Abstract
Endocytosis and trafficking of receptors and nutrient transporters are dependent on an acidic intra-endosomal pH that is maintained by the vacuolar H+-ATPase (V-ATPase) proton pump. V-ATPase activity has also been associated with cancer invasiveness. Here, we report on a new V-ATPase-associated protein, which we identified in insulin-like growth factor I (IGF-I) receptor-transformed cells, and which was separately identified in Caenorhabditis elegans as HRG-1, a member of a family of heme-regulated genes. We found that HRG-1 is present in endosomes but not in lysosomes, and it is trafficked to the plasma membrane upon nutrient withdrawal in mammalian cells. Suppression of HRG-1 with small interfering RNA causes impaired endocytosis of transferrin receptor, decreased cell motility, and decreased viability of HeLa cells. HRG-1 interacts with the c subunit of the V-ATPase and enhances V-ATPase activity in isolated yeast vacuoles. Endosomal acidity and V-ATPase assembly are decreased in cells with suppressed HRG-1, whereas transferrin receptor endocytosis is enhanced in cells that overexpress HRG-1. Cellular uptake of a fluorescent heme analogue is enhanced by HRG-1 in a V-ATPase-dependent manner. Our findings indicate that HRG-1 regulates V-ATPase activity, which is essential for endosomal acidification, heme binding, and receptor trafficking in mammalian cells. Thus, HRG-1 may facilitate tumor growth and cancer progression.
Keywords: Diseases/Cancer, Diseases/Cancer/Transformation, Hormones/Growth Factors, Hormones/Insulin, Metabolism/Glucose, Signal Transduction/Protein Kinases, Insulin-Like Growth Factor I, Vacuolar ATPase
Introduction
Cancer cell invasiveness is associated with increased expression of components of the endocytic trafficking machinery (1, 2), and altered trafficking of growth factor receptors and integrins may have a profound effect on tumor growth and invasive potential (3, 4). Cancer cells are also highly dependent on a continuous supply of nutrients and micronutrients. These are acquired through transporters that may be either channels or receptors that deliver their cargo via endocytosis (4–6). The expression levels and trafficking of nutrient transporters are tightly regulated by growth factors, especially through activity of the insulin/IGF-I3-activated PI3K/mammalian target of rapamycin signaling pathway (7). Both overexpression of nutrient transporters and decreased degradation in lysosomes have been linked to cellular transformation (7, 8).
An increasingly acidic lumenal pH gradient from early to late endosomes is required for receptor trafficking and is maintained by the V-ATPase proton pump acting in concert with other pH regulatory channels (9, 10). The V-ATPase consists of two multisubunit domains. The V1 domain has eight subunits (A–H), is located on the cytoplasmic side of membranes, and hydrolyzes ATP. The V0 domain has six subunits (a, d, e, c, c′, and c″), is embedded in membranes, and translocates protons. Activity of the complex is regulated in at least three ways as follows: by reversible domain assembly, which is dependent on glucose and PI3K signaling; by the RAVE regulatory protein complex in yeast; and by targeting of subunits to specific membranes (10–13). In mammals, further regulation may be attributed to subunit isoforms that target V-ATPase to specific tissues or organelles (9, 10).
The V-ATPase has been detected at the plasma membrane of cancer cells (14, 15), which is thought to favor invasiveness, although the c subunit from the V0 domain has been associated with promoting cellular transformation (16). Very little is known about the regulation of V-ATPase function in epithelial cancer cells. Nonetheless, it is reasonable to propose that it may be influenced by the insulin/IGF-I/PI3K signaling pathway, which has increased activity in many cancers. PI3K has been shown to be essential for V-ATPase assembly in renal epithelial cells (12).
In an effort to identify IGF-I-regulated proteins that are important for cancer progression, we isolated a series of genes that are differentially expressed in nontransformed cells (R−, derived from the IGF-IR knock-out mouse) and highly transformed cells (R+ and R− cells that overexpress the IGF-IR) (17–19). Among these was a gene encoding an endosomal protein that we found to associate with the endosomal V-ATPase and to regulate V-ATPase activity in endosomes. As this study was completed, the same gene was identified to encode a heme-binding protein necessary for heme homeostasis in Caenorhabditis elegans and for erythropoiesis and development in zebrafish (20). Heme transport into and within cells may involve endocytosis and trafficking of heme transporters/receptors (21–23). Overall, our data indicate that HRG-1 is the first known heme-binding protein that regulates function of the V-ATPase in endosome acidification and trafficking of receptors essential for cell metabolism.
MATERIALS AND METHODS
General Reagents and Antibodies
IGF-I was from PeproTech Inc. (Rocky Hill, NJ), Concanamycin A, bafilomycin A, nocodazole, propidium iodide, goat serum, crystal violet, leupeptin, E-64, FITC-transferrin and FITC-dextran 40 kDa (FD40), DMEM, glucose-free DMEM, sodium azide, 2-deoxy-d-glucose, nigericin, NaCl and all other salts and reagents were from Sigma unless otherwise stated. LY294002, rapamycin, and PD89059 were from Calbiochem. Alexa-488 transferrin, LysoTracker Red, and LysoSensor Green were from Molecular Probes (Eugene, OR). Antibodies used were as follows: anti-EEA1 (BD Transduction Laboratories); anti-transferrin receptor and anti-Rab11 antibody (Zymed Laboratories Inc.); anti-β-actin and Rab7 (Sigma); anti-LAMP1 and anti-HA (clone 16B12) (Covance); anti-His (Qiagen, UK); anti-c subunit (Chemicon); and anti-A1 subunit (Santa Cruz Biotechnology). Generation of mouse monoclonal V-ATPase anti-A subunit antibody was described previously (24).
Cloning of Human HRG-1 cDNA
cDNA encoding the human HRG-1 gene sequence was obtained from the I.M.A.G.E. Consortium. The open reading frame (nucleotides 98–538) was amplified by PCR using primers to incorporate XhoI restriction sites at the 5′ and 3′ ends and ligated into the pcDNA3-HA vector. For cloning into the yeast expression vector, the human HRG-1 coding sequence was digested with XhoI and SmaI restriction enzymes and ligated into XhoI/SmaI-digested pGBK-T7 vector.
HRG-1 Polyclonal and Monoclonal Antibody Generation
A peptide corresponding to amino acid residues 131–146, both inclusive (HRYRADFADISIL SDF), of the human HRG-1 protein sequence was conjugated to keyhole limpet hemocyanin for inoculation of rabbits (Davids Biotechnologie, Germany). The generated antibody was affinity-purified with immobilized peptide. Alternatively, the peptide CHRYRADFADISILSD (amino acids 130–145) conjugated to keyhole limpet hemocyanin was used to immunize BALB/c mice and subsequent generation of hybridoma cell lines producing HRG-1 monoclonal antibodies (GenScript). The generated antibody was affinity-purified with immobilized protein A. Specificity was confirmed by peptide competition assays. Both antibodies were used at a dilution of 1:250 for immunofluorescence and immunoblotting.
Cell Culture and Transfection
R− cells are an embryonic fibroblast cell line derived from igf-1r−/− mice, and the R+ cell line is R− cells stably overexpressing the IGF-IR (19). R+, R−, HeLa, HEK-293T, normal rat kidney, and MCF-7 cells were maintained in DMEM supplemented in 1 mm l-glutamine, 10% fetal bovine serum (FBS), and 5 mg/ml penicillin and streptomycin antibiotics. For IGF-I stimulation, R+ and MCF-7 cells were starved of serum for 4 h before stimulation with 100 ng/ml IGF-I for the indicated times. HEK-293T cells were transfected using the calcium phosphate transfection method. MCF-7/HA-HRG-1 pools were generated by transfecting MCF-7 cells using Lipofectamine followed by selection for 3 weeks with 1 mg/ml neomycin. Protein expression was confirmed by Western blot and immunofluorescence.
Immunofluorescence
Cells were seeded on serum-coated 10-mm glass coverslips and allowed to attach. The cells were washed with phosphate-buffered saline (PBS), fixed with 3.7% formaldehyde in PBS, and permeabilized with 0.1% Triton X-100 in PHEM (60 mm PIPES, 25 mm HEPES, 10 mm EGTA, 2 mm MgCl2, pH 6.9) for 5 min. Prior to incubation with primary antibody, the cells were blocked with 5% goat serum in PBS. Primary antibodies were diluted in 5% goat serum/PBS, and incubations were performed for 1 h at room temperature, after which the cells were washed with PHEM and incubated with Cy2- or Cy3-conjugated secondary antibodies (Jackson ImmunoResearch, Soham, Cambridgeshire, UK) together with Hoechst before examination with a Nikon T600 fluorescent microscope. Images were captured under ×100 objective using a SPOT CCD camera. Confocal sections were obtained using an Olympus Fluoview FV1000 confocal laser scanning biological microscope (software FV10-ASW version 1.6). Serial z-sections were obtained every 1 μm (usually 8–10 sections/field) to generate representative composite images, and cross hairs through zoom images correspond to z sections depicted on x and y axis, respectively.
Antibody Uptake by Live Cells
MCF7/HA-HRG-1 growing on coverslips cells were incubated overnight with either HA or HRG-1 antibodies diluted in complete or serum-free medium. Cells were then fixed in 4% paraformaldehyde, permeabilized, and incubated with Cy2- or Cy3-conjugated secondary antibodies together with Hoechst for 1 h. As a positive control Fo2 antibody staining, cells seeded at the same time were fixed, permeabilized, and incubated with primary antibody for 1 h followed by incubation with secondary antibodies as before.
Analysis of Plasma Membrane Expression of HRG-1 by Flow Cytometry
Cells were cultured in complete media and then serum-starved for 2 h with or without 20 μm nocodazole for the last 30 min of starvation. Cells were lifted with PBS/EDTA (2 mm) and washed with PBS. Cell pellets were resuspended in 100 μl of anti-HRG-1 antibody diluted in 5% goat serum, 0.1% NaN3/PBS and incubated on ice for 1 h. Samples were washed twice with PBS and incubated with Cy2-conjugated secondary antibody for 1 h on ice. Samples were washed with PBS and resuspended in 500 μl of PBS, and fluorescence intensity was analyzed by flow cytometry.
Western Blotting and Immunoprecipitation
Whole cell lysates were prepared in RIPA lysis buffer (150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mm Tris, pH 8.0, 1 mm phenylmethylsulfonyl fluoride, 1 μm pepstatin, 2 mg/ml aprotinin, 1 μm NaVO4) and analyzed by Western blotting. Primary antibodies were diluted in 5% milk/TBS-Tween 20 (TBS-T) and incubated with membranes at 4 °C overnight. Membranes were washed with TBS-T, incubated with horseradish peroxidase-conjugated secondary antibody (DakoCytomation Denmark, Glostrup, Denmark) at room temperature for 1 h, and detected using ECL. Alternatively, detection and quantification of protein were performed using Odyssey Image Scanner System (Licor Biosciences, Cambridge, UK) using IRdye-conjugated secondary antibodies (Licor) and the Odyssey quantification software.
For immunoprecipitation of His-c subunit, 800 μg of transfected HEK-293T lysates were precleared with protein G-Sepharose beads (GE Healthcare) at 4 °C for 1 h. The lysate was incubated with anti-His antibody at 4 °C overnight, and protein G beads were then added for 1 h at 4 °C with gentle rotation. Immunocomplexes were analyzed by Western blotting using anti-His and anti-HA antibodies. For immunoprecipitation of endogenous V-ATPase A subunit, HA-HRG-1-transfected HEK-293T lysates were precleared as for His-c subunit immunoprecipitations. The lysates were then incubated with the monoclonal anti-A subunit antibody (25) at 4 °C overnight, and immune complexes were analyzed by Western blotting.
Small Interfering RNA (siRNA)
To suppress HRG-1 expression specifically, two siRNAs targeting the human HRG-1 mRNA were obtained from Dharmacon (Lafayette, CO). The sequences of the siRNA oligonucleotides were as follows: siHRG-11 5′-UGGUGACGCACGUGAUGUA-3′ (nucleotides 158–176 of HRG-1 coding sequence) and siHRG-12 5′-GCACGUGAUGUACAUGCAA-3′ (nucleotides 165–183). HeLa and MCF-7 cells were transfected with 10 nm siRNA oligonucleotides using the Oligofectamine transfection reagent (Invitrogen), as per manufacturer's instructions. A nontargeting oligonucleotide from Ambion (Cambridgeshire, UK) was used as a negative control. Suppression of HRG-1 expression was examined 24–96 h after transfection by Western blotting with the anti-HRG-1 polyclonal antibody.
Yeast Two-hybrid Screen
The MATCHMAKER yeast two-hybrid system (Clontech) was used to identify HRG-1-interacting proteins. The pGBKT7-HRG-1 recombinant yeast expression vector, containing the GAL4 DNA binding domain, was used as bait to screen a human fetal brain library, constructed in the pACT2 vector, containing the GAL4 DNA activation domain. Selection of colonies containing putative interacting proteins was carried out by plating onto synthetic dropout media, as per the manufacturer's protocol. Library plasmid DNA was extracted, and the inserts were sequenced.
Cell Viability and Migration Assays
siRNA-transfected HeLa cells were seeded at a density of 3 × 104 cells/well in triplicate wells of a 24-well tissue culture plate 24 h after transfection. Cell viability in DMEM, 10% FBS was assessed every 24 h by the uptake of propidium iodide using flow cytometry on a FACSCalibur instrument (BD Biosciences).
To measure cell migration, siRNA-transfected HeLa cells were starved of serum for 4 h before harvesting with trypsin/EDTA, pelleting, and resuspending in DMEM. Cell suspensions were diluted to a final concentration of 1.8 × 104 cells/100 μl of DMEM and loaded into the upper wells of a Boyden Transwell chamber (Corning Glass) in triplicate. Lower wells were loaded with DMEM, 10% FBS. After 4 h at 37 °C, chambers were disassembled, and cells that had migrated through the membrane were fixed with methanol, stained with 0.1% crystal violet, washed extensively with water, and air-dried. The membranes were then examined by phase contrast microscopy under ×10 magnification using a Nikon TE300 microscope. Three fields from each of the membranes were counted.
Transferrin Receptor-mediated Endocytosis Assays and Transferrin-Alexa488 Binding Assay
Twenty four hours after siRNA transfection, HeLa cells were starved of serum for 30 min before being pulsed with either 25 μg/ml transferrin Alexa Fluor488 (Molecular Probes, Eugene, OR) or transferrin-FITC (Sigma) in 10% FBS/DMEM for 15 min at 37 °C, 5% CO2. The extent of transferrin uptake was determined by fluorescence intensity in the FL1H channel using the FACSCalibur instrument.
To assess transferrin-Alexa488 binding, cells were serum-starved for 1 h, then transferred to ice, and washed with cold 0.1% BSA/PBS. Cells were incubated with 25 μg/ml transferrin Alexa Fluor488 diluted in 0.1% BSA/PBS on ice for 90 min. The cells were washed three times with cold 0.1% BSA/PBS, trypsinized, washed once more, and resuspended in 2% FBS/PBS before FACS analysis. For TfR/Tf-Alexa488 internalization assays, after the 90-min binding of Tf-Alexa488, prewarmed complete media were added to the cells, and they were left at 37 °C for the indicated periods of time. The cells were then transferred to ice, washed with cold BSA/PBS, and the noninternalized receptors removed by a 5-min acid wash (0.2 m acetic acid, 0.2 m NaCl). The cells were then washed and harvested for FACS analysis.
Endosomal pH Estimation
We followed the protocol described by Nilsson et al. (26), with some modifications. Twenty four hours after siRNA transfection, MCF-7 or HeLa cells were seeded in 6-well plates. The next day, the media were replaced with complete media containing 0.5 mg/ml FITC-dextran 40 kDa (FD40), with or without 100 nm of either concanamycin A or bafilomycin A, and incubated at 37 °C with 5% CO2 for 3 h. The samples were then washed twice with PBS, trypsinized, transferred to FACS tubes, centrifuged and then washed with HBSS (Invitrogen) and centrifuged again. Cell pellets were kept on ice and resuspended in HBSS immediately before analysis by FACS. The mean fluorescence detected in FL1 and FL2 channels was quantified, and the FL1/FL2 ratio was calculated for each sample. To create the calibration curve, cells were loaded with FD40 and treated as described above, but the cell pellets were resuspended in a series of citric acid/potassium phosphate (C/K) buffers based on McIlvaine tables, with pH ranging from 4.0 to 6.0, and containing 50 mm sodium azide, 50 mm 2-deoxy-d-glucose, and 10 μm nigericin to equilibrate the intracellular pH with the pH of the C/K buffers. The cells were incubated for 10 min on ice, and the FL1 and FL2 mean fluorescence values were quantified. The FL1/FL2 ratio of these samples was then correlated with the pH value of the solutions used to equilibrate the cells, and the resulting equation was used to estimate the endosomal pH value of the samples.
Lysosomal pH Estimation
Cells transfected with siRNAs were transferred to complete media with no phenol red (Sigma) containing either 1 μm LysoSensor Green for 1 h or 250 nm LysoTracker Red for 2 h. Control cells were also subsequently incubated with 100 nm concanamycin A or bafilomycin A. Cells were washed once with PBS; fresh complete media without phenol red were added to each well, and the fluorescence was analyzed in a FLEXstation II fluorimeter (Molecular Devices). Afterward, the plates were stained with crystal violet and scanned in the Odyssey Infrared system to estimate cell number. The fluorescence data were then normalized to the cell staining data to control for cell density.
V-ATPase Activity Assays
The yeast strain YPH500 was transformed with human HRG-1 in the pGBK-T7 vector (HRG-1) or the pGBK-T7 empty vector. Vacuolar membranes were isolated as described previously (27) followed by SDS-PAGE and Western blotting using the monoclonal antibodies 10D7 (24) against subunit a, 8B1-F3 (24) against subunit A, or antibody against HRG-1. ATPase activity was measured using a coupled spectrophotometric assay in the presence or absence of 1 μm concanamycin, as described previously (27). ATP-dependent proton transport was measured using the fluorescence probe 9-amino-6-chloro-2-methoxyacridine in the presence or absence of 1 μm concanamycin, as described previously (27).
V-ATPase Assembly Assay
siNeg- and siHRG-1-transfected MCF7 cells were maintained in complete media for 24 h post-transfection, starved from serum and glucose for 4 h, and then stimulated with 10 mm glucose for 15 min. The cells were transferred to ice, and the cytosolic fraction was purified. Cells were resuspended in cold STE buffer (250 mm sucrose, 10 mm HEPES, 1 mm EDTA, pH 7.6) supplemented with protease and phosphatase inhibitors and lysed mechanically by passing them through a 26-gauge needle. The lysates were centrifuged at 1200 × g for 10 min at 4 °C to obtain the post-nuclear supernatant, containing cytosol and cellular membranes. Equal volumes of post-nuclear supernatant were centrifuged at 100,000 × g for 1 h to produce the S-100/cytosolic fraction and a membrane pellet. Equal volumes of cytosolic fractions were loaded to a protein gel, and the amount of A1 present in these fractions was quantified by Western blotting. Actin was used as loading control for the cytosolic fraction and the A1/actin ratio was used as measurement of the amount of A1 present in the cytosol of the cells.
ZnMP Uptake Assays
We performed these assays as described previously (20) with some modifications. Control cells and cells overexpressing the HRG-1 protein were grown to confluence and incubated with 5 μm ZnMP (Frontier Scientific, Lancashire, UK) for 15 min in uptake medium (25 mm HEPES, pH 7.4, 130 mm NaCl, 10 mm KCl, 1 mm CaCl2, 1 mm MgSO4, and 2.5 μm BSA). ZnMP uptake was stopped by placing cells on ice and washing with ice-cold 5% BSA in uptake medium to remove surface binding. Cells were then washed three times with cold PBS, trypsinized, washed, and resuspended with HBSS, and the fluorescence intensity of ZnMP was measured by flow cytometry (FACSCalibur) in the FL3 channel. Where indicated, cells were pretreated with 100 nm bafilomycin A for 1 h prior to the uptake. For studies in cells with suppressed HRG-1 expression, cells were transfected with siNeg and siHRG-1 oligonucleotides and 36 h later were incubated with ZnMP as described.
Hemin-Agarose Pulldown Assays
The hemin-agarose pulldown assay was performed as described by Rajagopal et al. (20) using MCF-7/HA-HRG-1 cells lysed in MS buffer (210 mm mannitol, 70 mm sucrose, 10 mm HEPES) at pH 5.0, 6.0, or 7.5. We used 300 nmol of hemin-agarose (Sigma) per assay (equivalent to 50 μl of hemin-agarose slurry) and 50 μl of Sepharose 4B slurry as negative control, with 300 μg of lysate.
RESULTS
HRG-1 mRNA and Protein Are Induced by IGF-I
The mouse slc48a1/hrg-1 gene (NCBI accession number NM_026353) is expressed as two transcripts of 2.5 and 1.6 kb that are more abundant in R+ cells than in R− cells (Fig. 1A). The 2.5-kb variant has a larger 3′-untranslated region, but both species are predicted to encode a 16.5-kDa protein. The hrg-1 1.6-kb mRNA variant is more predominantly expressed in adult mouse tissues than the 2.5-kb variant, with the highest expression in liver and kidney, followed by heart and brain (Fig. 1B). Immunofluorescence microscopy analysis using an antibody raised against the C terminus of the protein demonstrated that HRG-1 expression is higher in R+ than in R− cells (Fig. 1C). The hrg-1 1.6-kb mRNA (Fig. 1D) and HRG-1 protein (Fig. 1E) are induced in response to IGF-I stimulation of R+ cells. Induction of mRNA was not greatly altered by inhibition of PI3K or extracellular signal-regulated kinase (ERK) signaling pathways (data not shown), suggesting it is not entirely due to IGF-I-mediated activation of these pathways.
FIGURE 1.
HRG-1 is a new IGF-I-responsive gene/protein. A, Northern blot with RNA from R+ and R− cells hybridized with hrg-1 and igf-1r probes and actin as loading control. B, Northern blot showing HRG-1 mRNA expression in a murine multiple tissue RNA blot (Clontech). Actin is shown as loading control. S.muscle, skeletal muscle. C, R+ and R− cells were stained for endogenous HRG-1, using Cy3-conjugated secondary antibody, and analyzed by epifluorescence microscopy. Nuclei were visualized by Hoechst stain. D, HRG-1 mRNA is up-regulated by IGF-I. R+ cells were serum-starved for 4 h and stimulated with 100 ng/ml IGF-I for the indicated times before total RNA extraction for Northern blot analysis. The RNA blot was probed for hrg-1, and igf-1r levels were used as loading control. E, HRG-1 protein is induced by IGF-I. R+ cells were treated as in D, and HRG-1 expression was analyzed by Western blot using polyclonal anti-HRG-1 antibody. Actin was used as a loading control.
HRG-1 Is Located in Endosomes and Is Trafficked to the Plasma Membrane
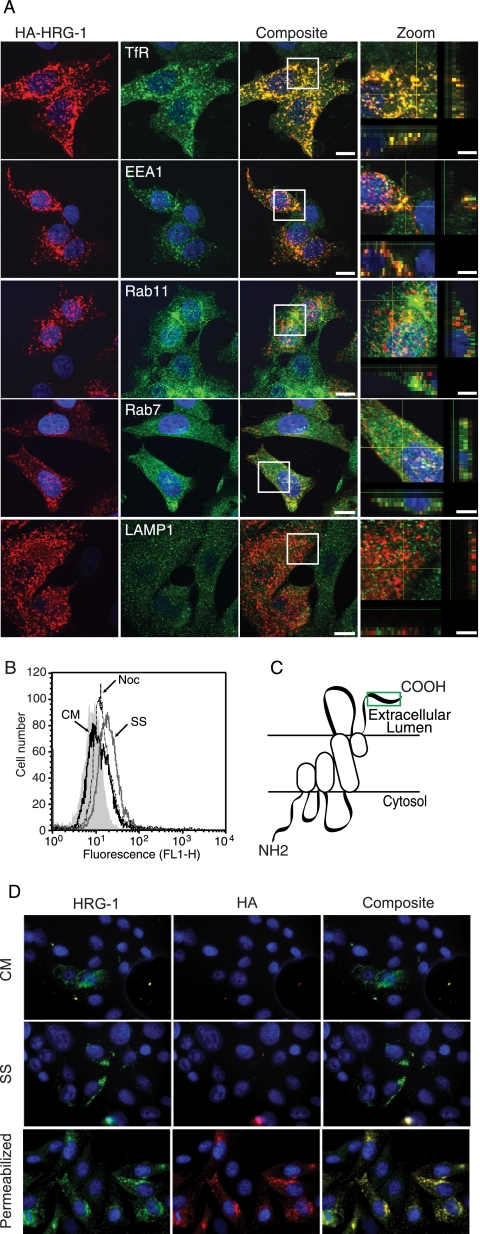
The subcellular location of HRG-1 was investigated by immunofluorescence using the anti-HRG-1 antibody. HRG-1 is expressed in several cell lines with a punctated cytoplasmic distribution resembling endosomes (supplemental Fig. S1). The colocalization of HRG-1 with endosomal compartments was investigated by confocal microscopy in MCF-7 cells expressing HA-tagged HRG-1. HA-HRG-1 strongly colocalizes with the TfR and partially colocalizes with EEA1 (early endosomal autoantigen-1), Rab11, and Rab 7 (Fig. 2A). This indicates that HRG-1 is located in early, recycling, and late endosomes. However, HA-HRG-1 does not colocalize with LAMP-1, which implies no significant expression in lysosomes.
FIGURE 2.
HRG-1 is expressed throughout endosomal compartments and traffics to the plasma membrane. A, HRG-1 colocalization with endosomal markers. MCF7/HA-HRG-1 cells were costained with anti-HA antibody (red) and antibodies against TfR, EEA1, Rab11, Rab7, and LAMP1 (green) and analyzed by confocal microscopy. Representative composite images for markers of each of the vesicular structures and merges with zoom are shown. Cross hairs through zoom images correspond to z sections depicted on the x and y axis, respectively. Original magnification is ×60, and bars represent 5 or 10 μm (zoom images). B, HRG-1 expression at the plasma membrane was detected by flow cytometry with anti-HRG-1 antibody on HeLa cells that were maintained in complete media (CM, black line), serum-starved (SS, dark gray line) for 2 h, or serum-starved for 1.5 h and incubated with 20 μm nocodazole (Noc, dashed-line) for a further 30 min in serum-free media. Control cells stained with secondary antibody alone are indicated by solid gray histogram. C, schematic representation of the predicted transmembrane topology of the HRG-1 protein taking into consideration the orientation of the N and C termini. The epitope (peptide) used to raise anti-HRG-1 antibodies is indicated in green. D, anti-HRG-1 antibody becomes internalized. MCF-7/HA-HRG-1 cells were incubated with either HRG-1 (green) or HA (red) antibodies overnight, in complete media (CM) or serum-free media (SS), and then fixed permeabilized and stained with secondary antibodies. The pictures show a typical example of HRG-1 staining due to HRG-1 antibody uptake by live cells. As positive control for antibody staining, cells were fixed, permeabilized, and stained with HA and HRG-1 antibodies (bottom panels).
The distribution of HRG-1 throughout the endosomal compartments prompted us to investigate whether HRG-1 is trafficked to the plasma membrane. Using the polyclonal anti-HRG-1 antibody, HRG-1 was detected by flow cytometry at the plasma membrane of live HeLa cells (Fig. 2B, black line). Serum starvation increased HRG-1 expression at the plasma membrane (Fig. 2B, gray line), but this translocation was prevented by nocodazole, which disrupts microtubule polymerization necessary for endosome trafficking (Fig. 2B, dashed line). These results indicate that the C terminus of HRG-1 is located on the extracellular/lumenal side of the membrane and that HRG-1 traffics to the membrane upon nutrient withdrawal (Fig. 2C).
To further investigate the relative orientation of the N and C termini of HRG-1 on cell membranes, we incubated live MCF-7/HA-HRG-1 cells with either anti-HRG-1 antibody (epitope on C terminus) or anti-HA antibody (epitope on N terminus) in complete media or under serum starvation conditions. As can be seen in Fig. 2D, live MCF-7/HA-HRG-1 cells internalize the HRG-1 antibody in both conditions, but there is no visible internalization of the HA antibody. Staining of permeabilized cells showed that both antibodies recognized HA-HRG-1 protein. These data demonstrate that HRG-1 is present at the plasma membrane and that the C terminus of the protein (HRG-1 antibody epitope) resides at the external face of the plasma membrane and in the endosome lumen; the N terminus (HA antibody epitope) is located on the cytosolic side.
Analysis of the HRG-1 protein sequence with MemBrain software (28) suggests two full transmembrane helices and two half-transmembrane helices. However, taken together with the data indicating orientation of the N and C termini on opposite sides of the membrane, we propose that the protein may contain one transmembrane helices and three half-transmembrane helices, as illustrated in the model in Fig. 2C.
HRG-1 Is Required for Receptor Endocytosis, Cell Migration, and Survival
As HRG-1 is an endosomal protein, we next investigated whether HRG-1 expression is important for endocytosis, and thus cell survival or migration, by suppressing its expression with siRNA in HeLa cells. Two hrg-1-specific siRNAs, siHRG1 and siHRG2, effectively suppress expression of HRG-1 in HeLa cells compared with control siRNA (Fig. 3A). HeLa cultures are viable at 48 h post-transfection but exhibit substantial cell death by 72 and 96 h (Fig. 3B and supplemental Fig. S2). Based on these observations, all cellular assays were performed between 24 and 48 h to preclude effects of impaired viability. SiHRG1 and siHRG2 reduced HeLa cell migration in Transwell assays by 40 and 50%, respectively, compared with controls (Fig. 3C).
FIGURE 3.
HRG-1 suppression decreases receptor-mediated endocytosis, cell viability, and migration. A, HeLa cells were transfected with 10 nm HRG-1-specific siRNAs or control siRNA for 24, 35, and 48 h, and HRG-1 protein levels were measured by Western blot analysis. B, viability of the siRNA-transfected cultures was quantified as the percentage of propidium iodide (PI)-positive cells by flow cytometry. Results represent one of three independent experiments with similar results. C, cell migration assays were performed on siRNA-transfected HeLa cells 24 h post-transfection and following 4 h of serum starvation. Cell suspensions were loaded into the upper wells of Boyden Transwell chambers and were left to migrate for 4 h. The graph represents the mean ± S.D. number of cells that had traversed the membrane from n = 9. *, p > 0.05 using test. D, receptor-mediated endocytosis was measured in siRNA-transfected HeLa cells 36 h post-transfection in triplicate wells. Cells were serum-starved for 30 min and then incubated with 25 μg/μl of Alexa-488-labeled transferrin (Tf-Alexa488) for 15 min at 37 °C. The amount of Tf-Alexa488 uptake was quantified by flow cytometry. The data are presented as the percentage of Tf-Alexa uptake relative to control siNeg transfectants (100%). Results represent one of four independent experiments with similar results. **, p < 0.01 using t test. E, surface TfR levels were assessed in siRNA-transfected HeLa cells 24 h post-transfection in triplicate wells. Cells were starved for 1 h, transferred to ice, and incubated with 25 μg/μl Tf-Alexa488 in cold PBS, 0.1% BSA for 90 min. The cells were washed extensively and analyzed by FACS. The data are presented as the percentage of surface-bound Tf-Alexa488 relative to siNeg levels (100%). Results are representative of three independent experiments. *, p < 0.05 using t test.
Endocytosis was assessed by measuring the uptake of fluorescently labeled transferrin (Tf-Alexa488) via the constitutively recycled TfR. Uptake of Tf-Alexa488 was reduced by ∼20 and 40%, respectively, in siHRG1 and siHRG2-transfected HeLa cells when compared with controls (Fig. 3D). This correlated with a reduction of ∼15 and 30%, respectively, in Tf-Alexa488 binding to the cells (Fig. 3E), which indicates decreased plasma membrane TfR levels. Similar effects on TfR endocytosis and expression were observed in MCF-7 cells with suppressed HRG-1 expression (supplemental Fig. S3).
HRG-1 Associates with the V-ATPase and Enhances Its Activity in Yeast
To investigate HRG-1 function in the endocytic pathway, we searched for HRG-1-interacting proteins using a yeast two-hybrid screen. The most frequently isolated clone from a human library encoded the 16-kDa c subunit of the V-ATPase (ATP6V0C), a component of the V0 domain. This interaction was confirmed with coexpressed HA-HRG-1 and His-c subunit in HEK-293T cells, with the interaction more pronounced in serum-starved cells (Fig. 4A). Colocalization of HA-HRG-1 with endogenous c subunit was also observed by immunofluorescence (Fig. 4B). The c subunit is known to interact with proteins other than V-ATPase components (29, 30), so we also tested whether HRG-1 associates with the V-ATPase holoenzyme. HA-HRG-1 could be coimmunoprecipitated with endogenous A subunit (cytosolic V1 domain) (Fig. 4C), indicating that HRG-1 associates with the assembled V-ATPase holoenzyme.
FIGURE 4.
HRG-1 associates with the V-ATPase proton pump and enhances V-ATPase activity in yeast. A, HRG-1 interacts with V-ATPase c subunit. HEK-293T cells were cotransfected with HA empty vector and His-c subunit or HA-HRG-1 and His-c subunit. Transfected cells were maintained in complete media (CM), starved of serum (SS) for 20 h, or serum-starved (20 h) and stimulated with 100 ng/ml IGF-I. His-c subunit was immunoprecipitated (IP) using anti-His antibody, and coimmunoprecipitation of HA-HRG-1 was detected using anti-HA antibody. B, HRG-1 localizes with V-ATPase c subunit. HeLa cells transiently expressing HA-HRG-1 were cultured in complete media (CM) or serum-starved for 20 h (SS), costained with anti-HA antibody (red) or anti-c subunit antibody (green), and analyzed by epifluorescence microscopy. Merged images are shown, together with a zoom image of the insets. Arrows mark cells with overlapping fluorescence. C, HRG-1 interacts with V-ATPase holoenzyme. HEK-293T cells were transfected with HA (empty vector) or HA-HRG-1. Endogenous A subunit (A sub) was immunoprecipitated, and coprecipitation of HA-HRG-1 was examined by Western blot with anti-HA antibody. D, HRG-1 enhances V-ATPase activity in yeast. The yeast strain YPH500 was transformed with pGBK-T7-HRG-1 or pGBK-T7 vector alone. Top, vacuolar membranes were isolated, followed by Western blotting to detect subunit a (V0 domain), subunit A (V1 domain), and HRG-1. Bottom, vacuolar membranes were assayed for concanamycin A-sensitive ATPase activity (black bars) and concanamycin A-sensitive, ATP-dependent proton transport (white bars) as described in Ref. 27. Activities are expressed relative to vacuolar membranes isolated from the YPH500 strain expressing the pGBK-T7 vector alone. Error bars represent S.D. between vacuole preparations.
To investigate whether HRG-1 influences V-ATPase activity, we expressed it in yeast, which lack an HRG-1 orthologue. Vacuolar membranes purified from yeast overexpressing HRG-1 contain subunit a (V0 domain), subunit A (V1 domain), and HRG-1 (Fig. 4D). HRG-1-expressing vacuoles displayed an increase of 40% in concanamycin-sensitive ATPase activity (Fig. 4D, gray bars) and of 35% in concanamycin-sensitive proton transport compared with vector-expressing vacuoles (Fig. 4D, white bars). This demonstrates that HRG-1 is targeted to yeast vacuoles and that it enhances V-ATPase function.
HRG-1 Suppression Decreases V-ATPase Activity and Alters Endosomal pH with Consequences for Cell Survival and Transferrin Receptor Trafficking
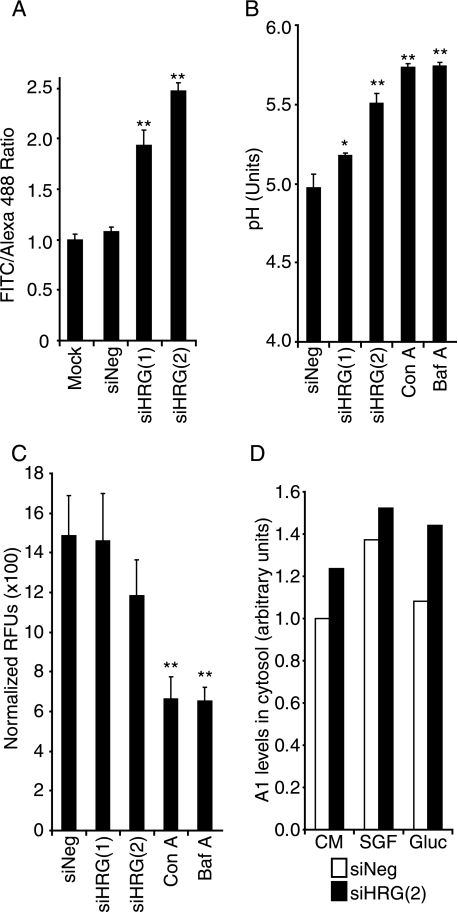
We next investigated whether HRG-1 expression is necessary for V-ATPase to function as a regulator of endosomal and lysosomal pH in mammalian cells. Early endosomal pH was assessed using transferrin-FITC (pH-dependent fluorescence) and transferrin-Alexa488 (pH-independent fluorescence) following uptake in parallel cultures for 15 min. An increase in Tf-FITC/Tf-Alexa488 fluorescence ratio indicates increased early endosomal pH, which reflects decreased V-ATPase activity. In siHRG1 and siHRG2 HeLa cells, the FITC/Alexa488 ratio was 2- and 2.5-fold higher, respectively, than in control cells (Fig. 5A), demonstrating that suppression of HRG-1 decreases the acidity of early endosomes. Average endosomal pH was calculated after uptake of FITC-dextran (FD40) for 3 h by using a calibration curve generated from cells prebuffered to defined pH values (supplemental Fig. S4). The endosomal pH of siHRG-1/HeLa cells increased from 0.2 to 0.5 pH units, comparable with the pH increase elicited by the V-ATPase inhibitors bafilomycin or concanamycin A (Fig. 5B). As HRG-1 does not colocalize with the lysosomal marker LAMP1 (Fig. 2A), we wanted to determine whether HRG-1 suppression affects lysosomal pH using the acidotropic lysosome dyes LysoTracker Red and LysoSensor Green. No significant difference in fluorescence was observed in cells with HRG-1 suppressed compared with controls (Fig. 5C and supplemental Fig. S5), which indicates that lysosomal pH is not altered by suppression of HRG-1. A similar decrease in endosomal acidity was observed in siHRG-1b/MCF-7 cells (supplemental Fig. S6).
FIGURE 5.
HRG-1 influences endosomal pH and V-ATPase assembly. A, HRG-1 suppression increases early endosome pH. Transferrin uptake was analyzed in siRNA-transfected HeLa cells by flow cytometry. The mean fluorescence of Tf-FITC (pH-sensitive) was compared with the mean fluorescence of Tf-Alexa488 (pH-insensitive), considering FITC/Alexa488 ratio in Mock cells (no oligonucleotide) as 1. Results represent one of three independent experiments with similar results. B, HRG-1 suppression increases average endosomal pH. HeLa cells were transfected as in Fig. 3A, and the endosomal pH value was estimated using FITC-dextran uptake, as described under “Materials and Methods.” The calibration curve of pH values correlated with the FL1/FL2 ratio is shown in supplemental Fig. S5. The data are presented as estimated endosomal pH values for siRNA-transfected HeLa cells. The V-ATPase inhibitors concanamycin A (Con A) and bafilomycin A (Baf A)were used as controls for V-ATPase inhibition. Error bars are S.D. for data from three independent experiments. C, HRG-1 suppression does not affect lysosomal pH. HeLa cells were transfected as in Fig. 3A. 36 h post-transfection, control cells were incubated with 250 nm LysoTracker Red for 2 h, and cell fluorescence was quantified by fluorimetry. The data are presented as relative fluorescence units (RFUs) normalized to cell numbers. Error bars are S.D. for data from three independent experiments. D, HRG-1 suppression decreases V-ATPase holoenzyme assembly. V-ATPase assembly in siRNA-transfected MCF-7 cells was assessed by quantifying cytosolic levels of A1 subunit. Cells were kept in complete media (CM), starved of serum and glucose for 4 h (SGF), or serum/glucose-starved, and then stimulated with 10 mm glucose for 15 min (Gluc). The cytosolic fraction of the cells was purified, and the presence of A1 was quantified by Western blot using actin as loading control. Data are presented as relative levels of cytosolic A1 in each of the conditions analyzed. This experiment is representative of three independent experiments with similar results.
Next we addressed the mechanism by which HRG-1 regulates V-ATPase activity. Because HRG-1 is not an integral component of the V-ATPase, we hypothesized that it may modulate V-ATPase holoenzyme assembly. V-ATPase disassembles when glucose and serum are withdrawn, and it re-assembles after glucose re-addition in a PI3K-dependent manner (12), so we asked if HRG-1 influences glucose-induced re-assembly of the V-ATPase. We quantified the amount of free cytosolic A1 subunit (V1 domain) as a measure of disassembled V-ATPase (method validation shown in supplemental Fig. S7). Cells with suppressed HRG-1 cultured in complete medium consistently had higher levels of cytosolic A1 subunit compared with controls, which indicates decreased V-ATPase assembly. Moreover, glucose re-addition did not reduce cytosolic A1 levels in serum- and glucose-starved siHRG2 cells (Fig. 5D). Therefore, glucose-induced re-assembly of the V-ATPase is impaired when HRG-1 is suppressed. Taken together, our data indicate that HRG-1 participates in V-ATPase activity regulation but only in the endocytic compartments in which HRG-1 is expressed (endosomes but not lysosomes).
Overexpression of HRG-1 Enhances TfR Internalization
Our findings that HRG-1 is expressed at high levels in transformed cells and cancer cell lines and that it enhances V-ATPase function in yeast suggest it may modulate endocytosis of receptors in mammalian cells. To test this, MCF-7 cell lines overexpressing HA-HRG-1 were assessed for endocytosis. In MCF-7/HA-HRG-1 cells total cellular levels of TfR protein were 20% higher than in controls (Fig. 6A), which suggests reduced degradation of TfR. Internalization rates of Tf-Alexa488 at 5 and 10 min were enhanced by ∼15% in HA-HRG-1 cells compared with controls, although cell surface TfR expression was equivalent (Fig. 6B). Our results indicate that HRG-1 enhances transferrin uptake via receptor endocytosis.
FIGURE 6.
HRG-1 overexpression regulates receptor trafficking. A, MCF-7 cells stably overexpressing HA-HRG-1 were assessed for cellular levels of TfR by Western blotting. The data represent the average of TfR levels from three experiments (n = 3; *, p < 0.05 using t test). B, TfR internalization was compared in MCF-7/HA and MCF-7/HA-HRG-1 cells. Cells were serum-starved for 1 h, followed by incubation at 4 °C with 25 μg/μl Tf-Alexa488 for 90 min in triplicate wells. Prewarmed media were added to the cells, which were transferred to 37 °C to induce TfR/Tf-Alexa488 internalization. At the indicated time points, cells were transferred to ice. Noninternalized ligand/receptors were removed by an acid wash, and internalized TfR/Tf-Alexa488 was quantified by FACS. The data are represented as the percentage of internalized receptor compared with cells maintained at 4 °C whereby receptor internalization is inhibited. *, p < 0.05 using t test. Panel shows relative cell surface TfR levels in MCF-7/HA and MCF-7/HA-HRG-1 cells, determined by FACS quantification of surface-bound Tf-Alexa488 as described for Fig. 3E.
HRG-1 Enhances Heme Transport in a V-ATPase-dependent Manner
After we had already characterized HRG-1 function in V-ATPase regulation, we found that the same gene was described by Rajagopal et al. (20) as a heme-binding protein (HRG-1) essential for erythropoiesis in zebrafish. Heme trafficking is a pH-dependent process, so we first investigated heme uptake in HeLa cells with suppressed HRG-1. We found that siHRG2/HeLa cells display ∼30% less uptake of the fluorescent heme analogue ZnMP than control cells (Fig. 7A). Because ZnMP fluorescence intensity depends on pH (31), we performed ZnMP uptake assays and measured ZnMP fluorescence after equilibrating the cells to pH 5.5. The proportion of change between siNeg and siHRG2 cells was the same as in cells without pH equilibration (Fig. 7B), indicating that the differences in ZnMP fluorescence in siHRG2 cells were due to decreased uptake and were not an artifact due to altered endosomal pH.
FIGURE 7.
HRG-1 enhances heme uptake in a V-ATPase-dependent manner. A, HRG-1 suppression reduces heme uptake. ZnMP uptake was measured by flow cytometry in siRNA-transfected HeLa cells 36 h post-transfection. The data are presented as the percentage of ZnMP uptake relative to control siNeg transfectants (100%). B, cells were transfected and treated as in A, but the cells were either resuspended in an isotonic buffer (HBSS) or in a pH-equilibrating buffer to adjust intracellular pH to 5.5 prior to flow cytometry analysis of ZnMP uptake. The data are presented as the percentage of ZnMP uptake relative to control siNeg transfectants (100%). C, HRG-1 overexpression enhances heme uptake. ZnMP uptake was measured by flow cytometry in MCF-7/HA-HRG-1 cells. The data are presented as the percentage of ZnMP uptake relative to control HA empty vector cells (100%). D, HRG-1 enhancement of heme uptake depends on V-ATPase activity. ZnMP uptake was measured by flow cytometry in MCF-7/HA-HRG-1 cells pretreated with vehicle (DMSO) or 100 nm bafilomycin A (Baf A). The data are presented as the percentage of ZnMP uptake relative to control HA empty vector cells (100%). In all panels, error bars are S.D. for data from three independent experiments. *, p < 0.05 using t test; **, p < 0.01 using t test. E, HA-HRG-1 binds to hemin-agarose at an acidic pH. MCF-7/HA-HRG-1 cell lysates were incubated with either Sepharose 4B as negative control (−) or hemin-agarose (H) at different pH values, and the bound proteins were analyzed by immunoblotting. Top panel shows one-tenth of inputs used in the assay as loading controls. Bottom panel shows HA-HRG-1 and endogenous HRG-1 bound to hemin-agarose detected using monoclonal anti-HRG-1 antibody.
We also assessed ZnMP uptake in cells overexpressing MCF-7/HA-HRG-1. These cells exhibited nearly 2-fold increases in ZnMP uptake compared with control cells (Fig. 7C), which is in agreement with published data for HRG proteins (20). To determine whether the increase in ZnMP uptake in HA-HRG-1 cells was linked with the effects of HRG-1 on V-ATPase activity, the ZnMP uptake assays were performed in the presence of the V-ATPase inhibitor bafilomycin A. In untreated cells, HA-HRG-1 expression increased ZnMP uptake by 2-fold, but pretreatment with bafilomycin A reduced this substantially (Fig. 7D). These results indicate that function of HRG-1 in promoting heme transport is dependent on V-ATPase activity.
We next investigated how the subcellular location and trafficking of HRG-1 may affect heme binding. HRG-1 binding to heme was previously shown to be stronger at pH 6.0 than at pH 8.5, whereas binding of heme to other HRG proteins was pH-independent (20). We performed hemin-agarose pulldown assays at pH 5.0, 6.0, and 7.5, representing average pH in late endosomes, early/recycling endosomes, and extracellular media, respectively. We found that both HA-HRG-1 and endogenous HRG-1 bound the strongest to heme at pH 5.0 (Fig. 7E), which is consistent with a role for HRG-1 in heme transport in late endosomes rather than at the cell surface.
DISCUSSION
There is considerable evidence to indicate that the endocytic machinery and trafficking of nutrients is de-regulated in cancer cells to support the transformed phenotype (reviewed in Refs. 6, 8). Here, we have shown that the endosomal protein HRG-1/SLC48A1 is a new IGF-I-regulated protein that may enhance the proliferative and invasive phenotype of cancer cells through its effects on endosomal acidification and receptor trafficking. HRG-1 was also previously described as a heme-binding protein (20) with conserved function. Thus, HRG-1 has a unique role in regulating pH-dependent endocytic trafficking and micronutrient uptake necessary for cellular metabolism. We initially identified HRG-1 because it had increased expression in cells transformed due to IGF-IR overexpression. As IGF-I signaling facilitates the growth, migration, and invasiveness of many tumors (32–34), we propose that HRG-1 is a micro-nutrient (heme) transporter that may enhance tumor growth and cancer progression. When HRG-1 is suppressed, V-ATPase activity is decreased; endosomal acidity is reduced, and receptor endocytosis is impaired. In contrast, overexpression of HRG-1 enhances receptor endocytosis. Thus, increased HRG-1 expression could have a profound effect on the growth and migratory potential of cancer cells by regulating the endocytosis and degradation of receptors that promote signaling for survival, growth, and migration.
The tissue distribution of HRG-1 mRNA (high in liver and kidney) is consistent with a dual role for HRG-1 in regulating V-ATPase activity as well as in heme intracellular transport. Both liver and kidney are known sites of heme transport, and HRG-1, HCP-1, and ABCB6 (other heme transporters) mRNAs are all expressed in those tissues (35, 36). In the kidney, the V-ATPase also plays a major role in acidification and bicarbonate absorption by nephrons (37).
The HRG protein family has four orthologues (HRG1–4) in C. elegans with different cellular locations and abilities to transport heme (20). It is interesting that HRG-1 is not present in yeast and that only the HRG-1 orthologue is present in mammalian cells. This suggests that the protein may have evolved for a broader function in mammalian cells involving the integration of heme transport with endosome acidification. Indeed, we have shown that HRG-1 not only binds heme and regulates its uptake, but we also have shown that HRG-1 has a direct effect on V-ATPase activity and glucose-induced re-assembly of the holoenzyme. Moreover, the dynamic interaction observed with the c subunit and trafficking to the plasma membrane suggests a role for HRG-1 in assembly or targeting of the V0 domain to specific membranes. Regulation of V-ATPase assembly is not completely understood but has previously been shown to involve several different proteins, including the protein complex RAVE that regulates assembly of the V1 domain in yeast (13), and chaperones for V0 domain assembly (9).
Our observations that HRG-1 is associated with early and late endosomes but not lysosomes and that HRG-1 suppression impairs endosome but not lysosome acidification support the specific role proposed by others for V-ATPase activity in trafficking from early to late endosomes (29). Inhibition of V-ATPase with bafilomycin or loss of a pH-dependent interaction of the V-ATPase with Arf6 and ARNO can specifically impair trafficking from early to late endosomes, which subsequently leads to impaired endocytosis (29). Here, we have shown that the defects in V-ATPase activity due to HRG-1 suppression also lead to impaired TfR endocytosis. The fact that HRG-1 is trafficked to the plasma membrane also suggests a possible role in chaperoning heme or transferrin-associated iron uptake by cells. This is supported by our observations on ZnMP uptake and TfR trafficking in cells with HRG-1 suppressed or overexpressed.
As HRG-1 regulates endosomal pH, it also has the potential to influence in a number of ways iron and heme availability within cells. HRG-1 suppression may diminish the internalization of iron through the TfR, and it could also affect the release of endocytosed iron from transferrin due to an increased endosomal pH. As a result, there would be a reduced iron supply for de novo heme biosynthesis. Although HRG-1 is present at the cell surface, it binds heme more efficiently at low pH than at pH 7.5, so it is not likely to transport heme directly across the plasma membrane (our data and see Ref. 20) but rather to mediate transport inside late endosomes. The increase in ZnMP uptake observed in cells overexpressing HRG-1 may therefore be due to enhanced internalization and/or trafficking of a plasma membrane heme transporter, which would rely on efficient V-ATPase activity.
Our findings indicate that the subcellular distribution of HRG-1 is slightly different from that reported previously (20). We found that HRG-1 is located at the plasma membrane and throughout the endosomal compartments but not in lysosomes, whereas Rajagopal et al. (20) described HRG-1 as a lysosomal protein with no apparent localization at the cell surface. These different observations may be related to the expression constructs and epitope tags used. We detected endogenous protein where possible and used an expression construct with an N-terminal HA tag, whereas the other study used a C-terminal cyan fluorescent protein fusion. We also assessed colocalization in lysosomes with endogenous LAMP-1 protein rather than overexpressed LAMP-1. Our observation that cells with reduced HRG-1 exhibit impaired acidification of endosomes but not lysosomes is also consistent with the lack of expression in lysosomes.
In summary, we propose that HRG-1-mediated regulation of endosomal pH through the V-ATPase is essential for function of the endocytic pathway. This in turn will have an impact on the ability of the cells to acquire nutrients, to mediate signaling in response to growth factor receptor activation, and to internalize and traffic integrins and other proteins, which is necessary for cell survival, migration, and proliferation.
Supplementary Material
Acknowledgments
We are grateful to Prof. Mary McCaffrey and research group for advice on membrane trafficking, to our colleagues in the Cell Biology Laboratory for helpful discussions, and to Kurt Tidmore for assistance with illustrations.
This work was supported by Science Foundation Ireland, the Irish Research Council for Science, Engineering, and Technology, and the Health Research Board.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
- IGF-I
- insulin-like growth factor I
- IGF-IR
- IGF-I receptor
- V-ATPase
- vacuolar H+-ATPase
- siRNA
- small interfering RNA
- Tf
- transferrin
- TfR
- transferrin receptor
- HA
- hemagglutinin
- PI3K
- phosphatidylinositol 3-kinase
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- BSA
- bovine serum albumin
- PBS
- phosphate-buffered saline
- FITC
- fluorescein isothiocyanate
- FACS
- fluorescence-activated cell sorter
- HBSS
- Hanks' balanced salt solution
- PIPES
- 1,4-piperazinediethanesulfonic acid
- ZnMP
- zinc mesoprotoporphyrin IX.
REFERENCES
- 1.Cheng K. W., Lahad J. P., Kuo W. L., Lapuk A., Yamada K., Auersperg N., Liu J., Smith-McCune K., Lu K. H., Fishman D., Gray J. W., Mills G. B. (2004) Nat. Med. 10, 1251–1256 [DOI] [PubMed] [Google Scholar]
- 2.Wang W., Goswami S., Lapidus K., Wells A. L., Wyckoff J. B., Sahai E., Singer R. H., Segall J. E., Condeelis J. S. (2004) Cancer Res. 64, 8585–8594 [DOI] [PubMed] [Google Scholar]
- 3.Caswell P. T., Norman J. C. (2006) Traffic 7, 14–21 [DOI] [PubMed] [Google Scholar]
- 4.Miaczynska M., Pelkmans L., Zerial M. (2004) Curr. Opin. Cell Biol. 16, 400–406 [DOI] [PubMed] [Google Scholar]
- 5.Caswell P., Norman J. (2008) Trends Cell Biol. 18, 257–263 [DOI] [PubMed] [Google Scholar]
- 6.Mosesson Y., Mills G. B., Yarden Y. (2008) Nat. Rev. Cancer 8, 835–850 [DOI] [PubMed] [Google Scholar]
- 7.Edinger A. L. (2007) Biochem. J. 406, 1–12 [DOI] [PubMed] [Google Scholar]
- 8.Ganapathy V., Thangaraju M., Prasad P. D. (2009) Pharmacol. Ther. 121, 29–40 [DOI] [PubMed] [Google Scholar]
- 9.Forgac M. (2007) Nat. Rev. Mol. Cell Biol. 8, 917–929 [DOI] [PubMed] [Google Scholar]
- 10.Marshansky V., Futai M. (2008) Curr. Opin. Cell Biol. 20, 415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lafourcade C., Sobo K., Kieffer-Jaquinod S., Garin J., van der Goot F. G. (2008) PLoS ONE 3, e2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sautin Y. Y., Lu M., Gaugler A., Zhang L., Gluck S. L. (2005) Mol. Cell Biol. 25, 575–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smardon A. M., Kane P. M. (2007) J. Biol. Chem. 282, 26185–26194 [DOI] [PubMed] [Google Scholar]
- 14.Hinton A., Sennoune S. R., Bond S., Fang M., Reuveni M., Sahagian G. G., Jay D., Martinez-Zaguilan R., Forgac M. (2009) J. Biol. Chem. 284, 16400–16408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez-Zaguilan R., Lynch R. M., Martinez G. M., Gillies R. J. (1993) Am. J. Physiol. 265, C1015–C1029 [DOI] [PubMed] [Google Scholar]
- 16.Goldstein D. J., Finbow M. E., Andresson T., McLean P., Smith K., Bubb V., Schlegel R. (1991) Nature 352, 347–349 [DOI] [PubMed] [Google Scholar]
- 17.Loughran G., Healy N. C., Kiely P. A., Huigsloot M., Kedersha N. L., O'Connor R. (2005) Mol. Biol. Cell 16, 1811–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loughran G., Huigsloot M., Kiely P. A., Smith L. M., Floyd S., Ayllon V., O'Connor R. (2005) Oncogene 24, 6185–6193 [DOI] [PubMed] [Google Scholar]
- 19.Sell C., Dumenil G., Deveaud C., Miura M., Coppola D., DeAngelis T., Rubin R., Efstratiadis A., Baserga R. (1994) Mol. Cell Biol. 14, 3604–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rajagopal A., Rao A. U., Amigo J., Tian M., Upadhyay S. K., Hall C., Uhm S., Mathew M. K., Fleming M. D., Paw B. H., Krause M., Hamza I. (2008) Nature 453, 1127–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamza I. (2006) ACS Chem. Biol. 1, 627–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krishnamurthy P., Xie T., Schuetz J. D. (2007) Pharmacol. Ther. 114, 345–358 [DOI] [PubMed] [Google Scholar]
- 23.West A. R., Oates P. S. (2008) J. Gastroenterol. Hepatol. 23, 150–158 [DOI] [PubMed] [Google Scholar]
- 24.Arai H., Berne M., Terres G., Terres H., Puopolo K., Forgac M. (1987) Biochemistry 26, 6632–6638 [DOI] [PubMed] [Google Scholar]
- 25.Arai H., Berne M., Forgac M. (1987) J. Biol. Chem. 262, 11006–11011 [PubMed] [Google Scholar]
- 26.Nilsson C., Kågedal K., Johansson U., Ollinger K. (2003) Methods Cell Sci. 25, 185–194 [DOI] [PubMed] [Google Scholar]
- 27.Vasilyeva E., Forgac M. (1996) J. Biol. Chem. 271, 12775–12782 [DOI] [PubMed] [Google Scholar]
- 28.Shen H., Chou J. J. (2008) PLoS ONE 3, e2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hurtado-Lorenzo A., Skinner M., El Annan J., Futai M., Sun-Wada G. H., Bourgoin S., Casanova J., Wildeman A., Bechoua S., Ausiello D. A., Brown D., Marshansky V. (2006) Nat. Cell Biol. 8, 124–136 [DOI] [PubMed] [Google Scholar]
- 30.Lee I., Skinner M. A., Guo H. B., Sujan A., Pierce M. (2004) J. Biol. Chem. 279, 53007–53014 [DOI] [PubMed] [Google Scholar]
- 31.Kepczyński M., Ehrenberg B. (2002) Photochem. Photobiol. 76, 486–492 [DOI] [PubMed] [Google Scholar]
- 32.Lopez T., Hanahan D. (2002) Cancer Cell 1, 339–353 [DOI] [PubMed] [Google Scholar]
- 33.Mauro L., Salerno M., Morelli C., Boterberg T., Bracke M. E., Surmacz E. (2003) J. Cell Physiol. 194, 108–116 [DOI] [PubMed] [Google Scholar]
- 34.Samani A. A., Yakar S., LeRoith D., Brodt P. (2007) Endocr. Rev. 28, 20–47 [DOI] [PubMed] [Google Scholar]
- 35.Krishnamurthy P. C., Du G., Fukuda Y., Sun D., Sampath J., Mercer K. E., Wang J., Sosa-Pineda B., Murti K. G., Schuetz J. D. (2006) Nature 443, 586–589 [DOI] [PubMed] [Google Scholar]
- 36.Shayeghi M., Latunde-Dada G. O., Oakhill J. S., Laftah A. H., Takeuchi K., Halliday N., Khan Y., Warley A., McCann F. E., Hider R. C., Frazer D. M., Anderson G. J., Vulpe C. D., Simpson R. J., McKie A. T. (2005) Cell 122, 789–801 [DOI] [PubMed] [Google Scholar]
- 37.Nakhoul N. L., Hamm L. L. (2002) J. Nephrol. 15, Suppl, 5, S22–S31 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.