Abstract
Acetyl xylan esterase (EC 3.1.1.72) is a member of a set of enzymes required to depolymerize hemicellulose, especially xylan that is composed of a main chain of β-1,4-linked xylopyranoside residues decorated with acetyl side groups. Fibrobacter succinogenes S85 Axe6B (FSUAxe6B) is an acetyl xylan esterase encoded in the genome of this rumen bacterium. The enzyme is a modular protein comprised of an esterase domain, a carbohydrate-binding module, and a region of unknown function. Sequences that are homologous to the region of unknown function are paralogously distributed, thus far, only in F. succinogenes. Therefore, the sequences were designated Fibrobacter succinogenes-specific paralogous module 1 (FPm-1). The FPm-1s are associated with at least 24 polypeptides in the genome of F. succinogenes S85. A bioinformatics search showed that most of the FPm-1-appended polypeptides are putative carbohydrate-active enzymes, suggesting a potential role in carbohydrate metabolism. Truncational analysis of FSUAxe6B, together with catalytic and substrate binding studies, has allowed us to delineate the functional modules in the polypeptide. The N-terminal half of FSUAxe6B harbors the activity that cleaves side chain acetyl groups from xylan-like substrates, and the binding of insoluble xylan was determined to originate from FPm-1. Site-directed mutagenesis studies of highly conserved active-site residues in the esterase domain suggested that the esterase activity is derived from a tetrad composed of Ser44, His273, Glu194, and Asp270, with both Glu194 and Asp270 functioning as helper acids, instead of a single carboxylate residue proposed to initiate catalysis.
The development of strategies for biomass conversion to fuels (biofuels) is a subject of keen interest as we search for energy resources alternative to fossil fuels (39). Plant cell matter accounts for 150 to 200 billion tons of biomass on our planet annually (31). It is technically possible, but economically far from realization, to convert plant cell wall to biofuels (41). Thus, currently, plant cell wall utilization as a source of biofuels is mostly at the laboratory scale, although there is a great impetus to move production to the industrial scale.
The main components of the plant cell wall are cellulose, hemicellulose, and lignin. These components form complex structures that provide the plant with physical strength (42). Biologically, there are two major steps in the production of alcohols from plant-based feedstock. The first step is an enzymatic hydrolysis of the plant cell wall components to fermentable sugars, and the second step is fermentation of the resultant sugars into alcohols. A major limitation of the process is the lack of highly efficient biocatalysts required for the first step. However, it is known that microbes, either as individuals or consortia, that harbor genes encoding enzymes that hydrolyze plant cell wall polysaccharides abound in nature. Research efforts directed at deepening knowledge of how multiple enzymes participate synergistically to degrade the plant cell wall will accelerate the capacity to achieve the goal of converting biomass to biofuels on a large scale (12, 27). However, improvement of “enzyme cocktails” developed for depolymerization of lignocellulosic biomass will be dependent on a better understanding of the structure/function of individual enzymes that together constitute the arsenal of enzymes (hydrolyzome) used by naturally occurring organisms known to be highly efficient in plant cell wall degradation.
Ruminant animals harbor a variety of plant cell wall-degrading bacteria in their first stomach or rumen (26). These animals digest forages with the aid of a microbial consortium that is able to metabolize plant cell wall polysaccharides to short-chain fatty acids, the main energy source for the ruminant host. Fibrobacter succinogenes is a ubiquitous rumen bacterium and has been estimated in previous reports to occupy 0.1% to 1.0% of the microbial population in the cattle rumen, based on the quantification of 16S rRNA genes as a marker (25, 43). F. succinogenes is a significant cellulolytic rumen bacterium, and it has the ability to grow on crystalline cellulose as a sole source of carbon and energy (17). Additionally, it has been demonstrated that this bacterium can solubilize hemicelluloses, although it only partially utilized the constituent monosaccharides released (34). As further evidence, F. succinogenes failed to grow on xylose (33), a constituent of most hemicelluloses. Since F. succinogenes is a highly versatile microbe capable of degrading both cellulose and hemicellulose, strains of this bacterium are attractive models to study natural strategies for efficient deconstruction of plant cell wall polysaccharides.
Through analysis of the genome sequence of F. succinogenes S85, a gene cluster that encodes more than 10 hemicellulose-targeting enzymes was identified. Most of the enzymes in the cluster are modular polypeptides, a common feature in many carbohydrate-active enzymes. Kam and coworkers (23) previously identified two acetyl xylan esterases (Axe6A and Axe6B) in this cluster and predicted that each gene encoded a polypeptide composed of two domains: an esterase catalytic domain and a family 6 carbohydrate-binding module (CBM6). Whereas Axe6A was fairly well characterized, difficulties in expression of recombinant Axe6B restricted its characterization (23). In this report, overproduction of recombinant F. succinogenes S85 Axe6B (FSUAxe6B) is demonstrated, and furthermore, it is shown that rather than having two domains, the polypeptide harbors three domains composed of an esterase, CBM6, and a region of unknown function. Bioinformatics analysis suggested that the unknown domain observed in FSUAxe6B is, so far, distributed only in F. succinogenes S85; thus, it was designated F. succinogenes-specific paralogous module 1 or FPm-1. Twenty-four polypeptides, with the majority containing glycoside hydrolase family motifs and CBMs, were found to harbor this peptide at the extreme C-terminal region. In addition to assigning a carbohydrate binding function to FPm-1, critical residues that confer esterase activity to the N-terminal half of FSUAxe6B were also identified through site-directed mutagenesis.
MATERIALS AND METHODS
Strains, media, and growth conditions.
Fibrobacter succinogenes subsp. succinogenes S85 was obtained from a culture collection at the Department of Animal Sciences, University of Illinois at Urbana-Champaign (35). F. succinogenes S85 was grown in a synthetic medium (40) under anaerobic conditions. Escherichia coli JM109 and E. coli BL21(DE3) CodonPlus RIPL-competent cells were purchased from Stratagene (La Jolla, CA). Gene manipulation and plasmid construction were performed with E. coli JM109. E. coli BL21(DE3) CodonPlus RIPL was used for gene expression. The E. coli cells were grown aerobically at 37°C in Luria-Bertani (LB) medium supplemented with appropriate antibiotics.
Gene cloning, expression, and protein purification.
F. succinogenes S85 was grown for 2 days, cells were harvested, and the genomic DNA was extracted using a DNeasy tissue kit (Qiagen, Hilden, Germany). The genes of wild-type (WT) FSUAxe6B and its truncational mutant proteins (TM1, TM2, TM3, and TM4) were amplified from the genomic DNA by PCR using Prime STAR HS DNA polymerase (Takara Bio, Otsu, Japan). Since it is expected that the functional FSUAxe6B protein produced by F. succinogenes cells will lack the signal peptide, which is required for secretion, the WT protein refers to the polypeptide with the signal peptide removed by PCR amplification. The forward and reverse primers used for the PCR were engineered to incorporate the NdeI and XhoI restriction sites, respectively. The primer pairs used for amplifying the WT protein and its truncated derivatives TM1, TM2, TM3, and TM4 were F1/R1, F1/R2, F1/R3, F2/R1, and F2/R2, respectively (Table 1; see also Fig. 2A). The amplified fragments were cloned into the pGEM-T vector (Promega, Madison, WI) and subcloned into a modified pET-28a expression vector (Novagen, San Diego, CA) that was engineered by replacing the kanamycin resistance gene with that for ampicillin resistance (7). For the construction of the TM5 expression vector, the Ek/LIC cloning kit was utilized (Novagen). The TM5 gene was amplified from the genomic DNA with the primers, F1′ and R1′ (Table 1; see also Fig. 2A). Both ends of the amplified gene fragment were digested, in the presence of dATP, with the 3′ to 5′ exonuclease activity of the T4 DNA polymerase. The resultant fragment was annealed to the pET-46 Ek/LIC vector. All genes encoding derivatives of FSUAxe6B were nucleotide sequenced to confirm the integrity of the coding sequence after cloning into the expression vector. The gene expression vectors for FSUAxe6B or its truncated derivatives were introduced individually into E. coli BL21(DE3) CodonPlus RIPL-competent cells and grown in 10 ml of LB medium with ampicillin (100 μg/ml) and chloramphenicol (50 μg/ml) at 37°C overnight. Each culture was transferred to a fresh LB medium (1 liter) with the same antibiotics and grown until the optical density at 600 nm reached approximately 0.4. For each culture, the temperature for culturing was then decreased to 16°C, and isopropyl β-d-thiogalactopyranoside at a final concentration of 0.1 mM was added to the medium to induce production of the target protein. After 14 h, cells were harvested by centrifugation (5,000 rpm, 4°C, 15 min), and resuspended in 50 ml of lysis buffer (50 mM Tris-HCl, pH 7.5, 300 mM NaCl, 20 mM imidazole). Cells were disrupted by using an EmulsiFlex C-3 cell homogenizer (Avestin Inc., Ottawa, Canada), and the lysate was clarified by centrifugation (15,000 rpm, 4°C, 30 min). The supernatant was filtered through a 0.22-μm-pore-size Durapore membrane (Millipore, Bedford, MA). The filtrate was applied to a HisTrap FF 5-ml column (GE Healthcare, Piscataway, NJ), and unbound proteins were washed with 20-column volumes of lysis buffer. The bound proteins were eluted with elution buffer (50 mM Tris-HCl, pH 7.5, 300 mM NaCl, 250 mM imidazole), and the buffer was exchanged to 50 mM Tris-HCl, pH 7.5, and 300 mM NaCl by use of a desalting column (HiPrep 26/10 desalting; GE Healthcare). The latter buffer served as the storage buffer. All columns used in the protein purification steps were fitted to an ÄKTAxpress system (GE Healthcare).
TABLE 1.
Primers used in this study
| Primer | Sequence | Expt |
|---|---|---|
| F1 | 5′-CATATGGCTCCGAACCCGAACTTCCATATCTACATTGC-3′a | Cloning |
| F2 | 5′-CATATGGGCCCGTACACGGACCCGATTGAAATCCCTGGCAAG-3′a | Cloning |
| F1′ | 5′-GACGACGACAAGATGGGAATCAAGAATATCCGC-3′b | Cloning |
| R1 | 5′-CTCGAGTTATTCATGTATCACCACCTTTTTTG-3′a | Cloning |
| R2 | 5′-CTCGAGCTATCCAATCGGCGGCTGAGCGCTGATTTCCTTGAATTC-3′a | Cloning |
| R3 | 5′-CTCGAGCTAGCCATATTCCTCGGGCGGTTCATCCGGAACCGTAG-3′a | Cloning |
| R1′ | 5′-GAGGAGAAGCCCGGTTATTCATGTATCACCACCTTTTTTG-3′b | Cloning |
| S44G | 5′-CATTGCTTATGGGCAGGGTAACATGGCGGGCAACGGC-3′c | Mutagenesis |
| E194N | 5′-CATCTTCCACCAGGGCAACAGTGACGGTACCGATGC-3′c | Mutagenesis |
| E194A | 5′-CATCTTCCACCAGGGCGCAAGTGACGGTACCGATGC-3′c | Mutagenesis |
| D270N | 5′-GCAGGGTAACGGCAAGAATCCGTACCACTTTGGCCG-3′c | Mutagenesis |
| D270A | 5′-GCAGGGTAACGGCAAGGCTCCGTACCACTTTGGCCG-3′c | Mutagenesis |
| H273Q | 5′-CGGCAAGGATCCGTACCAGTTTGGCCGTGCGGGC-3′c | Mutagenesis |
Nucleotides incorporated for restriction enzyme digestion are underlined.
Nucleotides incorporated for exonuclease digestion are underlined.
Nucleotides corresponding to the substituted amino acids are underlined.
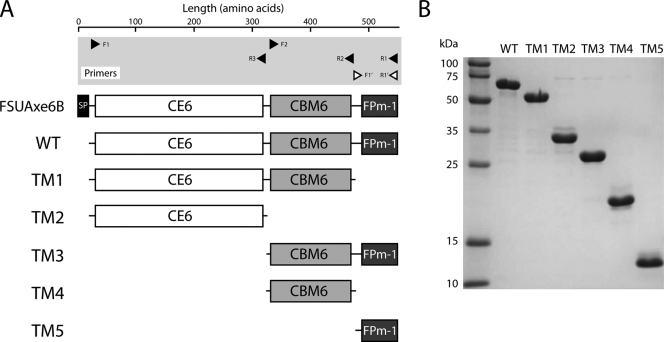
FIG. 2.
Truncational mutant proteins of FSUAxe6B. (A) Schematic representation of translated FSUAxe6B, the mature protein (WT), and its truncational mutant proteins (TM1 to TM5). The DNA primer sequences used for the amplification of the genes are described in Table 1. The arrowheads display the 5′ → 3′ direction of oligonucleotides. Primers with closed arrowheads contain restriction sites at the 5′ ends for insertion of their PCR products into a modified pET-28a vector. The primers with open arrowheads harbor nucleotide sequences that allow their PCR products to be digested by the exonuclease activity of T4 DNA polymerase prior to annealing to a pET-46 Ek/LIC vector. (B) SDS-PAGE image of purified WT and truncational proteins. Purified protein (2.5 μg) were loaded on the 12.5% polyacrylamide gel and stained with Coomasie brilliant blue G-250.
Bioinformatic analysis.
The genome sequence of F. succinogenes S85 was determined by the North American Consortium for Genomics of Fibrolytic Ruminal Bacteria in collaboration with the Institute for Genomic Research (TIGR) (FibRumba database; http://www.jcvi.org/rumenomics). A functional domain search was performed to determine the protein family and domain organization using the Pfam search server (http://www.sanger.ac.uk/Software/Pfam) and NCBI BLAST server (http://www.ncbi.nlm.nih.gov/BLAST). Prediction of lipoproteins and signal peptides were performed by using the LipoP 1.0 server (http://www.cbs.dtu.dk/services/LipoP). The secondary structure of FSUAxe6B was predicted by using the Advanced Protein Secondary Structure Prediction Server (http://imtech.res.in/raghava/apssp). Multiple amino acid sequences were aligned with ClustalW (http://www.ebi.ac.uk/clustalw). Protein Data Bank files were visually analyzed by the UCSF Chimera molecular graphics program (http://www.cgl.ucsf.edu/chimera).
Binding of insoluble polysaccharides.
Oat-spelt xylan (OSX) and Avicel PH-101 as ligands were purchased from Sigma-Aldrich (St. Louis, MO). Since OSX contains some soluble components, the soluble fraction was excluded as follows. One gram of OSX was stirred in 100 ml of distilled water for 12 h. After centrifugation (4,000 × g, 10 min, room temperature), the precipitate was further washed with 100 ml of distilled water and centrifuged (4,000 × g, 10 min, room temperature). The insoluble fraction was lyophilized and then ground into small particles in a mortar, producing insoluble OSX (is-OSX). Qualitative binding assessment between proteins and ligands was carried out as follows: one ml of 2 μM proteins in 50 mM Tris-HCl, pH 7.5, containing 300 mM NaCl (buffer A) was mixed with 20 mg of insoluble polysaccharide. The reaction mixture was gently mixed at 4°C for 1 h. Then, the insoluble polysaccharide was precipitated by centrifugation (13,000 rpm, 4°C, 1 min). The supernatant, including unbound protein, was resolved on a 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel. Blanks, for excluding the possibility of precipitation or adsorption of the protein to the tube during reaction, were prepared by incubating the protein without insoluble polysaccharide in the reaction buffer. Depletion binding isotherms were derived for quantitatively assessing the binding capacity of the protein for insoluble polysaccharide. The bicinchoninic acid protein assay kit (Thermo Scientific, Rockford, IL) was used for the quantification of proteins. One ml of various concentrations of proteins in buffer A was added to 20 mg of is-OSX and incubated with gentle mixing at 4°C. The supernatant after centrifugation (13,000 rpm, 4°C, 1 min) was used for the quantification of the unbound (free) protein. Total protein was measured after incubating protein without is-OSX under the same conditions. Bound protein was calculated by subtracting the free protein from the total protein.
For the determination of the binding constant between the protein and ligand, the Michaelis/Langmuir equation was applied. The equation is as follows: qad/ q = Kp × qmax/(1 + Kp × q), where qad is the amount of bound protein (nmol of proteins per g of is-OSX), q is the free protein in buffer (μM), Kp is the dissociation constant (μM), and qmax is the maximum amount of bound protein to ligand (28). GraphPad Prism version 5.01 (GraphPad Software, San Diego, CA) was utilized for the calculation of the binding parameters.
Enzyme assays and steady-state kinetics.
Acetyl xylan esterase activity was assayed using tetra-acetyl-xylopyranoside (Toronto Research Chemicals Inc., Ontario, Canada) for all proteins in this study, and the released acetic acid was measured using an acetic acid detection kit (Megazyme, Bray, Ireland) according to the manufacturer's instructions. The reduction of NADH was monitored continuously at an absorbance of 340 nm using a Synergy 2 microplate reader (BioTek, Winooski, VT) using the path length correction feature. All assays were carried out at 37°C. Five microliters of 1 μM enzyme and 20 μl of R2 enzyme solution (containing acetate kinase, pyruvate kinase, and d-lactose dehydrogenase in 100 mM Tris-HCl, pH 7.4, and 3 mM MgCl2) were thoroughly mixed. The tetra-acetyl-xylopyranoside was prepared in 290 μl of R1 solution (NADH, ATP, phospho-enol-pyruvate, and pyruvate). The concentrations of the ingredients in the R1 and R2 solutions were predetermined by the manufacturer (Megazyme). Both solutions were incubated separately at 37°C for 3 min to allow equilibration and then mixed to start the reaction. For active-site mutants with lower activity, the kinetic parameters were determined at a concentration of 10 μM for the E194N protein and 2 μM for the E194A, D270N, and D270A proteins. Initial rates were plotted against the tetra-acetyl-xylopyranoside concentrations, and the kinetic parameters were determined by the Michaelis-Menten equation utilizing GraphPad Prism version 5.01.
Site-directed mutagenesis.
Site-directed mutagenesis was carried out using the QuikChange multisite-directed mutagenesis kit (Stratagene), according to the manufacturer's instructions. Primers used in the site-directed mutagenesis study are presented in Table 1.
Circular dichroism (CD) spectra.
Determination of CD spectra for the FSUAxe6B WT protein and its site-directed mutant proteins was carried out using a J-815 CD spectropolarimeter (Jasco, Tokyo, Japan). Protein samples were prepared at a concentration of 0.1 mg/ml in 20 mM phosphate (NaH2PO4) buffer (pH 7.5) (24). For the measurements, a quartz cell with a path length of 0.1 cm was utilized. CD scans were carried out at 25°C from 190 nm to 260 nm at a speed of 50 nm/min with a 0.1-nm wavelength pitch, with five accumulations. Data files were analyzed on the DICHROWEB on-line server (http://www.cryst.bbk.ac.uk/cdweb/html/home.html) using the CDSSTR algorithm with reference set 4, which is optimized at 190 nm to 240 nm (29).
RESULTS
Domain organization of FSUAxe6B and proteins harboring FPm-1.
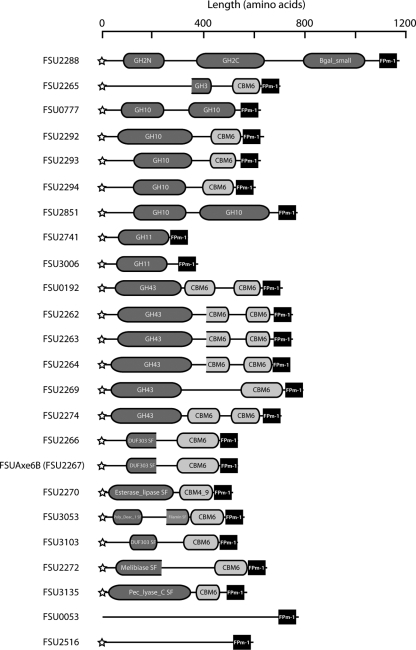
Based on the amino acid sequence identity, carbohydrate esterases (CEs) have been classified into 16 families (CE1 to CE16) according to the CAZy database (http://www.cazy.org/). A domain of FSUAxe6B, from amino acid position 30 to position 329, showed 46% identity to the polypeptide sequence of F. succinogenes acetyl xylan esterase Axe6A, a member of CE6 family. Therefore, FSUAxe6B was predicted to be a member of the CE6 family. Further analysis suggested that FSUAxe6B is a modular protein composed of the CE6 domain, a CBM6, and a C-terminal domain of unknown function. Acetyl xylan esterase is one of a set of enzymes that is required for xylan hydrolysis to its components. This enzyme cleaves ester bonds that link acetyl side groups to the β-1,4-linked xylopyranoside backbone of xylan, and members of CBM6 are known to bind to a variety of substrates (9, 20, 21, 36, 44). Although the likelihood that the CBM6 may include the region demarcated as harboring the unknown function was initially considered, this made the FSUAxe6B CBM unusually long. The GenBank database was searched to determine whether the sequence of unknown function occurs in other polypeptides, especially CBM6 proteins, already reported from other organisms. Interestingly, the results yielded no polypeptide with obvious similarity in amino acid sequence to this region. On the other hand, a search of the genome database of F. succinogenes S85 suggested that 23 other proteins harbor amino acid sequences that are similar to this C-terminally located domain of FSUAxe6B. These sequences were, therefore, designated Fibrobacter succinogenes-specific paralogous module 1 (FPm-1). Figure 1 shows the domain organizations of proteins harboring FPm-1 sequences. Most of these proteins, except for FSU0053, include signal peptides for secretion, suggesting that they function either extracellularly or in the periplasmic space. Among these 24 proteins, 15 proteins harbor glycosyl hydrolase (GH) family domains, which included a GH family 2 protein (GH2) (FSU2288), a GH3 protein (FSU2265), 5 GH10 proteins (FSU0777, FSU2292, FSU2293, FSU2294, and FSU2851), 2 GH11 proteins (FSU2741 and FSU3006), and 6 GH43 proteins (FSU0192, FSU2262, FSU2263, FSU2264, FSU2269, and FSU2274). Additionally, five of the proteins (FSU2266, FSU2267 [FSUAxe6B], FSU2270, FSU3053, and FSU3103) are (putative) esterases, while one of the gene products was predicted to be a melibiase (FSU2272) and another was similar to a pectate lyase (FSU3135). On the other hand, no conserved domains in FSU0053 and FSU2516 were found, although the BLAST search suggested that these proteins may contain pectate lyase activity. It is interesting that each of the proteins belongs to protein families that are related to hemicellulose or pectin metabolism. Seventeen of the proteins (FSU0192, FSU2262, FSU2263, FSU2264, FSU2265, FSU2266, FSU2267, FSU2269, FSU2270, FSU2272, FSU2274, FSU2292, FSU2293, FSU2294, FSU3053, FSU3103, and FSU3135) harbored, in addition to FPm-1, either single or double CBM domains, further suggesting that the FPm-1 sequence plays roles in the recognition or catalysis of certain carbohydrates. The FPm-1 domains were consistently located at the C-terminal end of these proteins, and CBMs, when present, were located N terminal to the FPm-1 domains. It was also noted that seven proteins (FSU0053, FSU0777, FSU2288, FSU2516, FSU2741, FSU2851, and FSU3006) that have the FPm-1 domains did not have identifiable CBM sequences, suggesting that FPm-1 is likely functionally independent of the CBM.
FIG. 1.
Domain architectures of proteins harboring FPm-1 in Fibrobacter succinogenes S85. Proteins harboring the FPm-1 domain were obtained through a search of the genome database of Fibrobacter succinogenes S85 (http://biocyc.org/FSUC59374/NEW-IMAGE?type=GENOME-OVERVIEW&object=FSU0013&chromosome=FS-CHROM-S85-FIBROBACTER). The presence of signal peptides was determined by the LipoP server and marked as stars at the N terminus of the protein architectures. Domain organizations were predicted using a BLAST protein search.
FSUAxe6B truncational derivatives.
To delineate and investigate the modules present in FSUAxe6B for functional role assignments, a gene truncation strategy was utilized. To create the truncated proteins, glycines in loop regions were selected as the terminal amino acids of the constructs. Based on the secondary structure analysis, five truncational derivatives of the polypeptide, as shown in Fig. 2A, were made. The construct TM1 (CE6 plus CBM6) was designed to investigate the contribution of FPm-1 to the WT protein in terms of its catalytic (esterase) and carbohydrate binding activities. Likewise, TM2 (CE6) was constructed for identifying the role of putative CBM6 on the two potential functions of the protein. TM3 (CBM6 plus FPm-1), TM4 (CBM6), and TM5 (FPm-1) were constructed for direct determination of the functions of the putative CBM6 and FPm-1 domains. All truncated derivatives of FSUAxe6B were successfully expressed in E. coli as soluble proteins and purified to near homogeneity (Fig. 2B).
Steady-state kinetic analysis of the FSUAxe6B WT and its truncational derivatives.
In order to obtain the basic catalytic information of FSUAxe6B, steady-state kinetic analysis was performed. Using tetra-acetyl-xylopyranoside as a substrate yielded a typical Michaelis-Menten plot, and a kcat value of 15 s−1 and a Km value of 0.08 mM were determined for this substrate (Table 2). Kinetic analysis of the two truncational mutant proteins, TM1 and TM2, which harbored the CE6 domain, were carried out. The TM1 protein exhibited a kcat value of 15 s−1 and a Km value of 0.09 mM, resulting in a kcat/Km value of 170 s−1 mM−1 (Table 2). Likewise, the kinetic parameters for the TM2 protein were 13 s−1 (kcat) and 0.07 mM (Km), resulting in a kcat/Km value of 190 s−1 mM−1 (Table 2). These values were quite similar to those of the WT protein, indicating that the CBM6 domain and FPm-1 domain of FSUAxe6B have no obvious effect on the esterase activity, at least with the substrate used in the present experiment. Also, the activity of TM2 delineated the catalytic region of FSUAxe6B.
TABLE 2.
Kinetic parameters for FSUAxe6B WT and its truncational mutantsa
| Protein | kcat (s−1) | Km (mM) | kcat/Km (s−1 mM−1) |
|---|---|---|---|
| WT | 15 ± 0.3 | 0.08 ± 0.01 | 190 ± 24 |
| TM1 | 15 ± 0.2 | 0.09 ± 0.01 | 170 ± 19 |
| TM2 | 13 ± 0.4 | 0.07 ± 0.01 | 190 ± 27 |
Data are shown as means ± standard errors.
Binding studies of FSUAxe6B and its truncational derivatives.
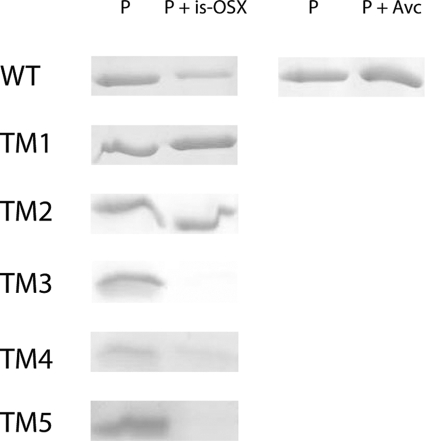
In order to investigate the carbohydrate binding activity of FSUAxe6B, Avicel (crystalline cellulose) and is-OSX, as substrates, were tested. The WT protein did not show any binding activity to Avicel. However, it showed binding activity for is-OSX (Fig. 3). Furthermore, to identify the location of the FSUAxe6B domains involved in the binding of is-OSX, the truncational derivatives (TM1 to TM5) were used for the binding assays. The qualitative binding assays demonstrated that although TM1 and TM2 have no discernible affinity for is-OSX, TM3, TM4, and TM5 bound to this substrate (Fig. 3). Taken together, these results indicate that the binding activity of FSUAxe6B for is-OSX is located in the TM5 peptide or the region designated an unknown domain.
FIG. 3.
Qualitative polysaccharide binding studies of the FSUAxe6B WT and its truncational mutants. is-OSX or Avicel PH-101 (Avc) was incubated with 2 μM protein. Lane P represents the same amount of protein incubated in the same buffer, but without substrate. The supernatants after incubation of the proteins with is-OSX or Avc were loaded on SDS-PAGE as P + is-OSX and P + Avc, respectively. In each case, except for TM5, 10 μl of solution with or without substrate for the WT, TM1, TM2, TM3, and TM4 were loaded for the SDS-PAGE analysis. The supernatants of the TM5 protein were concentrated up to 10 times, and then 10 μl of the solution was loaded on SDS-PAGE for visualization.
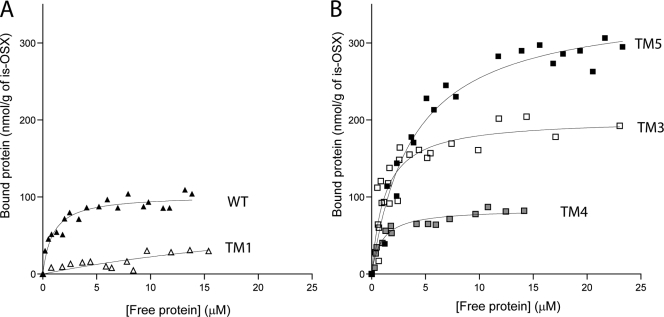
To ascertain these results and to quantify the binding capacity of the WT and its truncational mutants (TM1 to TM5) for is-OSX, binding isotherms were determined for these proteins. Figure 4A shows the binding isotherms for the WT protein and the truncated derivative lacking only FPm-1 (TM1). The truncation of FPm-1 from the WT protein led to a dramatic reduction of binding activity for the TM1 mutant, suggesting that the FPm-1 domain is key for binding to is-OSX (Fig. 4A), as was also observed with the qualitative binding assay (Fig. 3). Figure 4B shows the binding isotherms for the truncated derivatives that lacked the CE6 catalytic domain. The dissociation constant (Kp) of TM5 was 0.26 μM, which is much lower than that of TM4 (Kp = 1.1 μM). These values showed that the FPm-1 domain (TM5) exhibited binding activity for is-OSX that was much higher than that of the CBM6 domain (TM4), directly indicating that the FPm-1 domain is the true contributor to the binding of is-OSX (Fig. 4B and Table 3). Furthermore, the possibility that the TM4 and TM5 domains are one functional domain for binding was investigated. To test this hypothesis, TM3, which is composed of TM4 and TM5, was tested as well (Fig. 4B). The TM3 protein displayed a Kp value of 0.83 μM (Table 3), which is higher than that of TM5, indicating that the binding activity of FSUAxe6B is due mainly to the TM5 domain. Interestingly, the WT exhibited a dissociation constant of 1.1 μM (Fig. 4A and Table 3), which is much higher than that of TM5.
FIG. 4.
Quantitative studies of the binding of the FSUAxe6B WT and its truncational mutants to is-OSX. is-OSX (20 mg) was mixed with various concentrations of proteins, and the binding activities were estimated as described in Materials and Methods. The graphs depict the binding isotherms between bound proteins (nmol/g of is-OSX) and free proteins (μM). (A) The binding isotherms (closed triangles) for the WT and TM1, the protein with FPm-1 deleted (open triangles). (B) The binding isotherms for TM3 (open squares), TM4 (gray squares), and TM5 (closed squares) are shown. The binding constants for the wild type and its truncated mutants are presented in Table 3.
TABLE 3.
Binding parameters of FSUAxe6B WT and its truncated mutants for is-OSXa
| Protein | Kp (μM) | qmax (nmol protein/g is-OSX) |
|---|---|---|
| WT | 1.1 ± 0.2 | 100 ± 4 |
| TM3 | 0.83 ± 0.2 | 200 ± 10 |
| TM4 | 1.1 ± 0.2 | 84 ± 3 |
| TM5 | 0.26 ± 0.04 | 350 ± 10 |
Data are shown as means ± standard errors.
Multiple sequence alignment of FPm-1 sequences.
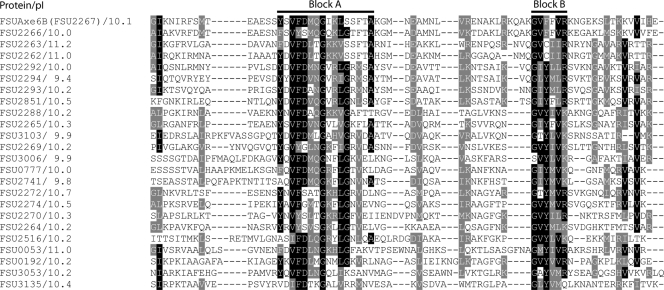
The 24 FPm-1 sequences were aligned using ClustalW (Fig. 5). The alignment revealed two conserved regions (block A and block B). Aromatic residues (tryptophan, tyrosine, and phenylalanine) in CBMs generally play a critical role in binding by forming hydrophobic stacking interactions with sugars in the carbohydrate polymer (5). Five relatively conserved aromatic residues (one tyrosine residue and two phenylalanine residues in block A and two phenylalanine residues in block B) were observed. Another interesting characteristic of the FSUAxe6B protein is the differences of the isoelectric points (pIs) of its different modules. The pI of TM2 (esterase domain), TM4 (CBM6), and TM5 (FPm-1) were 5.2, 4.6, and 10.1, respectively. The high pI of TM5 is due to the high proportion of positively charged amino acid residues in its sequence. Consistent with this observation, the other FPm-1 peptides (Fig. 5) also have high pI values, ranging from 9.4 for FSU2294 to 11.2 for FSU2263.
FIG. 5.
Multiple amino acid sequence alignment among FPm-1 domains in Fibrobacter succinogenes S85. Amino acid sequences of FPm-1 homologs in Fibrobacter succinogenes S85 were aligned utilizing ClustalW. The output files were entered into the BoxShade version 3.21 program (http://www.ch.embnet.org/software/BOX_form.html), with the fraction of sequences that must agree for shading set at 0.5. The conserved amino acids are shaded black, and similar amino acids are shaded gray. The pIs of the FPm-1 peptides are shown with protein identification numbers.
Determination of active-site residues in FSUAxe6B.
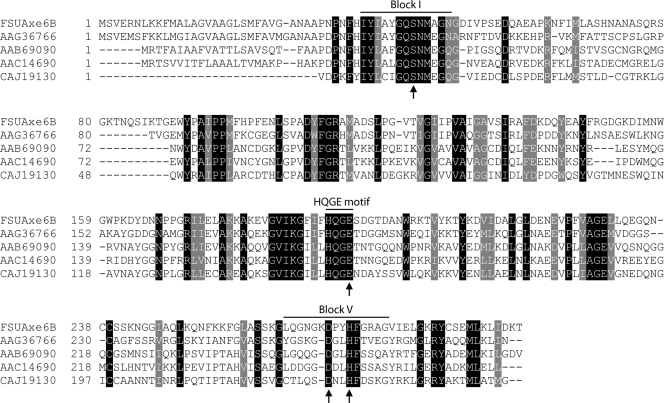
In previous studies of acetyl xylan esterases, the deacetylation mechanism of xylan was proposed (18, 19). The catalysis starts with an aspartate acting as a helper acid and forms a hydrogen bond with histidine, leading to an increase in the pKa of its imidazole nitrogen. This allows the histidine to become a strong general base, removing a proton from the hydroxyl group of serine. The deprotonated serine serves as a nucleophile and attacks the carbonyl carbon of the acetyl group. This mechanism allows the replacement of aspartate by a glutamate. Indeed, a catalytic triad formed by serine, histidine, and glutamate has been identified for the CE6 family protein R.44 from an uncultured rumen microbe (30). The three residues (Ser14, His231, and Glu152) reside in highly conserved regions in the CE6 family proteins (Fig. 6; see also Fig. S1 in the supplemental material). A comparison between the amino acid sequence of FSUAxe6B with that of biochemically characterized CE6 proteins was carried out. The results showed that these amino acids are completely conserved (Fig. 6) in the F. succinogenes protein. Following the previous study (30), the serine at position 44 of FSUAxe6B (S44), the glutamate at position 194 (E194), and the histidine at position 273 (H273) were mutated to glycine, asparagine, and glutamine, respectively. As expected, the S44G and H273Q mutations abolished detectable activities (Table 4). However, the E194N mutant exhibited detectable catalytic activity. Thus, a detailed kinetic analysis, which determined kcat and Km values of 2.8 s−1 and 7 mM, respectively, for E194N, was conducted. The catalytic efficiency (kcat/Km) of this mutant was 0.40 s−1 mM−1, which is considerably lower than that of the WT protein (190 s−1 mM−1). These results indicated that the glutamate at position 194 (E194) is contributing largely to catalysis. The possibility that the replaced asparagine formed a hydrogen bond with histidine by way of its carbonyl group was considered. Thus, to ascertain that the E194 is a member of the catalytic residues, it was substituted with alanine (E194A). Surprisingly, E194A also displayed some catalytic activity. The kcat and Km values of this mutant were 2.9 s−1 and 0.2 mM, respectively, resulting in a kcat/Km value of 14 (Table 4), which is also lower than that of the WT (kcat/Km = 190).
FIG. 6.
Amino acid sequence alignment of the FSUAxe6B esterase domain and similar domains from carbohydrate esterase family 6 (CE6) proteins. Amino acid sequences of FSUAxe6B and biochemically characterized CE6 proteins from Fibrobacter succinogenes (GenBank accession no. AAG36766), Neocallimastix patriciarum (GenBank accession no. AAB69090), Orpinomyces sp. PC-2 (GenBank accession no. AAC14690), and an unidentified microorganism (GenBank accession no. CAJ19130) were aligned utilizing ClustalW. The output files were entered into the BoxShade version 3.21 program (http://www.ch.embnet.org/software/BOX_form.html), with the fraction of sequences that must agree for shading set at 1.0. The conserved amino acids were shaded black, and similar amino acids were shaded gray. Arrows indicate the catalytic residues identified in this study for FSUAxe6B. An expanded alignment is shown in Fig. S1 in the supplemental material.
TABLE 4.
Kinetic parameters for FSUAxe6B WT and its site-directed mutantsa
| Protein | kcat (s−1)b | Km (mM) | kcat/Km (s−1 mM−1) |
|---|---|---|---|
| WT | 15 ± 0.3 | 0.08 ± 0.01 | 190 ± 24 |
| S44G | ND | ||
| E194N | 2.8 ± 0.3 | 7 ± 1 | 0.40 ± 0.07 |
| E194A | 2.9 ± 0.1 | 0.2 ± 0.02 | 14 ± 2 |
| D270N | 2.0 ± 0.1 | 0.2 ± 0.03 | 10 ± 2 |
| D270A | 1.8 ± 0.03 | 0.2 ± 0.01 | 9.0 ± 0.5 |
| H273Q | ND | ||
| E194A/D270A | ND |
Data are shown as means ± standard errors.
ND, no activity was detected.
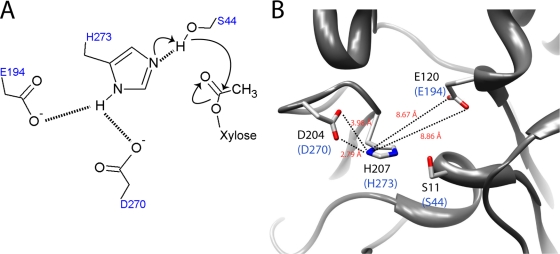
Since mutating E194, located in the vicinity of the catalytic pocket, did not completely abolish catalysis, a search was done for another residue that could serve as the helper acid in the catalysis. To facilitate the search, FSUAxe6B was modeled after the three-dimensional structure of a Clostridium acetobutylicum putative acetyl xylan esterase (Protein Data Bank accession number 1ZMB), the most similar protein structure available in the database. The residues, S44, E194, and H273 in FSUAxe6B, are completely conserved in 1ZMB (Fig. 7B). Furthermore, a potential helper acid, an aspartate with 3.39 Å as the mean value (distance) between its ionized group and the nitrogen of the imidazole group in H273, was located. This aspartate is also conserved in FSUAxe6B (D270) (Fig. 7B). Interestingly, the D270A and D270N mutants of FSUAxe6B displayed catalytic activities against tetra-acetyl-xylopyranoside. Thus, the D270A mutant, which showed catalytic properties similar to those of the D270N mutant, exhibited kcat and Km values of 1.8 s−1 and 0.2 mM, respectively. The kcat/Km value of D270A is therefore 9.0 (Table 4). These kinetic parameters were comparable to those of the E194A mutant. Since no other potential helper acid could be identified, an E194A/D270A double mutant was created. The activity of this mutant was completely abolished (Table 4), suggesting that both E194 and D270 contribute to catalysis, both residues perhaps acting as helper acids.
FIG. 7.
Active-site residues of FSUAxe6B. (A) Predicted reaction mechanism of FSUAxe6B. The two residues E194 and D270 form hydrogen bonds (dotted lines) with H273, leading to an increase in the pKa of its imidazole nitrogen. H273 as a strong general base removes a proton from the hydroxyl group of serine. The deprotonated serine serves as a nucleophile and attacks the carbonyl carbon of the acetyl group. (B) The three-dimensional structure illustrating the predicted active-site residues of FSUAxe6B (Fig. 7A) in a putative acetyl xylan esterase from Clostridium acetobutylicum (Protein Data Bank accession number 1ZMB). The side chains in the C. acetobutylicum protein are presented in the model. The corresponding residues in FSUAxe6B are shown in blue letters in closed brackets.
The CD spectrum analyses for the WT protein and the mutants were carried out to investigate the structural effects of the amino acid substitutions (Table 5). Among the mutant proteins, D270N and D270A showed secondary structural compositions similar to that of the WT protein. Also, other than the percentage of β-sheets, which was slightly increased, the parameters for the H273Q mutant were not very different from that of the WT protein. On the other hand, some increases in the α-helix structure were observed for S44G (17% compared with 14% for the WT). The percentages of α-helices increased slightly and the percentages of β-sheets decreased slightly for the E194N, E194A, and E194A/D270A double mutants compared to the wild type. The corresponding amino acid residues of S44 and E194 in FSUAxe6B are both located in an α-helix structure in the putative acetyl xylan esterase from Clostridium acetobutylicum (Protein Data Bank accession number 1ZMB) (Fig. 7B), and this might be the reason why the proportion of α-helical structures in FSUAxe6B was slightly increased when the residues were mutated. Of much interest are the two mutants E194A and D270A, originally selected as potential helper acids during catalysis. The D270A mutant has almost no detectable structural difference from the WT, and although it dramatically decreased esterase activity, it failed to abolish catalytic activity. The E194A mutant, on the other hand, exhibited some structural differences compared with the WT but was not very different from the D270A mutant in terms of its catalytic activity. Fascinatingly, however, a double mutant of the two residues E194A/D270A failed to exhibit detectable activity, suggesting that both residues may be critical to catalysis.
TABLE 5.
CD spectra for FSUAxe6B WT and its site-directed mutantsa
| Protein | α-Helix (%) | β-Sheet (%) | β-Turn (%) | Unordered (%) |
|---|---|---|---|---|
| WT | 14 ± 0 | 32 ± 1 | 23 ± 0 | 29 ± 1 |
| S44G | 17 ± 1 | 31 ± 1 | 23 ± 1 | 29 ± 0 |
| E194N | 19 ± 1 | 27 ± 2 | 24 ± 0 | 30 ± 1 |
| E194A | 17 ± 1 | 30 ± 0 | 23 ± 0 | 30 ± 0 |
| D270N | 15 ± 1 | 31 ± 2 | 24 ± 1 | 29 ± 1 |
| D270A | 14 ± 0 | 32 ± 1 | 23 ± 0 | 30 ± 0 |
| H273Q | 13 ± 0 | 34 ± 1 | 23 ± 0 | 30 ± 1 |
| E194A/D270A | 17 ± 0 | 29 ± 0 | 24 ± 0 | 31 ± 1 |
Data are presented as means ± standard deviations.
DISCUSSION
The gram-negative rumen bacterium Fibrobacter succinogenes S85 is estimated to have 104 putative glycoside hydrolases, 4 polysaccharide lyases, and 14 carbohydrate esterases from its complete genome information (16). It is clear that this bacterium has well-developed machinery that is devoted to plant cell wall degradation. The abundant carbohydrate-active enzymes, along with the modular protein structures, likely endow F. succinogenes S85 with the flexibility to survive on diverse polysaccharides and also to compete in the rumen environment. An example of these versatile proteins is the modular protein FSUAxe6B fully characterized in this study. F. succinogenes S85 Axe6A, a protein similar to FSUAxe6B, was shown to possess esterase activity and also to bind to Avicel cellulose, beech-wood xylan, and to a lesser extent is-OSX (23). A similar characterization for Axe6B was restricted by low expression of recombinant Axe6B. Of interest from the previous study is the prediction based on protein modeling and conservation of key amino acids that the CBM6 of Axe6B was different from that of Axe6A (23). This report presents overexpression, delineation of modules, and biochemical characterization of each module in FSUAxe6B to show that, indeed, the polypeptide is composed of a family 6 acetyl xylan esterase domain, a CBM6, and an unknown domain, to which a function is now assigned.
Biochemical analysis utilizing the truncational mutants of FSUAxe6B revealed the function of the C-terminal unknown domain as a novel CBM. Although it is currently found in F. succinogenes S85, it is anticipated that this module may be found, in the future, in some other organisms. FPm-1 clearly bound to is-OSX (Fig. 3 and 4). CBMs are protein folds that recognize specific polysaccharides and are often linked to a catalytic glycoside hydrolase domain through flexible loops (5). Many CBMs have been identified experimentally and classified into 54 families based on similarity of amino acid sequence, and some members from different families, such as families 2 (3), 4 (1), 6 (15, 23), 13 (6), 22 (8), 35 (4), 37 (45), and 54 (13) are known to bind to insoluble β-1,4-xylan. The F. succinogenes S85 FPm-1 is proposed as a novel CBM family because there is no characterized CBM that shares homology with its sequence.
There is a proposal to classify CBMs into three groups (type A, type B, and type C) based on their structures and functionalities (5). Type A CBMs are defined as surface binding, and they bind to insoluble cellulose and/or chitin crystals. FPm-1 preferred insoluble xylan, harboring heterogeneous amorphous structure (12), to crystalline cellulose (Fig. 3). On the other hand, type B and type C CBMs are peptides that are able to bind to soluble polysaccharides using a cleft in their structure. Although it was shown that FPm-1 of FSUAxe6B binds to is-OSX, the binding experiments with isothermal titration calorimetry suggested that the module does not bind to soluble substrates such as xylobiose, xylopentaose, and soluble arabinoxylan (data not shown). Thus, currently it is not possible to assign FPm-1 to any of the proposed groups of CBMs.
In CBMs, the common binding mechanism is interaction between aromatic amino acids and the carbohydrates as ligand. The amino acid sequence of FPm-1 in FSUAxe6B showed the presence of a single tyrosine residue, five phenylalanine residues, and no tryptophan (Fig. 5). Alanine scans for these aromatic residues did not abolish the binding capacity of TM5 for is-OSX (data not shown), suggesting that the binding mechanism reported to be mediated by these residues is not critical for FPm-1 or that multiple aromatic residues are involved in the interactions with substrate. Testing of the latter hypothesis will require creating multiple mutations in FPm-1 in the future.
Since the initial report of a C-terminal basic domain (BTD), specific to enzymes in F. succinogenes (32), many BTDs in this strain have been reported (22, 37, 38). To date, the role of the BTDs remain unknown. From the data on FPm-1s (Fig. 1), all identifiable homologs of this domain are located at the C terminus of the individual proteins. In addition, they are likely to display basic features (Fig. 5) at neutral pH, as is generally found in the rumen environment. Thus, similar to the BTDs, the FPm-1s are C-terminally located and also have basic properties. The FPm-1s, therefore, share some common features or properties with the BTDs. Interestingly, the amino acid sequences of hitherto reported BTDs are different from those of the FPm-1s identified in this study. While, the function of the BTDs is unknown, in the present report, a carbohydrate binding property is demonstrated for a member of the FPm-1s. It would be interesting to determine whether this property of the FPm-1s is shared with the BTDs.
It is also of interest that domains that share similar properties with FPm-1s have been observed with proteins from the gram-positive rumen bacterium Ruminococcus albus. The so-called X domains were first reported as C-terminal modules in the cellulose-binding proteins Cel9B and Cel48A through proteomic analysis (11). The domains exhibited a wide binding specificity for ligands and are currently classified as CBM family 37. This CBM family has members reported from only R. albus (45). Recently, a CBM37 domain was demonstrated to be crucial for binding to the bacterial cell surface (14). Similar to the FPm-1 domains in F. succinogenes, the ∼100 C-terminal amino acid sequences (CBM37s) in Cel5G, Cel9C, and Cel48A have the following high pIs: 9.78 (Cel5G), 9.59 (Cel9C), and 9.70 (Cel48A), respectively. The gram-negative F. succinogenes and gram-positive R. albus are two of the major microbes that adhere to and degrade insoluble polysaccharides in the rumen (16). It is hypothesized that the CBM37s and the FPm-1s share some common function, such as an electrostatic interaction between peptides and cell wall surface, and testing of this hypothesis is anticipated in the future.
Many CBM6s have been characterized, and their ligand specificities have been shown (9, 20, 21, 36, 44). Based on information derived from a previous study (23), the binding sites of biochemically and structurally characterized CBM6s of Cellvibrio mixtus endoglucanase 5A (21, 36) and Clostridium thermocellum xylanase 11A (9) are not conserved in the FSUAxe6B CBM6. Some affinity was detectable between the CBM6 domain (TM4) and is-OSX. However, the activity was much lower than that of the FPm-1 domain (TM5). Although CBM6 of FSUAxe6B is likely to bind to a specific carbohydrate or may exhibit other functional roles for efficient catalysis, it was not possible to assign a clear role to it in the current study.
The GDS(L) esterase/lipase family possesses a catalytic serine in the conserved motif GDS(L), and it was suggested that this protein family employs a catalytic triad formed by a serine in the block I consensus sequence and a histidine and an aspartate in the block V consensus sequence (2, 10). Although the carbohydrate esterase 6 (CE6) family is a member of the GDS(L) esterase/lipase family, it was recently demonstrated that the glutamate in the HQGE motif of block III is the sole catalytic helper acid in the R.44 protein (30). In the present study, to determine whether this finding is applicable to FSUAxe6B, a member of the CE6 family, site-directed mutagenesis studies of the esterase were carried out. The serine, as a nucleophile in block I, and the histidine, as a base to deprotonate the hydroxyl group of the serine in block V, were identified (Fig. 6 and 7 and Table 4). However, analysis of mutants with a single mutation (E194N, E194A, D270N, and D270A) and a mutant with double mutations (E194A/D270A) suggested that E194 and D270 may both be important for catalysis, potentially serving as dual helper acids, instead of the single helper acid proposed to function in the deacetylation mechanism described above. The two carboxylates are highly conserved among CE6 family proteins (see Fig. S1 in the supplemental material), and this may be a common catalytic mechanism in this family. Axe6A, with a 61% amino acid sequence similarity to the catalytic domain of Axe6B, exhibited some similarity of kinetic data to Axe6B (Km value of 0.08 mM and 0.06 mM for Axe6A and Axe6B, respectively), although the Vmax values for the two proteins were quite different (23). Mutational analysis of the predicted catalytic residues of Axe6A should yield interesting data.
Although extensive research has yielded deep insight into the strategies used by microbes to release nutrients from plant matter for fermentation, it is likely that there is yet fascinating biology and chemistry to be learned in this field. This may especially be true for specialist plant cell wall-degrading bacteria, such as F. succinogenes, from which a new CBM, FPm-1, is described in addition to a potentially new mechanism for esterase activity that employs a catalytic tetrad instead of a catalytic triad.
Supplementary Material
Acknowledgments
We gratefully thank Dylan Dodd and Shinichi Kiyonari of the Energy Biosciences Institute for scientific discussions and Brad Jelinek and Charles Hespen of University of Illinois for technical assistance.
This research was supported by a grant from the Energy Biosciences Institute.
Footnotes
Published ahead of print on 6 November 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abou Hachem, M., E. Nordberg Karlsson, E. Bartonek-Roxå, S. Raghothama, P. J. Simpson, H. J. Gilbert, M. P. Williamson, and O. Holst. 2000. Carbohydrate-binding modules from a thermostable Rhodothermus marinus xylanase: cloning, expression and binding studies. Biochem. J. 345:53-60. [PMC free article] [PubMed] [Google Scholar]
- 2.Akoh, C. C., G. C. Lee, Y. C. Liaw, T. H. Huang, and J. F. Shaw. 2004. GDSL family of serine esterases/lipases. Prog. Lipid Res. 43:534-552. [DOI] [PubMed] [Google Scholar]
- 3.Black, G. W., G. P. Hazlewood, S. J. Millward-Sadler, J. I. Laurie, and H. J. Gilbert. 1995. A modular xylanase containing a novel non-catalytic xylan-specific binding domain. Biochem. J. 307:191-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolam, D. N., H. Xie, G. Pell, D. Hogg, G. Galbraith, B. Henrissat, and H. J. Gilbert. 2004. X4 modules represent a new family of carbohydrate-binding modules that display novel properties. J. Biol. Chem. 279:22953-22963. [DOI] [PubMed] [Google Scholar]
- 5.Boraston, A. B., D. N. Bolam, H. J. Gilbert, and G. J. Davies. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382:769-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boraston, A. B., P. Tomme, E. A. Amandoron, and D. G. Kilburn. 2000. A novel mechanism of xylan binding by a lectin-like module from Streptomyces lividans xylanase 10A. Biochem. J. 350:933-941. [PMC free article] [PubMed] [Google Scholar]
- 7.Cann, I. K. O., S. Ishino, M. Yuasa, H. Daiyasu, H. Toh, and Y. Ishino. 2001. Biochemical analysis of replication factor C from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 183:2614-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charnock, S. J., D. N. Bolam, J. P. Turkenburg, H. J. Gilbert, L. M. A. Ferreira, G. J. Davies, and C. M. G. A. Fontes. 2000. The X6 “thermostabilizing” domains of xylanases are carbohydrate-binding modules: structure and biochemistry of the Clostridium thermocellum X6b domain. Biochemistry 39:5013-5021. [DOI] [PubMed] [Google Scholar]
- 9.Czjzek, M., D. N. Bolam, A. Mosbah, J. Allouch, C. M. G. A. Fontes, L. M. A. Ferreira, O. Bornet, V. Zamboni, H. Darbon, N. L. Smith, G. W. Black, B. Henrissat, and H. J. Gilbert. 2001. The location of the ligand-binding site of carbohydrate-binding modules that have evolved from a common sequence is not conserved. J. Biol. Chem. 276:48580-48587. [DOI] [PubMed] [Google Scholar]
- 10.Dalrymple, B. P., D. H. Cybinski, I. Layton, C. S. McSweeney, G. P. Xue, Y. J. Swadling, and J. B. Lowry. 1997. Three Neocallimastix patriciarum esterases associated with the degradation of complex polysaccharides are members of a new family of hydrolases. Microbiology 143:2605-2614. [DOI] [PubMed] [Google Scholar]
- 11.Devillard, E., D. B. Goodheart, S. K. Karnati, E. A. Bayer, R. Lamed, J. Miron, K. E. Nelson, and M. Morrison. 2004. Ruminococcus albus 8 mutants defective in cellulose degradation are deficient in two processive endocellulases, Cel48A and Cel9B, both of which possess a novel modular architecture. J. Bacteriol. 186:136-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dodd, D., and I. K. O. Cann. 2009. Enzymatic deconstruction of xylan for biofuel production GCB Bioenergy. 1:2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dvortsov, I. A., N. A. Lunina, L. A. Chekanovskaya, W. H. Schwarz, V. V. Zverlov, and G. A. Velikodvorskaya. 2009. Carbohydrate-binding properties of a separately folding protein module from β-1,3-glucanase Lic16A of Clostridium thermocellum. Microbiology 155:2442-2449. [DOI] [PubMed] [Google Scholar]
- 14.Ezer, A., E. Matalon, S. Jindou, I. Borovok, N. Atamna, Z. Yu, M. Morrison, E. A. Bayer, and R. Lamed. 2008. Cell surface enzyme attachment is mediated by family 37 carbohydrate-binding modules, unique to Ruminococcus albus. J. Bacteriol. 190:8220-8222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandes, A. C., C. M. G. A. Fontes, H. J. Gilbert, G. P. Hazlewood, T. H. Fernandes, and L. M. A. Ferreira. 1999. Homologous xylanases from Clostridium thermocellum: evidence for bi-functional activity, synergism between xylanase catalytic modules and the presence of xylan-binding domains in enzyme complexes. Biochem. J. 342:105-110. [PMC free article] [PubMed] [Google Scholar]
- 16.Flint, H. J., E. A. Bayer, M. T. Rincon, R. Lamed, and B. A. White. 2008. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat. Rev. Microbiol. 6:121-131. [DOI] [PubMed] [Google Scholar]
- 17.Forsberg, C. W., B. Crosby, and D. Y. Thomas. 1986. Potential for manipulation of the rumen fermentation through the use of recombinant DNA techniques. J. Anim. Sci. 63:310-325. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh, D., M. Erman, M. Sawicki, P. Lala, D. R. Weeks, N. Li, W. Pangborn, D. J. Thiel, H. Jörnvall, R. Gutierrez, and J. Eyzaguirre. 1999. Determination of a protein structure by iodination: the structure of iodinated acetylxylan esterase. Acta Crystallogr. D Biol. Crystallogr. 55:779-784. [DOI] [PubMed] [Google Scholar]
- 19.Hakulinen, N., M. Tenkanen, and J. Rouvinen. 2000. Three-dimensional structure of the catalytic core of acetylxylan esterase from Trichoderma reesei: insights into the deacetylation mechanism. J. Struct. Biol. 132:180-190. [DOI] [PubMed] [Google Scholar]
- 20.Henshaw, J., A. Horne-Bitschy, A. L. van Bueren, V. A. Money, D. N. Bolam, M. Czjzek, N. A. Ekborg, R. M. Weiner, S. W. Hutcheson, G. J. Davies, A. B. Boraston, and H. J. Gilbert. 2006. Family 6 carbohydrate binding modules in β-agarases display exquisite selectivity for the non-reducing termini of agarose chains. J. Biol. Chem. 281:17099-17107. [DOI] [PubMed] [Google Scholar]
- 21.Henshaw, J. L., D. N. Bolam, V. M. R. Pires, M. Czjzek, B. Henrissat, L. M. A. Ferreira, C. M. G. A. Fontes, and H. J. Gilbert. 2004. The family 6 carbohydrate binding module CmCBM6-2 contains two ligand-binding sites with distinct specificities. J. Biol. Chem. 279:21552-21559. [DOI] [PubMed] [Google Scholar]
- 22.Iyo, A. H., and C. W. Forsberg. 1996. Endoglucanase G from Fibrobacter succinogenes S85 belongs to a class of enzymes characterized by a basic C-terminal domain. Can. J. Microbiol. 42:934-943. [DOI] [PubMed] [Google Scholar]
- 23.Kam, D. K., H. S. Jun, J. K. Ha, G. D. Inglis, and C. W. Forsberg. 2005. Characteristics of adjacent family 6 acetylxylan esterases from Fibrobacter succinogenes and the interaction with the Xyn10E xylanase in hydrolysis of acetylated xylan. Can. J. Microbiol. 51:821-832. [DOI] [PubMed] [Google Scholar]
- 24.Kelly, S. M., T. J. Jess, and N. C. Price. 2005. How to study proteins by circular dichroism. Biochim. Biophys. Acta 1751:119-139. [DOI] [PubMed] [Google Scholar]
- 25.Koike, S., and Y. Kobayashi. 2001. Development and use of competitive PCR assays for the rumen cellulolytic bacteria: Fibrobacter succinogenes, Ruminococus albus and Ruminococcus flavefaciens. FEMS Microbiol. Lett. 204:361-366. [DOI] [PubMed] [Google Scholar]
- 26.Krause, D. O., S. E. Denman, R. I. Mackie, M. Morrison, A. L. Rae, G. T. Attwood, and C. S. McSweeney. 2003. Opportunities to improve fiber degradation in the rumen: microbiology, ecology, and genomics. FEMS Microbiol. Rev. 27:663-693. [DOI] [PubMed] [Google Scholar]
- 27.Kumar, R., S. Singh, and O. V. Singh. 2008. Bioconversion of lignocellulosic biomass: biochemical and molecular perspectives. J. Ind. Microbiol. Biotechnol. 35:377-391. [DOI] [PubMed] [Google Scholar]
- 28.Kyriacou, A., R. J. Neufeld, and C. R. Mackenzie. 1988. Effect of physical parameters on the adsorption characteristics of fractionated Trichoderma reesei cellulase components. Enzyme Microb. Technol. 10:675-681. [Google Scholar]
- 29.Lobley, A., L. Whitmore, and B. A. Wallace. 2002. DICHROWEB: an interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics 18:211-212. [DOI] [PubMed] [Google Scholar]
- 30.López-Cortés, N., D. Reyes-Duarte, A. Beloqui, J. Polaina, I. Ghazi, O. V. Golyshina, A. Ballesteros, P. N. Golyshin, and M. Ferrer. 2007. Catalytic role of conserved HQGE motif in the CE6 carbohydrate esterase family. FEBS Lett. 581:4657-4662. [DOI] [PubMed] [Google Scholar]
- 31.Lykov, O. P. 1994. Selection of raw material for basic organic synthesis. Chem. Technol. Fuels Oils 30:302-309. [Google Scholar]
- 32.Malburg, L. M., Jr., A. H. Iyo, and C. W. Forsberg. 1996. A novel family 9 endoglucanase gene (celD), whose product cleaves substrates mainly to glucose, and its adjacent upstream homolog (celE) from Fibrobacter succinogenes S85. Appl. Environ. Microbiol. 62:898-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matte, A., C. W. Forsberg, and A. M. Verrinder Gibbins. 1992. Enzymes associated with metabolism of xylose and other pentoses by Prevotella (Bacteroides) ruminicola strains, Selenomonas ruminantium D, and Fibrobacter succinogenes S85. Can. J. Microbiol. 38:370-376. [DOI] [PubMed] [Google Scholar]
- 34.Miron, J., and D. Ben-Ghedalia. 1993. Digestion of cell-wall monosaccharides of ryegrass and alfalfa hays by the ruminal bacteria Fibrobacter succinogenes and Butyrivibrio fibrisolvens. Can. J. Microbiol. 39:780-786. [DOI] [PubMed] [Google Scholar]
- 35.Montgomery, L., B. Flesher, and D. Stahl. 1988. Transfer of Bacteroides succinogenes (Hungate) to Fibrobacter gen. nov. as Fibrobacter succinogenes comb. nov. and description of Fibrobacter intestinalis sp. nov. Int. J. Syst. Bacteriol. 38:430-435. [Google Scholar]
- 36.Pires, V. M. R., J. L. Henshaw, J. A. M. Prates, D. N. Bolam, L. M. A. Ferreira, C. M. G. A. Fontes, B. Henrissat, A. Planas, H. J. Gilbert, and M. Czjzek. 2004. The crystal structure of the family 6 carbohydrate binding module from Cellvibrio mixtus endoglucanase 5A in complex with oligosaccharides reveals two distinct binding sites with different ligand specificities. J. Biol. Chem. 279:21560-21568. [DOI] [PubMed] [Google Scholar]
- 37.Qi, M., H. S. Jun, and C. W. Forsberg. 2008. Cel9D, an atypical 1,4-β-d-glucan glucohydrolase from Fibrobacter succinogenes: characteristics, catalytic residues, and synergistic interactions with other cellulases. J. Bacteriol. 190:1976-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qi, M., H. S. Jun, and C. W. Forsberg. 2007. Characterization and synergistic interactions of Fibrobacter succinogenes glycoside hydrolases. Appl. Environ. Microbiol. 73:6098-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rubin, E. M. 2008. Genomics of cellulosic biofuels. Nature 454:841-845. [DOI] [PubMed] [Google Scholar]
- 40.Scott, H. W., and B. A. Dehority. 1965. Vitamin requirements of several cellulolytic rumen bacteria. J. Bacteriol. 89:1169-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Somerville, C. 2007. Biofuels. Curr. Biol. 17:R115-R119. [DOI] [PubMed] [Google Scholar]
- 42.Somerville, C., S. Bauer, G. Brininstool, M. Facette, T. Hamann, J. Milne, E. Osborne, A. Paredez, S. Persson, T. Raab, S. Vorwerk, and H. Youngs. 2004. Toward a systems approach to understanding plant cell walls. Science 306:2206-2211. [DOI] [PubMed] [Google Scholar]
- 43.Stevenson, D. M., and P. J. Weimer. 2007. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl. Microbiol. Biotechnol. 75:165-174. [DOI] [PubMed] [Google Scholar]
- 44.van Bueren, A. L., C. Morland, H. J. Gilbert, and A. B. Boraston. 2005. Family 6 carbohydrate binding modules recognize the non-reducing end of β-1,3-linked glucans by presenting a unique ligand binding surface. J. Biol. Chem. 280:530-537. [DOI] [PubMed] [Google Scholar]
- 45.Xu, Q., M. Morrison, K. E. Nelson, E. A. Bayer, N. Atamna, and R. Lamed. 2004. A novel family of carbohydrate-binding modules identified with Ruminococcus albus proteins. FEBS Lett. 566:11-16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.