape has been suggested to play the major role in species-specific recognition of thioredoxins (13). Surface analysis of Trx and TrxR from both E. coli and D. radiodurans Trx systems suggests an important role for shape in recognition. The two E. coli TrxR residues that fit into a hydrophobic pocket on E. coli Trx have corresponding residues in D. radiodurans TrxR (F148 and F149, respectively). However, one of the corresponding D. radiodurans TrxR residues (F149) points in the opposite direction from that of E. coli TrxR, which suggests some differences between the shapes of the binding pockets of E. coli and D. radiodurans Trx proteins (Fig. 6). All eight residues that form the Trx-binding groove on the E. coli TrxR surface are identical to those of D. radiodurans TrxR, except the conserved substitution of R130 (E. coli TrxR) with K137 (D. radiodurans TrxR). This substitution also contributes to differences in the shape of the Trx-binding site. Gap volume index calculations also suggest that homologous TrxR/Trx systems show more complementarity than heterologous systems. The gap volume index for the E. coli TrxR/D. radiodurans Trx complex is higher (3.37 Å) than that of the E. coli TrxR/E. coli Trx complex (2.50 Å); likewise, the gap volume index for the D. radiodurans TrxR/E. coli Trx complex is higher (2.79 Å) than that of the D. radiodurans TrxR/D. radiodurans Trx complex (0.20 Å).
FIG. 6.
Superimposed TrxR structures from E. coli (red) and D. radiodurans (blue), showing some key residues that are important for binding to thioredoxin. The figure was generated by PYMOL (9).
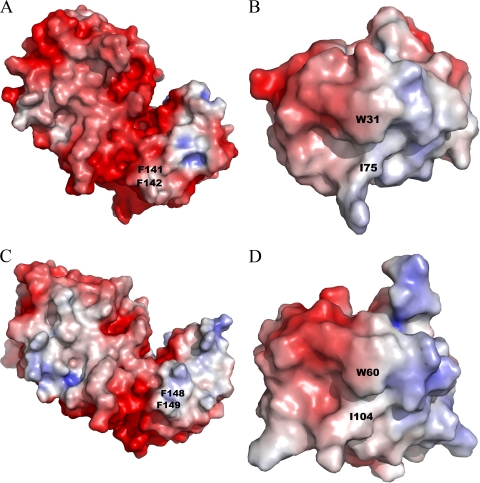
A comparison of the overall surface charge distributions shows some charge complementarity between Trx and TrxR. The surface surrounding the Trx-TrxR complex interface is both positively and negatively charged in Trx and in TrxR. The interface itself is mostly nonpolar in both Trx and TrxR. This complementarity is seen in both the E. coli and D. radiodurans thioredoxin systems, although the surface around the Trx-binding site of E. coli TrxR is slightly more negative than that of D. radiodurans TrxR (Fig. 7). This difference in surface charge may also play a role in the species-specific recognition of Trx.
FIG. 7.
Electrostatic potentials of TrxR and Trx from E. coli (A and B, respectively) and D. radiodurans (C and D, respectively), calculated by APBS software (4). Positively and negatively charged areas are colored in red and in blue and contoured at −5 kBTe−1 and +5 kBTe−1, respectively. Key residues of the TrxR-Trx complex interface are shown on the diagram. The overall shapes of the Trx-binding sites differ between the two TrxR proteins, but there are only small differences in the electrostatic potentials. The figures were generated with PYMOL (9) using E. coli TrxR, E. coli Trx, and D. radiodurans TrxR coordinates (PDB accession no. 1TDE, 2TRX, and 2Q7V, respectively). The D. radiodurans Trx figure was generated using the D. radiodurans Trx homology model.
Acknowledgments
We thank Tiffany Dickinson, Danée Klassen, and Ohmin Kwon from Evan Hardy Collegiate for their contributions to this project. This crystallography work was based upon research conducted at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source, which is supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health.
This work was funded by an NSERC grant (RGPIN-250238) to D.A.R.S. S.A.B. and V.P. were both funded through the NSERC USRA program.
Footnotes
Published ahead of print on 20 November 2009.
REFERENCES
- 1.Akif, M., K. Suhre, C. Verma, and S. C. Mande. 2005. Conformational flexibility of Mycobacterium tuberculosis thioredoxin reductase: crystal structure and normal-mode analysis. Acta Crystallogr. D Biol. Crystallogr. 61:1603-1611. [DOI] [PubMed] [Google Scholar]
- 2.Alkhalfioui, F., M. Renard, and F. Montrichard. 2007. Unique properties of NADP-thioredoxin reductase C in legumes. J. Exp. Bot. 58:969-978. [DOI] [PubMed] [Google Scholar]
- 3.Baker, L. M. S., A. Raudonikiene, P. S. Hoffman, and L. B. Poole. 2001. Essential thioredoxin-dependent peroxiredoxin system from Helicobacter pylori: genetic and kinetic characterization. J. Bacteriol. 183:1961-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker, N. A., D. Sept, S. Joseph, M. J. Holst, and J. A. McCammon. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 98:10037-10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, R., L. Li, and Z. Weng. 2003. ZDOCK: an initial-stage protein-docking algorithm. Proteins 52:80-87. [DOI] [PubMed] [Google Scholar]
- 6.Cox, M. M., and J. R. Battista. 2005. Deinococcus radiodurans—the consummate survivor. Nat. Rev. Microbiol. 3:882-892. [DOI] [PubMed] [Google Scholar]
- 7.Daly, M. J. 2009. A new perspective on radiation resistance based on Deinococcus radiodurans. Nat. Rev. Microbiol. 7:237-245. [DOI] [PubMed] [Google Scholar]
- 8.Daly, M. J., E. K. Gaidamakova, V. Y. Matrosova, A. Vasilenko, M. Zhai, A. Venkateswaran, M. Hess, M. V. Omelchenko, H. M. Kostandarithes, K. S. Makarova, L. P. Wackett, J. K. Fredrickson, and D. Ghosal. 2004. Accumulation of Mn(II) in Deinicoccus radiodurans facilitates gamma-radiation resistance. Science 306:1025-1028. [DOI] [PubMed] [Google Scholar]
- 9.DeLano, W. L. 2002. The PyMol molecular graphics system. DeLano Scientific, San Carlos, CA.
- 10.Dennis, R. J., E. Micossi, J. McCarthy, E. Moe, E. J. Gordon, S. Kozielski-Stuhrmann, G. A. Leonard, and S. McSweeney. 2006. Structure of the manganese superoxide dismutase from Deinococcus radiodurans in two crystal forms. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 62:325-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 12.Gouet, P., X. Robert, and E. Courcelle. 2003. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31:3320-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gustafsson, T. N., T. Sandalova, J. Lu, A. Holmgren, and G. Schneider. 2007. High-resolution structures of oxidized and reduced thioredoxin reductase from Helicobacter pylori. Acta Crystallogr. D Biol. Crystallogr. 63:833-843. [DOI] [PubMed] [Google Scholar]
- 14.Holmgren, A. 1979. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J. Biol. Chem. 254:9627-9632. [PubMed] [Google Scholar]
- 15.Holmgren, A., and F. J. Morgan. 1976. Enzymic reduction of disulfide bonds by thioredoxin. The reactivity of disulfide bonds in human choriogonadotropin and its subunits. Eur. J. Biochem. 70:377-383. [DOI] [PubMed] [Google Scholar]
- 16.Keegan, R. M., and M. D. Winn. 2008. MrBUMP: an automated pipeline for molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 64:119-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi, I., T. Tamura, H. Sghaier, I. Narumi, S. Yamaguchi, K. Umeda, and K. Inagaki. 2006. Characterization of monofunctional catalase KatA from radioresistant bacterium Deinococcus radiodurans. J. Biosci. Bioeng. 101:315-321. [DOI] [PubMed] [Google Scholar]
- 18.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. Mcgettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 19.Laskowski, R. A. 1995. SURFNET: a program for visualizing molecular surfaces, cavities, and intermolecular interactions. J. Mol. Graph. 13:323-330. [DOI] [PubMed] [Google Scholar]
- 20.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 21.Lennon, B. W., C. H. Williams, Jr., and M. L. Ludwig. 2000. Twists in catalysis: alternating conformations of Escherichia coli thioredoxin reductase. Science 289:1190-1194. [DOI] [PubMed] [Google Scholar]
- 22.Levin-Zaidman, S., J. Englander, E. Shimoni, A. K. Sharma, K. W. Minton, and A. Minsky. 2003. Ringlike structure of the Deinococcus radiodurans genome: a key to radioresistance? Science 299:254-256. [DOI] [PubMed] [Google Scholar]
- 23.Liu, H., K. Woznica, G. Catton, A. Crawford, N. Botting, and J. H. Naismith. 2007. Structural and kinetic characterization of quinolinate phosphoribosyltransferase (hQPRTase) from homo sapiens. J. Mol. Biol. 373:755-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luthman, M., and A. Holmgren. 1982. Rat liver thioredoxin and thioredoxin reductase: purification and characterization. Biochemistry 21:6628-6633. [DOI] [PubMed] [Google Scholar]
- 25.Makarova, K. S., L. Aravind, Y. I. Wolf, R. L. Tatusov, K. W. Minton, E. V. Koonin, and M. J. Daly. 2001. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 65:44-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mattimore, V., and J. R. Battista. 1996. Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J. Bacteriol. 178:633-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miranda-Vizuete, A., A. E. Damdimopoulos, J. Gustafsson, and G. Spyrou. 1997. Cloning, expression, and characterization of a novel Escherichia coli thioredoxin. J. Biol. Chem. 272:30841-30847. [DOI] [PubMed] [Google Scholar]
- 28.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 29.Mustacich, D., and G. Powis. 2000. Thioredoxin reductase. Biochem. J. 346:1-8. [PMC free article] [PubMed] [Google Scholar]
- 30.Navarro, J. A., F. K. Gleason, M. A. Cusanovich, J. A. Fuchs, T. E. Meyer, and G. Tollin. 1991. Kinetics of electron transfer from thioredoxin reductase to thioredoxin. Biochemistry 30:2192-2195. [DOI] [PubMed] [Google Scholar]
- 31.Obiero, J., S. A. Bonderoff, M. M. Goertzen, and D. A. R. Sanders. 2006. Expression, purification, crystallization and preliminary X-ray crystallographic studies of Deinococcus radiodurans thioredoxin reductase. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 62:757-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perrakis, A., T. K. Sixma, K. S. Wilson, and V. S. Lamzin. 1997. wARP: improvement and extension of crystallographic phases by weighted averaging of multiple-refined dummy atomic models. Acta Crystallogr. D Biol. Crystallogr. 53:448-455. [DOI] [PubMed] [Google Scholar]
- 33.Poole, L. B., A. Godzik, A. Nayeem, and J. D. Schmitt. 2000. AhpF can be dissected into two functional units: tandem repeats of two thioredoxin-like folds in the N-terminus mediate electron transfer from the thioredoxin reductase-like C-terminus to AhpC. Biochemistry 39:6602-6615. [DOI] [PubMed] [Google Scholar]
- 34.Poole, L. B., C. M. Reynolds, Z. A. Wood, P. A. Karplus, H. R. Ellis, and M. Li Calzi. 2000. AhpF and other NADH:peroxiredoxin oxidoreductases, homologues of low M(r) thioredoxin reductase. Eur. J. Biochem. 267:6126-6133. [DOI] [PubMed] [Google Scholar]
- 35.Reynolds, C. M., J. Meyer, and L. B. Poole. 2002. An NADH-dependent bacterial thioredoxin reductase-like protein in conjunction with a glutaredoxin homologue form a unique peroxiredoxin (AhpC) reducing system in Clostridium pasteurianum. Biochemistry 41:1990-2001. [DOI] [PubMed] [Google Scholar]
- 36.Reynolds, C., D. Damerell, and S. Jones. 2009. ProtorP: a protein-protein interaction analysis server. Bioinformatics 25:413-414. [DOI] [PubMed] [Google Scholar]
- 37.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779-815. [DOI] [PubMed] [Google Scholar]
- 38.Schulz, G. E., R. H. Schirmer, W. Sachsenheimer, and E. F. Pai. 1978. The structure of the flavoenzyme glutathione reductase. Nature 273:120-124. [DOI] [PubMed] [Google Scholar]
- 39.Seo, H. J., and Y. N. Lee. 2006. Occurrence of thioredoxin reductase in Deinococcus species, the UV resistant bacteria. J. Microbiol. 44:461-465. [PubMed] [Google Scholar]
- 40.Song, D., R. Chen, C. Shao, L. Wu, and Z. Yu. 2000. Effect of N+ beam exposure on superoxide dismutase and catalase activities and induction of Mn-SOD in deinococcus radiodurans. Plasma Sci. Technol. 2:491. [Google Scholar]
- 41.Stefanková, P., D. Perecko, I. Barak, and M. Kollarova. 2006. The thioredoxin system from Streptomyces coelicolor. J. Basic Microbiol. 46:47-55. [DOI] [PubMed] [Google Scholar]
- 42.Thelander, L. 1967. Thioredoxin reductase. Characterization of a homogenous preparation from Escherichia coli B. J. Biol. Chem. 242:852-859. [PubMed] [Google Scholar]
- 43.Tian, B., Z. Xu, Z. Sun, J. Lin, and Y. Hua. 2007. Evaluation of the antioxidant effects of carotenoids from Deinococcus radiodurans through targeted mutagenesis, chemiluminescence, and DNA damage analyses. Biochim. Biophys. Acta 1770:902-911. [DOI] [PubMed] [Google Scholar]
- 44.Vagin, A., and A. Teplyakov. 1997. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30:1022-1025. [Google Scholar]
- 45.Waksman, G., T. S. R. Krishna, C. H. Williams, Jr., and J. Kuriyan. 1994. Crystal structure of Escherichia coli thioredoxin reductase refined at 2 Å resolution. Implications for a large conformational change during catalysis. J. Mol. Biol. 236:800-816. [PubMed] [Google Scholar]
- 46.White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson, R. J. Dodson, D. H. Haft, M. L. Gwinn, W. C. Nelson, D. L. Richardson, K. S. Moffat, H. Qin, L. Jiang, W. Pamphile, M. Crosby, M. Shen, J. J. Vamathevan, P. Lam, L. McDonald, T. Utterback, C. Zalewski, K. S. Makarova, L. Aravind, M. J. Daly, K. W. Minton, R. D. Fleischmann, K. A. Ketchum, K. E. Nelson, S. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]