Abstract
Bacterial persistence in the environment and in the infected host is often aided by the formation of exopolymer-enclosed communities known as biofilms. Heterogeneous gene expression takes place in microcompartments formed within the complex biofilm structure. This study describes cell differentiation within an isogenic bacterial cell population based on the example of biofilm formation by Salmonella enterica serovar Typhimurium. We analyzed the expression of the major biofilm regulator CsgD at the single-cell level with a chromosomal CsgD-green fluorescent protein (GFP) translational fusion. In individual cells, CsgD-GFP expression is mostly found in the cytoplasm. Quantitative expression analysis and results from three different models of S. Typhimurium biofilms demonstrated that CsgD is expressed in a bistable manner during biofilm development. CsgD expression is, however, monomodal when CsgD is expressed in larger amounts due to a promoter mutation or elevated levels of the secondary signaling molecule c-di-GMP. High levels of CsgD-GFP are associated with cellular aggregation in all three biofilm models. Furthermore, the subpopulation of cells expressing large amounts of CsgD is engaged in cellulose production during red, dry, and rough (rdar) morphotype development and in microcolony formation under conditions of continuous flow. Consequently, bistability at the level of CsgD expression leads to a corresponding pattern of task distribution in S. Typhimurium biofilms.
The key to the ecological success of bacteria as the main colonizers of the planet is their remarkable phenotypic plasticity. Bacteria are capable of profoundly modulating their gene expression patterns in response to environmental stimuli. Thus, most bacteria alternate between two distinct modes of growth: as free-living cells or as members of surface-attached and exopolymer-embedded communities known as biofilms (6, 21, 48). Biofilms have a complex architecture consisting of many microenvironments with diverse environmental conditions. These microenvironments provide ecological niches where individual cells execute specialized genetic programs and aid the establishment of highly heterogeneous cell populations (25, 50).
Associated with medical settings, biofilm formation is a virulence factor in chronic infections and contributes to the environmental persistence and transfer of pathogenic bacteria between hosts (9, 54). Resistance to antimicrobials, ability to escape from the action of the immune system, and resistance to environmental assaults are properties that make biofilm formation an important problem in clinical settings (9, 26).
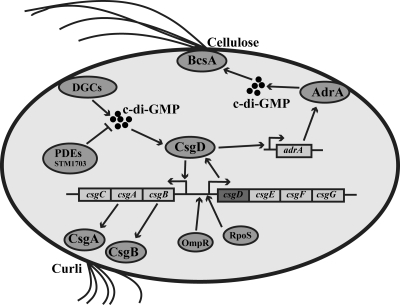
Biofilm formation is an integral part of the life cycle of the food-borne pathogen Salmonella enterica and has an impact on host colonization, environmental persistence, and transmission (30, 31, 36, 52). The rdar (red, dry, and rough) morphotype is a biofilm behavior of Salmonella enterica serovar Typhimurium (32, 34). The rdar morphotype is characterized, among other factors, by the production of an adhesive extracellular matrix consisting of curli fimbriae (previously called thin aggregative fimbriae [agf]) and cellulose (Fig. 1) (34, 55). CsgD, a master regulator of the rdar morphotype, positively regulates the synthesis of both matrix components (37). CsgD directly activates transcription of the csgBA operon, which encodes structural components of curli fibers (2, 18, 34). The effect of CsgD on cellulose production proceeds through transcriptional activation of the diguanylate cyclase AdrA, which generates c-di-GMP, an allosteric activator of cellulose synthase (39, 44, 51, 55). Expression of CsgD requires the stress sigma factor RpoS and is tightly controlled by environmental conditions and global regulatory proteins, such as the response regulator OmpR and the novel second messenger c-di-GMP (Fig. 1) (14, 15, 23, 34). An unusually large region upstream and downstream of the csgD promoter has been implicated in the control of csgD expression through binding of several regulatory proteins (3, 12, 13, 22); however, single point mutations in the csgD promoter region can lead to enhanced semiconstitutive and temperature-independent expression of CsgD, which does not require the stress sigma factor RpoS (38). Many of the links between environmental cues and CsgD expression are still unclear and are likely to involve both transcriptional and posttranscriptional control.
FIG. 1.
Regulatory network of CsgD-mediated biofilm formation in S. Typhimurium UMR1. CsgD expression requires the stress sigma factor RpoS and the response regulator OmpR (34). CsgD is required for the expression of the csgBA operon, encoding structural subunits of curli fimbriae (19), and adrA, encoding a diguanylate cyclase required for the activation of cellulose biosynthesis, on agar plates (55). CsgD expression can be positively modulated by c-di-GMP, the steady-state level of which is determined by the activity of c-di-GMP-specific phosphodiesterases, such as STM1703 and diguanylate cyclases (23, 43).
Adhesion of Salmonella to abiotic surfaces is important for biofilm formation in industrial settings. Adherence to glass and polystyrene has several components in common with the rdar morphotype, including positive regulation by CsgD and the expression of curli and cellulose as extracellular matrix components (29, 37, 38, 55). Formation of a pellicle at the air-liquid interface is yet another model for Salmonella biofilms that requires a set of genes similar to the one involved in rdar morphotype expression (37, 47).
In this study, we addressed the pattern of expression of a master biofilm regulator, CsgD, at the single-cell level. To this end, we visualized the expression of a chromosomal CsgD-green fluorescent protein (GFP) translational fusion in three different models of S. Typhimurium biofilms: rdar morphotype, biofilm formation in steady-state liquid culture, and biofilm formation in continuous-flow culture. Expression of CsgD in all three models of S. Typhimurium biofilm is not uniform but is subject to a bistable switch. This bistable CsgD expression corresponds to the pattern of task distribution in the biofilms, whereby the cells involved in producing the cellulose matrix or building up the biofilm structure express CsgD. However, monophasic expression of CsgD is observed at higher overall CsgD expression levels created by a csgD promoter mutation or epigenetically by elevated c-di-GMP concentrations, suggesting that CsgD expression in S. Typhimurium strain UMR1 is adapted to give rise to phenotypic variability to fine tune the highly delicate balance between cell communities and single cells.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. For cloning purposes, Escherichia coli and Salmonella enterica serovar Typhimurium were grown on Luria-Bertani (LB) agar. When appropriate, antibiotics were included at the following concentrations: 100 μg ml ampicillin−1 and 20 μg ml chloramphenicol−1. In the cases where expression of the rdar morphotype was analyzed, the strains were grown on LB agar plates without salt at 28°C after being precultured overnight on LB agar at 37°C. “Spot” cultures were inoculated by spotting 10 μl preculture containing 109 CFU/ml onto LB agar without salt. In this case, the preculture was grown in LB broth without salt at 37°C overnight.
TABLE 1.
Strains of S. Typhimurium (ATCC 14028) used in this work
| Strain | Genotype | Reference |
|---|---|---|
| UMR1 | ATCC 14028-1s Nalr | 34 |
| MAE14 | UMR1 csgBA101::Kmr | 37 |
| MAE18 | UMR2 csgBA101::Kmr | 34 |
| MAE50 | UMR1 ΔcsgD101 | 37 |
| MAE52 | UMR1 PcsgD101 | 38 |
| MAE171 | MAE52 ΔbcsA102 | 55 |
| MAE119 | MAE51 zxx::gfp | 55 |
| MAE222 | UMR1 bcsA101::MudJ | 55 |
| MAE265 | UMR1 ΔcsgD bcsA101::MudJ | 23 |
| MAE282 | UMR1 STM1703::Cmr | 23 |
| MAE299 | UMR1 ΔbcsA102 | This study |
| MAE318 | UMR1 ompR43::MudJ | 13 |
| MAE775 | UMR1 ΔbcsA102 csgBA::Km | This study |
| MAE776 | UMR1 Φ(csgD-gfp) ΔbcsA102 csgBA::Km | This study |
| MAE777 | MAE52 ΔbcsA102 csgBA::Km | This study |
| MAE778 | MAE52 Φ(csgD-gfp) ΔbcsA102 csgBA::Km | This study |
| MAE779 | UMR1 Φ(csgD-gfp) ompR43::MudJ | This study |
| MAE780 | UMR1 Φ(csgD-gfp) STM1703::Cmr | This study |
| MAE851 | UMR1 Φ(csgD-gfp)Hyb Nalr Cmr | This study |
For phenotypic evaluation of the rdar morphotype, the strains were grown on LB agar plates without salt supplemented with Congo red (40 μg ml−1) and Coomassie brilliant blue (20 μg ml−1).
Microaerophilic conditions were obtained by filling 60% of the volume of flasks with medium (150 rpm).
Construction of strains.
To construct the CsgD-GFP translational fusion, the gene for GFP was fused to the csgD open reading frame in its native chromosomal locus with the help of Lambda Red recombination technology (7). The stop codon of csgD was replaced by a DNA sequence containing the gene encoding the GFP variant GFP+ (42) followed by a chloramphenicol resistance cassette amplified from plasmid pZEP08 (20). Primers were designed with 40-bp overhangs homologous to the 3′ end of csgD precisely up- and downstream of the csgD stop codon and to delete the stop codon of csgD and the start codon of gfp with the following nucleotide sequences: 5′-CTCTGCTGCTACAATCCAGGTCAGATAGCGTTTCATGGCCAATAACTGCCTTAAAAAAATTA-3′ (For.CsgD.Gfp) and 5′-CACCCAGGCAGTTTCATGGGCAAACGATAATCTCAGGCGGAGTAAAGGAGAAGAACTTTT-3′ (Rev.CsgD.Gfp) (the 40 bp containing overhangs are underlined). Candidate clones were screened by colony PCR for the insertion of gfp at the 3′ end of csgD using the primers listed in Table 2. The final construct was confirmed by sequencing. CsgD-GFP fusions were constructed in the UMR1 background (strain MAE851 [UMR1 csgD-gfp]) and the MAE52 background (MAE778 [MAE52 ΔbcsA ΔcsgBA csgD-gfp]). If required, the CsgD-GFP fusion was transduced to other strain backgrounds using chloramphenicol resistance as a selection marker. The csgD-gfp junction was confirmed by PCR.
TABLE 2.
Primers used in this work
| Primer | Sequence |
|---|---|
| Primers used to construct the translational CsgD-GFP fusion | |
| For.CsgD.Gfp | CTCTGCTGCTACAATCCAGGTCAGATAGCGTTTCATGGCCAATAACTGCCTTAAAAAAATTA |
| Rev.CsgD.Gfp | CACCCAGGCAGTTTCATGGGCAAACGATAATCTCAGGCGGAGTAAAGGAGAAGAACTTTT |
| Primers used to verify constructed mutants | |
| bcsAN1(KpISDstart) | AAAAAGGTACCAAGGAGGGCCTGCGATGAGCGCCCTTTCCCGGTGGCTGC |
| bcsAC2(Xb) | GCCGGTCTAGAAATCATTGTTGAGCCTGAGCCATAACCCGATCC |
| agfC | CGAGGATCCGGCCATTGTTGTGATAAA |
| agfBD | ACGAAAGCTTGCACTGCTGTGGGTTG |
| 1703_control_rev | AAATTGATTGTTGTCGGGAGT |
| ycir_KO_check | GATGTCATTGATGTCACTATTG |
| Control_ompR_F | GCTGCTGTTAAATATGCTTTGT |
| In MudJ | CTAGAGTGAAACGCTTTCGC |
| Stop.CsgD.Gfp | ATCCTCAATAAGTTACGTATT |
| RevGfp606 | TTCGAAAGGGCAGATTGTGT |
| Primers used for quantitative real-time RT-PCR | |
| qCsgD ffw | ACGCTACTGAAGACCAGGAAC |
| qCsgD rev | GCATTCGCCACGCAGAATA |
| qadrA ffw | GGCCATTAAATTAGCGGAAC |
| qadrA rev | AATAAAATTTCCCAGTGGCG |
| qcsgB ffw | CCAACGATGCCAGTATATCG |
| qcsgB rev | TGGCCTTATTTCCAGAACCT |
| qrecA FFW | GGCGAAATCGGCGACTCT |
| qrecA REV | CATACGGATCTGGTTGATGAAAATC |
An in-frame deletion in the cellulose synthase gene, bcsA, was created using plasmid pXZO1 as described previously (33, 55). Phage transduction was carried out using phage P22 HT105/1 int-201 (41). The constructed strains were verified by PCR with the control primers listed in Table 2.
Western blot analysis.
Bacteria were grown on LB agar plates without salt at 28°C. Five mg (wet weight) of cells was harvested, resuspended in 200 μl sample buffer, and incubated at 95°C for 10 min. The total protein content was analyzed by Coomassie blue staining after gel separation. Equal amounts of total protein were separated by SDS-PAGE (12% resolving gel and 4% stacking gel) and transferred to a polyvinylidene difluoride membrane (Immobilon P; Millipore). Detection of CsgD was carried out as described previously (37) using a polyclonal anti-CsgD peptide antibody (1:5,000) and horseradish peroxidase-conjugated goat anti-rabbit IgG (1:2,000; Jackson ImmunoResearch Laboratories Inc.). GFP was detected with a horseradish peroxidase-conjugated polyclonal anti-GFP antibody (1:1,000; Affinity Bioreagents). RpoS was detected with a monoclonal anti-RNAP sigmas IgG (1:5,000; Neoclone) and horseradish peroxidase-conjugated anti-mouse IgG (1:2,000; Sigma). Chemiluminescence (Lumi-Light WB substrate; Roche) was recorded using the Las-1000 system (Fujifilm) and quantified with ImageJ software (http://rsb.info.nih.gov/ij/).
Assay for in vivo protein stability of CsgD and CsgD-GFP.
The UMR1 wild type and the isogenic strain MAE851 expressing the CsgD-GFP fusion protein were grown in liquid cultures in LB without salt at 28°C under microaerophilic conditions, as described above. After 16 to 22 h of incubation, the optical density at 600 nm (OD600) was measured to ensure that the two cultures contained the same amount of cells. Chloramphenicol was added to the cultures at a final concentration of 200 μg ml−1, and 1-ml samples were removed at the indicated time points. The culture medium was removed by centrifugation, and the cells were resuspended in sample buffer (10 μl per 1 mg [wet weight]). CsgD and RpoS were detected by Western blot analysis as described above.
RNA extraction.
Extraction of total RNA was performed using the SV Total RNA Isolation System (Promega) with minor modifications. Prior to RNA extraction, bacterial cells were incubated in ice-cold 5% (vol/vol) phenol-95% (vol/vol) ethanol to stabilize the RNA (≥30 min on ice). The samples were centrifuged at 4,000 rpm for 10 min, and the supernatant was discarded. Subsequently, the pellet was resuspended in 100 μl lysis buffer (50 mg/ml lysozyme in Tris-EDTA [TE] buffer) and incubated for 5 min at room temperature. The subsequent steps of RNA purification were performed according to the manufacturer's instructions, including an on-column DNase I treatment. The quality of RNA samples was assessed via gel electrophoresis. RNA concentrations were determined using the NanoDrop System (Thermo Scientific), and RNA samples were stored at −70°C.
cDNA synthesis and quantitative real-time RT-PCR.
For quantitative real-time reverse transcription (RT)-PCR, S. Typhimurium cells were grown in liquid LB medium (without NaCl) at 28°C and 150 rpm for 24 h, and RNA was extracted as described previously. The expression of target genes was determined by two-step real-time RT-PCR using Power SYBR green PCR Master Mix (Applied Biosystems) and the 7500 Real-Time PCR System (Applied Biosystems). First-strand cDNA synthesis from total RNA was performed with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) following the manufacturer's instructions. Relative transcript abundance was determined by the 2−ΔΔCT method (27) using 7500 SDS Software v1.3.1 (Applied Biosystems). The recA gene was used as an endogenous control for internal normalization. Experiments were performed as biological triplicates using the mean expression from quadruplicates per real-time PCR assay relative to a calibrator value (UMR1).
Flow cell experiments.
Biofilms were cultivated at ambient temperature in three-channel flow cells (5) with an individual-channel flow of 8 ml h−1 of M9/glucose minimal medium (the dimensions of an individual cell were 1 by 4 by 40 mm). The substratum for biofilm attachment was a glass coverslip (Menzel-Glaser; thickness, 0.13 to 0.16 mm). Each of the flow channels was inoculated with 200 μl culture grown overnight in LB medium without salt at 37°C and adjusted to an OD600 of 0.04 in M9/glucose minimal medium. In order to allow the initial attachment of bacteria, the flow was resumed 1 h after inoculation.
Fluorescence microscopy.
Cells were grown on LB agar without salt or in LB broth without salt at 28°C. Living cells were mounted in LB broth without salt on agarose-coated slides (1% agarose [Sigma] in H2O). Cellulose staining was performed by immersing living cells in LB broth without salt containing 0.001% calcofluor (Fluorescent Brightener 28; Sigma) on agarose-coated slides. (A fresh stock of 1% calcofluor in 20% glycerol containing 25 mM NaOH was prepared prior to each experiment.) The cell membranes of bacteria were stained with FM4-64 (Molecular Probes/Invitrogen) diluted in phosphate-buffered saline (PBS) to a final concentration of 2 μg/ml.
Fluorescence images were acquired using a Nikon ECLIPSE E400 microscope equipped with a Plan Apo 100×/1.4 oil objective and a cooled Hamamatsu charge-coupled device (CCD) camera. Biofilms growing in the flow cell system were visualized using a Laser Confocal System based on Nipkow spinning-disk technology. Z stacks were acquired with the UltraViewers Fret OH confocal imaging system (Perkin Elmer). Cells in the biofilm were stained with the Live/Dead Baclight bacterial viability kit (Invitrogen) according to the manufacturer's instructions. Digital image processing was carried out using ImageJ software (http://rsb.info.nih.gov/ij/) and Adobe Photoshop.
FACS.
Bacterial cells were scraped off LB agar plates without salt, resuspended in fixation buffer (4% formaldehyde in PBS), and incubated for 2 h at room temperature in the dark. The cells were washed, diluted in PBS, and directly subjected to fluorescence-activated cell sorting (FACS) analysis using a Becton Dickinson FACScan apparatus. Data were captured and further analyzed using Cell Quest Pro software version 6.0. Strain MAE119, which constitutively expresses GFP, served as a positive control (55). Strain UMR1 ΔompR csgD-gfp, which does not express CsgD-GFP (see Results) (Fig. 1) (38), served as a negative control. These strains were used to calibrate the FACS instrument and enabled us to identify nonfluorescent (CsgD-OFF) and fluorescent (CsgD-ON) events. Gates identifying the bacterial population were applied in an identical manner for all strains and time points. The figures were prepared for publication using Cell Quest Pro 6.0 and Adobe Photoshop.
RESULTS
CsgD-GFP translational fusion: functionality, expression, and protein stability.
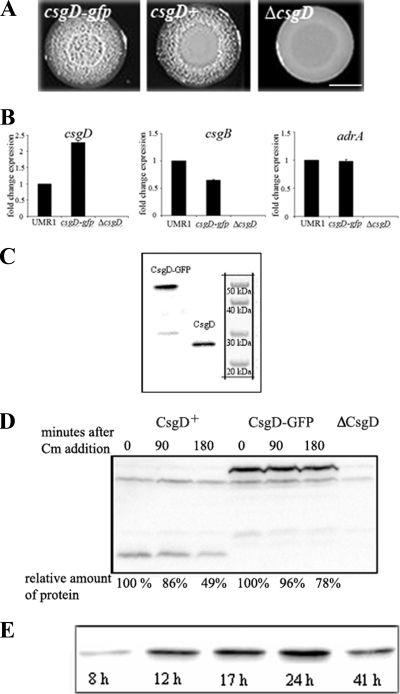
We constructed a C-terminal translational fusion of GFP+ to CsgD (CsgD-GFP) in its native chromosomal locus in S. Typhimurium UMR1, yielding the strain MAE851. The CsgD-GFP fusion protein was functional, as judged by the ability of strain MAE851 to express the rdar morphotype on LB plates without salt supplemented with Congo Red. The color and appearance of the strain expressing the CsgD-GFP fusion protein on Congo Red plates indicated production of curli and cellulose in comparison to the wild-type UMR1 (Fig. 2A) (34). Thus, the phenotype suggested that CsgD remained functional when tagged with GFP. The functionality of the CsgD-GFP fusion protein was confirmed by expression analysis of the CsgD-controlled genes csgB and adrA (Fig. 2B).
FIG. 2.
Functionality and expression of the CsgD-GFP fusion protein. (A) Colony appearance of the strains MAE851 (csgD-gfp), UMR1 (wild-type; csgD), and MAE50 (ΔcsgD) after 48 h of growth on LB agar without salt supplemented with Congo Red. Bar, 5 mm. (B) Expression of csgD, adrA, and csgB in S. Typhimurium UMRI (wild type), MAE851 (csgD-gfp), and MAE50 (ΔcsgD). Quantitative real-time RT-PCR was performed to compare the expression of csgD and the CsgD-regulated genes csgB and adrA. RNA was isolated from cells grown for 24 h at 28°C in liquid LB medium without NaCl. One representative experiment is shown. (C) Detection of CsgD by Western blotting in cells from strains MAE851 (csgD-gfp) and UMR1 (wild type; csgD) grown on LB agar without salt for 24 h. (D) Stability of CsgD and CsgD-GFP. Shown is detection of CsgD by Western blot analysis from cells treated with chloramphenicol to inhibit protein synthesis. Cells were removed from the culture for analysis after the indicated time points. (E) Growth phase-dependent expression of CsgD in strain MAE851 (csgD-gfp) as analyzed by Western blotting in cells grown on LB agar without salt. Cell lysates were made at the indicated time points.
In order to assess the integrity of the CsgD-GFP fusion protein, immunoblot analysis was performed. A major band of 51 kDa, corresponding in size to the intact CsgD-GFP fusion, was detected with either α-CsgD (Fig. 2C) or α-GFP antibody (data not shown). These results showed that the CsgD-GFP fusion protein was stable. Therefore, the GFP fluorescence observed by subsequent microscopic analysis derived from the CsgD-GFP fusion protein.
Quantification of CsgD expression in UMR1 and MAE851 showed that the CsgD-GFP fusion protein in strain MAE851 and the native CsgD in the wild-type UMR1 were present in approximately equal amounts (Fig. 2C and data not shown). The protein stabilities of CsgD-GFP and CsgD were measured in vivo after translation was blocked with chloramphenicol. Native CsgD had a half-life of 180 min, while the half-life of the CsgD-GFP fusion protein was somewhat longer (Fig. 2D). As a positive control for proteolysis, we found that the half-life of RpoS in the same samples was shorter than 15 min (data not shown), which is consistent with previous reports (16). Thus, CsgD and CsgD-GFP are stable proteins.
The temporal expression of the CsgD-GFP fusion was analyzed in comparison to that of the native CsgD by immunoblotting. The overall expression of both CsgD and CsgD-GFP was low at 8 h of growth on LB agar without salt and steadily increased until 24 h, when the maximum expression was observed. At 41 h of growth, the protein levels of both CsgD-GFP and native CsgD had decreased. Thus, the temporal pattern of CsgD-GFP expression followed that of the untagged CsgD in the wild-type UMR1 (Fig. 2E and data not shown) (45).
CsgD-GFP is localized in the cytoplasm but is localized to the cell membrane in aging parts of the colonies.
The expression of the CsgD-GFP fusion protein was visualized at optimal expression levels in live cells of strain MAE851, i.e., after 22 h of growth on LB agar without salt (Fig. 3A). In an individual cell, the fluorescent signal from CsgD-GFP was evenly dispersed in the cytoplasm. Microscopic observation of cells grown for 48 h, however, indicated that CsgD-GFP localized to the membrane in a small fraction of the cells (less than 0.1%) (data not shown).
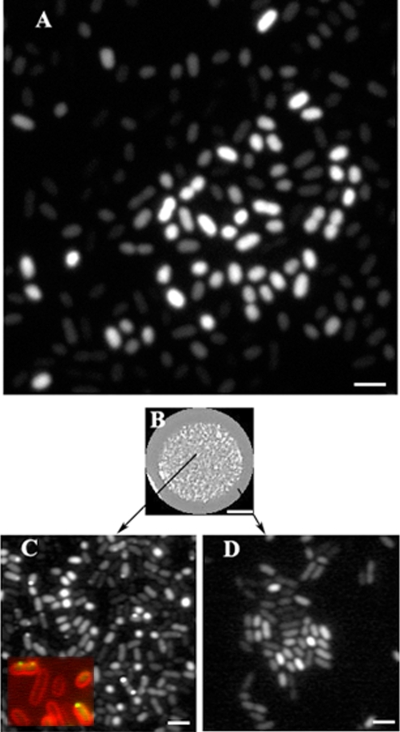
FIG. 3.
Visualization of CsgD-GFP in cells grown on agar plates. (A) Fluorescence micrograph (confocal) of CsgD-GFP-expressing strain MAE851 after 22 h of growth on LB agar without salt. Bar, 2 μm. (B) The CsgD-GFP-expressing strain MAE851 was grown as a giant “spot” colony on LB agar without salt for 72 h. Bar, 3 mm. (C and D) GFP fluorescence micrographs of bacteria that were scraped off the agar surface either from the inner (i.e., aging) parts (C) or from the outermost (i.e., young) parts (D) of the same “spot” colony. (Inset in C) Membrane localization of CsgD. Green, CsgD-GFP; red, membrane stain FM4-64. Bar, 2 μm.
In order to address the effect of colony age on CsgD-GFP localization in more detail, we grew the cells in “spot” cultures on LB agar without salt (Fig. 3B). These cultures represent colonies growing by radial expansion of a drop of inoculum containing a defined number of cells. Therefore, cells situated in the middle parts of such a colony are older than the cells in the outermost parts. Inspection of the inner (i.e., older) parts of a “spot” colony after 72 h of growth revealed membrane foci of CsgD-GFP of variable size in approximately 10% of the cells (Fig. 3C). In contrast, in the outermost (and youngest) part of the colony, CsgD-GFP fluorescence was evenly dispersed in the cytoplasm of cells (Fig. 3D).
Interestingly, on the population level, the amounts of fluorescence showed great cell-to-cell variation. Quantification of pixel brightness showed that the difference in fluorescence intensity could be greater than 20-fold (data not shown). Consequently, the cells could be divided into two populations with respect to their CsgD expression, one population with CsgD expression on and one population with CsgD expression off. In other words, these observations suggested that the expression of CsgD during rdar morphotype development follows a bistable pattern.
Expression of CsgD-GFP during rdar morphotype development.
To investigate whether a bistable pattern of CsgD expression is present throughout the growth phase, we visualized CsgD-GFP in living cells from different stages of rdar morphotype development on LB agar without salt. Cells were scraped off the agar surface at the following time points after inoculation: 8 h, 12 h, 17 h, 24 h, and 41 h (representative examples are shown in Fig. 4). At the single-cell level, microscopic observation revealed CsgD-GFP fluorescence throughout the cytoplasm at all time points. As an exception, CsgD-GFP was found to localize to the membrane in a minute fraction of the cells at 41 h of growth (data not shown).
FIG. 4.
Visualization of CsgD-GFP during rdar morphotype development. Strain MAE851 (csgD-gfp) was grown on LB agar without salt at 28°C for 8 h (A), 12 h (B), 24 h (C), or 41 h (D). Bacteria were carefully scraped off the agar surface and mounted on agarose-coated slides in LB without salt. (B and C) Two distinct cell populations characterized by high or low intensity of GFP fluorescence are evident. Red, membrane stain FM4-64. Bar, 2 μm.
At the level of the total cell population, a differential pattern of CsgD-GFP expression was established over time. At 8 h, only a few individual cells expressed large amounts of CsgD-GFP, while the majority of the cells did not express CsgD-GFP (Fig. 4A). After 12 h, and more pronounced after 24 h of growth, the proportion of cells showing CsgD-GFP expression had increased (Fig. 4B and C). At 41 h of rdar morphotype development, the population of cells with bright CsgD-GFP signal decreased in number (Fig. 4D).
Next, we correlated CsgD expression with the formation of multicellular communities of cells. At 8 h of growth, expression of CsgD-GFP was observed in single individual cells (Fig. 4A). At later time points, however, expression of CsgD-GFP was associated with cell aggregation, consistent with earlier reports that CsgD controls the synthesis of extracellular matrix components in rdar morphotype expression (38). At 12 and 24 h, cells expressing CsgD-GFP fusion protein were most frequently part of large aggregates or microcolonies that could not be disrupted easily (Fig. 4B and C). However, at 41 h, the CsgD-expressing cells were not as tightly associated with aggregates (Fig. 4D). Cell aggregates of comparable sizes were absent from the cultures of a csgD deletion mutant grown under identical conditions (reference 55 and data not shown). This is in agreement with our previous results demonstrating that CsgD is required for cellular aggregation during rdar morphotype development (55).
Cellulose production is confined to the population of cells with high levels of CsgD expression.
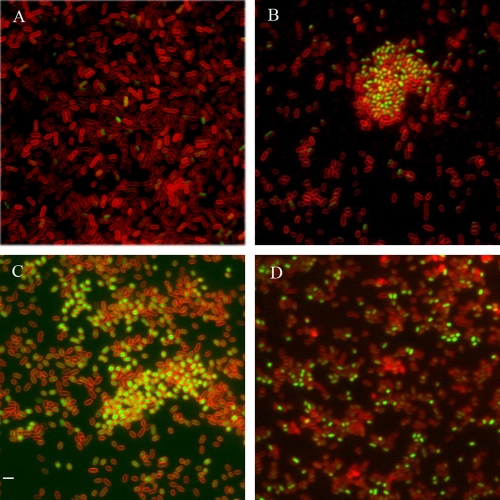
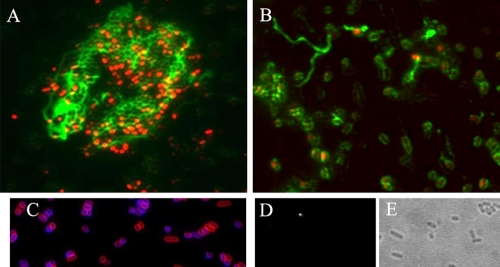
We have previously shown that one of the functions of CsgD is to promote cellulose production through transcriptional activation of the diguanylate cyclase AdrA (44, 55). We hypothesized that the bistable pattern of CsgD expression would translate into a corresponding pattern of cellulose synthesis. To test that hypothesis, we stained surface-grown cells expressing CsgD-GFP with calcofluor, a dye previously demonstrated to specifically stain cellulose in S. Typhimurium under these growth conditions (55). We observed that cellulose production was mainly associated with the cell aggregates containing CsgD-GFP fluorescence (Fig. 5A). In many cases, the production of cellulose polymers could be traced down as produced by individual cells with strong GFP signal (Fig. 5B). As a negative control, bacteria from the csgD deletion mutant MAE50 were stained with calcofluor. Minute amounts of cellulose were detected around the peripheries of some cells (Fig. 5C), while the long and branched cellulose filaments characteristic of the wild-type UMR1 were absent in the ΔcsgD mutant. It has been shown that CsgD-independent cellulose biosynthesis can occur (11, 47). The absence of detectable calcofluor fluorescence from cultures of the cellulose synthase knockout mutant confirmed that calcofluor bound cellulose specifically (Fig. 5D and E).
FIG. 5.
Cellulose production in the subpopulation of cells with high levels of CsgD. Detection of cellulose production by calcofluor staining was performed with the following strains: MAE851 (csgD-gfp) (A and B; red, GFP fluorescence; green, calcofluor), MAE50 (ΔcsgD) (C; red, membrane staining with FM4-64; blue, calcofluor), and MAE222 (ΔbcsA) (D [calcofluor] and E [corresponding phase-contrast image]). Bar, 2 μm.
Bistable expression of CsgD-GFP in biofilm formed in steady-state liquid culture.
In addition to agar plates, CsgD also controls biofilm formation in liquid culture (38). Strain MAE851, expressing the CsgD-GFP fusion protein, was grown under microaerophilic conditions shown to be optimal for biofilm formation (15). Forty-eight hours postinoculation, biofilm formation was evidenced by a bacterial pellicle attached to the walls of the flask and by the presence of cell aggregates at the bottom of the flask. The culture flask was left standing on the bench for 30 min in order to let cell aggregates sediment to the bottom. Samples for microscopic observation were taken from the pellicle (see Fig. S1, upper panel, in the supplemental material), from the liquid part of the culture (see Fig. S1, middle panel, in the supplemental material), and from the cell aggregates at the bottom (see Fig. S1, bottom panel, in the supplemental material). The cells from the pellicle and cell aggregate fractions of the culture expressed CsgD-GFP in almost all cells. In contrast, the liquid part of the culture contained mainly the subpopulation of cells that did not express CsgD-GFP (see Fig. S1, middle panel, in the supplemental material). The cells with CsgD-OFF were single cells, while the cells with CsgD-ON showed a tendency to aggregate. A control culture of the csgD deletion mutant grown under identical conditions consisted mainly of planktonic cells (data not shown).
Bistable expression of CsgD in flow cell-formed biofilms.
The flow cell model of biofilm formation provides the opportunity to monitor biofilm development unperturbed and in real time under continuous-flow conditions. To establish the role of CsgD during biofilm formation in the flow cell, the wild-type UMR1 and the isogenic csgD deletion mutant MAE50 were compared. In this way, initial attachment to the glass surface at 2 h postinoculation and biofilm formation after 48 h of cultivation in the flow cell system were assessed. We found that the csgD deletion mutant MAE50 was impaired in initial attachment, with the number of cells attaching to the glass surface being 60% of the number of wild-type UMR1 cells attached (data not shown). Furthermore, the UMR1 wild type formed three-dimensional biofilms with a high degree of confluence after 48 h of cultivation in the flow cell system (Fig. 6A). In contrast, the csgD deletion mutant formed dispersed microcolonies with occasional three-dimensional biofilm structures, which were much smaller in diameter than in the wild-type UMR1 and were disconnected from each other (Fig. 6B). We concluded from these observations that CsgD promotes primary attachment to the glass surface and is required for the formation of mature biofilms.
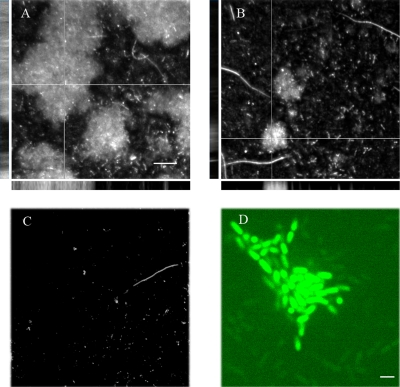
FIG. 6.
Role and expression of CsgD in flow cell biofilms. (A and B) Confocal micrographs of 48-h-old biofilms of wild-type UMR1 (A) and isogenic strain MAE50 (ΔcsgD) (B). The large panels represent top-down views of the biofilms, and the side panels show orthogonal projections (XZ and YZ) taken along the thick and the thin lines, respectively. The cells were stained with Live/Dead stain. Bars, 20 μm. (C) The double mutant MAE265 (ΔcsgD ΔbcsA) did not form biofilm. The cells were stained with Live/Dead stain. Bar, 20 μm. (D) Confocal micrograph of 24-h-old biofilm of strain MAE851 (CsgD-GFP). Two subpopulations of cells are evident, one, with high GFP fluorescence, involved in building up the biofilm structure, and one subpopulation of individual cells with low GFP signal. Size bar, 2 μm.
In order to test whether CsgD-independent biofilm formation is still dependent on cellulose production, we used a mutant with both csgD and the gene for cellulose synthase, bcsA, deleted. The latter mutant was severely impaired in primary attachment (10% of the wild-type value) and formed only tiny microcolonies of 10 to 15 loosely attached cells after 48 h of incubation (Fig. 6C), similar to the phenotype of the bcsA deletion strain (36). Thus, the production of cellulose is partially uncoupled from CsgD expression during initial attachment to the glass surface and subsequent biofilm formation in the flow cell system.
The CsgD-GFP-expressing strain MAE851 formed biofilms with confluence and complexity indistinguishable from those of the wild-type UMR1 (data not shown). CsgD-GFP fluorescence could be observed around 24 h of cultivation in the flow cell (Fig. 6D) and was not visible in mature biofilms. In 24-hour-old biofilms, high-intensity GFP signals were predominantly associated with the firmly attached cells, which were arranged as microcolonies (Fig. 6D). On the other hand, the single and motile cells contained low levels of CsgD-GFP fluorescence. In summary, two distinct subpopulations of cells were present within flow cell-grown biofilms in respect to their CsgD-GFP contents, and the subpopulation of cells expressing CsgD-GFP was more often involved in building up the three-dimensional biofilm structures.
Quantification of CsgD-GFP expression during rdar morphotype development.
FACS analysis was used in order to quantify the expression of CsgD-GFP in individual cells. Production of biofilm matrix components by the CsgD-GFP-expressing strain UMR1, however, leads to tightly connected cell populations (37), which hinders quantitative FACS analysis of single cells. Therefore, a csgD-gfp strain lacking the extracellular matrix components curli fimbriae (through deletion of the genes encoding structural components of curli fimbriae, csgBA) and cellulose (through deletion of the gene encoding the cellulose synthase, bcsA) was constructed. Such a strain was previously shown to consist of individual cells (37). As a control experiment, the impact of the expression of the extracellular matrix components cellulose and curli fimbriae on CsgD expression was analyzed by Western blotting. At 8 h of growth, the expression of CsgD-GFP in the ΔbcsA ΔcsgBA deletion mutant was low and almost undetectable by Western blot analysis, a situation like that in the wild-type UMR1 (Fig. 7A). In agreement with the observation made in the UMR1 background, only a few cells expressed large amounts of CsgD-GFP in the ΔbcsA ΔcsgBA deletion mutant, as observed by fluorescence microscopy (compare Fig. 7B with Fig. 4A). After 24 h of growth, the expression of CsgD-GFP was significantly increased in the mutant deficient in extracellular matrix components and was approximately two times higher than the wild-type UMR1 (Fig. 7A). The upregulated CsgD expression observed in the wild-type UMR1 expressing native CsgD upon deletion of the matrix components is consistent with this observation (Fig. 7A). Nevertheless, fluorescence microscopy analysis revealed that the cell population of the ΔbcsA ΔcsgBA deletion strain showed an on/off expression pattern of CsgD-GFP expression similar to that of the wild-type UMR1 (compare Fig. 4 and 7B).
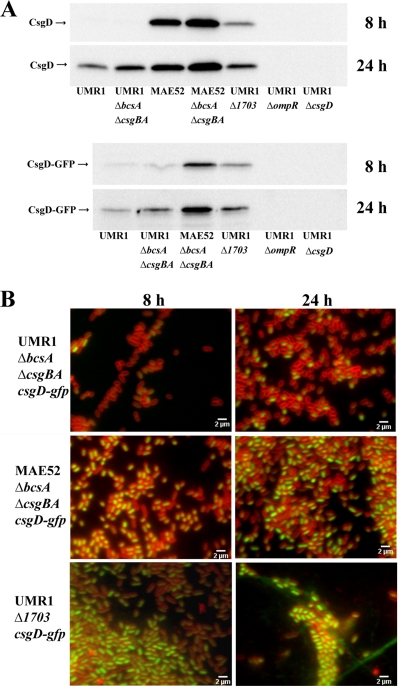
FIG. 7.
(A) CsgD and CsgD-GFP expression of S. Typhimurium UMR1 and derivatives. UMR1 showed detectable CsgD expression at 24 h, but not at 8 h. MAE52 displayed high CsgD expression at 8 h and 24 h. UMR1 ΔbcsA ΔcsgBA and MAE52 ΔbcsA ΔcsgBA showed upregulation of CsgD expression at all time points compared to the respective parental strains, UMR1 and MAE52. Strain UMR1 Δ1703 displayed upregulation of CsgD expression compared to wild-type UMR1, although the upregulation in the MAE52 strain was more pronounced. The relative expression of CsgD-GFP was similar to the relative expression of CsgD for all strains. CsgD and CsgD-GFP expression was not observed in UMR1 ΔompR. UMR1 ΔcsgD served as a negative control. (B) Fluorescence microscopy studies of CsgD-GFP expression in S. Typhimurium UMR1 and mutants with altered CsgD expression. Strain UMR1 ΔbcsA ΔcsgBA csgD-gfp, deficient for the production of the extracellular matrix components cellulose and curli fimbriae, exhibits a time-dependent bistable expression pattern of CsgD-GFP comparable to that of wild-type strain UMR1 csgD-gfp (Fig. 4). In strain MAE52 ΔbcsA ΔcsgBA csgD-gfp, the majority of cells expressed high levels of CsgD-GFP at 8 and 24 h. Strain UMR1 Δ1703 csgD-gfp expressed high levels of CsgD-GFP. Extensive cell aggregation and production of extracellular matrix components can be observed in strain UMR1 Δ1703 csgD-gfp. The microscopic observations are consistent with the Western blot analysis shown in panel A. Green, CsgD-GFP fluorescence; red, fluorescence of membrane stain FM4-64. Bars, 2 μm.
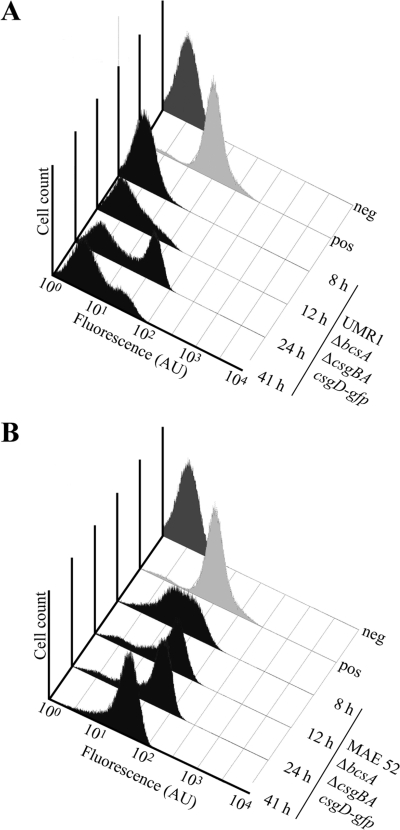
Subsequently, the fluorescence intensity of UMR1 ΔbcsA ΔcsgBA csgD-gfp cells was measured over a period of 41 h by FACS analysis (Fig. 8A). After 8 h of growth, the majority of cells did not show detectable CsgD-GFP expression and only 2% of the cells were positive for CsgD expression. At 12 h of growth, 12% of the cells were positive for CsgD-GFP expression. At 24 h of growth, the histogram showed two distinct peaks with a maximum of 52% of all analyzed cells expressing CsgD-GFP. At 41 h of development, the number of CsgD-GFP-expressing cells decreased, reaching 28%. In summary, the FACS analysis supported the observations made by fluorescence microscopy and demonstrated that CsgD-GFP is expressed in a bistable manner.
FIG. 8.
FACS analysis of CsgD-GFP fluorescence detected in strains UMR1 ΔbcsA ΔcsgBA csgD-gfp (A) and MAE52 ΔbcsA ΔcsgBA csgD-gfp (B) after 8 h, 12 h, 24 h, and 41 h of growth (black shading). CsgD-GFP was virtually unexpressed in UMR1 ΔbcsA ΔcsgBA csgD-gfp at 8 and 12 h of growth. At 24 and 41 h of growth, a bistable expression pattern was evident in strain UMR1 ΔbcsA ΔcsgBA csgD-gfp. Due to the elevated CsgD-GFP expression of strain MAE52 ΔbcsA ΔcsgBA csgD-gfp, the majority of cells were positive for CsgD-GFP expression at all time points. A bistable expression pattern was not observed. Dark gray, the control strain UMR1 ΔompR csgD-gfp, expressing no CsgD-GFP; light gray, the positive control (strain MAE119). The x axis shows arbitrary units (AU) of fluorescence in a logarithmic scale. The y axis shows the cell count in the cell population-representing gate, where total numbers ranged between a minimum of 58,000 and a maximum of 89,500 events.
Mechanism(s) of bistable CsgD-GFP expression.
A bistable expression pattern of a gene/protein can be observed when the overall expression of a gene or protein is low in combination with positive feedback regulation (46). In order to investigate the impact of enhanced CsgD expression on the bistable expression pattern, CsgD expression was investigated by FACS analysis in strain MAE52, a strain with 3-fold-higher CsgD expression than UMR1 (38) (Fig. 8B). Again, a ΔbcsA ΔcsgBA strain of MAE52 csgD-gfp was constructed. By Western blot analysis, high levels of CsgD-GFP were detected at 8 h and 24 h in the MAE52 mutant deficient for cellulose and curli fimbriae expression compared to the corresponding UMR1 derivative (Fig. 7A). As we were not able to construct a csgD-gfp fusion in the MAE52 background, the impact of deletion of cellulose and curli fimbriae on the CsgD-GFP level was investigated by comparing native CsgD levels in the parental strain, MAE52, and its ΔbcsA ΔcsgBA derivative (Fig. 7A). In the two strains, CsgD was expressed at high levels at 8 and 24 h compared to UMR1. As in the UMR1 background, CsgD expression was higher in the mutant deficient for cellulose and curli expression than in the MAE52 parent strain.
Fluorescence microscopy studies suggested that the high levels of CsgD-GFP expression detected by Western blot analysis after 8 h of growth was due to a high number of cells expressing the protein (Fig. 7B). At 24 h of growth, the majority of cells were detected to fluoresce brightly (Fig. 7B). FACS analyses revealed that at 8 h of growth, 69% of cells were positive for CsgD-GFP expression (Fig. 8B). However, in contrast to the UMR1 derivative, a single broad peak with a continuous distribution of CsgD expression was observed. At 12 h of growth, the peak narrowed, with the maximum, 80% of all cells, positive for CsgD-GFP expression. Narrowing of the peak continued over time, with up to 91% of all cells expressing CsgD-GFP at 41 h of growth. It is important to note that a bistable expression pattern was not detected at any time point, suggesting that the bistable regulation of CsgD is dependent on the presence of the wild-type csgD promoter and is generated at the level of transcriptional regulation and/or elevated expression.
Impact of the c-di-GMP signaling pathway on CsgD expression.
The c-di-GMP signaling system is known to dynamically regulate CsgD expression (Fig. 1) (23, 43). c-di-GMP levels and CsgD expression are low in the wild-type UMR1 (43), but enhanced c-di-GMP levels and CsgD expression can be created by deletion of c-di-GMP-specific phosphodiesterases, like STM1703, responsible for the degradation of the c-di-GMP pools dedicated to CsgD regulation (Fig. 1) (43). In order to investigate the influence of elevated c-di-GMP levels on CsgD-GFP expression, strain UMR1 csgD-gfp ΔSTM1703 was constructed. Deletion of STM1703 led to upregulation of CsgD expression at 8 and 24 h of growth (compare CsgD and CsgD-GFP levels of wild-type UMR1 and UMR1 ΔSTM1703 in Fig. 7A) (45). In addition, CsgD-GFP expression was analyzed by fluorescence microscopy (Fig. 7B). At 8 h of growth, high expression levels of CsgD-GFP were observed in many cells. At 24 h of growth, the majority of cells were brightly fluorescent due to upregulated CsgD-GFP expression. CsgD-GFP-expressing cells were part of biofilm communities. These biofilm-forming cells were surrounded by long fibers of extracellular matrix, presumably cellulose. Only a minority of cells did not express CsgD and did not take part in biofilm communities (Fig. 7B). Although we did not quantify CsgD-GFP expression in the STM1703 mutant, we concluded from the results obtained by fluorescence microscopy that high c-di-GMP levels lead to growth phase-independent monophasic expression of CsgD.
DISCUSSION
In this work, we studied the spatial and temporal patterns of expression of the major biofilm regulator CsgD in S. Typhimurium biofilms at the single-cell level. To this end, we constructed a functional CsgD-GFP translational fusion expressed from the native chromosomal locus of csgD under its natural promoter. We used fluorescence microscopy to analyze the expression of CsgD in single living cells and FACS analysis to demonstrate bimodal expression of CsgD. To our knowledge, this is the first report that deals with direct visualization of the expression of a major biofilm regulator.
Using the wild-type strain UMR1, a strain with a CsgD expression level representative of S. Typhimurium (35), we found bimodal expression of CsgD-GFP within the total population of cells in three different models of S. Typhimurium biofilm formation, the rdar morphotype, biofilm formation in steady-state culture, and continuous-flow (flow cell-grown) biofilms. In all three biofilm models, two subpopulations of cells could be distinguished based on their CsgD contents: a high-expressing (CsgD-ON) and a low-expressing (CsgD-OFF) population. Consequently, the csgD promoter activity is adapted so that stochastic fluctuations in the expression of the promoter give rise to phenotypic variability.
Phenotypic variation is frequently found in natural systems in bacteria and spans from nutrient utilization through competence to virulence gene expression (46). Cells in biofilms are considered to be especially prone to phenotypic variation, as the biofilm consists of spatially restricted microenvironments, resulting in heterogeneous populations that differ, e.g., with respect to antibiotic susceptibility, motility, and production of extracellular matrix components (4, 17, 24).
What is the adaptive advantage conferred by bistable CsgD expression? Conventionally, it is considered that biofilm formation is an energetically costly process through, e.g., the production of a complex exopolymer matrix. Only a subpopulation of cells is responsible for production of extracellular matrix components, which can subsequently be shared by the whole population (1), thus minimizing the cost and maximizing the benefits of the collaborative effort to make a biofilm. Consequently, we observed cells in the colony expressing the rdar morphotype that did not express the biofilm regulator CsgD but were still part of large cellular aggregates (Fig. 3D and 4B). The energy theory arises from the observation that the capability to express multicellular behavior is easily lost under laboratory conditions (8, 34). Although there is no doubt that production of extracellular matrix is energetically expensive, another reason for phenotypic heterogeneity can be to maintain the developmental potential of the population. This becomes evident in flow cell biofilms, where CsgD-ON cells form microcolonies whereas CsgD-OFF cells are single, motile cells (Fig. 7D). In Pseudomonas aeruginosa, motility has been shown to contribute to the establishment of a mature biofilm architecture (24). In addition, generation of heterogeneity in a clonal population is a strategy to maximize the chances for survival in a constantly changing environment. Consistent with this hypothesis, cells expressing CsgD have been demonstrated to be more resistant to disinfectants and long-term desiccation (15, 40, 47, 53).
The subpopulation of cells expressing large amounts of CsgD was associated with cellular aggregation in rdar morphotype expression and biofilm formation in liquid culture, formed microcolonies in the flow cell system, and was more often engaged in cellulose production during rdar morphotype development. Consequently, bimodal CsgD expression leads to the establishment of phenotypic diversity during biofilm development. White and coworkers have recently shown that transcriptional fusions to the promoters of adrA (encoding a diguanylate cyclase required for the activation of cellulose biosynthesis) and csgB (encoding a curli structural subunit) are expressed primarily by the subpopulation of aggregated cells (52). In this paper, we inferred for the first time the direct correlation between expression of the biofilm regulator CsgD and the phenotypic output, namely, the production of cellulose.
In this study, the role of CsgD during biofilm formation in a continuous-flow model, the flow cell system, was investigated. We found that CsgD is critical for the formation of extended mature biofilm structures while it is dispensable for the establishment of isolated microcolonies. Furthermore, by using a double mutant with both csgD and the gene coding for the cellulose synthase deleted, we showed that, as previously reported, biofilm formation is entirely dependent on expression of the cellulose synthase under the experimental conditions used. Furthermore, we could conclude from these results that production of cellulose is partially uncoupled from CsgD expression during flow cell biofilm formation. There have been reports about CsgD-independent cellulose synthesis, which is controlled by the expression of the GGDEF domain protein STM1987 and other unknown GGDEF domain proteins (11, 28, 47). It will be of interest to determine the factors involved in control of cellulose synthesis in the flow cell system. Another question for the future is the molecular mechanism through which CsgD affects the three-dimensional structure of the flow cell-formed biofilms. We can exclude a possible role for curli fimbriae in this process, since we have previously shown that a curli-deficient mutant (ΔcsgA) of S. Typhimurium colonizes the flow cell in a manner indistinguishable from the wild-type UMR1 (36).
The observed bistable expression of the CsgD protein raised a number of questions about the molecular mechanisms involved in this phenomenon. In a genetic regulatory network, bistability can be generated by low-level noisy gene expression amplified by net positive feedback (10, 49). In E. coli, an (indirect) positive feedback loop has been implicated in the control of CsgD expression. CsgD activates the expression of the IraP protein, which counteracts the proteolytic degradation of the RpoS sigma factor required for CsgD expression. Increased RpoS stability results in enhanced csgD transcription, thus establishing an autoactivation loop for CsgD expression (16). However, CsgD autoactivation has not been observed in S. Typhimurium, as inactivation of csgD does not diminish the amount of mRNA of the csgDEFG operon (37). On the other hand, the bistable expression of CsgD that we observed can be the result of bimodal expression of an upstream regulator. A likely candidate to mediate bistable expression of CsgD is the stress sigma factor RpoS; however, it is unknown whether the activity of RpoS (18, 34) shows bistability.
Clearly, however, csgD promoter activity is tuned to give rise to bimodal expression. We observed that this highly delicate expression of CsgD can be modulated by genetic changes and epigenetic regulation. A single-base-pair insertion in the spacer region of the CsgD promoter, which led to approximately 3-fold upregulation of CsgD expression throughout the growth phase (38), virtually abolished the bistable expression of CsgD. On the other hand, CsgD expression can be modulated epigenetically by the secondary messenger c-di-GMP. High c-di-GMP levels led to elevated CsgD expression throughout the growth phase, practically abolishing biphasic CsgD expression. It has to be noted that the levels of CsgD expression in strain UMR1 are representative of the S. Typhimurium strain population (35). Therefore, it can be concluded that most strains of S. Typhimurium show bistable expression of biofilm formation. This bistable expression of CsgD enables S. Typhimurium to readily adapt to changing environmental conditions, like upcoming stress or a host environment.
Supplementary Material
Acknowledgments
We thank Tabea Hörnlen for experimental contributions to the project.
This work was supported by research grants from Carl Tryggers Stiftelse (CTS-07:306), the KI Infection Biology Network, the Swedish Research Council (621-2004-3979 and 621-2007-6509), the European Commission under contract number MEST-CT-2004-008475, and Längmanska Kulturfonden to U.R.
Footnotes
Published ahead of print on 6 November 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Branda, S. S., F. Chu, D. B. Kearns, R. Losick, and R. Kolter. 2006. A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 59:1229-1238. [DOI] [PubMed] [Google Scholar]
- 2.Brombacher, E., C. Dorel, A. J. Zehnder, and P. Landini. 2003. The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology 149:2847-2857. [DOI] [PubMed] [Google Scholar]
- 3.Brown, P. K., C. M. Dozois, C. A. Nickerson, A. Zuppardo, J. Terlonge, and R. Curtiss III. 2001. MlrA, a novel regulator of curli (AgF) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium. Mol. Microbiol. 41:349-363. [DOI] [PubMed] [Google Scholar]
- 4.Chai, Y., F. Chu, R. Kolter, and R. Losick. 2008. Bistability and biofilm formation in Bacillus subtilis. Mol. Microbiol. 67:254-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen, B. B., C. Sternberg, J. B. Andersen, R. J. Palmer, Jr., A. T. Nielsen, M. Givskov, and S. Molin. 1999. Molecular tools for study of biofilm physiology. Methods Enzymol. 310:20-42. [DOI] [PubMed] [Google Scholar]
- 6.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidson, C. J., A. P. White, and M. G. Surette. 2008. Evolutionary loss of the rdar morphotype in Salmonella as a result of high mutation rates during laboratory passage. ISME J. 2:293-307. [DOI] [PubMed] [Google Scholar]
- 9.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubnau, D., and R. Losick. 2006. Bistability in bacteria. Mol. Microbiol. 61:564-572. [DOI] [PubMed] [Google Scholar]
- 11.Garcia, B., C. Latasa, C. Solano, F. Garcia-del Portillo, C. Gamazo, and I. Lasa. 2004. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol. Microbiol. 54:264-277. [DOI] [PubMed] [Google Scholar]
- 12.Gerstel, U., A. Kolb, and U. Römling. 2006. Regulatory components at the csgD promoter—additional roles for OmpR and integration host factor and role of the 5′ untranslated region. FEMS Microbiol. Lett. 261:109-117. [DOI] [PubMed] [Google Scholar]
- 13.Gerstel, U., C. Park, and U. Römling. 2003. Complex regulation of csgD promoter activity by global regulatory proteins. Mol. Microbiol. 49:639-654. [DOI] [PubMed] [Google Scholar]
- 14.Gerstel, U., and U. Römling. 2003. The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium. Res. Microbiol. 154:659-667. [DOI] [PubMed] [Google Scholar]
- 15.Gerstel, U., and U. Römling. 2001. Oxygen tension and nutrient starvation are major signals that regulate agfD promoter activity and expression of the multicellular morphotype in Salmonella typhimurium. Environ. Microbiol. 3:638-648. [DOI] [PubMed] [Google Scholar]
- 16.Gualdi, L., L. Tagliabue, and P. Landini. 2007. Biofilm formation-gene expression relay system in Escherichia coli: modulation of sigmaS-dependent gene expression by the CsgD regulatory protein via sigmaS protein stabilization. J. Bacteriol. 189:8034-8043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haagensen, J. A., M. Klausen, R. K. Ernst, S. I. Miller, A. Folkesson, T. Tolker-Nielsen, and S. Molin. 2007. Differentiation and distribution of colistin- and sodium dodecyl sulfate-tolerant cells in Pseudomonas aeruginosa biofilms. J. Bacteriol. 189:28-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammar, M., A. Arnqvist, Z. Bian, A. Olsen, and S. Normark. 1995. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 18:661-670. [DOI] [PubMed] [Google Scholar]
- 19.Hammar, M., Z. Bian, and S. Normark. 1996. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 93:6562-6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hautefort, I., M. J. Proenca, and J. C. Hinton. 2003. Single-copy green fluorescent protein gene fusions allow accurate measurement of Salmonella gene expression in vitro and during infection of mammalian cells. Appl. Environ. Microbiol. 69:7480-7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jefferson, K. K. 2004. What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 236:163-173. [DOI] [PubMed] [Google Scholar]
- 22.Jubelin, G., A. Vianney, C. Beloin, J. M. Ghigo, J. C. Lazzaroni, P. Lejeune, and C. Dorel. 2005. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol. 187:2038-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kader, A., R. Simm, U. Gerstel, M. Morr, and U. Römling. 2006. Hierarchical involvement of various GGDEF domain proteins in rdar morphotype development of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 60:602-616. [DOI] [PubMed] [Google Scholar]
- 24.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48:1511-1524. [DOI] [PubMed] [Google Scholar]
- 25.Lazazzera, B. A. 2005. Lessons from DNA microarray analysis: the gene expression profile of biofilms. Curr. Opin. Microbiol. 8:222-227. [DOI] [PubMed] [Google Scholar]
- 26.Lewis, K. 2001. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45:999-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 28.Monteiro, C., S. I. Wang, X. Kader, A. Bokranz, W. Simm, R. Nobles, M. Chromek, A. Brauner, R. M. Brown, Jr., and U. Römling. 2009. Characterisation of cellulose production in Escherichia coli Nissle 1917and its effect on bacterial-host interaction. Environ. Microbiol. 11:1105-1116. [DOI] [PubMed] [Google Scholar]
- 29.Prouty, A. M., and J. S. Gunn. 2003. Comparative analysis of Salmonella enterica serovar Typhimurium biofilm formation on gallstones and on glass. Infect. Immun. 71:7154-7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prouty, A. M., W. H. Schwesinger, and J. S. Gunn. 2002. Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect. Immun. 70:2640-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rabsch, W., H. L. Andrews, R. A. Kingsley, R. Prager, H. Tschape, L. G. Adams, and A. J. Baumler. 2002. Salmonella enterica serotype Typhimurium and its host-adapted variants. Infect. Immun. 70:2249-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Römling, U. 2005. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell Mol. Life Sci. 62:1234-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Römling, U. 2001. Genetic and phenotypic analysis of multicellular behavior in Salmonella typhimurium. Methods Enzymol. 336:48-59. [DOI] [PubMed] [Google Scholar]
- 34.Römling, U., Z. Bian, M. Hammar, W. D. Sierralta, and S. Normark. 1998. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180:722-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Römling, U., W. Bokranz, W. Rabsch, X. Zogaj, M. Nimtz, and H. Tschäpe. 2003. Occurrence and regulation of the multicellular morphotype in Salmonella serovars important in human disease. Int. J. Med. Microbiol. 293:273-285. [DOI] [PubMed] [Google Scholar]
- 36.Römling, U., D. Pesen, and S. Yaron. 2007. Biofilms of Salmonella enterica, p. 127-146. In M. Rhen (ed.), Salmonella: molecular biology and pathogenesis. Horizon Bioscience, Wymondham, United Kingdom.
- 37.Römling, U., M. Rohde, A. Olsen, S. Normark, and J. Reinkoster. 2000. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 36:10-23. [DOI] [PubMed] [Google Scholar]
- 38.Römling, U., W. D. Sierralta, K. Eriksson, and S. Normark. 1998. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28:249-264. [DOI] [PubMed] [Google Scholar]
- 39.Ryjenkov, D. A., R. Simm, U. Römling, and M. Gomelsky. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP. The PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 281:30310-30314. [DOI] [PubMed] [Google Scholar]
- 40.Scher, K., U. Römling, and S. Yaron. 2005. Effect of heat, acidification, and chlorination on Salmonella enterica serovar Typhimurium cells in a biofilm formed at the air-liquid interface. Appl. Environ. Microbiol. 71:1163-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmieger, H. 1972. Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 119:75-88. [DOI] [PubMed] [Google Scholar]
- 42.Scholz, O., A. Thiel, W. Hillen, and M. Niederweis. 2000. Quantitative analysis of gene expression with an improved green fluorescent protein. Eur. J. Biochem. 267:1565-1570. [DOI] [PubMed] [Google Scholar]
- 43.Simm, R., A. Lusch, A. Kader, M. Andersson, and U. Römling. 2007. Role of EAL-containing proteins in multicellular behavior of Salmonella enterica serovar Typhimurium. J. Bacteriol. 189:3613-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simm, R., M. Morr, A. Kader, M. Nimtz, and U. Römling. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123-1134. [DOI] [PubMed] [Google Scholar]
- 45.Simm, R., U. Remminghorst, I. Ahmad, K. Zakikhany, and U. Römling. 2009. A role for the EAL-like protein STM1344 in regulation of CsgD expression and motility. J. Bacteriol. 191:3829-3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smits, W. K., O. P. Kuipers, and J. W. Veening. 2006. Phenotypic variation in bacteria: the role of feedback regulation. Nat. Rev. Microbiol. 4:259-271. [DOI] [PubMed] [Google Scholar]
- 47.Solano, C., B. Garcia, J. Valle, C. Berasain, J. M. Ghigo, C. Gamazo, and I. Lasa. 2002. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43:793-808. [DOI] [PubMed] [Google Scholar]
- 48.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187-209. [DOI] [PubMed] [Google Scholar]
- 49.Veening, J. W., W. K. Smits, and O. P. Kuipers. 2008. Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 62:193-210. [DOI] [PubMed] [Google Scholar]
- 50.Vlamakis, H., C. Aguilar, R. Losick, and R. Kolter. 2008. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 22:945-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weinhouse, H., S. Sapir, D. Amikam, Y. Shilo, G. Volman, P. Ohana, and M. Benziman. 1997. c-di-GMP-binding protein, a new factor regulating cellulose synthesis in Acetobacter xylinum. FEBS Lett. 416:207-211. [DOI] [PubMed] [Google Scholar]
- 52.White, A. P., D. L. Gibson, G. A. Grassl, W. W. Kay, B. B. Finlay, B. A. Vallance, and M. G. Surette. 2008. Aggregation via the red, dry, and rough morphotype is not a virulence adaptation in Salmonella enterica serovar Typhimurium. Infect. Immun. 76:1048-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.White, A. P., D. L. Gibson, W. Kim, W. W. Kay, and M. G. Surette. 2006. Thin aggregative fimbriae and cellulose enhance long-term survival and persistence of Salmonella. J. Bacteriol. 188:3219-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winfield, M. D., and E. A. Groisman. 2003. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microbiol. 69:3687-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Römling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452-1463. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.