Abstract
The clinical efficacy of carbapenem antibiotics depends on their resistance to the hydrolytic action of β-lactamase enzymes. The structure of the class D β-lactamase OXA-1 as an acyl-complex with the carbapenem doripenem was determined to 1.4 Å resolution. Unlike most class A and class C carbapenem complexes, the acyl carbonyl oxygen in OXA-1/doripenem is bound in the oxyanion hole. Interestingly, no water molecules were observed in the vicinity of the acyl linkage, providing an explanation for why carbapenems inhibit OXA-1. The side-chain amine of K70 remains fully carboxylated in the acyl structure, and the resulting carbamate group hydrogen bonds to the alcohol of the 6α-hydroxyethyl moiety of doripenem. The carboxylate attached to the β-lactam ring of doripenem is stabilized by a salt-bridge to K212 and a hydrogen bond with T213, in lieu of the interaction with an arginine side-chain found in most other β-lactamase/β-lactam complexes (eg. R244 in the class A member TEM-1). This novel set of interactions with the carboxylate results in a major shift of the carbapenem's pyrroline ring compared to the structure of the same ring in meropenem bound to OXA-13. Additionally, bond angles of the pyrroline ring suggest that after acylation, doripenem adopts the Δ 1 tautomer. These findings provide important insights into the role that carbapenems may have in the inactivation process of class D β-lactamases.
Carbapenems are broad spectrum β-lactam antibiotics that represent one of the last lines of defense against severe bacterial infections. Doripenem (Figure 1A) was approved for use in the United States in 2007 with indications for urinary tract infections and complicated intra-abdominal infections (1). Though a broad spectrum antibiotic, it has shown higher efficacy than imipenem and meropenem in troublesome Gram-negative species such as Pseudomonas aeruginosa and Acinetobacter baumannii (1).
Figure 1.
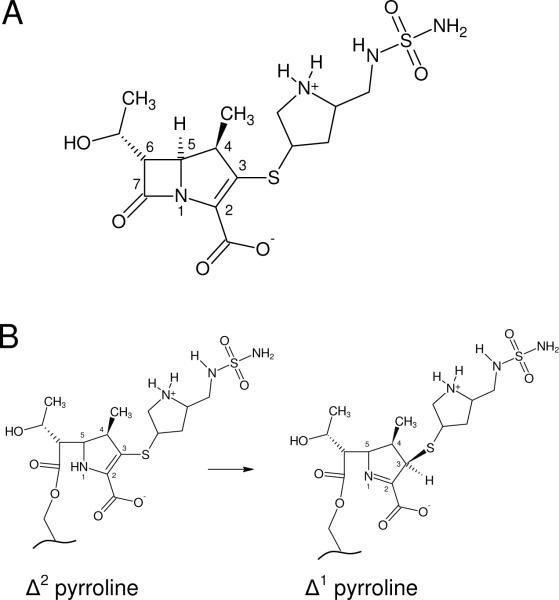
(A) The structure of doripenem. (B) Conversion of the Δ2 doripenem tautomer to the Δ1 form after acylation.
One of the defining features of carbapenems is the presence of a 6α-hydroxyethyl moiety in lieu of the more bulky 6β amide side-chains found in other β-lactam antibiotics. This unique structural element is thought to be responsible for resistance of these drugs to the hydrolytic action of some β-lactamases (2, 3). Classes A, C and D β-lactamases use a serine-nucleophile based covalent catalysis strategy (4). Interestingly, carbapenems are able to bind in the active site of these enzymes and can even proceed to a covalent acyl-intermediate with the active site serine (2, 3, 5). In many of these β-lactamases however, the water-dependent deacylation process is impaired. The end result is that carbapenems serve as effective irreversible inhibitors of many β-lactamases.
The interaction of carbapenems with β-lactamases has been studied extensively. X-ray crystallographic studies of imipenem and meropenem bound to class A (3, 6–9), class C (2) and class D (5) enzymes have yielded a wealth of structural detail about how the acyl-intermediate interacts with active site elements. In the case of most class A and class C enzymes, acylation is accompanied by a conformational change of the carbapenem species in which the acyl carbonyl is flipped ~ 180° away from the oxyanion hole (2, 3, 8). Removal of the carbonyl oxygen from the oxyanion hole explains the lack of carbapenem deacylation. This conformational flip was shown to be due to a steric clash between the hydroxyethyl group of the carbapenem and the side-chain of N132 in TEM-1 (7). The substitution of N132 by alanine in this enzyme allows the acyl-intermediate to rotate back into the oxyanion hole, though this alone is not enough to impart deacylation activity. The acyl carbonyl of meropenem is found in the oxyanion hole of one class A member, the BlaC β-lactamase from Mycobacterium tuberculosis (9). This may be explained by the fact that BlaC has a glycine at the position homologous to the asparagine typically found in class A and C enzymes.
Structures of OXA-13 with imipenem and meropenem represent the only class D β-lactamase/carbapenem structures determined so far (5). One feature unique to class D enzymes is the presence of a carboxylated lysine in the active site that plays the role of general base during the catalytic cycle (10–14). Interestingly, no carboxylation modification of the active site lysine was observed in either the apo or meropenem-bound structures of OXA-13 (5). However, the OXA-13 crystals were grown at pH 5.5, and pH values lower than 7 are deleterious toward lysine carboxylation in both OXA-10 (10) and OXA-1 (11). The ester carbonyl oxygen of meropenem is located in the oxyanion hole of OXA-13, so the lack of a properly positioned deacylating water molecule was cited as the cause of the inhibitory action of that drug.
One feature that is common to all β-lactamase/carbapenem structures determined so far is the formation of a salt-bridge between the carboxylate of the drug and a widely conserved arginine residue (e.g. R244 in TEM-1) (3). This arginine is not ubiquitous, however, so it is expected that those enzymes lacking this arginine (such as members of the class D β-lactamase OXA-1 subfamily) must use a different mechanism to attract and stabilize the carboxylate.
Another feature of the carbapenems is the presence of a double bond in the pyrroline ring which makes possible a tautomerization upon cleavage of the β-lactam amide bond (Figure 1B). Several studies have suggested that the Δ 1 pyrroline form of imipenem (bound to TEM-1) and meropenem (bound to SHV-1) is favored in the acyl-enzyme form (6, 8, 15). It has also been suggested that the two tautomeric states of the acyl-intermediate differ regarding their propensity for deacylation versus inhibition (6, 15).
Currently, the mechanism by which carbapenems inhibit class D β-lactamases remains unclear. What are the active site elements that determine whether the acyl carbonyl oxygen flips out of the oxyanion hole (as in TEM-1, SHV-1 and AmpC) or remains inside (as with OXA-13 and BlaC)? After acylation, are water molecules present in a position appropriate for deacylation? Is the active site carboxy-lysine involved in binding the drug, or is this modification lost upon acylation? Does tautomerization play any role in the inhibition process, as suggested for the class A enzymes? To explore these issues, we determined the crystal structure of the class D β-lactamase OXA-1 complexed with doripenem to 1.4 Å resolution. Beyond illuminating the interactions of carbapenems and class D β-lactamases, this structure is the first to show doripenem bound to a β-lactamase, and the first ligand-bound structure of any member of the OXA-1 subfamily.
Materials and Methods
Expression and Purification
E. coli cells expressing the blaoxa-1 gene were harvested and the enzyme was purified by carboxy-methyl cellulose CM-32 ion-exchange chromatography (13). OXA-1 purity was estimated to be >95% by SDS-PAGE. Pure enzyme was concentrated (Amicon Ultra) to the OXA-1 saturation limit (~6.0 mg/mL) in preparation for crystallization.
Crystal growth and structure determination
OXA-1 β-lactamase was crystallized as described previously (14). Briefly, a 10 μL drop containing 3.0 mg/mL OXA-1, 10% v/v polyethylene (PEG) 8000, 50 mM HEPES, pH 7.5 was equilibrated by hanging drop vapor diffusion over 20% v/v PEG 8000, 100 mM HEPES, pH 7.5, using microseeding techniques. Crystals appeared within 2–3 days at 25°C with maximal growth (0.05 × 0.05 × 0.15 mm) achieved after one week. Diffraction data were collected on the LS-CAT beamline (21-ID-D) at the Advanced Photon Source (Argonne, IL) at 100 K using a MarCCD detector. Prior to data collection the crystal was soaked in a solution containing > 20 mM doripenem for 3 min and then transferred into well buffer containing 25% 2-methyl-2,4-pentanediol (MPD) for 30 sec before flash-cooling in liquid nitrogen. A single crystal was used to collect both high and low resolution data sets.
Reflections were indexed, integrated, and scaled using HKL2000 (16). The space group was P21 with two OXA-1 monomers in the asymmetric unit. The structure of the OXA-1/doripenem complex was determined with Phaser (17) using the A monomer of apo OXA-1 (PDB entry 1M6K) as the initial phasing model (all water and ligand molecules removed). Refinement and electron density map calculations were performed with Refmac5 (18) in the CCP4 Program Suite (19). Manual rebuilding of the model was done with Coot (20). The final model of the OXA-1/doripenem complex contained 249 residues in each monomer, 434 water molecules, and one molecule of MPD. Doripenem was modeled into each active site based on initial Fo-Fc difference electron density maps and further refined with Refmac5. The coordinates for this structure have been deposited with the Protein Data Bank as 3ISG.
Molecular modeling
Starting coordinates for the semi-empirical molecular modeling of doripenem were obtained from an early model of the OXA-1/doripenem crystal complex. In this model, the doripenem molecule was built into the active site of OXA-1 as an acyl-enzyme intermediate covalently attached to the catalytic serine residue, Ser67. In preparation for modeling, the Cα atom of Ser67 was truncated to a methyl group, creating an ethyl ester linkage to the doripenem derivative. Using HyperChem (21), hydrogens were added to create the appropriate isomer and the structure was then geometry optimized using PM3, in gas phase, with a charge of −1.
Results
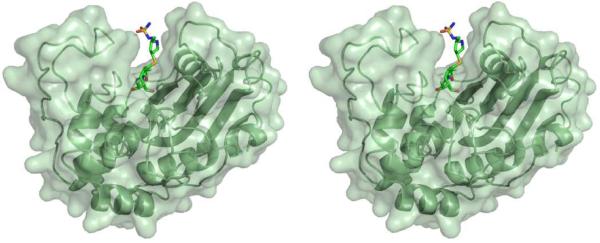
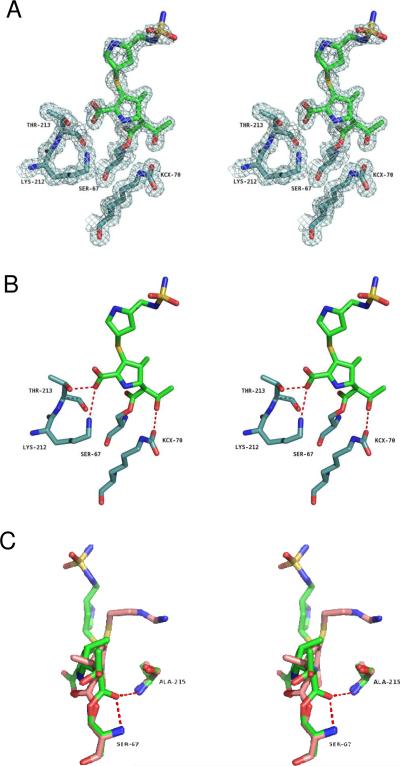
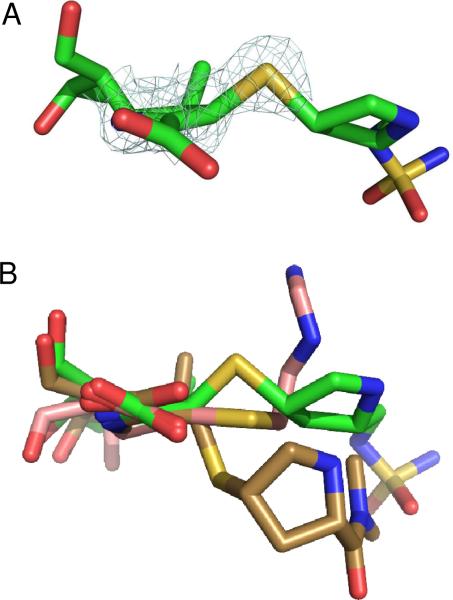
In an effort to understand the inhibition of class D β-lactamases by carbapenems, the structure of the complex of OXA-1 with doripenem was determined to 1.4 Å resolution (Figure 2). A comparison of the complex with the apo-enzyme structure shows strong similarity throughout the entire protein. Upon alignment of the two structures, an rmsd of 0.50 Å was determined for all Cα atoms. Initial Fo-Fc electron density maps contoured at 3.0 σ showed unambiguous density for the presence of doripenem bound in each of the OXA-1 active sites. Continuous electron density connects the carbonyl carbon atom (C7) of doripenem and the Oγ atom of S67 (Figure 3A). This serine is known to act as the catalytic nucleophile (13), indicating a covalent attachment between the drug and the enzyme. Additionally, clear density for all atoms of doripenem is observed, indicating that for the duration of the soak, no fragmentation of the drug occurs (Figure 3A). The final protein model was analyzed with the program Procheck (22), showing that 95% of the non-proline, non-glycine residues are in the preferred region of the Ramachandran plot (4% are in the additionally allowed region).
Figure 2.
Stereoview of the overall structure of OXA-1 bound to doripenem. This and all subsequent figures were created with PyMOL (29).
Figure 3.
Stereoview of the 2Fo-Fc electron density maps contoured at 1.0 σ surrounding doripenem and active site residues S67, KCX70, K212 and T213. Atoms are colored as follows: carbon atoms of OXA-1 are light blue, carbon atoms of doripenem green, nitrogens blue, oxygens red and sulfurs yellow. (B) Hydrogen bonding interactions (red dashes) between OXA-1 and doripenem. (C) Overlay of imipenem (salmon) in the active site of TEM-1 (PDB entry 1BT5) (3) and doripenem (green) in OXA-1, showing the different orientations of the carbonyl oxygen with respect to the oxyanion hole (right side). Residue numbers are for OXA-1.
The only significant effect of drug binding on the main-chain is seen in an approximately 1 Å movement of the loop formed by residues 94–103, a shift that slightly closes the active site. This loop forms the upper rim of the active site and is drawn toward the pyrrolidine/sulfonamide side-chain of doripenem when the drug is present.
It is highly notable that K70 is clearly carboxylated in the presence of doripenem (Figure 3A), and the resulting carbamate adopts essentially the same position observed in the apo structure (14). The presence of this modification contrasts with the OXA-13/meropenem complex, in which case the authors concluded the analogous lysine (K70) is not carboxylated (5). In contrast to the complexes of carbapenems with class A and class C β-lactamases, there is no water molecule within 5 Å of the carbamate of carboxy-K70 that could potentially act as the deacylating water. Instead, the atom OQ1 of the carbamate group makes a hydrogen bond to the hydroxyethyl group of doripenem (atom O62, 2.6 Å; Figure 3B). This hydrogen bond likely prevents the recruitment of the deacylating water, providing a satisfying explanation for the inhibitory nature of doripenem and other carbapenems towards OXA-1.
Another strong contrast between our structure and those of class A and C enzymes is the position of the acyl carbonyl. In the complexes of imipenem with TEM-1 (3) and AmpC (2), and meropenem with SHV-1 (8), this carbonyl is flipped 180° away from the two main chain amide nitrogens that comprise the oxyanion hole (Figure 3C). The carbonyl group of doripenem bound to OXA-1, on the other hand, forms two strong hydrogen bonds with the main-chain amides of S67 (2.7 Å) and A215 (2.8 Å). This is consistent with the position of meropenem in OXA-13, thus establishing a pattern for the mode of carbapenem binding to class D β-lactamases.
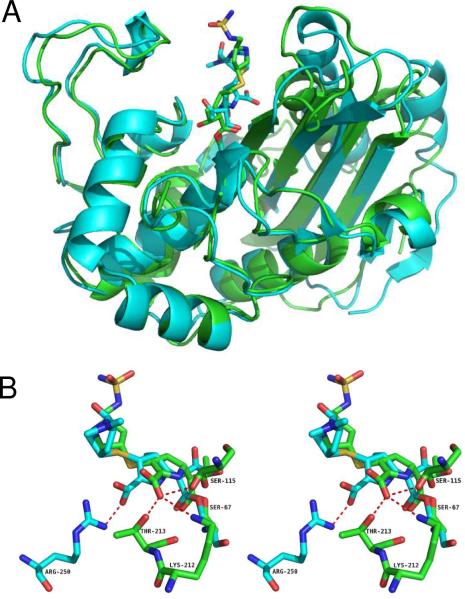
A unique feature of the OXA-1/doripenem complex is the interactions by which the carboxylate of the carbapenem is anchored to the enzyme. In most β-lactamase/β-lactam complexes, a highly conserved arginine (e.g. R244 in TEM-1, R349 in AmpC) forms a salt bridge to the carboxylate, either directly or through a bridging water molecule (2, 3, 5, 8). For apo OXA-1, Sun et al. noted the lack of any arginine near this area and suggested S258 may form a hydrogen bond with β-lactam substrates (14). Instead, the doripenem carboxylate hydrogen bonds to the side-chain of T213 and forms a salt bridge with K212 (Figure 3B). There is an additional hydrogen bond from the carboxylate to one of two alternative conformations of S115, a residue that has been proposed to shuttle a proton to the lactam nitrogen during catalysis (14). In order to contrast this novel mode of binding with the more common arginine interaction, we aligned OXA-1/doripenem with OXA-13/meropenem (5). The main-chains align with an rmsd of 2.65 Å (Figure 4A), with the entire ester linkage overlapping quite well (Figure 4B). However, the meropenem and doripenem structures greatly diverge starting with the pyrroline ring. Most notably, the interaction between the carboxylate and K212 causes a shift of the entire pyrroline ring back towards the helix comprising residues 108–114 (helix A4). The trajectory of the pyrrolidine ring (ie. the distal five-membered ring) and sulfonamide side-chain differs greatly from that of the similar side-chain of meropenem, with the sulfonamide approaching the loop comprising residues 97–99 (linking strands B5 and B6). The amine of the pyrrolidine ring, which is likely protonated at pH 7.5, makes a hydrogen bond to the carbonyl of L255, while the sulfonamide interacts with the carbonyl oxygens of S237 and G238 of a symmetrically related molecule.
Figure 4.
Superposition of the structures of OXA-13/meropenem (cyan) (5) with OXA-1/doripenem (green). (A) Overall structural similarity of the main-chains. (B) Comparison of the mode by which OXA-1 and OXA-13 stabilize the carbapenem carboxylate.
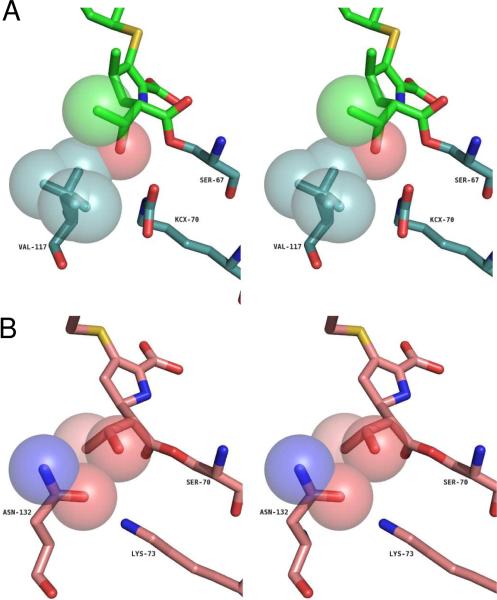
The side-chain of V117 packs tightly against the hydroxyethyl group of doripenem (Figure 5A), much as the homologous residue of TEM-1 (N132) does with the same group of imipenem (Figure 5B). Both the β-branched alkyl side-chain of V117 and the hydroxyethyl moiety of doripenem, however, show evidence of multiple rotational conformers, suggesting a degree of flexibility in this interaction.
Figure 5.
Comparison of the carbapenem hydroxyethyl moiety for (A) doripenem and V117 of OXA-1 and (B), imipenem and N132 of TEM-1 (3).
There has been much speculation about the role of carbapenem tautomerization in the mechanism of β-lactamase inhibition. While the positions of hydrogen atoms are not resolved in this structure, the resolution is sufficient to determine the geometry around the C3 carbon. In the intact drug, as well as the Δ2 tautomer produced just after acylation, the pyrroline double bond causes carbon C3 to be planar (Figure 1B). Isomerization to the Δ1 tautomer and the accompanying protonation of carbon C3 creates an additional tetrahedral center. The electron density of the sulfur attached to carbon C3 clearly shows that that atom rises significantly above the plane of the pyrroline ring (Figure 6A). This suggests that doripenem does in fact tautomerize after acylation, forming the R configuration of a novel chiral center at carbon C3. To determine the ideal geometry around the C3 carbon, we modeled an ethyl ester version of the acyl doripenem (no protein) using the program HyperChem (21). After semi-empirical geometry optimization using PM3, we found that the angle formed between the C3-S bond and the plane defined by atoms C2, C3 and C4 was 50°. The same angle in our structure was 31°, suggesting that protein interactions with the doripenem side-chain may prevent formation of the ideal tetrahedral geometry at C3. The presence of an R chiral center at this position contrasts to the geometry observed for meropenem bound to M. tuberculosis BlaC, in which the novel chiral center formed adopts the S configuration (9). Along with several structures modeled with an sp2 (planar) C3 atom (2, 3), these results highlight the diverse range of geometries that can be found in acyl-carbapenem structures (Figure 6B).
Figure 6.
The geometry of carbon C3 of doripenem bound to OXA-1. (A) Electron density showing that the location of the sulfur atom (S31) is above the plane of the pyrroline ring. (B) Overlay showing the different positions assumed by the sulfur in OXA-1/doripenem (R configuration; green), TEM-1/imipenem (planar; salmon) (3) and BlaC/meropenem (S configuration; brown) (9).
Discussion
The mode by which carbapenems inhibit β-lactamases is of great interest; a thorough understanding of their mechanism of action may aid in the design of more effective inhibitors. Moreover, understanding the lack of hydrolytic efficacy displayed by narrow spectrum enzymes like OXA-1 toward carbapenems should help explain how that inhibitory nature is overcome by carbapenemases. There are indeed many different possible explanations for the lack of carbapenem deacylation in class D β-lactamases, but analysis of the structure reported here points clearly in one direction.
When carbapenems acylate most class A and C β-lactamases, the ester carbonyl flips out of the oxyanion hole. This positively-charged cavity is known to stabilize the tetrahedral transition state, thus the conformational change provides a clear explanation for deacylation-deficiency. This flip does not occur in the OXA-1/doripenem complex, showing that class D β-lactamases are not inhibited by carbapenems in the same way as the other two classes. In TEM-1, this conformational change is caused by a steric clash between the 6α-hydroxyethyl moiety and the side-chain of N132. It is likely that the somewhat smaller side-chain of V117 in the homologous position of OXA-1 accounts at least partly for this major structural difference. It is tempting to speculate that substitution of asparagine (or an even larger residue) for this valine might cause a rearrangement similar to that seen for TEM-1, SHV-1 or AmpC.
Another potential inhibition mechanism that has been eliminated by this structure is deacylation-deficiency caused by decarboxylation of K70. We have recently shown that elimination of carboxy-K70 in OXA-1 by mutagenesis leads to an enzyme that accumulates acyl-enzyme intermediate of penicillin-class β-lactams (13). Moreover, the BlaR1 family of β-lactam sensor proteins are known to undergo decarboxylation of an active site carboxy-lysine after acylation, leading to the deacylation-deficiency property that is necessary for their function (23–26). The fact that we see unambiguous density for the carboxyl moiety of K70 in our ligand complex shows that this mechanism is not applicable here. The lack of lysine carboxylation in the OXA-13/meropenem structure (5), as noted earlier, is most likely an artifact of crystallization pH. The lack of a hydrogen bond between the (missing) carboxy-lysine of OXA-13 and the hydroxyethyl moiety of meropenem may contribute to the lateral shift of the ligand pyrroline ring (Fig 4B) compared to doripenem bound to OXA-1. The presence of R250 in OXA-13 and the lack of a similarly positioned arginine in OXA-1, however, also contribute to this lateral shift. Regardless of whether or not this shift reflects a true difference between carbapenems bound to different class D β-lactamases, the presence of the carboxy-lysine in OXA-1 argues that the doripenem structure we observe is the physiologically-relevant form.
In order to complete its full catalytic cycle, OXA-1 must recruit a water molecule to the vicinity of carboxy-K70 after acylation. The carbamate, acting as a general base, then activates it for attack on the ester bond. The fact that no water molecules are observed near the carbamate indicates that doripenem prevents recruitment of the deacylating water, providing a simple explanation for deacylation-deficiency/inhibition. The most likely source of interference in water recruitment is a hydrogen bond formed between the carbamate and the hydroxyethyl group of doripenem. In contrast, the hydroxyethyl group in both TEM-1/imipenem and AmpC/imipenem does not hydrogen bond to the putative general base (E166 in TEM-1; Y150 in AmpC), but rather interacts with an asparagine (N132 in TEM-1, N152 in AmpC). The general base in both enzymes is therefore able to recruit a water molecule, albeit one that is unable to complete the deacylation event. Because the asparagine of TEM-1 is replaced by a non-polar aliphatic residue in all class D enzymes, the interaction between the carbapenem hydroxyethyl group and the carboxy-lysine (and thus the mode of inhibition) is likely conserved throughout all of the class D enzymes.
The possibility of a tautomeric shift of the carbapenem after cleavage of the lactam bond has been postulated from the earliest description of carbapenem interactions with β-lactamases (6, 15, 27, 28). Biphasic hydrolysis profiles gave rise to the hypothesis that the two tautomers are deacylated at different rates. The Δ2 form, which is produced directly from lactam bond cleavage of the parental compound, deacylates rapidly (in this model). Alternatively, tautomerization to the Δ1 pyrroline leads to an acyl-penem conformation that is resistant to hydrolysis, causing inhibition. Isomerization of doripenem to the Δ1 tautomer is consistent with the spatial arrangement of the electron density around carbon C3 in our structure, though alternate techniques that are sensitive to tautomeric differences (i.e. Raman spectroscopy) will be needed for confirmation. One hypothesis suggests that tautomerization causes the carbapenem acyl carbonyl to flip out of the oxyanion hole in class A enzymes (6). Because we observe doripenem's acyl carbonyl in the oxyanion hole, this clearly cannot be the cause of slow deacylation in the case of OXA-1. In TEM-1, R244 has been suggested as a potential general acid for the protonation of imipenem C3 during the tautomerization (15). The side-chain of doripenem is exposed to solvent when bound to OXA-1, so the lack of a similarly positioned arginine in this enzyme is not problematic for tautomerization.
While the presence of two hydrogen bonds between doripenem and a symmetry-related OXA-1 molecule may impact the conformation of doripenem in the active site, we believe that the novel set of interactions we observe are likely present in the non-crystalline (ie. solution) structure. The two crystal lattice hydrogen bonds involve only the most distal portion of the doripenem side-chain (ie. the sulfonamide group). The position of the acyl carbonyl in the oxyanion hole and the position of the pyrroline ring and its carboxylate are stabilized by a large number of hydrogen bonds and/or ionic interactions within the same monomer unit. Also, the position of the pyrrolidine ring is stabilized by a hydrogen bond within the same protein unit (with the carbonyl of L255). Thus the two groups that most likely impact geometry of the C3 carbon and its attached sulfur atom (ie. the pyrroline and pyrrolidine rings) are anchored by polar groups within the same protein unit. Attempts to model the Δ2 tautomer (instead of the Δ1 form that we observe) result in the loss of the hydrogen bond with L255, and also in steric clashes between doripenem and the side-chains of M99 and A215.
In conclusion, the structure of OXA-1/doripenem reveals mechanisms of inhibition, carboxylate stabilization and tautomerization that differ from those observed in other β-lactamase/carbapenem complexes. This diversity reveals that subtle changes in active site architecture, even among highly similar β-lactamases, can result in major differences in ligand binding. As more structures become available in the future, these findings will contribute to the understanding of why some β-lactamases are inhibited by carbapenems, while others are able to hydrolyze them.
Table 1.
Crystallographic summary of the OXA-1/doripenem structure
| OXA-1/doripenem | |
|---|---|
| Cell constants (Å; °) | a=35.04 b=71.61 c=93.14; α=γ=90, β=99.18 |
| Spacegroup | P 21 |
| Resolution (Å) | 1.40 (1.45–1.40)a |
| Unique reflections | 86,546 |
| Total observations | 440,350 |
| Rmerge (%) | 7.4 (29.3) |
| Completeness (%)b | 96.6 (88.1) |
| <I>/<σI> | 13.5 (4.2) |
| Resolution range for refinement (Å) | 50–1.4 |
| Number of protein residues | 498 |
| Number of water molecules | 434 |
| rmsd bond lengths (Å) | 0.013 |
| rmsd bond angles (°) | 1.36 |
| R-factor (%) | 18.3 |
| Rfree (%)c | 20.6 |
| Average B-factor, protein atoms (Å2, molecule A) | 14.4 |
| Average B-factor, protein atoms (Å2, molecule B) | 12.7 |
| Average B-factor, doripenem atoms (Å2, molecule A) | 22.3 |
| Average B-factor, doripenem atoms (Å2, molecule B) | 22.9 |
| Average B-factor, water molecules (Å2) | 29.5 |
Values in parentheses are for the highest resolution shell.
Fraction of theoretically possible reflections observed.
Rfree was calculated with 5% of reflections set aside randomly.
Acknowledgments
This research was supported by National Institutes of Health grant 1R15AI082416-01 (D.A.L.). The Veterans Affairs Merit Review Program, Geriatric Research Education and Clinical Care VISN 10, and the National Institutes of Health (RO1 AI072219) supported R.A.B. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000817).
Abbreviations
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- MPD
2-methyl-2,4-pentanediol
- PDB
protein data bank
- rmsd
root mean square deviation
- KCX
carboxylysine residue
Footnotes
Coordinates have been deposited in the protein data bank (PDB) under accession # 3ISG
References
- (1).Pillar CM, Torres MK, Brown NP, Shah D, Sahm DF. In vitro activity of doripenem, a carbapenem for the treatment of challenging infections caused by gram-negative bacteria, against recent clinical isolates from the United States. Antimicrob. Agents Chemother. 2008;52:4388–4399. doi: 10.1128/AAC.00381-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Beadle BM, Shoichet BK. Structural basis for imipenem inhibition of class C β-lactamases. Antimicrob. Agents Chemother. 2002;46:3978–3980. doi: 10.1128/AAC.46.12.3978-3980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Maveyraud L, Mourey L, Kotra LP, Pedelacq J-D, Guillet V, Mobashery S, Samama J-P. Structural basis for clinical longevity of carbapenem antibiotics in the face of challenge by the common class A β-lactamases from the antibiotic-resistant bacteria. J. Am. Chem. Soc. 1998;120:9748–9752. [Google Scholar]
- (4).Fisher JF, Meroueh SO, Mobashery S. Bacterial resistance to β-lactam antibiotics: compelling opportunism, compelling opportunity. Chem. Rev. 2005;105:395–424. doi: 10.1021/cr030102i. [DOI] [PubMed] [Google Scholar]
- (5).Pernot L, Frenois F, Rybkine T, L'Hermite G, Petrella S, Delettre J, Jarlier V, Collatz E, Sougakoff W. Crystal structures of the class D β-lactamase OXA-13 in the native form and in complex with meropenem. J. Mol. Biol. 2001;310:859–874. doi: 10.1006/jmbi.2001.4805. [DOI] [PubMed] [Google Scholar]
- (6).Kalp M, Carey PR. Carbapenems and SHV-1 β-lactamase form different acyl-enzyme populations in crystals and solution. Biochemistry. 2008;47:11830–11837. doi: 10.1021/bi800833u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Wang X, Minasov G, Shoichet BK. Noncovalent interaction energies in covalent complexes: TEM-1 β-lactamase and β-lactams. Proteins. 2002;47:86–96. [PubMed] [Google Scholar]
- (8).Nukaga M, Bethel CR, Thomson JM, Hujer AM, Distler A, Anderson VE, Knox JR, Bonomo RA. Inhibition of class A β-lactamases by carbapenems: crystallographic observation of two conformations of meropenem in SHV-1. J. Am. Chem. Soc. 2008;130:12656–12662. doi: 10.1021/ja7111146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Hugonnet JE, Tremblay LW, Boshoff HI, Barry CE, 3rd, Blanchard JS. Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science. 2009;323:1215–1218. doi: 10.1126/science.1167498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Golemi D, Maveyraud L, Vakulenko S, Samama JP, Mobashery S. Critical involvement of a carbamylated lysine in catalytic function of class D β-lactamases. Proc. Natl. Acad. Sci. U. S. A. 2001;98:14280–14285. doi: 10.1073/pnas.241442898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Leonard DA, Hujer AM, Smith BA, Schneider KD, Bethel CR, Hujer KM, Bonomo RA. The role of OXA-1 β-lactamase Asp(66) in the stabilization of the active-site carbamate group and in substrate turnover. Biochem. J. 2008;410:455–462. doi: 10.1042/BJ20070573. [DOI] [PubMed] [Google Scholar]
- (12).Maveyraud L, Golemi-Kotra D, Ishiwata A, Meroueh O, Mobashery S, Samama JP. High-resolution X-ray structure of an acyl-enzyme species for the class D OXA-10 β-lactamase. J. Am. Chem. Soc. 2002;124:2461–2465. doi: 10.1021/ja016736t. [DOI] [PubMed] [Google Scholar]
- (13).Schneider KD, Bethel CR, Distler AM, Hujer AM, Bonomo RA, Leonard DA. Mutation of the active site carboxy-lysine (K70) of OXA-1 β-lactamase results in a deacylation-deficient enzyme. Biochemistry. 2009;48:6136–6145. doi: 10.1021/bi900448u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Sun T, Nukaga M, Mayama K, Braswell EH, Knox JR. Comparison of β-lactamases of classes A and D: 1.5-Å crystallographic structure of the class D OXA-1 oxacillinase. Protein Sci. 2003;12:82–91. doi: 10.1110/ps.0224303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Zafaralla G, Manavathu EK, Lerner SA, Mobashery S. Elucidation of the role of arginine-244 in the turnover processes of class A β-lactamases. Biochemistry. 1992;31:3847–3852. doi: 10.1021/bi00130a016. [DOI] [PubMed] [Google Scholar]
- (16).Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- (17).McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- (19).CCP4 (Collaborative Computational Project, Number 4) The CCP4 suite: programs for protein crystallography. Acta Crystallographica Section D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- (20).Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- (21).Hypercube, Inc. 1115 NW 4th Street, Gainesville, Florida 32601, USA: [Google Scholar]
- (22).Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- (23).Birck C, Cha JY, Cross J, Schulze-Briese C, Meroueh SO, Schlegel HB, Mobashery S, Samama JP. X-ray crystal structure of the acylated beta-lactam sensor domain of BlaR1 from Staphylococcus aureus and the mechanism of receptor activation for signal transduction. J. Am. Chem. Soc. 2004;126:13945–13947. doi: 10.1021/ja044742u. [DOI] [PubMed] [Google Scholar]
- (24).Cha J, Mobashery S. Lysine N(ζ)-decarboxylation in the BlaR1 protein from Staphylococcus aureus at the root of its function as an antibiotic sensor. J. Am. Chem. Soc. 2007;129:3834–3835. doi: 10.1021/ja070472e. [DOI] [PubMed] [Google Scholar]
- (25).Golemi-Kotra D, Cha JY, Meroueh SO, Vakulenko SB, Mobashery S. Resistance to β-lactam antibiotics and its mediation by the sensor domain of the transmembrane BlaR signaling pathway in Staphylococcus aureus. J. Biol. Chem. 2003;278:18419–18425. doi: 10.1074/jbc.M300611200. [DOI] [PubMed] [Google Scholar]
- (26).Thumanu K, Cha J, Fisher JF, Perrins R, Mobashery S, Wharton C. Discrete steps in sensing of β-lactam antibiotics by the BlaR1 protein of the methicillin-resistant Staphylococcus aureus bacterium. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10630–10635. doi: 10.1073/pnas.0601971103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Easton CJ, Knowles JR. Inhibition of the RTEM β-lactamase from Escherichia coli. Interaction of the enzyme with derivatives of olivanic acid. Biochemistry. 1982;21:2857–2862. doi: 10.1021/bi00541a008. [DOI] [PubMed] [Google Scholar]
- (28).Taibi P, Mobashery S. Mechanism of turnover of imipenem by the TEM β-lactamase revisited. J. Am. Chem. Soc. 2002;117:7600–7605. [Google Scholar]
- (29).DeLano WL. DeLano Scientific LLC; Palo Alto, CA, USA: 2008. [Google Scholar]