Abstract
This study was designed to discover blood biomarkers of cancer susceptibility using invasive multiple (n = 21), single primary breast cancer (n = 21), and control subjects (n = 20). Heparinized whole blood was incubated at 37 °C for 2 hours after 0–10 Gy of radiation, then cell cycle arrest marker CDKN1A and apoptosis marker BBC3 mRNA were quantified. This epidemiological study was practically feasible because radiation-induced mRNA was preserved for at least 1 day whenever blood was stored at 4 °C (r2 = 0.901). Moreover, blood could be stored frozen after radiation treatment (r2 = 0.797). Radiation-induced CDKN1A and BBC3 mRNA were dose dependent, and the degree of induction of CDKN1A was correlated with that of BBC3 (r2 = 0.679). Interestingly, multiple primary cases showed higher induction of CDKN1A mRNA than single primary and control groups, whereas BBC3 did not show such differences. The results suggested that cancer susceptibility represented by the multiple primary breast cancer cases was related to over-reaction of CDKN1A mRNA, not BBC3. The study also suggests that ex vivo gene expression analysis could potentially be used as a new tool in epidemiological studies for cancer and radiation sensitivity research.
Keywords: CDKN1A, BBC3, multiple primary cancer, DNA damage, cancer susceptibility Mitsuhashi et al
Introduction
An alteration of DNA functions either by mutation or epigenetic changes is considered to play a crucial role in cancer development. Each cell in our body frequently encounters various challenges of DNA alterations, but our cells repair these problems appropriately without inducing cancer or precancerous status in most cases. For sporadic cancer cases, agents causing DNA alterations may be too strong or persistent beyond such repair functions in the target cells, however, for high cancer risk individuals, it is reasonable to speculate that such repair function may be weak or altered congenitally or secondarily by acquired status. In fact, poor DNA-damage-response in ataxia telangiectasia, 1 is known to be associated with the development of cancer, and this hereditary cancer risk is related to the mutation of transcription factor tumor protein p53 (FLJ92943, LFS1, p53, TRP53) (TP53 is an offical name).2 Mutation of TP53 is also found frequently in many types of cancer cells,3 and single nucleotide polymorphism (SNP) of TP53 is now considered as one of risk factors of cancer susceptibility.4
The primary function of DNA is to transcribe appropriate mRNA. If DNA function is critically altered either by mutation or epigenetic changes, the resultant mRNA may be altered. Since TP53 is known to activate cyclin-dependent kinase inhibitor 1A (p21, Cip1) (CDKN1A is an official name)5 and BCL2 binding component 3 (JFY1, PUMA) (BBC3 is an official name) mRNAs,6–8 we thought in this study that the function of TP53 would be assessed quantitatively by measuring the induction of these genes. Although many hot spots of SNP were identified on TP53, each SNP is not always linked to the function of the gene. The mRNA assay has clear advantages over SNP because it is applicable as a screening tool beyond previously identified hot spots on TP53, and provides quantitative results.
Radiation-induced DNA damage was extensively studied using human peripheral blood lymphocytes, and various mRNAs were identified as potential biomarkers for cancer susceptibility as well as an assessment of individual radiation sensitivity.9–13 Our group has also been focused on the quantification of DNA-damage response using a unique method we developed,14 and found that CDKN1A and BBC3 were the most sensitive and universal marker mRNA for DNA damage in human whole blood.15,16 In this study, this assay was used to characterize cancer susceptibility, although we do not know the results in blood samples are representative to other tissues. We chose multiple primary cancer cases as a model of cancer susceptibility.
Materials and Methods
Subjects
Preliminary studies (Fig. 1) were carried out using blood samples collected from healthy adult donors (APEX Research Institute, Tustin, CA, institutional review board (IRB)-approved). Informed consent was obtained from each donor, and only limited demographic information (gender, age, and ethnicity only) was provided to us. For epidemiological studies (Figs. 2–4), the study protocol was approved by another IRB (University of California, Irvine (UCI)). We identified 38 breast cancer cases from the local cancer registry who were diagnosed with invasive multiple primary breast cancer (MP). The number of cases who were eligible and agreed to participate in the study was 21 cases. We then used their characteristics to choose single primary cases (SP) (n = 21) and unaffected control (UC) (n = 20) who were matched for age and ethnicity (Table 1). After informed consent was obtained, blood samples were collected by clinical nurses who went to their home for the blood collection and to fill out a questionnaire. Blood samples were drawn in 2 tubes, one for a complete blood count, and the other for mRNA analysis. Blood samples were immediately transferred at 4 °C to the laboratory. After radiation treatment (cesium-137, at Radiation Oncology, UCI) on the same day of blood draw, these samples were incubated at 37 °C for 2 hours, then stored frozen at −80 °C.
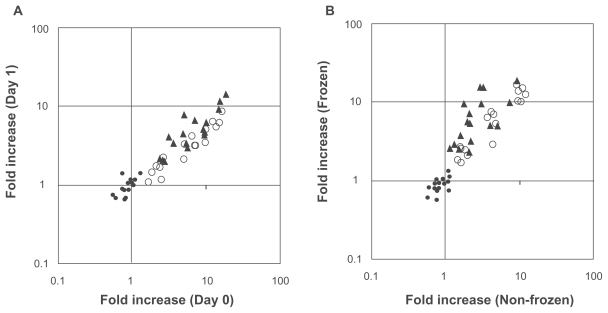
Figure 1. Validation of assay condition.
A) Blood storage at 4 °C. B) Applicability of frozen whole blood. Heparinized human blood samples were collected from 6 adult donors (APEX Research Institute), and were stored at 4 °C for 4–6 (Day 0), 24 (Day 1), or 48 hours (Day 2). Blood samples were then stimulated with 10, 1, 0.1 and 0 Gy of radiation (cesium-137) in triplicate, followed by the incubation at 37 °C for an additional 2 hours. One set of samples was stored at −80 °C, and the other set was processed immediately without freezing procedure. Frozen samples were analyzed 1 week later. ACTB (•), CDKN1A (○) and BBC3 (▴) mRNA were quantified as described in the Methods, and fold increase was calculated using the values of unexposed samples with 2 hours incubation. Blood volume used for each analysis was 150 μl (50 μl in triplicate), and each gene was amplified individually by TaqMan (CDKN1A and BBC3) or SYBR green (ACTB), respectively. Symbols are the mean value from each subject.
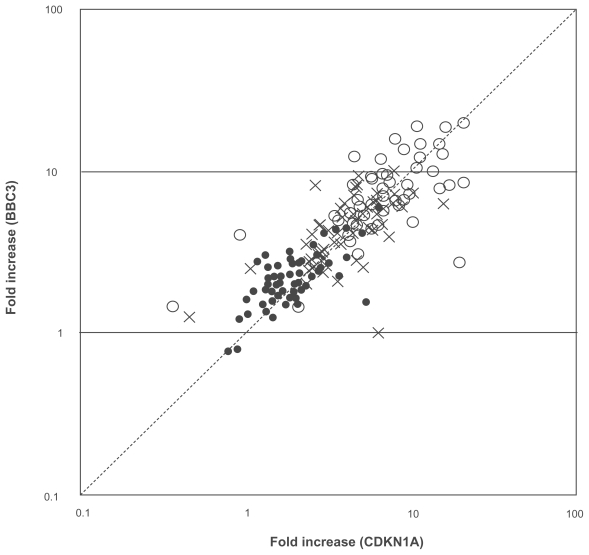
Figure 2. CDKN1A versus BBC3.
Heparinized human whole blood samples from MP (n = 21), SP (n = 21), and UC (n = 20) were stimulated with 0.1 (•), 1 (×), and 10 Gy (○) of radiation in quadruplicate, and incubated at 37 °C for 2 hours. CDKN1A (x-axis) and BBC3 (y-axis) mRNA were then quantified and fold increase was calculated as described in the Methods. Symbols are the mean value from each subject, and dashed line shows 45° angle.
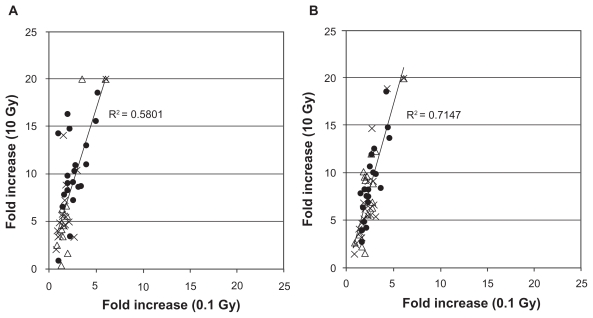
Figure 4. 0.1 Gy versus 10 Gy.
The data of CDKN1A and BBC3 at 0.1 Gy were compared with those of 10 Gy. A) CDKN1A, B) BBC3. •: MP,Δ: SP, ×: UC, respectively. Symbols are the mean value from each subject.
Table 1.
Demographic and tumor characteristics of recruited participants.
| MP*1 (n = 21) |
SP*2 (n = 21) |
UC*31 (n = 20) |
|
|---|---|---|---|
| N (%) | N (%) | N (%) | |
| Race/Ethnicity | |||
| NH White | 20 (95.2%) | 18 (85.7%) | 20 (100.0%) |
| H ispanic | 0 (0.0%) | 3 (14.3%) | 0 (0.0%) |
| Asian | 1 (4.8%) | 0 (0.0%) | 0 (0.0%) |
| Age Now | |||
| 40–49.9 | 2 (9.5%) | 1 (4.8%) | 3 (15.0%) |
| 50–59.9 | 6 (28.6%) | 8 (38.1%) | 9 (45.0%) |
| 60–69.9 | 10 (47.6%) | 10 (47.6%) | 3 (15.0%) |
| 70–79.9 | 3 (14.3%) | 2 (9.5%) | 5 (25.0%) |
| Age @ Invasive Breast Ca | |||
| 40–49.9 | 3 (14.3%) | 4 (19.0%) | |
| 50–59.9 | 7 (33.3%) | 13 (61.9%) | |
| 60–69.9 | 8 (38.1%) | 4 (19.0%) | |
| 70–79.9 | 3 (14.3%) | 0 (0.0%) | |
| Stage of Invasive Breast Ca | |||
| Localized | 13 (61.9%) | 14 (66.7%) | |
| Lymph Nodes | 6 (28.6%) | 7 (33.3%) | |
| Unknown | 2 (9.5%) | 0 (0.0%) | |
| Histology of Invasive Breast Ca | |||
| Papillary Ca | 1 (4.8%) | 0 (0.0%) | |
| Infiltrating Duct Ca | 12 (57.1%) | 17 (81.0%) | |
| Lobular Ca | 2 (9.5%) | 2 (9.5%) | |
| Duct./Lob. Ca | 3 (14.3%) | 2 (9.5%) | |
| Infilt Duct W/Other Ca | 3 (14.3%) | 0 (0.0%) | |
| Years bet Invasive Breast and Other Ca | |||
| 0–4.9 | 7 (33.3%) | ||
| 5–9.9 | 5 (23.8%) | ||
| 10–14.9 | 8 (38.1%) | ||
| 15–19.9 | 1 (4.8%) | ||
| Type Other Ca (5 have >1 other Ca) | |||
| Ovary | 2 (7.7%) | ||
| Lung | 2 (7.7%) | ||
| Breast | 4 (15.4%) | ||
| Endometrium | 5 (19.2%) | ||
| Thyroid | 1 (3.8%) | ||
| Rectum | 4 (15.4%) | ||
| Melanoma | 5 (19.2%) | ||
| Lymph Nodes | 2 (7.7%) | ||
| Kidney | 1 (3.8%) | ||
Multiple primary,
Single primary,
Unaffected control.
mRNA analysis
The mRNA and cDNA were prepared from whole blood as shown previously,14–18 where 50 μl of blood samples were dispensed into 96-well filterplates to collect leukocytes. Sixty μl of lysis buffer containing a cocktail of specific reverse primers was applied to the wells in the filterplate, and the resultant cell lysates were transferred to oligo(dT)-immobilized microplates (GenePlate, RNAture) for poly(A)+ mRNA purification. The cDNA was directly synthesized in 30 μl solutions at each well: specific primer-primed cDNA in the liquid phase and oligo(dT)-primed cDNA in the solid phase. Four μl cDNA solution was used for TaqMan PCR (Applied Biosystems),19 and SYBR green PCR (Bio-Rad).20 Each gene was amplified individually. The cycle threshold (Ct), which was the cycle of PCR to generate certain amounts of PCR products (fluorescence), was determined using analytical software (SDS, Applied Biosystems). The Ct values of radiation-treated triplicate (Fig. 1) or quadruplicate (Figs. 2–4) samples were subtracted by the mean Ct values of control samples (37 °C incubation without radiation treatment) to calculate ΔCt, and the fold increase was calculated as 2^(−ΔCt). Primers and TaqMan probes were published previously.14–17
Results
As shown in Figure 1A, radiation response was preserved for at least 1 day whenever whole blood was stored at 4 °C (r2 = 0.901, n = 54), although such function was reduced significantly after 2 days (data not shown). Furthermore, radiation-induced mRNA expression was maintained even when whole blood was stored frozen after radiation treatment (r2 = 0.797, n = 54) (Fig. 1B). These results made epidemiological studies feasible, because blood samples were collected one at a time over several months, then mRNA assay was performed simultaneously to minimize the cost and labor of the experiment as well as to avoid assay-to-assay variation. The fold increase of β-actin (ACTB) mRNA was all less than 1.5 (Figs. 1AB, •), suggesting that the experiments were performed appropriately. Time course and dose responses of radiation-induced CDKN1A and BBC3 were previously published.15
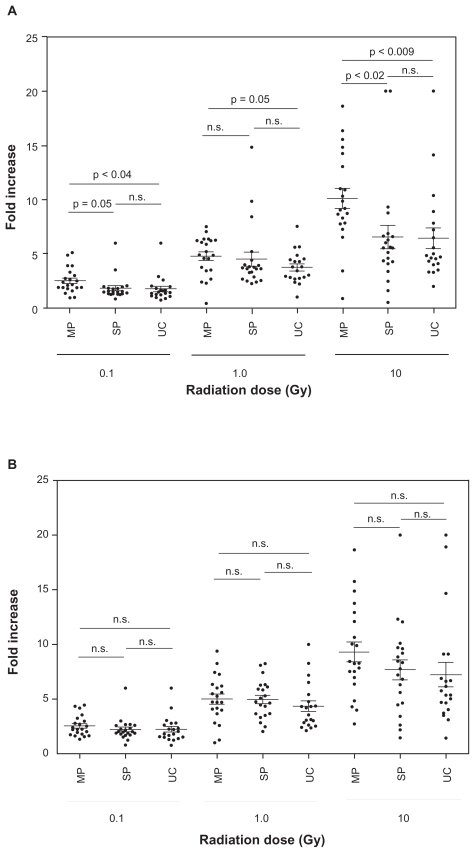
As shown in Figures 2 and 3, radiation induced both CDKN1A and BBC3 mRNA in a dose dependent manner, and the degree of induction of CDKN1A was correlated with that of BBC3 (r2 = 0.679, n = 62), although some discrepancy was found between 2 genes (Fig. 2). The leukocyte counts (6,300 ± 1,209 (MP), 5,580 ± 1,710 (SP), 5,230 ± 1,290 (UC), respectively) and demographic characteristics (Table 1) among 3 groups were not statistically significant. Although no significant difference was found among MP, SP, and UC for BBC3 mRNA (Fig. 3B), CDKN1A in MP was significantly higher than SP and UC in all doses of radiation (0.1, 1, and 10 Gy) (Fig. 3A), except MP versus SP at 1 Gy, where 3 individuals in SP showed very high fold increases. If these 3 individuals were excluded, it was statistically significant (p = 0.005). Interestingly, more than 2 fold increase of 0.1 Gy-induced CDKN1A was found in 3 (3/21 = 14.3%) and 4 (4/20 = 20%) individuals in SP and UC, respectively, whereas such population was significantly higher in MP (12/21 = 57.1%) (p = 0.004 (MP vs. SP), p = 0.01 (MP vs. UC) by χ2 test, respectively) (Fig. 3). Similarly at 1 Gy, 4 (SP) and 3 (UC) individuals showed more than 5 fold increase of CDKN1A mRNA, whereas it was 10 in MP (p = 0.05 (MP vs. SP), p = 0.02 (MP vs. UC), respectively) (Fig. 3). At 10 Gy, only 2 (SP) and 3 (UC) individuals showed more than 10 fold increase of CDKN1A mRNA, whereas it was 9 in MP (p = 0.01 (MP vs. SP), p = 0.05 (MP vs. UC), respectively) (Fig. 3). The immediate effect of chemotherapy may be excluded because blood was withdrawn at drug free period, and there was no significant difference between SP and UC at all doses of radiation (Fig. 3).
Figure 3. MP versus SP versus UC.
The same data shown in Figure 2 were expressed differently by comparing MP, SP, and UC for each dose of radiation for both CDKN1A A) and BBC3 B), respectively. Symbols are the mean value from each subject, and the bars indicate mean ± standard error of each group. Statistical differences (t-test) were also shown.
Furthermore, the data of 0.1 Gy of radiation were correlated with those of 10 Gy for both CDKN1A (r2 = 0.5801, n = 62) and BBC3 (r2 = 0.7147, n = 62), respectively (Fig. 4). Correlation between CDKN1A and BBC3 (Fig. 2) and 0.1 Gy and 10 Gy (Fig. 4) suggest that the variation of data are more likely to be derived from individual-to-individual variation, not assay techniques.
Discussion
Radiation-induced CDKN1A and BBC3 mRNA were well-known evidence, not new to this study. The uniqueness of this study was to characterize individual-to-individual variation of this response quantitatively, and found that MP showed higher induction of CDKN1A mRNA than SP and UC. This was confirmed by both t-test and non-parametric χ2 test. Although a variety of parameters is available for the assessment of cancer susceptibility or cancer risk, which includes family history, environmental exposure, diet, smoking, etc., we thought in this study that the patients having multiple primary cancers (not metastasis) (MP) in their life time would be considered as the cases of cancer susceptibility, especially when blood was tested after second or third cancers. The number of subjects in this study was limited, however, such small number was justified by the fact that we only found 38 MP in local cancer registry, and 21 subjects (21/38 = 55%) were successfully recruited. It was not easy to expand the territory beyond our local cancer registry, because we had to dispatch nurses to their home, and bring blood samples back to our laboratory on the same day for subsequent 2 hours’ incubation as described in Methods.
The measurement of mRNA from human whole blood requires multiple labor-intensive steps including leukocyte isolation, mRNA purification, and cDNA synthesis, and tiny variation in any step introduces a huge variation after exponential amplification of the target genes. Thus, the technical fluctuation of the assay itself must be substantially smaller than that of individual human variation. Although many studies in RT-PCR and DNA microarray demonstrated the results in duplicate or triplicate, the starting materials were often derived from a single source of blood. In order to draw statistical conclusion in each stimulation, the starting materials in our assay were triplicate-quadruplicate whole blood samples for both untreated control and treated samples.14–18 Our platform demonstrated much smaller variation than other conventional methods, leading to identify as small as 50% changes as significant results (see Fig. 3).15 As shown in Figure 1 and previous studies,14,18 synthetic poly(A)+RNA spiked into lysis buffer (external control) and reference gene β-actin (internal control) were rarely induced in our system, sugegsting that the false positive results are quite unlikely. Moreover, because each single blood sample becomes 16 aliquots (4 radiation doses × quadruplicate), high throughput 96-well format was fit nicely to manipulate 992 (62 subjects × 16) RNA preparations and cDNA syntheses, and 1,984 PCR (992 × 2 mRNAs).
Although the number of mRNA was limited to CDKN1A and BBC3 in this study, these 2 were selected as the most prominent and universal markers after screening experiments of 16 apoptosis-related mRNAs, including BAX, BAK1, BOK, BCL2L1, BAD, BID, BIK, BCL2L11, HRK, PMAIP1, BCL2, APAF1, SUMO1, DDIT3, BBC3, and CDKN1A.15 Furthermore, in addition to radiation, CDKN1A and BBC3 were used as chemosensitivity markers for aclarubicin, cytarabine, bleomycin, carboplatin, cisplatin, cyclophosphamide, daunorubicin, doxorubicin, fludarabine, idarubicin, methotrexate, pirarubicin, vinblastine, vincristine, vindesine, and etoposide.15,16
One of technical arguments regarding blood mRNA analysis was the stability of blood samples after blood draw. This is particularly important when blood is stimulated in vitro. In our previous study,14 we reported that the levels of mRNA were quite stable whenever blood samples were stored at 4 °C. As shown in Figure 1, we also confirmed that in vitro functionality in regard to radiation-induced CDKN1A and BBC3 mRNA was maintained for at least 1 day whenever blood was stored at 4 °C. Whole blood stimulation without isolation of mononuclear lymphocytes also made this epidemiological study practically feasible.
We initially hypothesized that cancer susceptibility represented by MP might be linked to hypo-functions of CDKN1A and/or BBC3, because of the knowledge of ataxia telangiectasia.1 However, surprisingly, the results were opposite as shown in Figure 3, and suggested that MP might be related to over-reaction of CDKN1A mRNA, not BBC3. Since CDKN1A is responsive for cell cycle arrest, which is a foundation of various DNA repair mechanisms, such overreaction of CDKN1A may increase the chance of false reaction, whereas BBC3-inducing cells will die by apoptosis without carrying damaged DNA to the daughter cells. Both CDKN1A and BBC3 mRNA are controlled by the transcription factor TP53,1,21 and in fact, the fold increase of these 2 mRNA was correlated to each other. The discrepancy of CDKN1A and BBC3 shown in MP may indicate an additional consideration into the TP53-CDKN1A-BBC3 axis. Our mRNA expression analysis should be supplemented in future studies by various downstream assays, such as protein synthesis of CDKN1A and BBC3, and detection of nuclear foci of the phosphorylated forms of histone H2AX (H2A histone family, member X) and ATM (ataxia telangiectasia mutated) kinase at sites of DNA double-strand breaks,22 although these assays are not immediately applicable to epidemiological studies.
Hyper-function of CDKN1A mRNA found in this study may be derived from SNP or epigenetic changes at the promoter regions of CDKN1A itself, TP53, or other related genes. Alternatively, it may be related to the strength or weakness of each individual’s antioxidant activity. Cancer susceptibility represented in MP may be developed secondarily by previous chemotherapy, not congenital factors. Thus, the present study is an interesting screening tool for various downstream molecular and biochemical pathways, and is applicable to both inherited and acquired status. SNP is not curable, however, we found that the results of this functional assay might be modified by exercise, dietary supplements, experimental drugs, etc. (data not shown). Although we do not know whether a couple of individuals in UC and SP who show hyper-function of CDKN1A will develop new or another cancer in their lifetime, the present study will be a clue for future cancer susceptibility research and prevention as well as radiation sensitivity.
Acknowledgements
We would like to thank Dr. MJ Tonkon, C. Fox, C. McGinty, and J. Haire (Apex Research Institute) for blood draw from healthy volunteers, Dr. Eugene Elmore (Radiation Oncology, UCI) for radiation treatment, and Cindy Yeh and Nancy Leighton (Department of Epidemiology, UCI) for their technical assistance during epidemiological studies. This study was funded by the grants from Hitachi Chemical Research Center, Inc., Lon V. Smith Foundation, and Hereditary Breast Cancer Research Project (NIH # CA-58860-12). MM declared the conflict of financial interest because he was an employee of Hitachi Chemical Research Center, Inc. Other authors declared no conflict of financial interest.
References
- 1.Paterson MC, Smith BP, Lohman PH, et al. Defective excision repair of gamma-ray-damaged DNA in human (ataxia telangiectasia) fibroblasts. Nature. 1976;260:444–7. doi: 10.1038/260444a0. [DOI] [PubMed] [Google Scholar]
- 2.Kastan MB, Zhan Q, el-Deiry WS, et al. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–97. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 3.Hernandez-Boussard T, Rodriguez-Tome P, Montesano R, et al. IARC p53 mutation database: a relational database to compile and analyze p53 mutations in human tumors and cell lines. International Agency for Research on Cancer. Hum Mutat. 1999;14:1–8. doi: 10.1002/(SICI)1098-1004(1999)14:1<1::AID-HUMU1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 4.Bond GL, Levine AJ. A single nucleotide polymorphism in the p53 pathway interacts with gender, environmental stresses and tumor genetics to influence cancer in humans. Oncogene. 2007;26:1317–23. doi: 10.1038/sj.onc.1210199. [DOI] [PubMed] [Google Scholar]
- 5.Han J, Flemington C, Houghton AB, et al. Expression of BBC3, a pro-apoptotic BH3-only gene, is regulated by diverse cell death and survival signals. Proc Natl Acad Sci U S A. 2001;98:11318–23. doi: 10.1073/pnas.201208798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu J, Zhang L, Hwang PM, et al. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell. 2001;7:673–82. doi: 10.1016/s1097-2765(01)00213-1. [DOI] [PubMed] [Google Scholar]
- 7.Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–94. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 8.Villunger A, Michalak EM, Coultas L, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science. 2003;302:1036–8. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- 9.Amundson SA, Do KT, Shahab S, et al. Identification of potential mRNA biomarkers in peripheral blood lymphocytes for human exposure to ionizing radiation. Radiat Res. 2000;154:342–6. doi: 10.1667/0033-7587(2000)154[0342:iopmbi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 10.Turtoi A, Brown I, Oskamp D, et al. Early gene expression in human lymphocytes after gamma-irradiation-a genetic pattern with potential for biodosimetry. Int J Radiat Biol. 2008;84:375–87. doi: 10.1080/09553000802029886. [DOI] [PubMed] [Google Scholar]
- 11.Paul S, Amundson SA. Development of gene expression signatures for practical radiation biodosimetry. Int J Radiat Oncol Biol Phys. 2008;71:1236–44. doi: 10.1016/j.ijrobp.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Badie C, Dziwura S, Raffy C, et al. Aberrant CDKN1A transcriptional response associates with abnormal sensitivity to radiation treatment. Br J Cancer. 2008;98:1845–51. doi: 10.1038/sj.bjc.6604381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turtoi A, Schneeweiss FH. Effect of (211)At alpha-particle irradiation on expression of selected radiation responsive genes in human lymphocytes. Int J Radiat Biol. 2009;85:403–12. doi: 10.1080/09553000902838541. [DOI] [PubMed] [Google Scholar]
- 14.Mitsuhashi M, Tomozawa S, Endo K, et al. Quantification of mRNA in whole blood by assessing recovery of RNA and efficiency of cDNA synthesis. Clin Chem. 2006;52:634–42. doi: 10.1373/clinchem.2005.048983. [DOI] [PubMed] [Google Scholar]
- 15.Mitsuhashi M, Endo K, Obara K, et al. Quantification of Drug-Induced mRNA in Human Whole Blood ex vivo. Clinical Medicine: Blood Disorders. 2008;1:1–11. (Open access at http://www.la-press.com/quantification-of-drug-induced-mrna-in-human-whole-blood-ex-vivo-a491) [Google Scholar]
- 16.Mitsuhashi M, Endo K, Obara K, et al. Ex vivo simulation of the action of antileukemia drugs by measuring apoptosis-related mRNA in blood. Clin Chem. 2008;54:673–81. doi: 10.1373/clinchem.2007.091975. [DOI] [PubMed] [Google Scholar]
- 17.Mitsuhashi M, Ogura M, Endo K, et al. Ex vivo induction of mRNA in human whole blood as a new platform of drug and dietary supplement development. Pharm Res. 2008;25:1116–24. doi: 10.1007/s11095-007-9510-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasegawa S, Kato H, Yamaguchi H, et al. Trastuzumab-induced CCL20 and interleukin-8 mRNA in human whole blood ex vivo. Invest New Drugs. 2009 Feb 10; doi: 10.1007/s10637-009-9223-y. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 19.Holland PM, Abramson RD, Watson R, et al. Detection of specific polymerase chain reaction product by utilizing the 5’----3’ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci U S A. 1991;88:7276–80. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morrison TB, Weis JJ, Wittwer CT. Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. Biotechniques. 1998;24:954–62. [PubMed] [Google Scholar]
- 21.Iyer NG, Chin SF, Ozdag H, et al. p300 regulates p53-dependent apoptosis after DNA damage in colorectal cancer cells by modulation of PUMA/p21 levels. Proc Natl Acad Sci U S A. 2004;101:7386–91. doi: 10.1073/pnas.0401002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson PF, Nham PB, Urbin SS, et al. Inter-individual variation in DNA double-strand break repair in human fibroblasts before and after exposure to low doses of ionizing radiation. Mutat Res. 2009 Nov 5; doi: 10.1016/j.mrfmmm.2009.10.013. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]