Abstract
Hematopoietic stem cells (HSCs) are thought to divide infrequently based on their resistance to cytotoxic injury targeted at rapidly cycling cells1, 2 and have been presumed to retain labels such as the nucleotide analogue 5-bromodeoxyuridine (BrdU). However, recently it has been demonstrated that BrdU-retention is neither sensitive nor specific for HSCs3. Here we show that transient, transgenic expression of a Histone2B (H2B)-Green Fluorescent Protein (GFP) fusion protein in mice allows superior labeling of HSCs and permits improved analysis of their turnover in combination with other markers. Mathematical modeling of H2B-GFP dilution in HSCs, identified with a highly stringent marker combination (L−K+S+CD48−CD150+)4, revealed unexpected heterogeneity in their proliferation rates and suggests that ~ 20% of HSCs turn over at an extremely low rate (≤ 0.8–1.8% per day). Prospective isolation and transplantation of L−K+S+CD48−CD150+ HSCs with different H2B-GFP levels revealed that higher H2B-GFP label retention correlates with superior long-term repopulation potential.
The hypothesis that stem cells are relatively slow-cycling predicts that they might be identified by labels that are diluted with division, such as BrdU 5, 6, tritiated thymidine7, or H2B-GFP 8, 9. Indeed, label-retaining cells in the skin are enriched for epithelial stem cells9. Label-retaining cells in the intestine 10, mammary gland 11, heart 12, and bone marrow 5, 6 have also been proposed as candidate stem cell populations. However, recent observations indicated that BrdU retention is neither sensitive nor specific for HSCs 3 and intestinal stem cells 13, unsettling the widely held notion that label retention is a general property of stem cells. One major limitation of using BrdU to study label retention is that the slow turnover of putative stem cells may not only prevent these cells from losing label with time, but may also prevent them from incorporating label. In addition, it is not possible to test the function of cells prospectively isolated based on their BrdU content. To circumvent these problems, we generated a mouse strain that allows for ubiquitous, doxycycline-inducible expression of an H2B-GFP fusion protein (Supplementary Fig. 1A–D). To explore its utility for marking HSCs, we induced a large cohort of mice with doxycycline for 6 weeks (pulse; starting at 4 – 8 weeks of age) and followed loss of fluorescence in the bone marrow (chase). As intended, fluorescence levels exceeding the background by several orders of magnitude were observed immediately after the pulse. However, in agreement with the expectation that most bone marrow cells turn over rapidly, >95% of the cells lost H2B-GFP expression after only 4 weeks of chase and <1% expressed significant levels of H2B-GFP after 6 months or more (6 months: 0.58%, standard deviation (SD) 0.46, n=6; 1 year: 0.39%, SD 0.05, n=2; 1.5 years: 0.31%, SD 0.1, n=3). These frequencies of label retaining cells were 1–2 orders of magnitude higher than the known frequencies of HSCs 4, 14 and the majority of them expressed markers of mature lymphoid (T-cell lineage: CD3, CD4, CD8, TCRβ; B-cell lineage: CD19, B220) or myeloid lineages (Gr1, Mac1, Ter119) (data not shown). Thus, similar to recent findings that demonstrate lack of specificity of BrdU retention for the identification of HSCs 3, H2B-GFP label retention is not specific for HSCs when used as a single parameter.
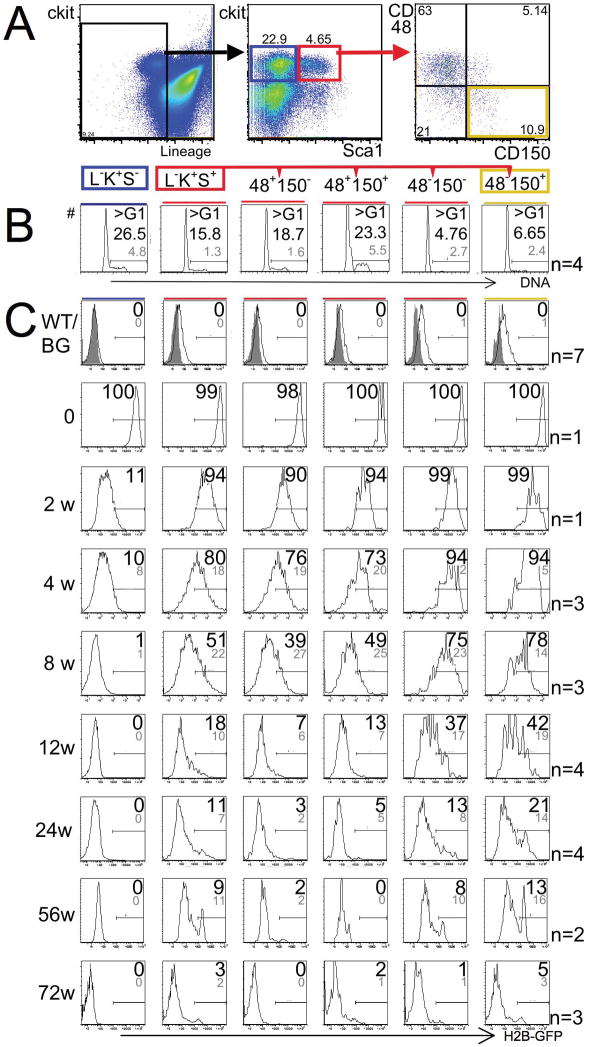
Next, we analyzed H2B-GFP in combination with well-established surface markers for progenitors and HSCs including lineage markers (L), c-Kit (K), Sca-1 (S), CD48, and CD150 4 (Fig. 1A) and compared proliferation rates of defined populations (Fig. 1B) with retention of H2B-GFP over time (Fig. 1C). Remarkably, all HSC/progenitor populations were quantitatively labeled immediately following the pulse (Figure 1C, second row “0”). Actively cycling myeloid progenitors (L−K+S−; Fig. 1A, middle panel, left frame, blue; Fig. 1B, left panel) lost the majority of H2B-GFP as soon as 2 weeks after the pulse and became entirely negative after ~8 weeks (Fig. 1C, left column). In contrast, a population enriched for HSCs (L−K+S+; Fig. 1A, middle panel, right frame, red), distinguished from progenitors by expression of Sca-1, cycled less actively (Fig. 1B, second panel from the left) and lost H2B-GFP much less rapidly (Fig. 1C; second column from left). Within the L−K+S+ population, absence of CD48 and presence of CD150 expression predict long-term repopulation potential 4 and both of these traits also predicted increased label retention. CD48-negative L−K+S+ cells cycled less than CD48-positive L−K+S+ cells (Fig. 1B, four right panels). Accordingly, CD48-positive L−K+S+ cells (Fig. 1C, two middle columns) lost H2B-GFP more rapidly than CD48-negative L−K+S+ cells (Fig. 1C, two right columns). CD150 expression was not associated with obvious differences in cycling rates within the CD48-negative L−K+S+ population (Fig. 1B, two right panels), but label retention was nevertheless slightly, but consistently, more pronounced in CD150-positive CD48−L−K+S+ cells (Fig. 1C, 2 right columns). Of note, ~20% of HSCs (L−K+S+CD48−CD150+) retained H2B-GFP after 24 weeks and ~5% of HSCs retained H2B-GFP after 72 weeks of chase (Fig. 1C, right column). These data compare favorably to BrdU-label retention where only ~2% of HSCs retained detectable label after ~17 weeks of chase in a recent study3. We confirmed by transplantation assays that long-term repopulation potential was strictly limited to L−K+S+CD48−CD150+ cells and demonstrated that L−K+S+CD48−CD150− cells harbored striking short-term repopulation potential (Supplementary Fig. 2, 3) 4, 15, 16. We speculate that the subtle nature of the difference in H2B-GFP retention between these two cell populations reflects that short-term HSCs are direct progeny of long-term HSCs, and that short-term HSCs preserve their low-proliferative state until they differentiate further into CD48-positive cells.
Figure 1. Immuno-phenotype predicts cell cycle-stage distribution and retention of H2B-GFP in early hematopoietic cells.
(A) Gating strategy for the analysis of progenitors and HSCs in the bone marrow. Left panel shows exclusion of dead (PI+) and lineage marker-expressing cells; previous forward and side scatter gates for exclusion of debris and enucleated red cells are not shown. Middle panel shows gates for committed myeloid progenitors (left, blue frame, Lineage (L) −, c-Kit (K)+, Sca1 (S) −) and enriched, but impure, HSCs (right, red frame, L−K+S+). Right panel shows further resolution of enriched HSC population with SLAM markers CD48 and CD150; highly purified HSCs are found in the right lower quadrant (yellow frame). (B) Cell-cycle stage distribution of early hematopoietic populations (as shown in A) assessed by visualizing DNA content with Hoechst 33342 dye. Shown are representative histograms; gates distinguish cells in G0/G1 from cells in S/G2/M phases of the cell cycle. Numbers show percentage of cells in S/G2/M (means: large black number; standard deviation: small grey number); difference between L−K+S−-cells (blue box) and L−K+S+-cells (red box) is significant (p=0.005); difference between CD150+CD48−L−K+S+- cells (yellow box) and CD150−CD48−L−K+S+- cells (second from right) is not significant (p=0.15); difference between CD150+CD48−L−K+S+- cells (yellow box) and either of the CD48+ populations (two middle panels) is significant p<0.001. (C) Expression of H2B-GFP in immuno-phenotypically defined early hematopoietic populations over time. In each column, analyses of a defined population (order as in B) are shown without pulse (first row), immediately after the pulse (second row), or at defined time points after the pulse (designated on the left in weeks (w), rows 3 – 9). Individual plots are representative histograms of H2B-GFP intensity on a logarithmic scale (x-axis covers 5 orders of magnitude). Large black numbers on individual plots show proportions (%) of H2B-GFP-positive cells above the arbitrary threshold. When multiple mice were analyzed, means are given and standard deviations are shown in smaller grey numbers (number of mice (n) per time point are shown on the right of the figure). Plots in first row show that H2B-GFP background (BG) is elevated in un-induced transgenic mice (black line, n=7, aged 2 to 15 months) as compared with wildtype (WT) mice (grey shaded area); arbitrary threshold was set above background as shown. Note: H2B-GFP is lost within 8 –12 weeks in progenitors (L−K+S−; first column from left, blue box) and is retained the most in highly purified hematopoietic stem cells (CD150+CD48−L−S+K+; first column from right, yellow box).
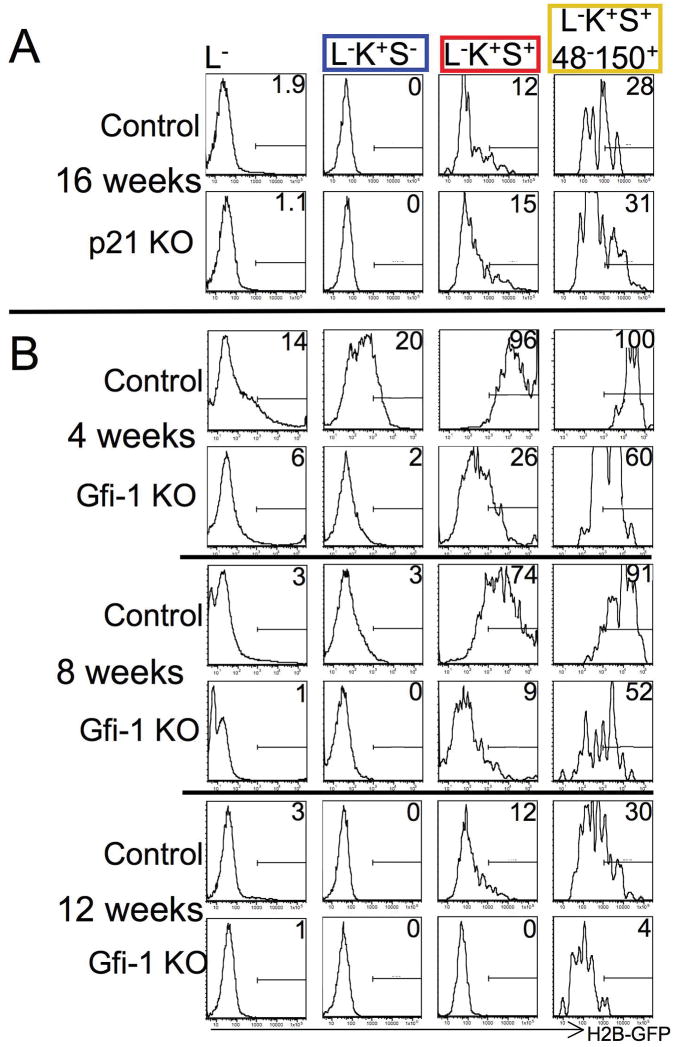
Figure 2. Accelerated H2B-GFP loss in Gfi-1−/− but not p21Cip1/Waf1−/− HSCs.
(A) No increased turnover of p21CIP1/WAF1−/− HSCs. Mice lacking p21CIP1/WAF1 and control mice were pulsed with doxycycline for 6 weeks. After 16 weeks of chase, H2B-GFP retention was analyzed in the entire lineage-marker-negative population (first column), committed myeloid progenitors (second column, blue frame), enriched but impure HSCs (third column, red frame), and highly purified HSCs (fourth column, yellow frame). (B) Increased turnover of Gfi-1−/− HSCs. Analysis after 4, 8, and 12 weeks of chase. Note loss of H2B-GFP in all early bone marrow populations, including HSCs, in Gfi-1−/− mice by 12 weeks. Letter/number-code for cell markers on top as in Fig. 1A. Representative data are shown. For each genotype, at least 4 mice were analyzed after 3 – 4 months of chase.
Similar to SLAM family markers CD48 and CD150, a number of other markers have been utilized to enrich for long-term repopulating HSCs within the L−K+S+ population, including CD34 14, Endothelial protein C receptor (EPCR, CD201) 17, Endoglin (CD105) 18, and Flt3 (CD135) 19. We found that all of these markers also enrich for H2B-GFP-retaining cells, albeit to varying degrees (Supplementary Fig. 4). Thus, our data establish a striking match between a history of low proliferation and markers for long-term repopulation potential in early hematopoietic cells. However, our findings contrast recent claims that the majority of HSCs (L−K+S+Flt3−) resides in a non-transplantable, quiescent “reserved” state that outnumbered “primed”, transplantable HSCs by ~ 5-fold 20. If such a “reserved” state existed, an inverse correlation between label retention and markers predicting long-term repopulation-potential within the L−K+S+-population should be expected, contrary to our findings. More directly, we failed to detect any correlation between H2B-GFP retention and staining with the N-cadherin antibody MNCD-2, the proposed marker for “reserved” L−K+S+Flt3−-HSCs (Supplementary Fig. 4F, 5) and, in agreement with others (Mark Kiel, Sean Morrison, personal communication), we demonstrated that MNCD-2 does not recognize N-cadherin in the analysis of bone marrow cells by flow-cytometry (Supplementary Fig. 6, 7).
Numerous molecular regulators have recently been implicated in restricting the proliferation of HSCs based on assessing cell cycle distribution or short-term BrdU incorporation assays (for review see21). Short-term BrdU incorporation assays are biased towards faster proliferating cells and analysis of cell cycle distribution using DNA and/or RNA stains may be flawed due to superimposing heterogeneous subpopulations. While our data prediced that H2B-GFP retention should be reduced in the presence of mutations that accelerate the turnover of HSCs, we failed to demonstrate accelerated loss of H2B-GFP in HSCs of p21Cip1/Waf1 knockout mice (Fig. 2A), contrasting previous conclusions based on experiments using less specific methods and markers 22. However, our data are consistent with a study that observed an impact of p21Cip1/Waf1 loss on HSCs only in stress conditions, but not in steady state 23 and with the observation that very pure HSCs express the cyclin-dependent kinase inhibitor p57KIP2 but not p21Cip1/Waf1 24. Together, these findings raise doubts on the widely held view that p21Cip1/Waf1 is a key regulator of quiescence in HSCs. In contrast, H2B-GFP loss in HSCs was indeed accelerated drastically in Gfi-1 knockout mice (Fig. 2B), confirming and extending previous investigations utilizing BrdU 25, 26. As the system described here initially labels all cells regardless of cell-cycle stage, our data unequivocally demonstrate that all lineage marker-negative cells are subject to faster turnover in the absence of Gfi-1 and rule out the possibility that sub-populations of Gfi-1−/−HSCs might be excluded from enhanced proliferation. Our data provide proof of principle that H2B-GFP retention analysis can refine the understanding of turnover of stem cells and progenitors in mutant mice.
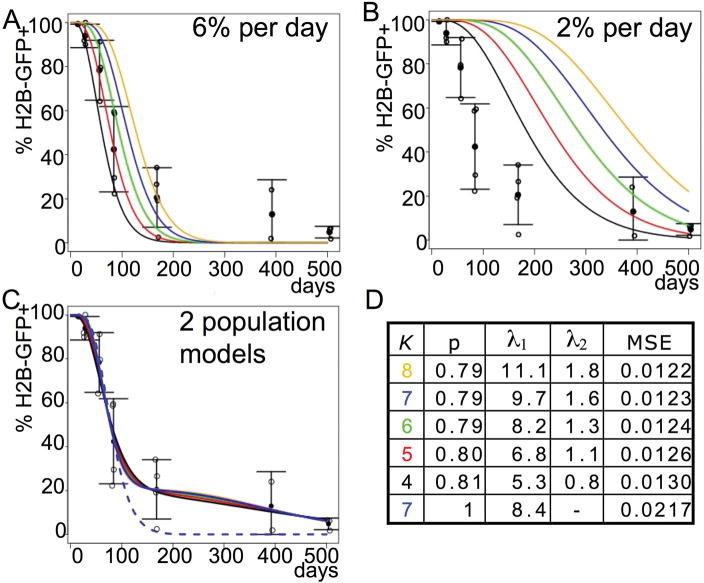
To assess the kinetics of HSC (L−S+K+CD48−CD150+) turnover, we generated mathematical models (Fig. 3). These models were created to reflect (i) the finding that HSCs distribute H2B-GFP equally to daughter cells (Supplementary Fig. 5), (ii) that the number of divisions necessary to dilute H2B-GFP from the levels at the end of the pulse to levels below the arbitrary threshold is unknown; we estimated that it took ~7–8 divisions to fall below the threshold, which corresponds to ~2 logs on the GFP intensity scale (Fig. 1C), however we cannot exclude non-proliferation associated losses over the long period of observation, so curves for 4–8 divisions are shown in Fig. 3, and (iii) the assumption that HSCs in the bone marrow are maintained in a steady state. Given a uniform HSC turnover rate of 6% per day 3, our model predicts that all HSCs will become H2B-GFP-negative before 300 days after the pulse (Fig. 3A). While this model roughly fits the experimental data for the first 100–200 days, it sharply underestimates the detected proportions of H2B-GFP positive cells at later time points suggesting a lower rate. Models obtained with a uniform division rate of 2% per day (Fig. 3B), which is lower than any rate previously estimated with BrdU incorporation (6% per day 3, 7.8% per day 19 ~3% per day 27, 28), are closer to the observed proportions at later time points, but underestimate H2B-GFP loss at early time points. Thus, our data cannot be easily reconciled with the previous assumption that all HSCs proliferate at similar rates. Next, we generated models assuming a larger population (~80%) cycling faster and a smaller population (~20%) cycling more slowly. Indeed, such two-component models appropriately predicted the observed proportions of GFP-positive cells over time (Fig. 3C,D) and showed significantly better fit to the experimental data (Fig. 3D). Depending on whether loss of detectable GFP was assumed to occur after 4 or 8 divisions, these models fit the data with a larger population cycling at 5.3%–11.1% per day and a slower population cycling at 0.8%–1.8% per day (Fig. 3D). It is likely that the actual proliferation rates of HSCs are even lower than our model suggests because we cannot estimate the losses of H2B-GFP resulting from protein turnover during the long observation period. Nevertheless, our findings are compatible with the conclusion from long-term (180 day) BrdU administration 19 that all HSCs do eventually turn over.
Figure 3. Mathematical modeling of H2B-GFP retention in HSCs.
(L−S+K+CD48−CD150+). (A, B) Models for H2B-GFP loss over time based on homogeneous HSC division rates of 6% per day (A) and 2% per day (B). Colored lines show models based on 4 (black), 5 (red), 6 (green), 7 (blue), or 8 (yellow) divisions for H2B-GFP to fall below the arbitrary detection threshold. Open circles represent the observed proportions of cells positive for H2B-GFP over time (derived from experiments shown in Fig. 1, LS+K+CD48−CD150+, first column from right). Closed circles show averages, error bars show standard deviations. Note: neither a high (6%/day) nor a low (2%/day) rate fits the data at both early and late time points. (C) Models based on mixture of two populations dividing at different rates. Color codes are as indicated above. Parameters for the models are shown in the adjacent table D. (D) K indicates the number of divisions until H2B-GFP falls below threshold (models for different K’s correspond to colored curves in C); P denotes the proportion of the larger population (rounded); λ1 denotes the proportion of cells cycling per day in the larger population (%), λ2 denotes the proportion of cells cycling in the smaller population. Shown are values of K and associated optimal estimates for P, λ1, λ2, and the mean squared error (MSE) for each model. Note: Curves for different Ks appear superimposed because P, λ1, and λ2 were varied to fit the data optimally for each K. The bottom row (dashed blue curve in C) is the optimal homogeneous model for K=7 exhibiting a substantial degradation of fit (higher MSE).
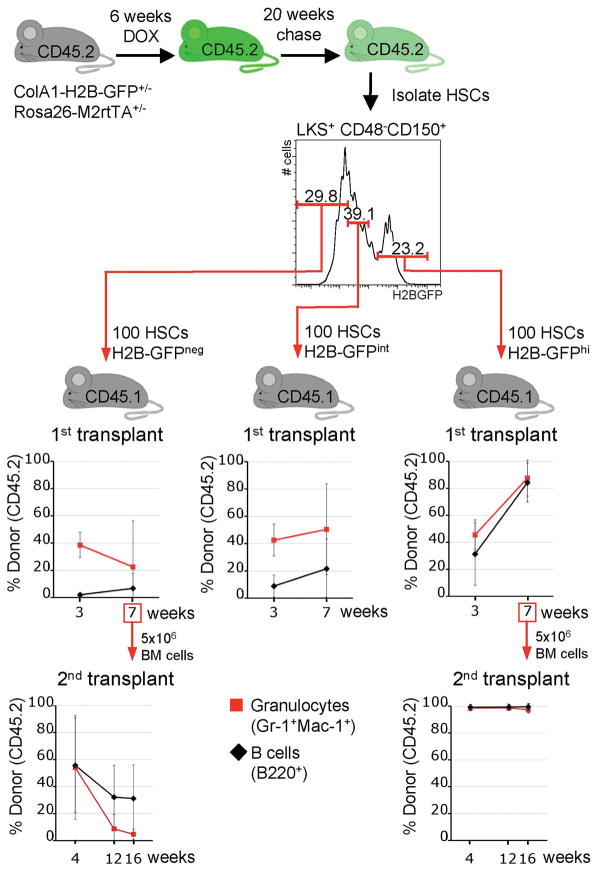
To test predictions of our model and to establish if the existence of different turnover rates amongst L−K+S+CD48−CD150+-defined HSCs is associated with functional differences, we prospectively isolated and transplanted L−K+S+CD48−CD150+-cells that had retained different levels of H2B-GFP after 5 months (Fig. 4). HSCs with the highest H2B-GFP content (~ top 20%) gave rise to lympho-myeloid hematopoiesis that improved with time (Fig. 4, upper right panel). In contrast, HSCs with reduced H2B-GFP content had diminished repopulation potential (Fig. 4, upper middle panel), and HSCs that had entirely lost H2B-GFP had markedly reduced repopulation potential (Fig. 4, upper left panel). These findings were confirmed in secondary transplants (Fig. 4, bottom panels). Of note, loss of H2B-GFP retention correlated with the acquisition of CD34 (Supplementary Fig. 6B), which is absent on mouse long-term HSCs14, and H2B-GFP retention correlated with high expression of EPCR (Supplementary Fig. 6C), an alternative marker for HSCs17. Addition of either of these markers to the L−K+S+CD48−CD150+ combination allowed for enhanced identification of label retaining cells (Supplementary Fig. 7), raising the possibility that more complex marker combinations might be utilized for the development of even more refined HSC identification protocols in the future.
Figure 4. H2B-GFP retention of L−K+S+CD48−CD150+-HSCs predicts function.
100 L−K+S+CD48−CD150+-defined HSCs from H2B-GFP transgenic mice (CD45.2) were transplanted into irradiated recipients (CD45.1) together with a small number of support bone marrow cells (CD45.1) to ensure survival. Three upper plots show the proportions of donor-derived B-cells and granulocytes in groups of recipient mice (primary recipients; H2B-GFPneg, left, n=4; H2B-GFPint, middle, n=3; H2B-GFPhi, right, n=2). Note that cells with the same immuno-phenotype, but distinct H2B-GFP content, harbor strikingly different repopulation potential. Bottom panels show donor-derived hematopoiesis in recipients of secondary transplants (performed 7 weeks after the primary transplant) from either the H2B-GFPneg group (left, n=5, donor was the most highly reconstituted primary transplant recipient) or the H2B-GFPhi group (right, n=5). Means and standard deviations (error bars) are shown.
Our study reveals that H2B-GFP label retention correlates well with long-term repopulation potential in the presence of highly selective markers for HSCs. However, we cannot exclude that the heterogeneity in proliferation and repopulation potential among L−K+S+CD48−CD150+-cells reflects its mixed composition of stem and progenitor cells. Thus, we cannot unambiguously conclude that the proliferation history of HSCs predicts their long-term repopulation potential or that there is an absolute restriction in the numbers of divisions that HSCs can undergo. However, it is tempting to speculate that HSCs “count” the number of divisions they undergo and shut down self-renewal programs after a threshold is reached. This may explain why the frequency of functional long-term repopulating HSCs among L−K+S+CD48−CD150+-cells diminishes as mice age and HSC proliferation increases (in 6 week old mice 1 in 2 phenotypic HSCs 4 and in 22 week old mice 1 in 7 HSCs repopulate long-term29). It may also explain why self-renewal eventually exhausts after serial transplantation, a setting where previous division has already been linked to loss of stem cell activity30, and why attempts at in vitro expansion of HSCs have met with limited success. It is unlikely that telomere attrition explains our observation because the number of divisions associated with functional differences of HSCs is low. Therefore, the molecular basis of proliferation-associated loss of self-renewal remains unknown. The experimental system introduced here opens the door to definitively clarify whether a proliferation clock of HSCs exists, define its molecular basis and, eventually, manipulate it to expand stem cells.
Methods
Flow cytometry was performed as previously described 25. Generation of mice, bone marrow transplantation, and mathematical modeling are described in supplementary “Full Methods” online.
Supplementary Material
Acknowledgments
We thank Laura Prickett and Kat Folz-Donahue from the HSCI Flow-core at MGH for their expert assistance, Geoffrey Wahl for providing the H2B-GFP cDNA, and Matthias Stadtfeld, Wenjun Guo, and Ben Wittner for valuable input. We thank Linheng Li for the kind gift of biotinylated MNCD-2. We thank Mark Kiel and Sean Morrison for sharing unpublished results and for the gift of excised bone marrow of conditional N-cadherin mutant mice. We thank Glenn Radice for permission to use bone marrow from conditional N-cadherin mutant mice. This work was supported by a contribution from the Ellison Foundation to MGH start-up funds for H.H., a seed grant by Harvard Stem Cell Institute to H.H., and by RO1CA122726. H.H. is the recipient of an ASH Scholar Award. K.H. was supported by the NIH Director’s Innovator Award, the Harvard Stem Cell Institute, the Kimmel Foundation, and the V Foundation. A.F. was the recipient of an MGH ECOR Fund for Medical Discovery Award. The authors do not have financial interests to disclose.
Footnotes
Note: Supplementary information is available on the Nature Biotechnology website.
References
- 1.Lerner C, Harrison DE. 5-Fluorouracil spares hemopoietic stem cells responsible for long-term repopulation. Exp Hematol. 1990;18:114–8. [PubMed] [Google Scholar]
- 2.Van Zant G. Studies of hematopoietic stem cells spared by 5-fluorouracil. J Exp Med. 1984;159:679–90. doi: 10.1084/jem.159.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kiel MJ, et al. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature. 2007;449:238–42. doi: 10.1038/nature06115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiel MJ, et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–21. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 5.Arai F, et al. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118:149–61. doi: 10.1016/j.cell.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–41. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 7.Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61:1329–37. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- 8.Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–85. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 9.Tumbar T, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–63. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potten CS, Hume WJ, Reid P, Cairns J. The segregation of DNA in epithelial stem cells. Cell. 1978;15:899–906. doi: 10.1016/0092-8674(78)90274-x. [DOI] [PubMed] [Google Scholar]
- 11.Welm BE, et al. Sca-1(pos) cells in the mouse mammary gland represent an enriched progenitor cell population. Dev Biol. 2002;245:42–56. doi: 10.1006/dbio.2002.0625. [DOI] [PubMed] [Google Scholar]
- 12.Urbanek K, et al. Stem cell niches in the adult mouse heart. Proc Natl Acad Sci U S A. 2006;103:9226–31. doi: 10.1073/pnas.0600635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 14.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–5. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 15.Akala OO, et al. Long-term haematopoietic reconstitution by Trp53−/−p16Ink4a−/−p19Arf−/− multipotent progenitors. Nature. 2008;453:228–32. doi: 10.1038/nature06869. [DOI] [PubMed] [Google Scholar]
- 16.Kiel MJ, Yilmaz OH, Morrison SJ. CD150- cells are transiently reconstituting multipotent progenitors with little or no stem cell activity. Blood. 2008;111:4413–4. doi: 10.1182/blood-2007-12-129601. author reply 4414–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balazs AB, Fabian AJ, Esmon CT, Mulligan RC. Endothelial protein C receptor (CD201) explicitly identifies hematopoietic stem cells in murine bone marrow. Blood. 2006;107:2317–21. doi: 10.1182/blood-2005-06-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen CZ, Li L, Li M, Lodish HF. The endoglin(positive) sca-1(positive) rhodamine(low) phenotype defines a near-homogeneous population of long-term repopulating hematopoietic stem cells. Immunity. 2003;19:525–33. doi: 10.1016/s1074-7613(03)00265-6. [DOI] [PubMed] [Google Scholar]
- 19.Cheshier SH, Morrison SJ, Liao X, Weissman IL. In vivo proliferation and cell cycle kinetics of long-term self-renewing hematopoietic stem cells. Proc Natl Acad Sci U S A. 1999;96:3120–5. doi: 10.1073/pnas.96.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haug JS, et al. N-cadherin expression level distinguishes reserved versus primed states of hematopoietic stem cells. Cell Stem Cell. 2008;2:367–79. doi: 10.1016/j.stem.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 21.Jude CD, Gaudet JJ, Speck NA, Ernst P. Leukemia and hematopoietic stem cells: balancing proliferation and quiescence. Cell Cycle. 2008;7:586–91. doi: 10.4161/cc.7.5.5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng T, et al. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science. 2000;287:1804–8. doi: 10.1126/science.287.5459.1804. [DOI] [PubMed] [Google Scholar]
- 23.van Os R, et al. A Limited role for p21Cip1/Waf1 in maintaining normal hematopoietic stem cell functioning. Stem Cells. 2007;25:836–43. doi: 10.1634/stemcells.2006-0631. [DOI] [PubMed] [Google Scholar]
- 24.Yamazaki S, et al. Cytokine signaling, lipid raft clustering, and HSC hibernation. Ann N Y Acad Sci. 2007;1106:54–63. doi: 10.1196/annals.1392.017. [DOI] [PubMed] [Google Scholar]
- 25.Hock H, et al. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 2004;431:1002–7. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- 26.Zeng H, Yucel R, Kosan C, Klein-Hitpass L, Moroy T. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. Embo J. 2004;23:4116–25. doi: 10.1038/sj.emboj.7600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bradford GB, Williams B, Rossi R, Bertoncello I. Quiescence, cycling, and turnover in the primitive hematopoietic stem cell compartment. Exp Hematol. 1997;25:445–53. [PubMed] [Google Scholar]
- 28.Sudo K, Ema H, Morita Y, Nakauchi H. Age-associated characteristics of murine hematopoietic stem cells. J Exp Med. 2000;192:1273–80. doi: 10.1084/jem.192.9.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yilmaz OH, Kiel MJ, Morrison SJ. SLAM family markers are conserved among hematopoietic stem cells from old and reconstituted mice and markedly increase their purity. Blood. 2006;107:924–30. doi: 10.1182/blood-2005-05-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ema H, et al. Quantification of self-renewal capacity in single hematopoietic stem cells from normal and Lnk-deficient mice. Dev Cell. 2005;8:907–14. doi: 10.1016/j.devcel.2005.03.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.