Abstract
Eukaryotic DNA replication uses kinase regulatory pathways to facilitate coordination with other processes during cell division cycles and response to environmental cues. At least two cell cycle-regulated protein kinase systems, the S phase-specific cyclin-dependent protein kinases (S-CDKs) and the Dbf4-Cdc7 kinase (DDK, the Dbf4-dependent protein kinase) are essential activators for initiation of DNA replication 1-5. While the essential mechanism of CDK activation of DNA replication in Saccharomyces cerevisiae has been established 6, 7, exactly how DDK acts has been unclear. Here we show that the N-terminal serine/threonine-rich domain (NSD) of Mcm4 plays both inhibitory and facilitating roles in DNA replication control and that the sole essential function of DDK is to relieve an inhibitory activity residing within the NSD. By combining an mcm4 mutant lacking the inhibitory activity with mutations that bypass the requirement for CDKs for initiation of DNA replication, we show that DNA synthesis can occur in G1 phase when CDKs and DDK are limited. However, DDK is still required for efficient S phase progression. In the absence of DDK, CDK phosphorylation at the distal part of the Mcm4 NSD becomes crucial. Moreover, DDK-null cells fail to activate the intra-S-phase checkpoint in the presence of hydroxyurea-induced DNA damage and are unable to survive this challenge. Our studies establish that the eukaryote-specific NSD of Mcm4 has evolved to integrate multiple protein kinase regulatory signals for progression through S phase.
In the early 1970s, studies on fusion of human cells suggested that DNA in G1 nuclei was competent for initiation of DNA replication, but G1 cells lacked an activator(s) that was present in S phase cells 8. The competent state has been defined as licensing of replication origins prior to S phase 1, 5, 9, 10. The process occurs at the M-phase exit through G1 phase, when a pre-replicative complex (pre-RC) forms at each origin. Pre-RC assembly begins with the binding of the Origin Recognition Complex (ORC), which recruits more protein factors, and ultimately completes with the loading the minichromosome maintenance (MCM) complex. Subsequently S phase-specific kinases, S-CDKs and DDK, activate this competent state by promoting assembly of the Cdc45-MCM-GINS (CMG) complex, the active replicative helicase 11-13. The minimal set of S-CDK targets essential for initiation of replication has been identified 6, 7. S-CDKs phosphorylate Sld2 and Sld3, enabling them to bind to Dbp11 6, 7, 14. Genetic and biochemical evidence suggested the MCM complex as one DDK target 3, 4. In budding yeast, DDK phosphorylates several MCM subunits and a mutation in MCM5, mcm5-bob1, can survive without DDK 15-19.
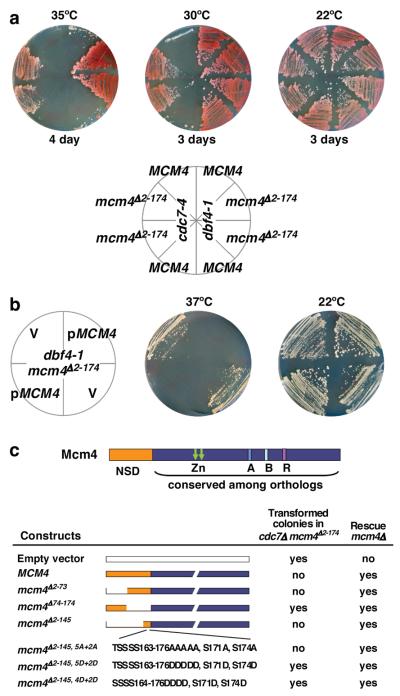
DDK binds to Mcm4 via a kinase-docking domain, allowing processive phosphorylation of multiple sites within the adjacent 174 amino acid NSD 18. Since deletion of NSD does not prevent cells from initiating DNA replication, it is likely that the role of NSD is regulatory. One hypothesis is that the NSD of Mcm4 blocks the activation of licensed origins and phosphorylation of the NSD by DDK alleviates the inhibition. To test this idea, we replaced the chromosomal MCM4 with mcm4Δ2-174, which lacks the entire NSD, in the temperature sensitive (ts) DDK mutants cdc7-4 and dbf4-1. Deletion of the Mcm4 NSD rescued the ts defect of cdc7-4 or dbf4-1 (Fig. 1a). Moreover, cdc7Δ mcm4Δ2-174 cells were viable (Fig. S1). The cdc7Δ mcm4Δ2-174 cells, however, grow slowly, likely due to (1) residues 2-174 harbor a domain needed for optimal MCM functions, or (2) DDK has another function in addition to its essential role in regulating Mcm4. Nevertheless, removing the Mcm4 NSD allows cells to bypass the essential function of DDK.
Figure 1. An inhibitory activity within the Mcm4 NSD is responsible for the dependency of cells on DDK for viability.
a, Yeast strains were grown on YPD plates at permissive and non-permissive temperatures (30°C and above for cdc7-4 and 35°C and above for dbf4-1). The mcm4Δ2-174 allele was introduced to cdc7-4 and dbf4-1 cells by two-step gene replacement. Shown are parental strains (top sectors) and three isolates of the second-step homologous recombination products. b, The dbf4-1 mcm4Δ2-174 cells transformed with empty vector (V) or vector carrying MCM4 were streaked on selective media and allowed to grow at 37°C or 22°C for 5 days. c, diagram of Mcm4 and summary of transformation assay and complementation of mcm4Δ by the same plasmid constructs (Fig. S2c). However, mcm4Δ2-145,5A+2A does exhibit growth defect even in the presence of DDK 18.
The ability of mcm4Δ2-174 to bypass DDK is recessive to MCM4. Re-introducing a MCM4 vector into dbf4-1 mcm4Δ2-174 allows cells to grow better at the permissive temperature (22°C), but they did not grow at 37°C, in contrast to the empty vector (Fig. 1b). Moreover, the MCM4 plasmid, unlike the empty vector, failed to yield transformed colonies in cdc7-4 mcm4Δ2-174 or cdc7Δ mcm4Δ2-174 cells, while CDC7 efficiently rescued cdc7Δ mcm4Δ2-174 cells (Fig. S2a and S2b). Together, these results suggest that the Mcm4 NSD contains an inhibitory activity that renders DDK essential for viability. Therefore, we used transformation of cdc7Δ mcm4Δ2-174 cells as an assay to map the inhibitory activity. While transformation of the mcm4Δ74-174 plasmid or empty vector yielded numerous colonies, transformation of plasmids carrying either MCM4, mcm4Δ2-73 or mcm4Δ2-145 produced none (Fig. 1c and S2c). Thus, the inhibitory activity resides within 74-174 of Mcm4 and residues 146-174 are sufficient for inhibiting transformation of cdc7Δ mcm4Δ2-174 cells. We have previously demonstrated DDK target sites within the 146-174 region 18. Here, we found that the phospho-mimetic mutation constructs (mcm4Δ2-145, 5D+2D or mcm4Δ2-145, 4D+2D) produced many transformed colonies whereas a construct that could not be phosphorylated by DDK (mcm4Δ2-145, 5A+2A) failed to produce transformants in cdc7Δ mcm4Δ2-174 cells (Fig. 1c and S2c). Thus, at least a portion of the Mcm4 NSD proximal to the DDK docking domain is inhibitory and phosphorylation of this region by DDK antagonizes the inhibitory effect.
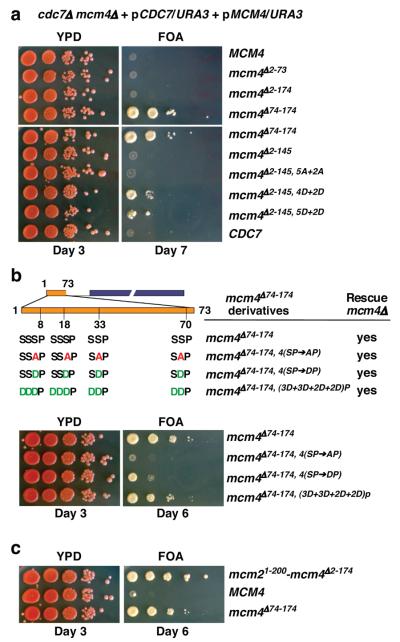
We tested those mcm4 alleles without the inhibitory activity for DDK bypass using a modified plasmid shuffle assay (Fig. 2a; see legend). This assay is stringent and relies on a CEN-based plasmid with a single replication origin to carry the tested allele. Thus, only those mcm4 alleles that can both fulfill the function of Mcm4 and bypass the requirement for DDK efficiently would allow the tester strain to survive on 5-Fluoroorotic Acid (5-FOA) media. Plasmids carrying mcm4Δ74-174, mcm4Δ2-145, 5D+2D and mcm4Δ2-145, 4D+2D allowed growth on 5-FOA media, indicating that these mutant alleles can cope with simultaneous loss of both MCM4 and CDC7 genes. In contrast, plasmids carrying CDC7, MCM4, mcm4Δ2-174, mcm4Δ2-73, mcm4Δ2-145 and mcm4Δ2-145,5A+2A scored negative in this assay. Although unphosphorylated mcm4Δ2-145 appears to be sufficient for exerting the inhibitory effect (Fig. 1c), the inhibitory domain extends beyond residues 146-174 because mcm4Δ147-174, mcm4Δ123-174 or mcm4Δ98-174 also fail to support DDK-independent growth (Fig. S3). Importantly, alanine substitution of 11 potential DDK phosphorylation sites within 74-174 in the full length NSD is lethal even in the presence of DDK (Fig. S4). It is possible that, when unphosphorylated, the proximal portion of the NSD exerts its inhibitory effect by imposing on the MCM complex a conformation that is not permissive for recruitment of activating factors, such as Cdc45 and GINS, and phosphorylation by DDK or removal of this domain allows the complex to assume a permissive state. The NSD inhibitory domain may also alter the MCM hexamer oligomeric state. Alternatively, the domain may directly block the access of activating factors.
Figure 2. The status of the Mcm4 N-terminus determines the efficiency of DDK-independent cell proliferation.
a-c, a modified plasmid shuffle assay was used to identify MCM4 alleles that allow cell growth in the absence of CDC7 and MCM4 when expressed from single-origin vectors. The tester yeast strain was constructed with both MCM4 and CDC7 genes deleted from its chromosomes while carrying both essential genes on plasmids with the URA3 gene that can be counter-selected in the presence of 5-FOA. The tester cells were transformed with indicated plasmids and assayed for growth on YPD and 5-FOA media. All mcm4 alleles used in the assay complement mcm4Δ. a, DDK-independent cell growth by the mcm4 mutants described in figure 1. b, Top panel, diagram of mcm4Δ74-174 derivatives with mutations at CDK phosphorylation sites and their preceding phospho-acceptor residues within the distal NSD (residues 2-73). Bottom panel, the ability of mcm4 alleles described above to support DDK-independent growth. c, DDK-independent cell growth supported by mcm21-200-mcm4Δ2-174.
The fact that mcm4Δ2-174 scored negative in the stringent assay for DDK-independent cell proliferation is consistent with our previous finding that it executes the function of MCM4 poorly 18. In contrast, mcm4Δ74-174 exhibited the best growth on 5-FOA media in the same experiment (Fig. 2a), suggesting that the distal part of the Mcm4 NSD (residues 2-73) plays a positive role in supporting DDK-independent growth. A shorter version (residues 2-37) could function similarly (Fig. S3). The distal NSD of Mcm4 is serine/threonine (S/T) rich and contains four CDK target (S/T-P) sites 20, all of which are preceded by additional S/Ts (Fig. 2b), which would become favorable phospho-acceptors for DDK upon priming phosphorylation 21, 22. Converting all four CDK sites to alanines within mcm4Δ74-174 (i.e. mcm4Δ74-174, 4(SP→AP)) had little effect on its ability to rescue mcm4Δ (Fig. 2b and S5). However, this mutant failed to bypass DDK. In contrast, phospho-mimetic substitution of these sites (i.e. mcm4Δ74-174, 4(SP→DP)) allowed DDK-independent growth and additional phospho-mimetic substitutions of all the preceding S/Ts further improved the growth (Fig. 2b). One caveat is that constitutive phosphorylation or phospho-mimetic substitution may compromise other aspects of Mcm4 function in DNA replication (see Fig. S10). As a result, the phospho-mimetic derivatives of mcm4Δ74-174 do not support growth better than mcm4Δ74-174 which is regulated by phosphorylation. Nevertheless, the positive function within the distal NSD may depend on phospho-regulation by CDK, and possibly by DDK. In the absence of DDK, CDK control of this region becomes essential. It remains to be addressed whether additional kinases also contribute to regulation of the Mcm4 NSD, which also contains multiple potential ATM/ATR target (S/T-Q) sites and many of these are also preceded by stretches of S/Ts.
Other MCM subunits such as Mcm2 and Mcm6 also have extended unstructured amino-terminal domains (NTDs) harboring DDK target sites (Fig. S6a). However, none of the N-terminal deletion mutants of MCM2 or MCM6 tested supported DDK-independent cell growth using analogous plasmid shuffle assays (Fig. S6 b and c). Thus, the inhibitory activity may be a unique feature of Mcm4. Yet, we have previously demonstrated that the NTD of Mcm2 can functionally replace the Mcm4 NSD in supporting normal cell proliferation and timely S phase progression 18. The mcm21-200-mcm4Δ2-174 fusion can function as an mcm4 allele that supports DDK-independent cell proliferation to the extent that surpasses mcm4Δ74-174 (Fig. 2c and S7a). Therefore, the Mcm2 NTD has a positive role in activating DNA replication. This region contains >30 % negatively charged aspartic acid (D) and glutamic acid (E) residues, reminiscent of phosphorylated S/Ts. Thus the Mcm2 NTD may act like a phosphorylated distal NSD of Mcm4.
DDK-bypass alleles of MCM4 were introduced into the endogenous locus in subsequent experiments. mcm4Δ74-174, mcm21-200-mcm4Δ2-174 and mcm5-bob1 cells grew at the same rate as the WT cells (Fig. S7a and S7b). The proliferation rates of cdc7Δ mcm4Δ74-174 and cdc7Δ mcm5-bob1 were comparable. Consistent with earlier observations (Fig. 2a and c), cdc7Δ mcm21-200-mcm4Δ2-174 cells proliferated faster than cdc7Δ mcm4Δ74-174 cells while cdc7Δ mcm4Δ2-174 cells proliferated more slowly (Fig. S7b). Cells without DDK grew slowly, entered S phase later and progressed through S phase at slower rates than their DDK positive counterparts (Fig. 3a). These results suggest that DDK has other non-essential roles in regulating S phase progression in addition to alleviating the inhibitory activity within the proximal NSD. For example, DDK may phosphorylate the distal NSD of Mcm4 or other substrates for efficient S phase progression.
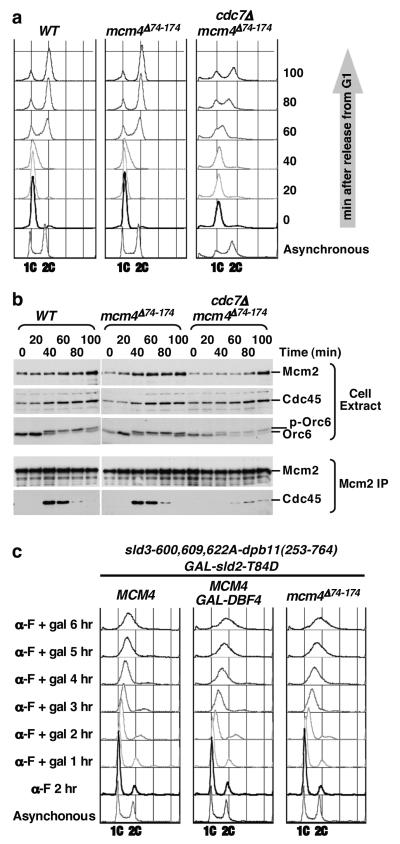
Figure 3. Removal of the N-terminal inhibitory domain of Mcm4 allows DDK-independent initiation of DNA replication and S phase progression.
a, Cell cycle progression of WT, mcm4Δ74-174 and cdc7Δ mcm4Δ74-174 cells. Cells were synchronized by G1 arrest and released into fresh YPD media at 25°C, collected at indicated times, and analyzed for DNA content by flow cytometry. b, Kinetics of Cdc45-MCM complex formation. Cell extracts were prepared from samples collected in a and subjected to immuno-precipitatation (IP) using monoclonal antibody against Mcm2. Cell extracts and IP were analyzed by immunoblotting. c, Flow cytometry analysis for DNA content in G1. CDK bypass cells containing SD fusion (sld3-600,609,622A-DBP11(253-764) and GAL-sld2-T84D, with or without additional modification (GAL-DBF4 or mcm4Δ74-174) were synchronized and held in G1 using α-factor (α-F) and galactose (gal) was added to induce expression of sld2-T84D and Dbf4.
One important consequence of DDK action during S phase is formation of a stable complex between Cdc45 and MCM at each origin as it is activated 18, 23-25. To determine if mcm4Δ74-174 can bypass the requirement of DDK for Cdc45-MCM complex formation, co-immunoprecipitation of Cdc45 with Mcm2 antibodies in cdc7Δ mcm4Δ74-174 cells, mcm4Δ74-174 and WT cells was examined. Cells were synchronized to allow progression through the cell cycle from G1 at 25°C (Fig. 3a). The Cdc45-MCM complex was detected at similar intensity and kinetics in WT and mcm4Δ74-174 cells, with a peak at ~40 minutes after G1 release (Fig. 3b). In the cdc7Δ mcm4Δ74-174 cells, the complex appeared at a later time (at ~60 minutes, peak at ~80 minutes) and at reduced levels. Nevertheless, these results demonstrated that eliminating the inhibitory domain in the Mcm4 NSD allows the Cdc45-MCM complex to form in the absence of DDK but DDK is still needed for timely Cdc45-MCM association under this bypass condition.
Recent studies reported conditions that allow yeast cells to replicate DNA in the absence of S-CDKs 6, 7. For example, the requirement for S-CDK for DNA synthesis can be bypassed by combining an sld3-dpb11 fusion (SD fusion) and over-expression of a phospho-mimetic sld2-T84D mutation 7. Under this S-CDK bypass condition, DDK is limiting for DNA replication and over-expression of DBF4 is necessary for extensive DNA synthesis in α-factor arrested, G1 cells. Instead of Dbf4 over-production, replacing the chromosomal MCM4 with mcm4Δ74-174 in this S-CDK bypass system allowed a similar extent of DNA replication in G1 (Fig. 3c and S8). We also observed a modest but consistent DDK-independent increase of DNA content in G1 by introducing mcm4Δ74-174 to a different S-CDK bypass condition 6 (Fig. S9). Furthermore, unlike another DDK bypass mutation mcm5-bob1, mcm4Δ74-174 does not exhibit synthetic lethality with the SD fusion. Thus, DDK bypass is not necessarily synthetic lethal with SD fusion as previously suggested 7. This result suggests DDK bypass by mcm4Δ74-174 is different from DDK bypass by mcm5-bob1. Moreover, accumulating biochemical evidence suggest that Mcm4, Mcm2 and Mcm6, but not Mcm5, are substrates of DDK 15, 17-19, 21, 22, 24, 26. Since MCM4 does not cause lethality in mcm5-bob1 cells lacking DDK, mcm5-bob1 is epistatic to MCM4 in this condition. Thus, DDK bypass by mcm5-bob1 is likely downstream of the inhibitory function of the Mcm4 NSD.
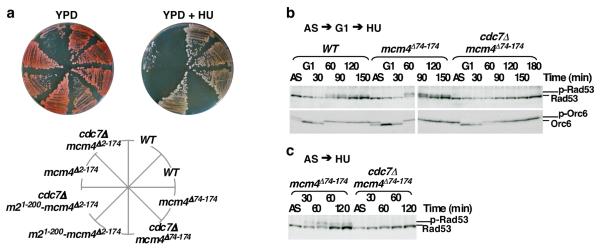
In the presence of DDK, mcm4Δ2-174 cells, but not mcm4Δ74-174 or mcm21-200-mcm4Δ2-174 cells, were sensitive to the ribonucleotide reductase inhibitor hydroxyurea (HU) (Fig. 4a), suggesting that the distal Mcm4 NSD or its functional equivalent (e.g. NTD of Mcm2) is required under DNA damaging conditions. Like cdc7Δ mcm5-bob1 cells 19, cdc7Δ mcm4Δ2-174, cdc7Δ mcm4Δ74-174 and cdc7Δ mcm21-200-mcm4Δ2-174 cells were not viable in HU (Fig. 4a). Thus, mcm4Δ74-174 and mcm21-200-mcm4Δ2-174 do not bypass the requirement of DDK for growth in the presence of HU. We examined checkpoint activation under synchronous G1 to HU release by monitoring Rad53 hyper-phosphorylation (Fig. 4 b and c). While checkpoint activation in MCM4 and mcm4Δ74-174 cells was efficient, Rad53 hyper-phosphorylation was not detectable in cdc7Δ mcm4Δ74-174 cells over the course of 3 hours in 200 mM HU. A similar defect in checkpoint activation in S phase was also found in cdc7Δ mcm5-bob1 cells 27. Although it is conceivable that insufficient initiation in cdc7Δ mcm4Δ74-174 cells would evade checkpoint 28, HU treatment of the asynchronous cdc7Δ mcm4Δ74-174 culture, which accumulates a large population of S phase cells, still failed to elicit robust Rad53 phosphorylation, unlike the response of the asynchronous mcm4Δ74-174 culture (Fig. 4c). Thus, it remains possible that DDK is required for the checkpoint response through Rad53 under replication stress (Fig. S11 and S12). Overall, our results demonstrate that mcm4Δ74-174 can bypass the requirement for DDK in an unperturbed S phase, but it cannot bypass the requirement for DDK in proper intra-S-phase checkpoint response.
Figure 4. Deletion of the N-terminal inhibitory domain of Mcm4 does not bypass the requirement for DDK for cell survival and checkpoint activation in the presence of HU.
a, Yeast strains of indicated genotypes at endogenous loci were grown on YPD media with or without 100 mM HU. b and c, Immunoblot analysis for Rad53 and Orc6 phosphorylation status in WT, mcm4Δ74-174 and cdc7Δ mcm4Δ74-174 cells. b, Protein samples were prepared from cells synchronized in G1 and released into 200 mM HU for indicated time. c, Protein samples were prepared from log-phase cells treated with HU for indicated time.
Cell fusion experiments suggested that “certain substances which are present in the S component probably migrate into G1 nucleus and cause initiation of DNA synthesis” 8. The results presented here for DDK and elsewhere for CDK 6, 7, 14 have uncovered essential targets for such activators that must act on the competent pre-RC. One surprising finding is that the essential DDK activity is to inhibit an intrinsic inhibitor of initiation of DNA replication. Our study reveals that the unstructured Mcm4 NSD is a multi-function domain that may serve to integrate various signals to regulate eukaryotic DNA replication.
Methods Summary
Yeast genetic methods and strain construction, cell extract preparation, immunoprecipitation, immunoblot analysis and antibodies are described in detail in Methods. Conditions of cell growth, cell cycle block and synchronization, and flow cytometry analysis are similar to published methods 6, 7, 18, 19.
Methods
Yeast strains
Yeast strains generated in this study were derived from W303-1a (MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1) and were described in the Supplementary Table, except for YS2041 (see below). A PCR-based gene deletion strategy was used for deletion of CDC7 and BAR1. The deletion cassettes cdc7Δ::KanMX6, cdc7Δ::HIS3 and bar1Δ::TRP1 are from the Yeast Knockout Collection (distributed by Open Biosystems), YB514 19 and y2007 7 were as described. All deletions were confirmed by PCR in combination with phenotypic assessment and gene complementation. A two-step gene replacement method was used to replace the endogenous MCM4 with mcm4 mutants. Plasmid constructs for two-step gene replacement were generated by sub-cloning the SphI/MluI fragments from pRS415-based constructs carrying Mcm4 N-terminal deletion mutants into the same restriction sites of a pRS306-based integration plasmid (a generous gift from J. J. Li) containing MCM4. CEN-based plasmid constructs carrying Mcm4 N-terminal deletion and site-directed mutants were created using fusion PCR strategy. The mcm4 mutant PCR products were digested with SphI/StuI and cloned into the same sites of pEM54.3 (pRS415/MCM4). Constructs were confirmed by DNA sequencing. Some constructs used in this work have been described in our previous work 18.
Plasmid Shuffle Assay
The tester strain YS2041 (MATa cdc7Δ::HIS3 mcm4Δ::TRP1 ade2 ura3 his3 leu2 trp1? + pRS416/CDC7 pRS416/MCM4) is a meiotic product of a diploid strain obtained by crossing YB514 (MATa ade2-101 ura3-52 lys2-801 his3Δ-200 leu2Δ-1 cdc7Δ::HIS3 + pRS416/CDC7) 19 and YS958 (MATα mcm4Δ::TRP1 ade2-1 can1-100 his3-11,-15 leu2-3,112 trp1-1 ura3-1 + pRS416/MCM4). Deletion of CDC7 and MCM4 were confirmed by selection for autotrophic traits, PCR, and Gene complementation. The pRS415-based test plasmids were used for transformation of YS2041. The transformed colonies were isolated and grown in SC-LEU media overnight, and 10-fold serial dilutions starting from 106 cells were spotted onto 5-FOA plates to select for loss of URA3 plasmids carrying CDC7 and MCM4. The same dilutions starting from 105 cells were spotted on YPD plates as control sets.
Cell Extract Preparation and Immunoprecipitation
Yeast cell pellet containing ~6 ×108 cells was resuspended in 150 μl of EB buffer containing 50 mM HEPES/KOH pH7.5, 100 mM KCl, 2.5 mM MgCl2, 2 mM NaF, 0.5 mM spermidine, 20 mM β-Glycerophosphate, 0.1 mM ZnSO4, 1 mM ATP, 1 mM DTT, 1 mM PMSF, Protease inhibitor tablets (EDTA free, Roche). An equal volume of chilled 0.5 mm zirconia/silia beads (BiosSpec Products, Inc.) was added to cell suspension and cells were lyzed by vortexing for 15 cycles of 30 sec on 30 sec off at maximal strength. The efficiency of cell breakage here was ~ 50 % after 12 cycles, as determined by visualization under a microscope. Cell lysates were collected in siliconized microfuge tubes after micro-centrifugation for 10 min at 12,000 rpm at 4°C. Pellets of cell debris and beads were resuspended with 400 μl of EBX buffer (EB Buffer with 0.25% Triton X-100) supplemented with 1 mM MnCl2 and 100 U/ml DNase I (Roche). The suspensions were allowed to mix for 30 min at 4°C. Supernatants were collected and pooled with previous lysates after centrifugation for 10 min at 12,000 rpm at 4°C. Pellets were resuspended again in the same buffer and allowed to mix for 15 min at 30°C, before supernatants were collected and pooled with earlier preps. Cell extracts from this preparation were analyzed for protein concentrations. Standard immunoprecipitation (IP) procedure were performed by mixing ~4 mg of total proteins and 2.5 μl of the mcm2-49 antiboy at 4°C for 30 min and precipitating the complex with GammaBind™G Sepharose (GE Healthcare) after washing extensively with EBX buffer.
TCA precipitation of yeast proteins
Yeast cell pellet containing ~5 × 107 cells was resuspended in 100 μl of TCA lysis buffer (1.85M NaOH and 7.4 % β-Mercaptoethanol), vortexed and left on ice. After 10 min, 100 μl of 20% TCA was added and gently mixed by inverting tubes. After incubation on ice for another10 min, pellets was collected by centrifugation at 13,000 rpm for 2 min, washed with 1ml ice-cold acetone and dried in vacuumed rotors. Dry pellets were resuspended carefully in 100 μl of 0.1 M NaOH. For analysis on SDS-PAGE, 100 μl of 2X sample buffer was added and samples were boiled for 10 minutes before loading.
Immunoblot analysis
Proteins from cell extract, IP or TCA precipitation were fractionated by SDS-10% PAGE and transferred to nitrocellulose membrane. Immunoblot analysis was performed as described previously using antibodies against Mcm2 (mcm2-39), Cdc45 (CS1485) and Orc6 (SB49) 18, 19. 12CA5 was used to detect 4HA-sld2-T84D and 2HA-dbf4 and 9E10 was used to detect sld2-11D-Myc. For detection of Rad53 from TCA precipitated yeast proteins, Rad53 (yC-19) antibody sc-6749 (Santa Cruz Biotechnology, Inc.) was used at 1:1000 dilution and TBS + 0.1% Tween 20 was used for preparing blocking and washing solutions.
Supplementary Material
Acknowledgements
We thank A. Stenlund and H.-G. Wendel for critical reading of the manuscript, J. Li, H. Araki and J. Diffley for plasmids and yeast strains. This work was supported by a grant from the US National Institute of General Medical Sciences.
Footnotes
Supplementary information is found in the attached material.
Competing Interests The authors have no competing interests
References
- 1.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 1.Diffley JF. Regulation of early events in chromosome replication. Curr Biol. 2004;14:R778–786. doi: 10.1016/j.cub.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 3.Masai H, Arai K. Cdc7 kinase complex: a key regulator in the initiation of DNA replication. J Cell Physiol. 2002;190:287–296. doi: 10.1002/jcp.10070. [DOI] [PubMed] [Google Scholar]
- 4.Sclafani RA. Cdc7p-Dbf4p becomes famous in the cell cycle. J Cell Sci. 2000;113(Pt 12):2111–2117. doi: 10.1242/jcs.113.12.2111. [DOI] [PubMed] [Google Scholar]
- 5.Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka S, et al. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- 7.Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- 8.Rao PN, Johnson RT. Mammalian cell fusion: studies on the regulation of DNA synthesis and mitosis. Nature. 1970;225:159–164. doi: 10.1038/225159a0. [DOI] [PubMed] [Google Scholar]
- 9.Forsburg SL. Eukaryotic MCM proteins: beyond replication initiation. Microbiol Mol Biol Rev. 2004;68:109–131. doi: 10.1128/MMBR.68.1.109-131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laskey RA, Fairman MP, Blow JJ. S phase of the cell cycle. Science. 1989;246:609–614. doi: 10.1126/science.2683076. [DOI] [PubMed] [Google Scholar]
- 11.Gambus A, et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 12.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell. 2006;21:581–587. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 14.Masumoto H, Muramatsu S, Kamimura Y, Araki H. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature. 2002;415:651–655. doi: 10.1038/nature713. [DOI] [PubMed] [Google Scholar]
- 15.Francis LI, Randell JC, Takara TJ, Uchima L, Bell SP. Incorporation into the prereplicative complex activates the Mcm2-7 helicase for Cdc7-Dbf4 phosphorylation. Genes Dev. 2009;23:643–654. doi: 10.1101/gad.1759609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardy CF, Dryga O, Seematter S, Pahl PM, Sclafani RA. mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc Natl Acad Sci U S A. 1997;94:3151–3155. doi: 10.1073/pnas.94.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lei M, et al. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 1997;11:3365–3374. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheu YJ, Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell. 2006;24:101–113. doi: 10.1016/j.molcel.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weinreich M, Stillman B. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. Embo J. 1999;18:5334–5346. doi: 10.1093/emboj/18.19.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Devault A, Gueydon E, Schwob E. Interplay between S-cyclin-dependent kinase and Dbf4-dependent kinase in controlling DNA replication through phosphorylation of yeast Mcm4 N-terminal domain. Mol Biol Cell. 2008;19:2267–2277. doi: 10.1091/mbc.E07-06-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho WH, Lee YJ, Kong SI, Hurwitz J, Lee JK. CDC7 kinase phosphorylates serine residues adjacent to acidic amino acids in the minichromosome maintenance 2 protein. Proc Natl Acad Sci U S A. 2006;103:11521–11526. doi: 10.1073/pnas.0604990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montagnoli A, et al. Identification of Mcm2 phosphorylation sites by S-phase-regulating kinases. J Biol Chem. 2006;281:10281–10290. doi: 10.1074/jbc.M512921200. [DOI] [PubMed] [Google Scholar]
- 23.Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 24.Masai H, et al. Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. J Biol Chem. 2006;281:39249–39261. doi: 10.1074/jbc.M608935200. [DOI] [PubMed] [Google Scholar]
- 25.Zou L, Stillman B. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science. 1998;280:593–596. doi: 10.1126/science.280.5363.593. [DOI] [PubMed] [Google Scholar]
- 26.Jiang W, McDonald D, Hope TJ, Hunter T. Mammalian Cdc7-Dbf4 protein kinase complex is essential for initiation of DNA replication. Embo J. 1999;18:5703–5713. doi: 10.1093/emboj/18.20.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogi H, Wang CZ, Nakai W, Kawasaki Y, Masumoto H. The role of the Saccharomyces cerevisiae Cdc7-Dbf4 complex in the replication checkpoint. Gene. 2008;414:32–40. doi: 10.1016/j.gene.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Shimada K, Pasero P, Gasser SM. ORC and the intra-S-phase checkpoint: a threshold regulates Rad53p activation in S phase. Genes Dev. 2002;16:3236–3252. doi: 10.1101/gad.239802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.