Abstract
To confirm and fine map previous reports of association, the Type I Diabetes (T1D) Genetics Consortium (T1DGC) assembled a large collection of DNA samples from affected sib-pair (ASP) families with T1D (5003 affected individuals) and genotyped polymorphic markers. One of these loci, involving the IL2RA gene, had been reported to be due to three independent effects. The T1DGC genotyped 69 single-nucleotide polymorphisms (SNPs) that span ~ 88 kb from the 5′ flanking to 3′ flanking region of the IL2RA locus. The most highly associated SNP reported earlier (ss52580101) was not included in the genotyping list; however, a 5-SNP (rs3134883, rs3118470, rs7072793, rs4749955 and rs12251307) haplotype (H5) was identified that strongly tagged its minor allele with r2 = 0.869 (95% CI, 0.850–0.885). This haplotype was significantly protective (P = 3.2 × 10−5) in the T1D ASP families, with an odds ratio virtually identical to that reported for ss52580101. The SNP marking the second independent locus, (rs11594656) showed no association in the T1DGC set and the third (rs2104286) could not be distinguished, by conditional regression, from H5. Instead, the most significant independent effect was detected from the 5′ flanking IL2RA SNP rs4749955, which remained significant after regression for H5. Thus, we confirm independent effects at the IL2RA locus.
Keywords: autoimmune disease, genetic susceptibility, IL2RA, linkage disequilibrium, single-nucleotide polymorphism, type I diabetes
Introduction
IL2RA encodes the α chain of the interleukin-2 (IL2) receptor complex, also known as CD25. IL2 is a powerful growth factor for lymphocytes1 and IL2Rα has a key function in mediating its effect in the form of a quaternary complex with IL2, IL2Rβ and γc.2 The expression of IL2RA is an important marker of CD4+CD25+ regulatory T cells (Treg), known to be crucial in maintaining immune homeostasis and inhibiting autoimmune disease.3,4
Type I diabetes (T1D) is due to T-cell-mediated autoimmune destruction of pancreatic β cells. An association of T1D with the IL2RA locus was first reported using a multilocus genetic association test.5 This association was replicated independently in a dataset of 949 European-descent nuclear family trios from Canada.6 Two intronic single-nucleotide polymorphisms (SNPs) in the 5′ region of the IL2RA gene, rs706778 and rs3118470, exhibited highly significant association with T1D. Recently, by a large-scale fine mapping of the IL2RA region based on a case–control sample from the UK, it was reported that the most significantly associated SNP was ss52580101 (rs41295061); this SNP also maps to the 5′ flanking region of IL2RA (−10 388 C/A).7 Further analysis identified an SNP (rs11594656) with an independent effect on risk that also is located in the 5′ flanking region of IL2RA.7
To evaluate the role of polymorphisms in the IL2RA gene on T1D risk, the Type I Diabetes Genetics Consortium (T1DGC) assembled a large collection of DNA samples from affected sib-pair (ASP) families with T1D and genotyped polymorphic markers. In this report, we analyze the T1DGC ASP families, and incorporate data of a CEU (Centre de l’Etude du Polymorphisme Humain (CEPH) Utah residents with ancestry from northern and western Europe) set of European CEPH individuals and a Canadian case–parent trio panel to provide the haplotype information. The purpose of this study was to confirm the IL2RA association with T1D and contribute to its fine mapping.
Results
A total of 69 SNPs in IL2RA were genotyped by the T1DGC in 2298 ASP families. The family collection contained 11159 individuals, 5003 of whom were affected. The samples were collected through an international effort, and most subjects were of European ancestry. The 69 SNPs spanned ~ 88 kb from the 5′ flanking to 3′ flanking region of the IL2RA gene and captured 93.7% of HapMap SNPs with minor allele frequency (MAF) ≥0.05 at r2>0.8 in the IL2RA region, and 45% of HapMap SNPs with 0.01 ≤MAF <0.05 (Supplementary Figure 1). The major T1D association (ss52580101) that was recently reported was not tagged by any genotyped SNP. The second independent T1D susceptibility marker (rs11594656) was included in the T1DGC SNPs.
To determine whether ss52580101 may be tagged by a haplotype, this SNP was genotyped in a CEU panel of 86 European CEPH individuals used in the HapMap project and in a Canadian case–parent trio collection from our previous mapping of IL2RA.6 Of the 69 SNPs proposed for genotyping, 66 SNPs were successfully genotyped on the Illumina platform and 65 SNPs were genotyped on the Sequenom platform. There were 62 SNPs genotyped by both platforms. Of the 11159 individuals from 2298 families, 1878 (16.8%) individuals had no Illumina genotyping data for any locus and 1799 (16.1%) individuals had no genotyping data by Sequenom. These were treated as missing samples. As shown in Supplementary Table 1, all SNPs genotyped by the Illumina and/or Sequenom platform had good call rates with a minimum 94.3%.
The genotype distribution of each Illumina SNP met Hardy–Weinberg Equilibrium assumptions; however, 8 of the 65 SNPs genotyped on the Sequenom platform had genotype distributions that failed to meet Hardy–Weinberg Equilibrium expectations. For the 62 SNPs genotyped by both methods, the concordance rates were high, with the exception of rs12722486 (concordance rate of 73.1%). For this SNP, the Sequenom genotypes were not in Hardy–Weinberg Equilibrium, suggesting that the discrepancy may be due to poor Sequenom genotyping quality or difficulty in the calling algorithm to assign genotypes for this marker.
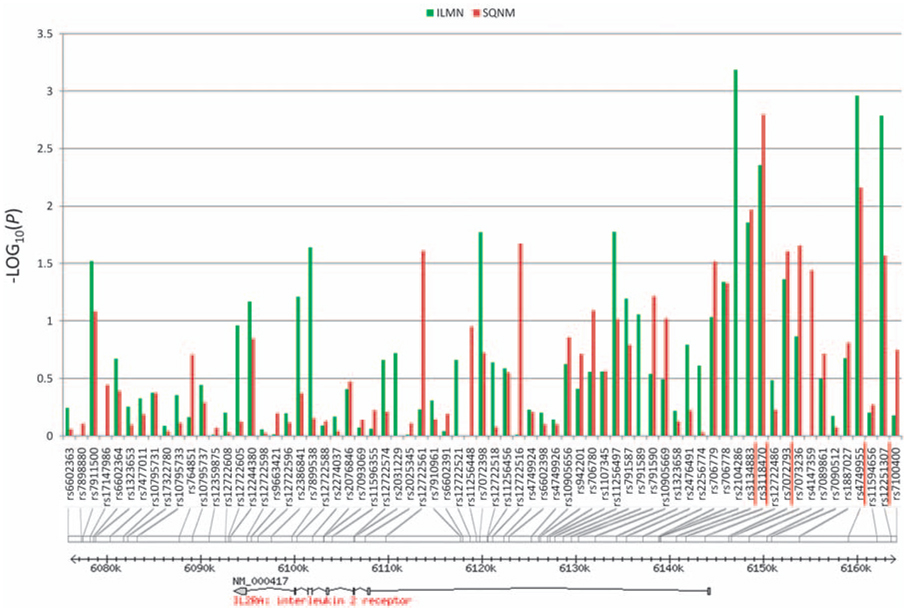
The T1D association with IL2RA SNPs is shown in Figure 1 (and Supplementary Table 1). The strongest T1D association occurs in SNPs at the 5′ end of the gene and flanking region. The linkage disequilibrium (LD) analysis of the T1D-associated SNPs is shown in Supplementary Figure 2. The most strongly associated marker reported earlier is ss52580101,7 but this was not genotyped in the T1DGC collection. Thus, ss52580101 was genotyped by our group in the 86 HapMap CEU collection and in 373 Canadian family trios to look for a tag haplotype. Five SNPs were identified (rs3134883, rs3118470, rs7072793, rs4749955 and rs12251307) in which the haplotype G-T-T-C-A was in perfect LD with the minor allele of ss52580101 (r2 = 1) in CEU. An expectation-maximization algorithm by the partition/ligation method8 was used for the haplotype identification and validated by phase reconstruction using PHASE.9 From the 120 CEU chromosomes (30 CEU pairs of parents), all the 8 chromosomes with the minor allele of ss52580101 have the haplotype G-T-T-C-A. Considering the rare allele frequency of ss52580101 and the relative small sample size of the HapMap CEU samples (n = 60), a subset of Canadian family trios was further investigated by phase reconstruction from our own genome-wide association scan data.10 By testing the LD in 373 complete family trios of European descent, the G-T-T-C-A was confirmed to be in strong LD with the minor allele of ss52580101 (r2 = 0.869, 95% CI 0.850–0.885).
Figure 1.
The 69 IL2RA SNPs for the T1D association assay. The five SNPs tagging the T1D-associated haplotype are red underlined. The peak of T1D association is from the 5′ end of the gene and flanking region.
As seen from the haplotype analysis of the T1DGC data (Table 1), the highest significance is from the protective haplotype H5, which is in perfect LD with the minor allele of ss52580101, and, in the Illumina genotyping data, has an estimated OR=0.699 (95% CI 0.602, 0.811). This result is extremely close to the effect size of ss52580101 in the previous family-based study (rapid response (RR) = 0.70 (95% CI 0.61–0.81)).7 Thus, the T1D association from ss52580101 reported earlier7 is confirmed by the T1DGC data. In contrast, the additional effect from rs11594656 SNP7 was not replicated by the T1DGC data. Neither the Illumina nor the Sequenom genotyping data show tendency of T1D association with this marker (Supplementary Table 1). For the T1D association of rs11594656, OR = 0.84 as reported earlier,7 the T1DGC has greater than 95% power to detect it at α = 0.05 level, assuming a multiplicative effect model with MAF = 0.252. Thus, there is failure to replicate this independent IL2RA SNP.
Table 1.
Haplotypic association analysis
| Haplotypea | Alias | Frequency | Illumina T:U | Illumina P value | Sequenom T:U | Sequenom P value |
|---|---|---|---|---|---|---|
| A-C-C-T-G | H1 | 0.309 | 1373:1244 | 0.012 | 1392:1264 | 0.013 |
| G-T-T-T-G | H2 | 0.249 | 1170:1147 | 0.634 | 1185:1177 | 0.875 |
| G-T-T-C-G | H3 | 0.237 | 1061:1077 | 0.732 | 1074:1114 | 0.386 |
| G-T-C-C-G | H4 | 0.090 | 482:533 | 0.112 | 479:539 | 0.057 |
| G-T-T-C-A | H5 | 0.067 | 320:432 | 4.41 × 10−5 | 331:415 | 2.10 × 10−3 |
| G-C-C-C-A | H6 | 0.018 | 109:103 | 0.680 | 115:95 | 0.173 |
| G-T-C-T-G | H7 | 0.016 | 102:73 | 0.026 | 104:74 | 0.025 |
The haplotype of five SNPs is rs3134883, rs3118470, rs7072793, rs4749955 and rs12251307. The under-transmitted H5 haplotype G-T-T-C-A is 1in strong linkage disequilibrium with the minor allele of ss52580101.
The haplotypic association was done based on the T1DGC data using the Haploview v4.0 software (www.broad.mit.edu/personal/jcbarret/haploview).
To search of other additional and independent IL2RA SNP effects, six T1D-associated SNPs with P < 0.05 in both the Illumina and the Sequenom data were examined, plus the most associated SNP (rs2104286) by the Illumina data. The haplotypes were phased using the expectation-maximization algorithm and transmission distortion of SNPs conditioning on the H5 haplotype was determined by the conditional extended TDT method.11 By the conditional analysis, an additional SNP effect was identified from the IL2RA 5′ flanking region. The SNP with greatest significance (P = 9.90 × 10−4) after regression for H5 was rs4749955 (Table 2).
Table 2.
Transmission of the T1D-associated SNPs conditioning on the H5 haplotype
| SNP | MAF | T1D association P |
P value conditional on the H5 haplotype |
|---|---|---|---|
| rs706778 | 0.439 (T) | 0.046 | 0.728 |
| rs2104286 | 0.231 (C) | 6.56 × 10−4 | 0.552 |
| rs3134883 | 0.322 (A) | 0.014 | 4.29 × 10−3 |
| rs3118470 | 0.342 (C) | 4.43 × 10−3 | 3.37 × 10−3 |
| rs7072793 | 0.443 (C) | 0.043 | 3.93 × 10−3 |
| rs4749955 | 0.426 (C) | 1.10 × 10−3 | 9.90 × 10−4 |
| rs12251307 | 0.099 (A) | 1.64 × 10−3 | 1.35 × 10−3 |
Abbreviations: MAF, minor allele frequency; SNP, single-nucleotide polymorphism; T1D, type I diabetes.
Results are based on the Illumina genotyping data. The conditioning analysis was done using the conditional extended TDT method,11 implemented in the UNPHASED software package (http://www.mrc-bsu.cam.ac.uk/personal/frank/software/unphased/).13 As shown by the conditioning analysis, five SNPs remain significant after conditioning on the H5 haplotype.
Discussion
The T1DGC data confirmed a substantial and statistically significant association (OR = 0.7) from the relatively uncommon ss52580101 SNP. This result is the same magnitude in the T1DGC as in the earlier published7 UK family-based dataset. An additional independent effect was observed but could not be mapped to the earlier reported IL2RA SNP, rs11594656.
A recently reported independent effect tagged by rs2104286 (the Group III SNPs)12 was not replicated either. Significant T1D association of rs2104286 was seen in the T1DGC data but when conditioned on H5, it lost significance (P = 0.552). Conversely, H5 retained significance when conditioned on rs2104286 (P = 6.14 × 10−4). For the independent effect of rs2104286, OR = 0.86 as reported,12 the T1DGC has greater than 95% power to detect it at α = 0.05 level, assuming a multiplicative effect model with MAF = 0.231. T1D association of rs2104286 was seen in the T1DGC data, but the effect can be explained by the LD with H5.
The best currently known marker for the independent and additional effect of IL2RA on T1D risk is rs4749955. The reason for the discrepancy in the second effect between the T1DGC data and the UK data7 is unclear. Both studies were adequately powered and genotyping error is unlikely, as rs11594656 had good call rate in both Illumina and Sequenom platforms. The results were highly consistent in both genotyping protocols. A cohort difference in allele frequencies or LD patterns is a possible explanation. Alternatively, population stratification in the UK case–control data could account for the difference, as the family-based T1DGC data are impervious to stratification. Further fine mapping in additional cohorts will be needed to define this effect, which illustrates the complexity of mechanisms underlying susceptibility to complex traits.
Functionally, the fine mapping of the T1D association to independent effects, all from the 5′ flanking region of IL2RA, suggests a regulatory effect. This region is also 5′ flanking to the neighboring gene RBM17, which could be the gene involved, a question that cannot be answered by further genetic dissection and requires functional studies.
Materials and methods
Subjects
The T1DGC collected DNA from 2298 ASP families containing 11 159 individuals (5003 affected) as part of an experiment to examine published reports of candidate gene associations with T1D. The samples were collected through an international effort, and most subjects were of European ancestry. Details of the samples and quality control and data collection can be found in the report of Brown et al.,13 this volume. For the haplotype analysis of the T1DGC data, the ss52580101 SNP was genotyped in a CEU set (86 European CEPH individuals in the HapMap project) and in a Canadian case–parent trio collection.
Genotyping
A total of 69 IL2RA tagging SNPs were genotyped by the T1DGC ASP family collection on two genotyping platforms (Illumina GoldenGate and Sequenom). For the haplotype analysis of the T1DGC data, the ss52580101 SNP was genotyped in an 86 individual CEU set and in a Canadian case–parent trio collection using the Acyclo-Prime-FP SNP Detection kit (PerkinElmer Inc., Boston, MA, USA). For the ss52580101, the call rate was 100% and Mendelian error rate was 0.1%. In the Canadian family trios, the phase reconstruction was based on our own genome-wide association scan data10 that was generated using the Illumina Infinium II HumanHap550 BeadChip technology (Illumina, Inc., San Diego, CA, USA).
Statistical analysis
Of the 11159 individuals from 2298 families, 1878 (16.8%) individuals had no Illumina genotyping data for any locus and 1799 (16.1%) individuals had no genotyping data by Sequenom. These were treated as missing samples and were not taken into account for the genotyping quality assessment and analyses of data from each platform. The overlap of missing data in the two platforms was 1477 samples. There were 34 non-Caucasian families (162 individuals) in the T1DGC collection; these were not included in the haplotype analysis.
T1D association was tested by the Family Based Association Test (FBAT) (http://www.biostat.harvard.edu/~fbat/fbat.htm).14 For haplotype identification, an expectation-maximization algorithm by the partition/ligation method8 was used, as implemented in the Haploview software (www.broad.mit.edu/personal/jcbarret/haploview). The results were validated using the PHASE program9 (http://stephenslab.uchicago.edu/software.html) by the method of phase reconstruction. To search of other additional and independent IL2RA SNP effects, haplotypes were phased using the expectation-maximization algorithm implemented in the PLINK software (http://pngu.mgh.harvard.edu/~purcell/plink/index.shtml). Transmission distortion of these SNPs conditioning on the H5 haplotype was investigated by the conditional extended TDT method,11 implemented in the UNPHASED software package (http://www.mrc-bsu.cam.ac.uk/personal/frank/software/unphased/).15
Supplementary Material
Acknowledgements
The Type I Diabetes Genetics Consortium (T1DGC) is funded by the NIH Grant U01-DK62418. This study was additionally funded by the Juvenile Diabetes Research Foundation International, Genome Canada and the Children’s Hospital of Philadelphia. We thank all the T1D families for their participation in the study. HQQ is supported by a fellowship from the Canadian Institutes of Health Research. Special thanks for helpful discussion and comments from Joanna Howson. Genotyping was performed at the Broad Institute Center for Genotyping and Analysis is supported by grant U54 RR020278 from the National Center for Research Resources.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Lowenthal JW, Zubler RH, Nabholz M, MacDonald HR. Similarities between interleukin-2 receptor number and affinity on activated B and T lymphocytes. Nature. 1985;315:669–672. doi: 10.1038/315669a0. [DOI] [PubMed] [Google Scholar]
- 2.Wang X, Rickert M, Garcia KC. Structure of the quaternary complex of interleukin-2 with its alpha, beta, and gammac receptors. Science. 2005;310:1159–1163. doi: 10.1126/science.1117893. [DOI] [PubMed] [Google Scholar]
- 3.Shevach EM. Certified professionals: CD4(+)CD25(+) suppressor T cells. J Exp Med. 2001;193:F41–F46. doi: 10.1084/jem.193.11.f41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willerford DM, Chen J, Ferry JA, Davidson L, Ma A, Alt FW. Interleukin-2 receptor [alpha] chain regulates the size and content of the peripheral lymphoid compartment. Immunity. 1995;3:521. doi: 10.1016/1074-7613(95)90180-9. [DOI] [PubMed] [Google Scholar]
- 5.Vella A, Cooper JD, Lowe CE, Walker N, Nutland S, Widmer B, et al. Localization of a type 1 diabetes locus in the IL2RA/CD25 region by use of tag single-nucleotide polymorphisms. Am J Hum Genet. 2005;76:773–779. doi: 10.1086/429843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qu H-Q, Montpetit A, Ge B, Hudson TJ, Polychronakos C. Toward further mapping of the association between the IL2RA locus and type 1 diabetes. Diabetes. 2007;56:1174–1176. doi: 10.2337/db06-1555. [DOI] [PubMed] [Google Scholar]
- 7.Lowe CE, Cooper JD, Brusko T, Walker NM, Smyth DJ, Bailey R, et al. Large-scale genetic fine mapping and genotype-phenotype associations implicate polymorphism in the IL2RA region in type 1 diabetes. Nat Genet. 2007;39:1074–1082. doi: 10.1038/ng2102. [DOI] [PubMed] [Google Scholar]
- 8.Qin ZS, Niu T, Liu JS. Partition-ligation-expectation-maximization algorithm for haplotype inference with single-nucleotide polymorphisms. Am J Hum Genet. 2002;71:1242–1247. doi: 10.1086/344207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hakonarson H, Grant SF, Bradfield JP, Marchand L, Kim CE, Glessner JT, et al. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007;448:591–594. doi: 10.1038/nature06010. [DOI] [PubMed] [Google Scholar]
- 11.Koeleman BPC, Dudbridge F, Cordell HJ, Todd JA. Adaptation of the extended transmission/disequilibrium test to distinguish disease associations of multiple loci: the Conditional Extended Transmission/Disequilibrium Test. Ann Hum Genet. 2000;64:207–213. doi: 10.1017/S0003480000008095. [DOI] [PubMed] [Google Scholar]
- 12.Maier LM, Lowe CE, Cooper J, Downes K, Anderson DE, Severson C, et al. IL2RA genetic heterogeneity in multiple sclerosis and type 1 diabetes susceptibility and soluble interleukin-2 receptor production. PLoS Genet. 2009;5:e1000322. doi: 10.1371/journal.pgen.1000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown WM, Pierce JJ, Hilner JE, Perdue LH, Lohman K, Lu L, et al. the Type I Diabetes Genetics Consortium. Overview of the Rapid Response data. Genes Immun. 2009;10 Suppl 1:S5–S15. doi: 10.1038/gene.2009.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype—phenotype associations. Eur J Hum Genet. 2001;9:301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- 15.Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol. 2003;25:115–121. doi: 10.1002/gepi.10252. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.