Abstract
The neuronal α4β2 nicotinic acetylcholine receptor (nAChR) is a potential molecular target for general anesthetics. It is unclear, however, whether anesthetic action produces the same effect on the open and closed channels. Computations parallel to our previous open channel study (J. Phys. Chem. B, 2009) were performed on the closed-channel α4β2 nAChR to investigate the conformation-dependent anesthetic effects on channel structures and dynamics. Flexible ligand docking and over 20-ns molecular dynamics simulations revealed similar halothane-binding sites in the closed and open channels. The sites with relatively high binding affinities (~−6.0 kcal/mol) were identified at the interface of extracellular (EC) and transmembrane (TM) domains or at the interface between α4 and β2 subunits. Despite similar sites for halothane binding, the closed-channel conformation showed much less sensitivity than the open channel to the structural and dynamical perturbations from halothane. Compared to the systems without anesthetics, the amount of water inside the pore decreased by 22% in the presence of halothane in the open channel, but only by 6% in the closed channel. Comparison of the non-bonded interactions at the EC/TM interfaces suggested that the β2 subunits were more prone than the α4 subunits to halothane binding. In addition, our data support the notion that halothane exerts its effect by disturbing the quaternary structure and dynamics of the channel. The study concludes that sensitivity and global dynamics responsiveness of α4β2 nAChR to halothane are conformation dependent. The effect of halothane on global dynamics of open-channel conformation might also account for the action of other inhaled general anesthetics.
Keywords: anesthesia mechanism, halothane, nicotinic acetylcholine receptor, alpha4beta2, membrane protein, molecular dynamic simulation
INTRODUCTION
Nicotinic acetylcholine receptors (nAChRs) belong to the Cys-loop receptor superfamily.1,2 The receptors in this superfamily consist of five subunits, which organize in the cell membrane to form pentameric ion channels. Each subunit has an extracellular (EC), a transmembrane (TM), and an intracellular (IC) domain. Agonist binding to the EC domain of these receptors triggers channel opening from a resting state and allows ions to flow through the membrane. Most general anesthetics noncompetitively inhibit nAChRs and decrease channel conductivities of nAChRs.3 Potentiating effects of some anesthetics and alcohols were also observed on nAChRs.4–7 Moreover, dual action of alcohols on neuronal nAChRs was reported.8 It is difficult to rationalize the varying, and sometimes opposing, responses of nAChRs to general anesthetics.
The neuronal α4β2 nAChR is one of the most abundant subtype nAChRs in the brain and was identified as a putative target for general anesthetics.9–13 As with other nAChRs, the molecular details regarding the interaction between anesthetics and the α4β2 nAChR have not been experimentally determined. A mechanistic understanding of how anesthetics altered the function of the α4β2 nAChR is still lacking. It is also unclear whether anesthetics exert their effects equally on the closed- and open-channel α4β2 nAChR or preferentially on one particular conformation. Answers to these questions are likely applicable not only to the α4β2 nAChR, but also to other receptors in the same superfamily.
Ideally, these questions are best answered with high-resolution structures of the α4β2 nAChR in the closed- and open-channel states. In reality, however, experimental structure determination of the α4β2 nAChR remains a great challenge. Based on the known structure of Torpedo nAChR14 and other related structural information,15–19 we recently generated in silico models of the closed- and open-channel α4β2 nAChR.20 In addition to providing valuable insights into plausible mechanisms of channel gating, these well-equilibrated structural models have provided a structural basis for investigating the α4β2 nAChR as a potential target of general anesthetics. More recently, using our open-channel structural model, we identified six amphiphilic interaction sites for the volatile anesthetic halothane through flexible ligand docking and subsequent 20-ns molecular dynamics simulations.21 The primary binding sites were found at the interface of extracellular (EC) and transmembrane (TM) domains (see Figure 1), where halothane perturbed conformations of the Cys loop, the β1–β2 loop, and the TM2–TM3 linker. Profound changes in residue flexibility and the concerted motions critical to protein function were observed through Gaussian network model analyses22,23 of the open-channel α4β2 nAChR structures at the end of 20-ns simulations in the absence or presence of halothanes, suggesting that the halothane effect on protein dynamics was on a global scale rather than confined to residues nearby the halothane-binding sites.21
Figure 1.
Top and side views of halothane binding sites in the closed-channel (A) and open-channel (B) α4β2 nAChR. The positions of halothane molecules at the beginning and end of the 20-ns MD simulations are highlighted in transparent and solid colors, respectively.
In the present study, we determined halothane binding and the binding-induced structural and dynamical changes in the closed-channel α4β2 nAChR using the same computational protocol applied previously to the open-channel conformation. A detailed comparison of halothane in two different channel states yielded the notion that the closed-channel α4β2 nAChR was less susceptible to anesthetic perturbation than the open-channel structure. The study suggested that the responsiveness of α4β2 nAChR to general anesthetics is conformation dependent.
METHODS
The system preparation of the closed-channel α4β2 nAChR was detailed in a previous publication.20 In brief, the initial structure of the closed-channel α4β2 nAChR was obtained through homology modeling using the cryo-EM structure of Torpedo nAChR (PDB Code: 2BG9)14 as a template. The closed channel was embedded in a pre-equilibrated ternary lipid bilayer of 162 POPC, 55 POPA, and 55 Cholesterol.24 The system also contained 33641 water molecules, 108 Na+, and 14 Cl−. The closed-channel structure had been equilibrated through a 11-ns molecular dynamics (MD) simulation before it was used for the current study, as in the case when halothane’s effect on the open-channel α4β2 nAChR was investigated.21
NAMD-2.625 and CHARMM-2726 force field were used for all MD simulations that were carried out on supercomputers at the Pittsburgh Supercomputer Center. The MD simulation protocols were the same as reported previously.21 Both control and halothane systems were simulated for over 20 ns under constant 1 atm pressure and 303 K temperature (NPT ensemble). Halothane binding sites in the closed channel were determined using the same strategy as used in the open channel.21 The majority of initial binding sites were identified through flexible docking using Autodock4,27 and the remaining sites were selected based on experimental suggestions.28 500 independent dockings were performed using a Lamarckian genetic algorithm with a grid spacing of 0.375Å. Twenty-four halothane molecules were included in a pre-equilibrated closed-channel system for a 15-ns MD simulation, from which 6 halothane-binding sites with relatively small halothane displacements during this period were chosen. Two new parallel 20-ns MD simulations were then restarted with the 6 chosen halothane molecules or without halothane; the two parallel systems are referred to as the halothane system and control system, respectively. The previously published halothane parameters29 were used for docking and MD simulations.
Halothane binding energy at each site in the closed-channel system was calculated using the Free Energy Perturbation (FEP)30,31 implemented in NAMD-2.6.25 The calculation protocol was identical to the one reported for the open channel.21 The same FEP calculation was also performed on a halothane molecule in a water box. The differences of the FEP outcomes in α4β2 nAChR and in water resulted in the halothane binding energies.
Gaussian network model (GNM)22,23 was used to analyze the halothane effect on the global dynamics of the closed-channel α4β2 nAChR. Each residue of the closed channel was represented by the Cα atom. A 10-Å cutoff was chosen. The five slowest modes of the GNM calculations on the structures after 20-ns MD simulations were included in the data analysis.
The pore radius was evaluated by the HOLE program.32 The average pore radius was calculated in a step size of 0.5 Å along the pore using 100 snapshots for each 1-ns simulation. The number of water molecules in the pore was counted along the channel axis (the Z coordinate) and averaged per 5-Å step over 1-ns simulation window.
The tilt angle of the second transmembrane domain (TM2) helix in each subunit was calculated using the same definition as reported previously.33 The Cα atoms of residue 245 to residue 267 were used for the calculations. A positive value of the radial tilt angle indicates that the extracellular end of the helix is further away from the channel axis compared to the intracellular end. A positive value of the lateral tilt angle indicates that the extracellular end of the helix has a counter-clockwise rotation viewed from the extracellular end of the pore. All the reported tilt angles for each system were obtained using the last 5-ns trajectory.
The cavities and preexisting binding pockets within the α4β2 nAChR were identified and measured using the online server CASTp with a probe radius of 1.4 Å.34
RESULTS AND DISCUSSION
Similarities and differences of halothane binding in the closed- and open-channel α4β2 nAChR
Figure 1A shows 6 halothane binding sites in the closed-channel α4β2 at the beginning and end of the 20-ns simulation. Some halothane molecules moved away from their initial positions more than others over the course of simulation. Displacements of halothane molecules in the 20-ns simulation ranged from 2 to 10 Å (see Fig. 1S in Supporting Information). The binding energy of individual halothane was determined through FEP calculations. Halothane binding energies and the corresponding disassociation constants are summarized in Table 1.
Table 1.
Halothane binding energies for the closed-channel α4β2 calculated from FEP along with the corresponding dissociation constants. The data for the open-channel system are also included for comparison.
| Site ID | Closed-channel α4β2 Conformation | Open-channel α4β2 Conformation | ||
|---|---|---|---|---|
| Binding Energy (kcal/mol) | Kd (mM)a | Binding Energy (kcal/mol) | Kd (mM)a | |
| 1 | −5.5 | 0.11 | −6.8 | 1.2 × 10−2 |
| 2 | −3.2 | 4.9 | −3.4 | 3.5 |
| 3 | −6.0 | 4.7 × 10−2 | −3.8 | 1.8 |
| 4 | −3.8 | 1.8 | −3.7 | 2.1 |
| 5 | −3.2 | 4.9 | −2.4 | 18.5 |
| 6 | −1.1 | 160.1 | −3.9 | 1.5 |
The apparent dissociation constant, R ln Kd, was calculated using the equation ΔΔG = T Kd, where R = 1.987 cal mol−1 K−1, T = 303 K, and ΔΔG is the calculated binding energy.
Some of the early observations of halothane binding in the open-channel system21 (Figure 1B) were also seen in the closed-channel system. First, halothane bound to multiple sites, located either within a subunit or between two subunits. Second, both systems have relatively high affinity binding sites with Kd values less than 0.2 mM, including halo-1 (halo-1open) in the open-channel system and halo-1 (halo-1closed) and halo-3 (halo-3closed) in the closed system. These sites were located either at the EC/TM interface or deep into the TM domain, and had comparable Kd values to those experimentally measured in the Torpedo nAChR (0.18 ± 0.04 mM).35 Third, lipids contributed to the high affinity binding sites. Both halo-1open21 and halo-3closed were adjacent to the lipid head groups. Similar to the lipids near halo-1open,21 the neighboring lipids of halo-3closed also experienced tilting and translation motions to optimize their interactions with halo-3closed. Finally, no halothane was found inside the channel, indicating that it is very unlikely for halothane to work as a channel blocker in the α4β2 nAChR.
A further inspection also revealed some differences in halothane binding in the closed- and open-channel systems. Halothane at the main immunogenic region (MIR) showed distinctly different binding affinity. Halo-6open had a Kd of 1.5 mM, the second highest affinity in the open-channel system.21 In contrast, Halo-6closed had the lowest binding affinity among all halothane sites. Its Kd value of ~160 mM suggested that halothane had virtually no binding at the MIR in the closed-channel α4β2 at anesthetizing halothane concentrations. Halo-5 near the agonist-binding site between the α4-1 and β2-3 subunits also behaved differently in the closed- and open-channel systems. The binding affinity of halo-5closed was almost 4 times greater than that of halo-5open, presumably due to a local protein structural deformation induced by nicotine binding in the open-channel system and unfavorable interactions between the bound nicotine and halo-5open. However, neither halo-5closed nor halo-5open had comparable binding affinities to those of agonists,36 such as nicotine and acetylcholine. This is consistent with an early finding that halothane unlikely acts as a competitive antagonist to inhibit the function of nAChR.37
It is worth noting that halothane can have different binding properties at analogous sites of the protein. Halo-1closed and halo-2closed were initially at equivalent inter-subunit sites of α4-1/β2-3 and α4-2/β2-2, respectively, but they migrated to different locations after 20-ns simulations, as displayed in Figure 1A. Two equivalent inter-subunit sites were not identical to halothane binding, largely due to asymmetric motions among subunits in a pentameric assembly.38 A substantial degree of asymmetry in the subunit motion exists not only in hetero-pentameric proteins, but also in homo-pentameric proteins, such as the α7 nAChR.39,40
Multiple halothane binding sites and similarly high binding affinity at the EC/TM interface and inter-subunit interface are the characteristics shared by both the closed- and open-channel α4β2 nAChR. In this regard, halothane binding in different conformational states was quite similar. This is not entirely unexpected. Early experiments even suggested that some anesthetics could have equal affinity for the open- and closed-channel states of nAChRs,4,5 though some anesthetics or long chain alcohols showed higher affinity for unclosed-channel states.41,42 It should be pointed out that similar anesthetic binding in different conformational states does not necessarily translate into similar anesthetic effects on protein structure and dynamics in the different states, as we have discovered in this study.
Halothane binding to the inter-subunit region induced less structural perturbation in the closed channel than in the open channel
Interface between two subunits often facilitates protein allosteric modulation. For instance, agonists of the nAChR bind to the interface of two subunits containing at least one α subunit to activate the channel.43–45 An interface of two subunits could also be the target for general anesthetics, as suggested by the photolabeling studies of [3H]azietomidate on the GABAA receptor46 and TDBzl-etomidate on the muscle-type nAChR.47
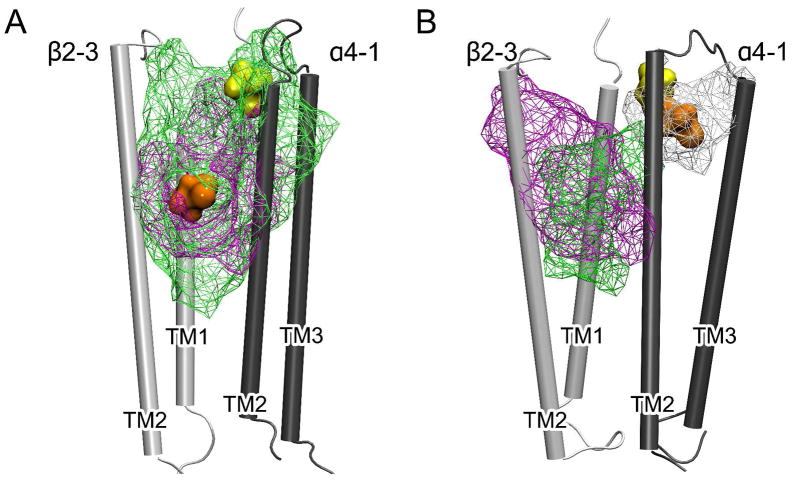
Halothane binding at the interface of α4 and β2 subunits was identified in both closed- and open-channel systems. The initial site of halo-1, obtained from docking, agreed with the halothane photolabeling on the Torpedo nAChR.28 At the end of 20-ns simulations, halo-1 resided at the interface of the α4-1 and β2-3 subunits, either facing the lumen of the pore in the closed-channel structure or facing the lipids in the open-channel structure, as shown in Figure 2. Irrespective of the absence or presence of halothane, a well-overlapped pocket in the closed channel (Figure 2A) suggests that the binding of halo-1closed to this pre-existing pocket caused negligible perturbation to the interface of the α4-1 and β2-3 subunits. The pocket was solvent accessible and filled with water in the control system. In the open-channel system, the corresponding pocket showed little change in shape and size after the 20-ns simulation in the absence of halothane. However, the pocket structure varied considerably when halothane molecules were added to the system, even though nearby halo-1open did not directly occupy this pocket (Figure 2B). It appears that the inter-subunit packing in the open-channel system is more susceptible to external perturbation, such as anesthetic binding.
Figure 2.
The cavities at the interface of β2-3 (white) and α4-1 (gray) subunits in the closed- (A) and open-channel (B) models. The cavities are viewed from inside the pore, presented in green wire frame for the control systems and in magenta and white wire frame for the halothane systems. For clarity, only parts of the α4-1 and β2-3 TM domains are shown. Halothane at the beginning and in the last 5-ns of the 20-ns simulations are shown in yellow and orange spheres, respectively. The structures averaged over either the first or the last 500-ps simulations were used for cavity identification.
The vulnerability of the inter-subunit pockets or residue packing in the open-channel α4β2 nAChR might result primarily from larger tilt angles between the TM helices at the subunit interface. The TM2 helices of α4-1 and β2-3 subunits in the closed channel were oriented almost in parallel (Figure 2A). In the open channel (Figure 2B), however, the TM2 helices of α4-1 and β2-3 had radial tilt angles of ~ 11° and ~ 7°, respectively, creating more space for more diversified side-chain movement. When halo-1open entered the inter-subunit site shown in Figure 2B, E266 in the TM2 of α4-1 moved closer to the TM1 of β2-3 and induced a large deformation to the original pocket. Although it has no direct interaction with E266 in the TM2 of α4-1, halo-1open can exert its effect to the pocket by interacting with P269 and S272, located at one and two helical turns above E266. Such quaternary structural changes in inter-subunit rearrangements might exist not only in the α4β2, but also in other Cys-loop receptors. Anesthetic binding to the inter-subunit space could either stabilize or destabilize the open-channel structure, appearing as potentiation or inhibition of ion channel functions,48,49 respectively. Further studies are required to confirm this prediction.
Halothane binding imposed less effect on the global dynamics of the closed channel than the open channel
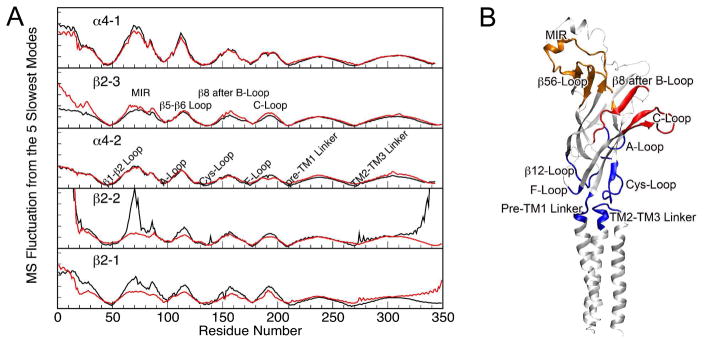
Gaussian network model is an effective tool for evaluating global dynamics of macromolecules.22,23 Using the GNM, we previously examined halothane modulation on the global dynamics of the open-channel α4β2 nAChR and observed halothane-induced mean-square fluctuation (MSF) changes.21 In comparison to the open channel, the closed channel seemed to experience less dynamical impact from halothane binding. As shown in Figure 3, observable changes in MSF in the absence or presence of halothane were limited only to a few regions in the EC domain of the closed-channel α4β2 nAChR. The minima of MSF from the slowest modes often correspond to the constrained residues critical for collective global motions.50,51 Five out of six minima (except the A loop) in the MSF plots resulted from linkers or loops located at the EC/TM domain interface (Figure 3B). It has been suggested by experiments that interactions among these linkers and loops play a key role in mediating signals of agonist binding to the channel opening.2,52–54 In the closed-channel model, all the minima remained almost the same in the absence or presence of halothane (Figure 3A), indicating negligible halothane effect on the global dynamics at the EC/TM domain interface. In contrast, several minima in the open-channel model, corresponding to the Cys loop, the F loop, the pre–TM1 linker, and the TM2–TM3 linker, exhibited considerable changes (Figure 2S).21 It appeared that halothane binding imposed much stronger effects on the global dynamics of the open-channel α4β2 nAChR than on that of the closed channel. Thus, the functional impact is more likely to the open-channel conformation rather than to the closed-channel structure of the receptor. Protein global dynamics are often directly related to protein’s biological functions.23,55,56 Therefore, the functional impact of halothane is likely to be more profound in the open-channel conformation than in the closed-channel conformation of the receptor. Indeed, previous experiments on various proteins also demonstrated the halothane effect varies over different conformations,57,58 and anesthetics preferentially inhibited the open-channel nAChR.41 However, our study indicates that halothane exerts its effect by influencing the global dynamics in the open-channel conformation, instead of blocking the open channel as suggested previously.41
Figure 3.
(A) Mean-square fluctuation (MSF) of individual subunits of the closed-channel α4β2 nAChR in the control (black) and halothane (red) systems. The data were generated using the five slowest normal modes of the GNM analysis on the pentameric structure at the end of the 20-ns simulations. MSF was reduced in certain regions of the extracellular (EC) domain in the presence of halothane. (B) A snapshot of the α4-2 subunit with color-coded maxima (red and orange) and minima (blue) of the MSF shown in (A).
β2 subunits showed greater susceptibility to halothane modulation than α4 subunits
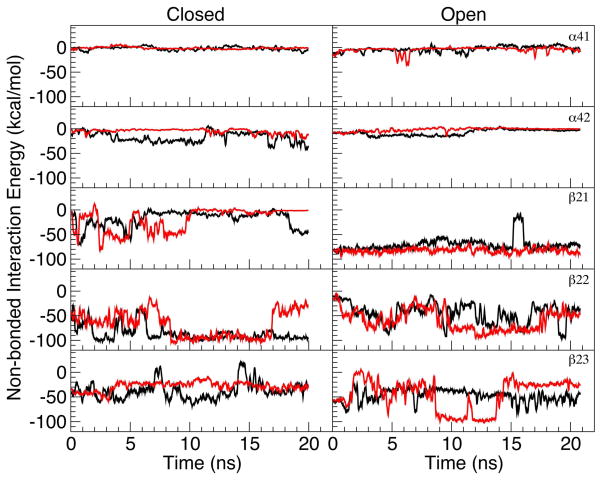
Previous experiments found that inhibition of nAChRs by anesthetics was subtype-dependent.9,59 Our simulation results support this notion. As shown in Figure 3A, almost all noticeable changes in MSF occurred in β2 subunits, even though overall β2 subunits had less direct exposure to halothane than α4 subunits. Greater susceptibility of β2 subunits to halothane modulation was also reflected in the energy profiles of the non-bonded interaction at the EC/TM interface of the α4β2 nAChR. The interaction energies, including van der Waals (vdW) and electrostatic energies, between the TM2–TM3 linker (S272 to L282 in the α4 subunits or S266 to L276 in the β2 subunits) and the β1–β2 loop (residues D47 to T57 in the α4 subunits or S44 to T54 in the β2 subunits) or other loops at the EC/TM interface were calculated for each subunit in the absence or presence of halothane. Figure 4 shows the non-bonded interaction energies between the TM2–TM3 linker and the β1–β2 loop in the closed- and open-channel α4β2 nAChR over the course of 20-ns simulations. The non-bonded interaction of the TM2–TM3 linker with the Cys loop is presented in Figure 3S. Weak non-bonded interaction energies were found consistently in the α4 subunits. Although the energy values fluctuated during 20-ns simulations, the β2 subunits demonstrated much stronger non-bonded interaction energies at the EC/TM interface than the α4 subunits. In the presence of halothane, near the end of the 20-ns simulations, non-bonded interactions in β2 subunits either retained the same levels as those in the control system or became much weaker, as evidenced in energy changing either close to zero or to less negative values in Figure 4. Separating the vdW and electrostatic energy terms revealed that electrostatics predominated the non-bonded interaction between the TM2–TM3 linker and the β1–β2 loop in the β2 subunits.
Figure 4.
The non-bonded interaction energies between the β1–β2 loop (residues D47 to T57 in the α4 subunits and S44 to T54 in the β2 subunits) and the TM2–TM3 linker (S272 to L282 in the α4 subunits and S266 to L276 in the β2 subunits) of the closed- (left panels) and open-channel (right panels) α4β2 nAChR in the absence (black) or presence (red) of halothane over the course of 20-ns MD simulations. Both electrostatics and van der Waals were accounted for the non-bonded interaction. Note that a more negative value indicates a stronger non-bonded interaction.
The charged residue pairs at the EC/TM interface of the β2 subunits are responsible for the distinct difference between the α4 and β2 subunits. The α4 subunits have no proper pairing of charged residues at the EC-TM interface, whereas the β2 subunits could form two salt bridges in the region.20 One pair is from R48 in the β1–β2 loop and D268 in the TM2–TM3 linker, the other is from D140 in the Cys loop and K274 in the TM2–TM3 linker. The large fluctuation of non-bonded interaction energies, as indicated in Figure 4, was mostly attributed to the formation and breakage of the salt bridges.
Halothane modulation of the pore region
Early experiments suggested that geometrical or electrostatic modifications in the lumen of the channel could alter the channel functioning.60,61 Water hydration in the pore is essential for channel conductance.62–65 Here, we evaluated the halothane modulation in the pore region of the α4β2 nAChR by assessing the changes in the pore radius and water population inside the pore in the presence or absence of halothane.
Figure 5A shows the differences in the averaged number of water and pore radius in the closed channel between the control and halothane systems in the first and last nanosecond of the 20-ns MD simulations. For comparison, the corresponding changes in the open channel are also presented in Figure 5B. In the halothane systems, the number of water molecules in the transmembrane region (−52.5Å < Z < −27.5Å) of the pore was reduced in both closed and open channels at the end of the 20-ns simulations, but the reduction was as much as 22% in the open channel, compared to only 6% in the closed channel. The pore radius of the transmembrane region also exhibited substantial decrease in the open channel, while only small fluctuation of decrease or increase was observed in the closed channel. The relative insensitivity of water population and pore radius to halothane modulation in the closed channel seems to be consistent with the aforementioned greater resistance of the closed channel to the halothane-induced changes in channel structure and dynamics.
Figure 5.
Comparisons of halothane-induced changes in the channel water (wide black bars and the left vertical axis) and the pore radius (thin green bars and the right vertical axis) for the closed-channel (A) and open-channel (B) systems. The number of water molecules was counted every 5 Å and the pore radius was calculated every 2.5 Å along the channel axis (Z-coordinate), and averaged over 100 snapshots in the first and the last 1-ns simulations. A positive value on the chart indicates more water molecules or larger pore radius in the halothane system than in the control system. The locations of pore lining residues along the channel axis are highlighted in the top panel.
The pore-water reduction in the presence of halothane, especially in the open channel, indicated an elevated barrier for water permeation. If liquid water encountered a high energetic barrier in the channel, so would ions.63 Thus, the pore-water reduction in the presence of halothane implies a possible reduction of ion permeation through the channel, which seems agree with the experimentally observed halothane inhibitory effect on channel conductance.59 It is worth noting that pore-water reduction in the closed channel was not always accompanied by pore-radius reduction. As suggested previously,62,63,66 in addition to pore radius, orientations of hydrophobic side chains and flexibility of pore-lining residues could also be the determinants of ion and water permeations. Without direct halothane binding to pore-lining residues to occlude the pore, the observed water reduction in Figure 5 could result from a combination of all these factors.
CONCLUSIONS
Our MD simulations revealed that the different susceptibilities of the open- and close-channel α4β2 nAChRs to anesthetics are largely due to the difference in channel global dynamics. The closed-channel conformation was much more resilient to the perturbation from halothane, whereas the open channel showed greater vulnerability in its structure and mobility toward halothane modulation. These results imply that halothane inhibition of the α4β2 nAChR may result from impairing the open-channel conformation instead of altering the closed-channel conformation. More profound anesthetic impact to the open-channel α4β2 nAChR agreed well with previous experimental and computational observations of other proteins, such as preferential inhibition of the open-channel conformation of the mouse muscle nAChR by octanol41 and more drastic changes in dynamics of the open-state potassium channel due to the presence of halothane molecules.67
The current study also offered a compelling explanation as to why the open- and closed- channel conformations responded differently to anesthetic binding. A more compacted structural fold of the closed channel, especially at the interface of EC/TM domains and the pore region, made the closed-channel conformation less susceptible to halothane modulation. Under the hypothesis that the “twist-to-open” motion is a key movement for nAChR to function,38 it follows that disturbance to the quaternary structure and its dynamics may render the altered open-channel conformation dysfunctional. Taking into account the fact that halothane has relatively low binding affinities to proteins; the accessibility to binding cavities is likely to be essential to anesthetic action. Volatile general anesthetics, with a size comparable to halothane molecule, are likely to target the EC/TM interface and the interface between two adjacent subunits, and to exert their effects by perturbing the dynamics and quaternary structural arrangement of α4β2 nAChRs.
Both the closed- and open-channel α4β2 nAChRs contained multiple halothane binding sites. A recent photolabeling of the Torpedo california nAChR by a photoactive analog of the general anesthetic etomidate also showed multiple sites.42 Thus, one can predict with confidence that more than one anesthetic binding site exists in each protein in the same family. In the case of α4β2 nAChR, since the majority of the high-affinity binding sites were either at the interface of EC/TM domains or at the interface between α4 and β2 subunits, the disruption to these interfaces seemed to be responsible for impairing normal functions of the receptor.
The β2 subunits exhibit greater susceptibility to halothane modulation than the α4 subunits. The unique electrostatic interaction at the EC/TM interface of the β2 subunits is, at least partially, responsible for the high sensitivity. These findings suggest a new topic for the future study that may shed light on why homo-pentameric α7 nAChR is much less sensitive to inhaled anesthetics than hetero-pentameric α4β2 nAChR.
Supplementary Material
Acknowledgments
This research was supported in part by the National Science Foundation through TeraGrid resources provided by the Pittsburgh Supercomputing Center. TeraGrid systems are hosted by Indiana University, LONI, NCAR, NCSA, NICS, ORNL, PSC, Purdue University, SDSC, TACC, and UC/ANL. This research was also supported by grants from the National Institutes of Health (R01GM066358, R01GM056257, R37GM049202, and T32GM075770).
Footnotes
Supporting Information Available
Three figures, demonstrating the halothane trajectories, mean-square fluctuation of individual subunits at the open-channel conformation, and halothane effects on the non-bonded interaction between the Cys loop and the TM2–TM3 linker of the α4β2 nAChR, are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Karlin A. Nat Rev Neurosci. 2002;3:102. doi: 10.1038/nrn731. [DOI] [PubMed] [Google Scholar]
- 2.Sine SM, Engel AG. Nature. 2006;440:448. doi: 10.1038/nature04708. [DOI] [PubMed] [Google Scholar]
- 3.Dilger JP. Br J Anaesth. 2002;89:41. doi: 10.1093/bja/aef161. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y, Dilger JP, Vidal AM. Mol Pharmacol. 1994;45:1235. [PubMed] [Google Scholar]
- 5.Dilger JP, Brett RS, Mody HI. Mol Pharmacol. 1993;44:1056. [PubMed] [Google Scholar]
- 6.Dilger JP, Brett RS, Lesko LA. Molecular pharmacology. 1992;41:127. [PubMed] [Google Scholar]
- 7.Hara K, Harris RA. Anesth Analg. 2002;94:313. doi: 10.1097/00000539-200202000-00015. [DOI] [PubMed] [Google Scholar]
- 8.Zuo Y, Aistrup GL, Marszalec W, Gillespie A, Chavez-Noriega LE, Yeh JZ, Narahashi T. Mol Pharmacol. 2001;60:700. [PubMed] [Google Scholar]
- 9.Flood P, Ramirez-Latorre J, Role L. Anesthesiology. 1997;86:859. doi: 10.1097/00000542-199704000-00016. [DOI] [PubMed] [Google Scholar]
- 10.Mori T, Zhao X, Zuo Y, Aistrup GL, Nishikawa K, Marszalec W, Yeh JZ, Narahashi T. Mol Pharmacol. 2001;59:732. doi: 10.1124/mol.59.4.732. [DOI] [PubMed] [Google Scholar]
- 11.Flood P, Role LW. Toxicol Lett. 1998;100–101:149. doi: 10.1016/s0378-4274(98)00179-9. [DOI] [PubMed] [Google Scholar]
- 12.Yamashita M, Mori T, Zhao X, Nagata K, Marszalec W, Yeh JZ, Narahashi T. International Congress Series. 2005;1283:243. [Google Scholar]
- 13.Yamashita M, Mori T, Nagata K, Yeh JZ, Narahashi T. Anesthesiology. 2005;102:76. doi: 10.1097/00000542-200501000-00015. [DOI] [PubMed] [Google Scholar]
- 14.Unwin N. J Mol Biol. 2005;346:967. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 15.Wilson GG, Karlin A. Neuron. 1998;20:1269. doi: 10.1016/s0896-6273(00)80506-1. [DOI] [PubMed] [Google Scholar]
- 16.Wilson G, Karlin A. Proc Natl Acad Sci U S A. 2001;98:1241. doi: 10.1073/pnas.031567798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. Nature. 2001;411:269. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- 18.Celie PH, van Rossum-Fikkert SE, van Dijk WJ, Brejc K, Smit AB, Sixma TK. Neuron. 2004;41:907. doi: 10.1016/s0896-6273(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 19.Hansen SB, Sulzenbacher G, Huxford T, Marchot P, Taylor P, Bourne Y. Embo J. 2005;24:3635. doi: 10.1038/sj.emboj.7600828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haddadian EJ, Cheng MH, Coalson RD, Xu Y, Tang P. J Phys Chem B. 2008;112:13981. doi: 10.1021/jp804868s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu LT, Willenbring D, Xu Y, Tang P. J Phys Chem B. 2009;113:12581. doi: 10.1021/jp9039513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bahar I, Atilgan AR, Erman B. Fold Des. 1997;2:173. doi: 10.1016/S1359-0278(97)00024-2. [DOI] [PubMed] [Google Scholar]
- 23.Yang LW, Liu X, Jursa CJ, Holliman M, Rader AJ, Karimi HA, Bahar I. Bioinformatics. 2005;21:2978. doi: 10.1093/bioinformatics/bti469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng MH, Liu LT, Saladino AC, Xu Y, Tang P. J Phys Chem B. 2007;111:14186. doi: 10.1021/jp075467b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. J Comput Chem. 2005;26:1781. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacKerell AD, Bashford D, Bellott, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. The Journal of Physical Chemistry B. 1998;102:3586. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 27.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. J Comput Chem. 1998;19:1639. [Google Scholar]
- 28.Chiara DC, Dangott LJ, Eckenhoff RG, Cohen JB. Biochemistry. 2003;42:13457. doi: 10.1021/bi0351561. [DOI] [PubMed] [Google Scholar]
- 29.Liu Z, Xu Y, Saladino AC, Wymore T, Tang P. J Phys Chem A. 2004;108:781. [Google Scholar]
- 30.Zacharias M, Straatsma TP, McCammon JA. The Journal of Chemical Physics. 1994;100:9025. [Google Scholar]
- 31.Chipot C, Pearlman DA. Molecular Simulation. 2002;28:1. [Google Scholar]
- 32.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. J Mol Graph. 1996;14:354. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 33.Cheng X, Ivanov I, Wang H, Sine SM, McCammon JA. Biophys J. 2007;93:2622. doi: 10.1529/biophysj.107.109843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. Nucleic Acids Res. 2006;34:W116. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eckenhoff RG. Proc Natl Acad Sci U S A. 1996;93:2807. doi: 10.1073/pnas.93.7.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holladay MW, Dart MJ, Lynch JK. J Med Chem. 1997;40:4169. doi: 10.1021/jm970377o. [DOI] [PubMed] [Google Scholar]
- 37.Rada EM, Tharakan EC, Flood P. Anesth Analg. 2003;96:108. doi: 10.1097/00000539-200301000-00023. [DOI] [PubMed] [Google Scholar]
- 38.Szarecka A, Xu Y, Tang P. Proteins. 2007;68:948. doi: 10.1002/prot.21462. [DOI] [PubMed] [Google Scholar]
- 39.Law RJ, Henchman RH, McCammon JA. Proc Natl Acad Sci U S A. 2005;102:6813. doi: 10.1073/pnas.0407739102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henchman RH, Wang HL, Sine SM, Taylor P, McCammon JA. Biophys J. 2003;85:3007. doi: 10.1016/S0006-3495(03)74720-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forman SA, Miller KW, Yellen G. Mol Pharmacol. 1995;48:574. [PubMed] [Google Scholar]
- 42.Chiara DC, Hong FH, Arevalo E, Husain SS, Miller KW, Forman SA, Cohen JB. Mol Pharmacol. 2009;75:1084. doi: 10.1124/mol.108.054353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galzi JL, Revah F, Bouet F, Menez A, Goeldner M, Hirth C, Changeux JP. Proc Natl Acad Sci U S A. 1991;88:5051. doi: 10.1073/pnas.88.11.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corringer PJ, Galzi JL, Eisele JL, Bertrand S, Changeux JP, Bertrand D. J Biol Chem. 1995;270:11749. doi: 10.1074/jbc.270.20.11749. [DOI] [PubMed] [Google Scholar]
- 45.Kotzyba-Hibert F, Grutter T, Goeldner M. Mol Neurobiol. 1999;20:45. doi: 10.1007/BF02741364. [DOI] [PubMed] [Google Scholar]
- 46.Li GD, Chiara DC, Sawyer GW, Husain SS, Olsen RW, Cohen JB. J Neurosci. 2006;26:11599. doi: 10.1523/JNEUROSCI.3467-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nirthanan S, Garcia G, 3rd, Chiara DC, Husain SS, Cohen JB. J Biol Chem. 2008;283:22051. doi: 10.1074/jbc.M801332200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arias HR, Kem WR, Trudell JR, Blanton MP. Int Rev Neurobiol. 2003;54:1. doi: 10.1016/s0074-7742(03)54002-8. [DOI] [PubMed] [Google Scholar]
- 49.Hemmings HC, Jr, Akabas MH, Goldstein PA, Trudell JR, Orser BA, Harrison NL. Trends Pharmacol Sci. 2005;26:503. doi: 10.1016/j.tips.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 50.Bahar I, Erman B, Jernigan RL, Atilgan AR, Covell DG. J Mol Biol. 1999;285:1023. doi: 10.1006/jmbi.1998.2371. [DOI] [PubMed] [Google Scholar]
- 51.Yang LW, Bahar I. Structure. 2005;13:893. doi: 10.1016/j.str.2005.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kash TL, Jenkins A, Kelley JC, Trudell JR, Harrison NL. Nature. 2003;421:272. doi: 10.1038/nature01280. [DOI] [PubMed] [Google Scholar]
- 53.Lee WY, Sine SM. Nature. 2005;438:243. doi: 10.1038/nature04156. [DOI] [PubMed] [Google Scholar]
- 54.Keramidas A, Kash TL, Harrison NL. J Physiol. 2006;575:11. doi: 10.1113/jphysiol.2005.102756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tama F, Sanejouand YH. Protein engineering. 2001;14:1. doi: 10.1093/protein/14.1.1. [DOI] [PubMed] [Google Scholar]
- 56.Ma J. Current protein & peptide science. 2004;5:119. doi: 10.2174/1389203043486892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eckenhoff RG, Tanner JW. Biophys J. 1998;75:477. doi: 10.1016/S0006-3495(98)77536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xi J, Liu R, Asbury GR, Eckenhoff MF, Eckenhoff RG. J Biol Chem. 2004;279:19628. doi: 10.1074/jbc.M313864200. [DOI] [PubMed] [Google Scholar]
- 59.Violet JM, Downie DL, Nakisa RC, Lieb WR, Franks NP. Anesthesiology. 1997;86:866. doi: 10.1097/00000542-199704000-00017. [DOI] [PubMed] [Google Scholar]
- 60.Galzi JL, Devillers-Thiery A, Hussy N, Bertrand S, Changeux JP, Bertrand D. Nature. 1992;359:500. doi: 10.1038/359500a0. [DOI] [PubMed] [Google Scholar]
- 61.Revah F, Bertrand D, Galzi JL, Devillers-Thiery A, Mulle C, Hussy N, Bertrand S, Ballivet M, Changeux JP. Nature. 1991;353:846. doi: 10.1038/353846a0. [DOI] [PubMed] [Google Scholar]
- 62.Liu Z, Xu Y, Tang P. J Phys Chem B. 2006;110:12789. doi: 10.1021/jp060688n. [DOI] [PubMed] [Google Scholar]
- 63.Beckstein O, Sansom MS. Physical biology. 2004;1:42. doi: 10.1088/1478-3967/1/1/005. [DOI] [PubMed] [Google Scholar]
- 64.Hummer G, Rasaiah JC, Noworyta JP. Nature. 2001;414:188. doi: 10.1038/35102535. [DOI] [PubMed] [Google Scholar]
- 65.Lynden-Bell RM, Jayendran CR. The Journal of Chemical Physics. 1996;105:9266. [Google Scholar]
- 66.Wang HL, Cheng X, Taylor P, McCammon JA, Sine SM. PLoS Comput Biol. 2008;4:e41. doi: 10.1371/journal.pcbi.0040041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vemparala S, Domene C, Klein ML. Biophys J. 2008;94:4260. doi: 10.1529/biophysj.107.119958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.